Summary
DNA double strand breaks (DSBs) initiate extensive local and global alterations in chromatin structure, many of which depend on the ATM kinase. Histone H2A ubiquitylation (uH2A) on chromatin surrounding DSBs is one example, thought to be important for recruitment of repair proteins. uH2A is also implicated in transcriptional repression; an intriguing yet untested hypothesis is that this function is conserved in the context of DSBs. Using a novel reporter that allows for visualization of repair protein recruitment and local transcription in single cells, we describe an ATM-dependent transcriptional silencing program in cis to DSBs. ATM prevents RNA polymerase II elongation dependent chromatin decondensation at regions distal to DSBs. Silencing is partially dependent on E3 ubiquitin ligases RNF8 and RNF168, while reversal of silencing relies on the uH2A deubiquitylating enzyme USP16. These findings give insight into the role of post-translational modifications in mediating cross talk between diverse processes occurring on chromatin.
Keywords: ATM, DNA repair, H2A ubiquitin, transcription
Introduction
Post-translational modifications (PTMs) on chromatin regulate diverse cellular processes such as transcription and mitosis. PTMs are also implicated in DNA double strand break (DSB) repair; the extensive DSB-induced phosphorylation of the histone variant H2AX (γH2AX) is a primary example (Rogakou et al., 1998). Emerging evidence implicates nondegradative ubiquitin signals as one such PTM at DSBs. These marks have been studied primarily as recognition signals for repair proteins (Kim et al., 2007; Sobhian et al., 2007; Wang et al., 2007). ATM (Ataxia Telangiectasia Mutated) kinase activity is a primary driving force for chromatin alterations emanating from DSB induction and these activities are in part thought to mediate ATM dependent suppression of genomic instability and carcinogenesis (Lavin, 2008). Recent studies have shed light on the ATM dependent molecular events that occur on chromatin adjacent to DSBs. Specifically, the E3 ubiquitin ligases RNF8 and RNF168 effect the formation of lysine 63-linked polyubiquitin (K63Ub) chains on damaged chromatin, including on histones H2A and H2AX, in an ATM dependent manner (Doil et al., 2009; Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007; Stewart et al., 2009; Wang and Elledge, 2007). The combined activity of these ligases is required for productive recruitment of repair proteins such as RAP80, BRCA1, and 53BP1.
Histone ubiquitylation is also linked to transcription (Zhang, 2003). Monoubiquitylation of histone H2A on lysine 119 is correlated with transcriptional repression (Wang et al., 2004). Notably, ubiquitylated H2A (uH2A) is observed at ionizing radiation-induced foci (IRIF). Like IRIF-associated K63Ub, uH2A enrichment at DSBs is RNF8/RNF168 dependent, though it is not known if K63Ub chains and uH2A represent the same functional signal at breaks. The commonality of uH2A in DNA repair and transcription suggests the intriguing possibility that DSB associated modifications signal to other processes on contiguous stretches of chromatin. Indeed, DSB-induced PTMs on chromatin may spread for hundreds of kilobases from sites of damage (Rogakou et al., 1999; Shroff et al., 2004), raising the possibility that they influence transcriptional regulatory elements at a distance.
To address this question, we developed a single cell assay by modifying a previously described transcriptional reporter system to allow simultaneous visualization of DNA damage responses and nascent transcription on a contiguous stretch of chromatin. Using this and other systems, we describe an ATM kinase-dependent silencing program that spreads across kilobases of chromatin in cis to DSBs to repress gene expression from a distant promoter. ATM activity is required to prevent transcription-associated large-scale chromatin decondensation, defining a novel role for this kinase in influencing chromatin dynamics within euchromatic environments. ATM-mediated silencing occurs, at least in part, through damage-responsive E3 ubiquitin ligases that catalyze the formation of uH2A, and is terminated by a deubiquitylating enzyme that opposes the actions of these ligases. We describe these findings and explore the mechanisms underlying this phenomenon.
Results
A novel reporter system reveals transcriptional silencing induced by DNA double strand breaks
To investigate how DSBs influence transcription, we developed a system that enables simultaneous visualization of transcription and DSB repair protein recruitment in single cells on a contiguous stretch of genomic DNA. This was achieved by modifying a previously described transcriptional reporter system (Janicki et al., 2004) with the capacity to create DSBs approximately 4-13 kb upstream of the promoter (Figure 1A). The reporter, integrated at a single site on chromosome 1p3.6 in the human osteosarcoma U2OS cell line, is visualized by expression of the red fluorescent mCherry protein fused to the lac repressor protein (mCherryLacI), which concentrates at the 256 copy lac operator array in the reporter. The operator sequences are separated from the promoter by approximately 4 kb of tandem tetracycline response elements (TREs), which bind a doxycycline-inducible transactivator. Upon doxycycline (dox) treatment, a minimal cytomegalovirus (CMV) promoter drives expression of a CFP-tagged peroxisomal targeting peptide (CFP-SKL), which accumulates in the cytoplasm. Nascent transcript is visualized by accumulation of yellow fluorescent protein tagged MS2 viral coat protein (YFP-MS2), which binds MS2 stem loop structures present as 24 repeats within the reporter transcript. Thus, chromosomal location, nascent transcript, and protein production are visualized in single cells.
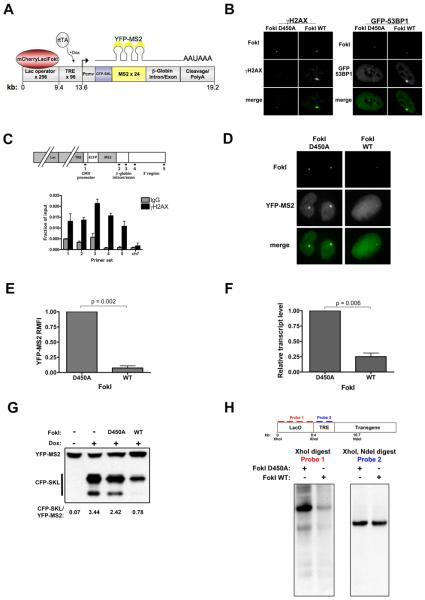
Figure 1. DSBs initiate local transcriptional silencing.
(A) Schematic of reporter locus. 256 lac operator repeats and a tetracycline response element (TRE) array are upstream of an inducible reporter gene that codes for the CFP-SKL protein and contains 24 repeats of the MS2 RNA stem loop sequence. The lac repressor is fused to mCherry and to the nuclease domain of the FokI endonuclease, enabling DSB induction within the 9 kb of lac operator repeats. Nascent transcript is visualized through binding of a YFP-MS2 protein to the MS2 stem loops. CFP-SKL accumulates in the cytoplasm upon induction of transcription. Not drawn to scale.
(B) Transfection of the wild type FokI nuclease fusion (FokI WT) but not the nuclease-deficient D450A mutant into U2OS cells containing the stably integrated reporter leads to local accumulation of damage response proteins γH2AX (left) and 53BP1 (right).
(C) Top – schematic of reporter locus and primers used for chromatin immunoprecipitation (ChIP).
Bottom – ChIP analysis demonstrates enrichment of γH2AX across reporter locus compared to IgG control. ch7 represents negative control locus on chromosome 7. Data are means + standard error of the mean (SEM) from two independent experiments.
(D) Nascent transcript (YFP-MS2) accumulation in cells expressing FokI D450A and FokI WT.
(E) Quantification of YFP-MS2 relative mean fluorescence intensity (RMFI) from (D) using ImageJ software (NIH). Data was derived from 3 independent experiments and is depicted relative to FokI D450A expressing cells. Error bars represent SEM.
(F) qRT-PCR for reporter mRNA levels in cells 48 hours after transduction with lentivirus expressing FokI WT or D450A. Transcription was monitored 3 hours after addition of 1 μg/ml doxycycline. Error bars represent SEM from 3 independent experiments.
(G) Representative immunoblot (IB) for CFP-SKL in cells expressing FokI WT or D450A after 4 hr induction with 1 μg/ml doxycycline. The ratio of CFP-SKL to YFP-MS2 signal density was quantified using ImageJ software (NIH).
(H) Top – schematic of Southern blot strategy. Bottom - Southern blot of genomic DNA from reporter cells expressing FokI WT or D450A digested with restriction enzymes XhoI, or XhoI and NdeI and probed for the indicated regions.
See also Figure S1.
We fused the nuclease domain of the FokI endonuclease to mCherryLacI, targeting nuclease activity to the lac operator repeats (Figure 1A). Expression of the wild-type FokI mCherryLacI fusion (FokI WT) in reporter cells resulted in a strong DNA damage response (DDR) at the reporter locus in over 90% of transfected cells, indicated by a striking local accumulation of γ2AX and GFP-tagged 53BP1, as well as several other repair proteins (Figures 1B, S1). In contrast, the nuclease-deficient D450A mutant of FokI fused to mCherryLacI (FokI D450A) (Waugh and Sauer, 1993) did not lead to a similar accumulation. This suggests FokI induced breaks recapitulate DNA recruitment processes found at IRIF. Moreover, by chromatin immunoprecipitation, γ2AX was significantly enriched throughout the reporter locus in FokI expressing cells, indicating induction of a chromatin mediated damage response (Figure 1C).
To assess the influence of DSBs on transcription occurring on chromatin contiguous to the site of damage, we first measured nascent transcript levels at the reporter after DSB induction. Dox-induced transcription in cells transfected with FokI D450A led to a strong local accumulation of nascent transcript (YFP-MS2), whereas little to no transcript was detected in cells expressing FokI WT (Figure 1D). YFP-MS2 fluorescence intensity analysis and quantitative reverse transcriptase PCR (qRT-PCR) revealed a four to ten-fold reduction in transcript levels in damaged cells compared to controls (Figure 1E, F). Consistent with a reduction in nascent transcript, western blot analysis demonstrated a strong reduction in reporter protein levels after DSB induction (Figure 1G).
We next tested the hypothesis that FokI-induced DSBs prevented transcription by simply degrading the TREs and transgene, or by inducing end resection through these regions. Southern blot analysis demonstrated that the TREs, promoter, and the CFP-SKL coding region were largely intact in FokI WT expressing cells (Figure 1H, right). In addition, efficient knockdown of the end resection factor CtIP, did not restore transcription (Figure S1), arguing against resection as the mechanism underlying DSB induced silencing. In contrast, a strong reduction in the lac operator region was observed after damage, indicative of extensive DSB induction by nuclease activity targeted to this region by the lac repressor (Figure 1H, left). Together, these data suggest DSBs do not themselves create a physical block to transcription, but rather initiate a signal that can inhibit transcription from a promoter at least 4 kb from the site of damage.
DSB induced silencing occurs in cis
To ascertain whether DSB-induced transcriptional silencing was a global response to DNA damage or a local phenomenon transmitted in cis to the lesion, we treated reporter cells with a low dose of ionizing radiation (IR), and analyzed YFP-MS2 accumulation at the locus. Three hours after 3 Gy of IR, there was a greater than 10-fold reduction in the mitotic (phospho-histone H3 positive) population of irradiated compared to control cells indicating a robust G2/M checkpoint (Figure 2A). However, this cell cycle checkpoint activation was not accompanied by transcriptional silencing, as reporter transcript and protein levels were comparable in both groups over three hours after damage (Figure 2A, S2). To address this question in an independent manner we assessed transcription in reporter cells that had been microirradiated with a 337 nm wavelength laser to create a linear array of DSBs through the nucleus. Damaged cells were visualized by immunofluorescence (IF) for γH2AX (Figure 2B, top). Consistent with results from IR-induced breaks, nascent transcript levels were comparable in damaged and undamaged cells (Figure 2B, bottom).
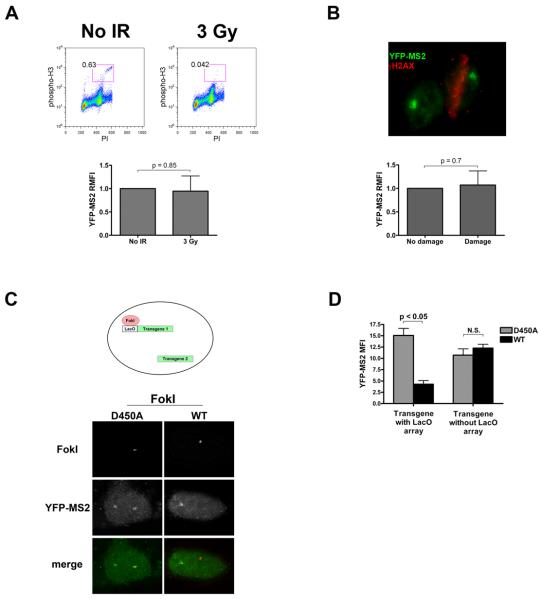
Figure 2. DSB induced silencing occurs in cis to DNA damage.
(A) Top – flow cytometric analysis of reporter cells mock treated (No IR) or treated with 3 Gy of ionizing radiation (IR), and analyzed for phospho-histone H3 and propidium iodide after 3 hours. Mitotic cells are contained within the red box and the percentage of mitotic cells indicated. Bottom – quantification of YFP-MS2 RMFI in mock and 3 Gy treated cells after 3 hour induction with 1 μg/ml doxycycline. The experiment was performed in triplicate and data represent mean + standard deviation (SD).
(B) Cells were induced with doxycycline for 3 hrs immediately after laser microirradiation. Top – representative image of microirradiated cell (right) identified by γH2AX (red) and a neighboring, undamaged cell. Nascent transcript is visible as YFP-MS2 in both cells. Bottom – Quantification of YFP-MS2 fluorescence intensity in damaged and undamaged cells. The experiment was performed in triplicate and data represent mean + SD.
(C) Top – schematic of two transgene containing reporter cells. Both transgenes contain identical promoters and regulatory elements, while only one contains the lac operator array. Bottom – representative images of two transgene reporter cells expressing FokI D450A (left) or WT (right), and YFP-MS2 accumulation at both transgenes.
(D) Quantification of YFP-MS2 intensity at both arrays as in (C). Data are presented as mean + SEM from two independent experiments.
See also Figure S2.
Recognizing the potential for differences in cellular responses to nuclease induced breaks in comparison to IR and laser induced damage, we developed a cell line to definitively test if FokI breaks transmitted silencing signals exclusively in cis to the site of damage. The original reporter cell line was engineered to contain a second reporter locus devoid of lac operator repeats (Figure 2C, top). The FokILacI fusion protein would consequently target DSBs to only one of the loci. Upon dox addition, we observed focal accumulation of YFP-MS2 at both loci in FokI D450A expressing cells. As predicted, FokI WT expression impaired transcription at the locus containing lac operator sequences, with no effect on the undamaged locus (Figures 2C, D). Collectively, these data strongly suggest DSBs initiate transcriptional repression that spreads in cis to the lesion. Consistent with this assertion, we observed mutually exclusive localization of IR induced 53BP1 foci and nascent RNA transcripts at endogenous sites in the genome as assessed by nuclear run-on experiments (Figure S2).
ATM is required for transcriptional silencing and accumulation of hypophosphorylated RNA polymerase II at DSBs
ATM has been implicated in multiple DNA damage responses, including global alterations to both chromatin structure and transcriptional programs (Bredemeyer et al., 2008; Goodarzi et al., 2008; Kim et al., 1999; Matsuoka et al., 2007; Ziv et al., 2006). Based on the varied roles ATM may play at DSBs – many of which remain unexplored – we asked if the kinase was involved in DSB-induced transcriptional silencing. A specific inhibitor of ATM kinase, KU55933, was administered at the time of dox addition, and nascent transcript levels assessed 3 hours later. Despite potent inhibition of ATM, KU55933 has little effect on the related PIKK kinases, ATR and DNA-PK (Hickson et al., 2004). Remarkably, ATM inhibition (ATMi) almost completely restored transcriptional activity in cells expressing FokI WT, but had no effect in control cells, as assessed by YFP-MS2 intensity and qRT-PCR (Figures 3A,B and S3). ATMi efficacy was documented by a decrease in phosphorylated H2AX after ionizing radiation (Fig. S3). Similar results were obtained with siRNA-mediated depletion of ATM (Figure 3B, right), supporting the specificity of the inhibitor. ATMi also rescued reporter protein levels in FokI WT expressing cells (Figure 3C). Conversely, pharmacologic inhibition of DNA-PK had no effect on transcriptional silencing (Figure S3).
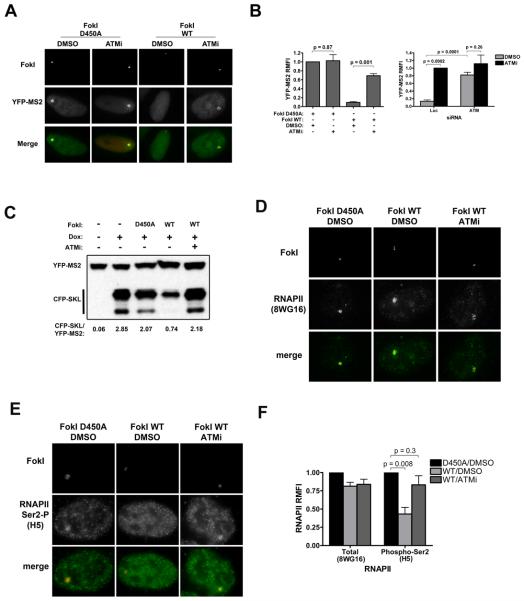
Figure 3. Transcriptional silencing at DSBs is dependent on ATM kinase activity.
(A) Reporter cells expressing FokI D450A or FokI WT were treated with 1μg/ml dox and either DMSO or ATMi (10 μM KU55933 ) for 3 hours before fixing for analysis of YFP-MS2 accumulation.
(B) Left - Quantification of YFP-MS2 RMFI from experiments as in (A). Error bars represent SEM from two to four independent experiments. Right – quantification of RMFI in cells expressing FokI WT, treated with siRNA against control or ATM, and treated with DMSO or ATMi. Error bars represent SEM from 2 independent experiments.
(C) Representative IB for CFP-SKL protein in D450A or FokI WT expressing cells treated with either DMSO or ATMi and 1μg dox for 4 hours. YFP-MS2 levels are used as a loading control.
(D) IF for total RNAPII (8WG16) was performed as indicated in cells treated with DMSO or with ATMi.
(E) IF was performed for each group as in (D) for actively elongating RNAPII with an antibody to RNAPII phosphorylated on Ser2 of the C-terminal domain (H5).
(F) Quantification of RMFI from experiments in panels D and E. Error bars represent SEM from at least three independent experiments.
See also Figure S3.
To gain insight into how DSBs mediate transcriptional silencing we examined RNA polymerase II (RNAPII) at the reporter. Due to the size and repetitive nature of the reporter, RNAPII accumulation is visible upon induction of transcription. Active transcription is associated with phosphorylation of Ser2 on the C-terminal domain (CTD) of the large subunit of RNAPII, whereas this site is predominately non-phosphorylated in non-elongating RNAPII complexes (Phatnani and Greenleaf, 2006). We observed no significant difference in RNAPII levels between D450A and WT FokI expressing cells using an antibody directed against hypophosphorylated RNAPII CTD repeats (8WG16) (Figure 3D,F), used as a proxy for total RNAPII (O'Brien et al., 1994). In contrast to unchanged total levels, upon DSB induction, we noted a specific loss of RNAPII phosphorylated on Ser2 of the CTD, which was largely reversed upon ATMi (Figure 3E,F), consistent with a resumption of transcription. Together, these data suggest that contrary to other forms of DNA damage, DSBs lead to a selective loss of actively elongating RNAPII, but do not affect total levels, supporting the concept that DSBs induce a unique form of transcriptional silencing distal to the site of DNA damage via an ATM-dependent stalling of RNAPII.
ATM kinase activity prevents transcriptional elongation dependent chromatin decondensation
Transcriptional activation is associated with chromatin decondensation in the vicinity of active genes; transcriptional reporter arrays represent a convenient tool to visualize this process. Though precise mechanisms remain to be identified, locus decondensation is known to rely on actively elongating RNAPII. Transcriptional inhibition with either the elongation inhibitor 5,6-Dichlorobenzimidazole 1-β-D-ribofuranoside (DRB) or with α-amanitin, which leads to RNAPII degradation, prevents locus decondensation (Muller et al., 2001). Therefore, we postulated that DSBs would prevent locus decondensation via stalling of RNAPII distal to the site of damage. To test this, we measured reporter locus area in cells expressing FokI D450A or WT before and after doxycyline addition. As anticipated, an approximately 5-fold increase in locus area was observed in D450A expressing cells, which was reversed by co-administration of DRB (Figure 4A, left and B). In control experiments, DRB treatment led to a selective loss of elongating RNAPII in comparison total RNAPII (Figure S4). FokI WT expressing cells qualitatively and quantitatively failed to demonstrate locus decondensation (Figure 4A, right and B).
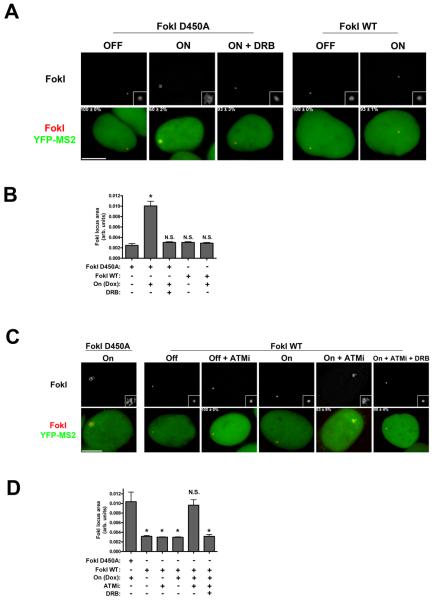
Figure 4. ATM prevents transcription-dependent large-scale chromatin decondensation at sites of DSBs.
(A) Representative images of mCherry-FokI loci in various transcriptional states with or without DSB induction. Numbers in bottom panels represent percentage of cells with the YFP-MS2 accumulation seen in the image + SEM. Insets in top panels are equivalent magnifications. Bar, 10 μM.
(B) Quantification of mCherry-FokI locus size from experiments as in (A). Representative cells were imaged for quantification based on YFP-MS2 signal. Bars represent mean + SEM from 3 independent experiments.
(C) Representative images of mCherry-FokI loci in various transcriptional states with or without DSB induction. Numbers in bottom panels represent percentage of cells with the YFP-MS2 accumulation seen in the image ± SEM. Insets in top panels are equivalent magnifications. Bar, 10 μM.
(D) Quantification of mCherry-FokI locus size from experiments as in (B). Representative cells were imaged for quantification based on YFP-MS2 signal. Bars represent mean + SEM from 3 independent experiments.
See also Figure S4.
Because ATM activity was necessary for RNAPII stalling and DSB induced silencing (Figure 3), we asked if ATM inhibition could restore locus decondensation in a transcription dependent manner. We monitored locus area in FokI WT expressing cohorts in which dox was co-administered with either DMSO or ATMi. We observed a striking 4-5 fold increase in locus area in ATMi treated cells, which was reversed by DRB (Figure 4C and D). Notably, ATMi did not increase locus size in the absence of dox, indicating a specific influence of ATM activity on large-scale chromatin decondensation in transcriptionally active regions.
Association of histone H2A ubiquitylation with ATM-dependent transcriptional silencing at DSBs
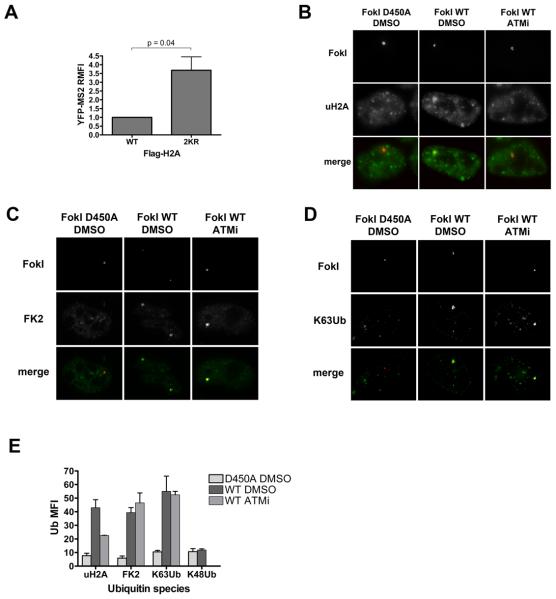
uH2A is closely linked to transcriptional repression (Wang et al., 2004); we therefore reasoned that uH2A might be involved in signaling between DNA damage and transcription responses. Because monoubiquitylation of H2A at lysine 119 is associated with transcriptional repression, we expressed a construct encoding either WT H2A or a version mutated at lysines 119 and 120 to arginines (2KR) in reporter cells along with FokI WT; stable incorporation into chromatin was monitored by IF (Figure S5). Transcription was partially restored in cells expressing the 2KR mutant compared to cells transfected with wild-type H2A (Figure 5A). These constructs had no significant effect on nascent transcript levels in control cells (Figure S5).
Figure 5. ATM-dependent transcriptional silencing is associated with H2A ubiquitylation.
(A) YFP-MS2 RMFI was quantified in cells expressing FokI WT and either Flag-H2A (WT) or a Flag-H2A allele mutated at lysines 119 and 120 to arginines (2KR). Error bars represent SEM from 4 independent experiments.
(B-D) Representative images of uH2A (B), conjugated polyubiquitin (FK2) (C), and K63Ub (D) in the presence of FokI D450A or FokI WT and DMSO or ATMi.
(E) Quantification of Ub MFI from experiments as in B-D. Error bars indicate SEM from three independent experiments.
See also Figure S5.
We next used antibodies against various ubiquitin chain topologies to further dissect the ubiquitin landscape at DSBs. We observed a striking accumulation of uH2A and conjugated polyubiquitin chains (FK2 antibody) at DSBs (Figure 5 B,C). Using antibodies specific for K63 and K48 conjugated polyubiquitin (Newton et al., 2008), we also detected an enrichment of K63Ub, but not K48Ub, consistent with previous reports (Figure 5D) (Doil et al., 2009; Sobhian et al., 2007; Stewart et al., 2009). ATM knockdown reduced the accumulation of either uH2A or K63Ub at FokI DSBs (Figure S5), indicating an ATM dependency for their genesis.
Total (FK2) and K63Ub enrichment at DSBs were both maintained during three hours of ATMi, while uH2A was strongly reduced (Figure 5E). Because ATM was inhibited after the induction of damage and establishment of ubiquitin signals at the locus, these data suggest the maintenance of K63Ub chains at DSBs is less sensitive to ATM inhibition than is uH2A maintenance. RAP80 persistence at DSBs was also insensitive to acute ATMi, consistent with the idea that RAP80 is recruited to K63Ub, rather than to uH2A (Figure S5). Conversely, uH2A levels correlate with transcriptional silencing and partially depend on ATM. These phenomena were not specific to the reporter system or cell type, as we observed similar results at IRIF in HeLa cells after ATMi (Figure S5). These data reveal a varied ubiquitin landscape at sites of damage, with multiple species of ubiquitin signals differing in their function as well as their reliance on different signaling pathways for maintenance.
USP16 is required for acute H2A deubiquitylation and transcriptional derepression at DSBs upon ATM inhibition
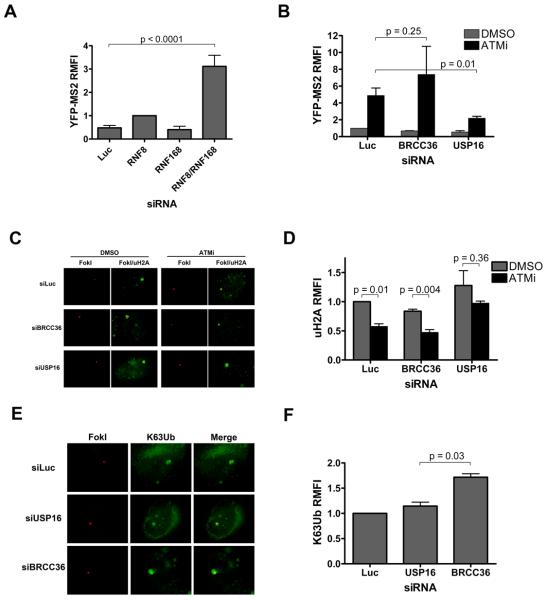
The cooperative E3 ligase activities of RNF8 and RNF168 are required for the synthesis of uH2A and K63Ub chains at sites of damage (Doil et al., 2009; Stewart et al., 2009). To assess possible roles for these E3 ligases in DSB-induced silencing, we analyzed YFP-MS2 intensity at the reporter locus following their depletion with siRNA. Whereas knockdown of either protein alone resulted in only small changes in accumulation of YFP-MS2, co-depletion led to a significant rescue of transcription (Figure 6A, S6). Moreover, expression of a dominant negative RNF8 allele partially reversed silencing (Figure S6). These data are consistent with cooperative roles for these E3 ligases in initiating and stabilizing ubiquitin signals at DSBs.
Figure 6. Opposing regulation of DSB induced transcriptional silencing by E3 ligases RNF8 and RNF168, and the deubiquitylating enzyme USP16.
(A) YFP-MS2 intensity analysis was performed 48-72 hours after transduction of cells with lentivirus expressing FokI WT and transfection with the indicated siRNAs. Error bars represent SEM from at least 3 independent experiments.
(B) YFP-MS2 accumulation was assessed in cells transduced with lentivirus expressing FokI WT and treated with siRNA against luciferase (Luc) or the deubiquitylating enzymes USP16 and BRCC36 (B36). Transcription was induced with 1 μg/ml doxycycline and either vehicle or 10 μM ATMi for 3 hours. Error bars represent SEM from at least 3 independent experiments.
(C) Representative images of uH2A (green) at FokI-induced DSBs (red) in cells transfected with siRNAs targeted to luciferase (Luc), USP16, or BRCC36 (B36) 48-72 hours prior to treatment with vehicle or 10 μM ATMi for 3 hours.
(D) Quantification of RMFI from experiments as in C. Error bars represent SEM from 3 independent experiments.
(E) Representative images of K63Ub at FokI-induced DSBs in cells transfected with the indicated siRNAs 48-72 hours prior to a 3 hour treatment with vehicle or 10 μM ATMi.
(F) Quantification of RMFI from experiments as in E. Error bars represent SEM from 2 independent experiments.
See also Figure S6.
The requirement of ATM for uH2A maintenance at DSBs suggests active deubiquitylation may occur in parallel to uH2A synthesis during the DSB response. A limited siRNA screen of deubiquitylating enzymes (DUBs) included BRCC36, a Zn2+-containing isopeptidase that exhibits K63 specific DUB activity (Cooper et al., 2009; Shao et al., 2009) and USP16, which belongs to a class of DUB enzymes that utilizes an active site cysteine nucleophile to cleave monoubiquitylated uH2A (Joo et al., 2007). Efficient knockdown of both DUBs was documented 48-72 hrs following siRNA transfection (Figure S6). Neither siRNA affected YFP-MS2 accumulation in the presence of DMSO (Figure 6B, grey bars). However, depletion of USP16 prevented ATMi from restoring transcription after DSB induction, while BRCC36 knockdown did not (Figure 6B, black bars). Similar data were obtained with three separate siRNAs directed against USP16, while expression of a siRNA resistant USP16 allele restored transcription after ATMi (Figure S6). Concordant with the maintenance of silencing, Usp16 knockdown prevented the decrease in uH2A at sites of damage upon ATMi (Figure 6C, D). Conversely, whereas USP16 knockdown did not affect K63Ub at sites of damage, depletion of BRCC36 resulted in an increase in K63Ub accumulation, consistent with its reported specificity (Figure 6E,F) (Cooper et al., 2009; Shao et al., 2009).
uH2A deubiquitylation is associated with rapid restoration of transcriptional capacity upon cessation of DNA damage
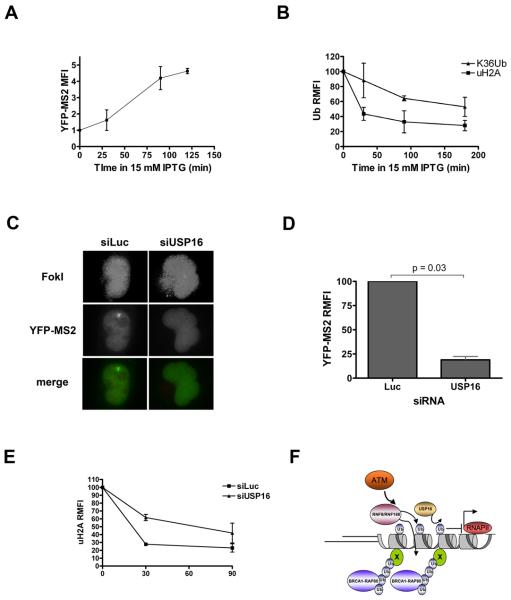
We next asked if repair processes could also reverse DSB associated uH2A and restore transcription. Reporter cells expressing FokI WT were treated with the allolactose analog IPTG (isopropyl-beta-D-thiogalactopyranoside) – which prevents association of the lac repressor with lac operator DNA -- to terminate nuclease action at the locus. IPTG treated cells showed a rise in YFP-MS2 intensity over two hours, indicating restoration of transcription, while both uH2A and K63Ub were noted to decrease at FokI-induced DSBs, notably, with differing kinetics (Figure 7A,B). Given that USP16 DUB activity provided a major contribution to uH2A lability in the presence of ATMi, we postulated that USP16 might be responsible for removal of uH2A during the repair process. This appears to be the case, as USP16 knockdown prevented nascent transcript accumulation and maintained uH2A levels at the locus after IPTG administration (Figure 7C-E). Together, these findings strongly implicate K63Ub and uH2A in distinct functional capacities at DSBs, with K63-Ub representing a recognition platform for various DNA repair complexes, and uH2A enforcing, at least in part, transcriptional silencing at contiguous stretches of chromatin (Figure 7F).
Figure 7. DSB repair leads to rapid transcriptional derepression that is dependent on USP16.
(A) Reporter cells expressing FokI WT were treated with 15 mM IPTG and a time course of nascent transcription was assessed by accumulation of YFP-MS2. Treatments were all timed to end after 3 hours of treatment with 1 μg/ml doxycycline. Error bars represent SD from duplicate experiments.
(B) Loss of DSB associated ubiquitin species was monitored following IPTG treatment by performing IF with antibodies to uH2A or K63Ub at breaks. Because IPTG was used, a mCherry transactivator was used to locate the locus. Error bars represent SD from duplicate experiments.
(C) Reporter cells were transfected with siRNA to luciferase (Luc) or USP16 and transduced with FokI expressing lentivirus 24 hours later. 48 hours after lentivirus transduction, cells were treated with 1 μg doxycycline and 15 mM IPTG for 3 hours. Representative images are shown. FokI images (top) are enhanced to demonstrate pan-nuclear localization upon IPTG treatment.
(D) Quantification of YFP-MS2 accumulation from experiments as in D. Error bars represent SEM from 2 independent experiments.
(E) Loss of uH2A at reporter locus over time in 15mM IPTG in cells expressing FokI WT and treated with control or siRNA against USP16. Because IPTG was used, a mCherry transactivator was used to locate the locus. Error bars represent SEM from 3 independent experiments.
(F) ATM dependent localization of RNF8 and RNF168 to DSBs facilitates K63-ubiquitylation of one or more as yet undefined substrates (X) as well as H2A ubiquitylation (gray cylinders with a single oval ubiquitin). K63Ub attracts the BRCA1-RAP80 complex, while uH2A mediates transcriptional silencing on chromatin spanning multiple kb in cis to the DSB. USP16 deubiquitylates uH2A to restore transcription following ATMi or cessation of damage.
Discussion
A novel system to visualize DSBs and local transcriptional activity reveals Double strand break Induced Silencing in Cis (DISC)
Despite an abundance of imaging based approaches to visualize either IR or laser induced DSBs within the genome, none provide the ability to view concomitant transcriptional responses. Elegant single cell systems have been reported that allow visualization of transcription in real-time (Darzacq et al., 2009; Janicki et al., 2004). Similar methodologies have been cleverly adapted to visualize responses to nuclease-induced breaks (Soutoglou et al., 2007; Soutoglou and Misteli, 2008), providing insights into DSB responses not attainable with other assays.
We have integrated these approaches and adapted a previously described transcriptional reporter array to induce DSBs at a defined genomic location upstream of an inducible promoter. This approach has enabled the discovery of a DSB induced transcriptional silencing program transmitted in cis through chromatin multiple kilobases distal to the site of damage (Fig. 7F). We refer to this phenomenon as DISC (Double strand break Induced Silencing in Cis). Silencing correlates with the spread of chromatin modifications emanating from DSBs on a contiguous stretch of chromatin (Figure 1C). This suggests DSB induced chromatin modifications are transmitted a considerable distance to influence other signaling processes.
Several features of this phenomenon distinguish it from other forms of DNA damage induced transcriptional repression. UV and other bulky adduct-induced DNA damage negatively influence transcription in cis to the DNA lesion – yet this is due to physical blockage, and has not been reported to occur at a distance from DSBs. In addition, these bulky lesions lead to ubiquitylation of RNAPII and subsequent proteasome-mediated degradation (Anindya et al., 2007; Bregman et al., 1996). Consequently, UV damage results in strongly reduced RNAPII after DNA damage. We observed neither a reduction of total RNAPII at FokI induced DSBs nor an accumulation of K48-linked ubiquitin chains, suggesting RNAPII is not targeted for degradation at FokI induced breaks. Moreover, unlike DNA damage from UV sources, IR does not result in RNAPII ubiquitylation (Bregman et al., 1996), suggesting different mechanisms of transcriptional inhibition exist at bulky adducts and DSBs. The reporter used in this study allows for substantial recruitment of RNAPII and therefore quantification by fluorescence imaging. It remains to be seen whether all promoters will recapitulate exactly the changes reported here for total and phospho-RNAPII.
IR induced DSBs have been reported to inhibit RNA Pol I dependent transcription of ribosomal RNA in the nucleolus (Kruhlak et al., 2007). Although dependent on ATM, this phenomenon also fundamentally differs from DISC. Low dose IR shut down RNA Pol I transcription in the nucleolus, but did not have an influence on RNAPII driven transcription at the YFP-MS2 reporter locus (Figure 2A). Moreover, RNAPI is not subject to the regulatory CTD phosphorylations that are perturbed in DSB-induced RNAPII stalling (Phatnani and Greenleaf, 2006). Together, these observations suggest the phenomenon described here defines a novel transcriptional silencing program.
ATM prevents large-scale chromatin decondensation at transcriptionally active loci in cis to DSBs
An ATM dependent decrease in actively elongating RNAPII was observed at the transcriptional reporter in the presence of DSBs (Figure 3E,F). Consistent with RNAPII stalling, DSBs prevented transcription dependent chromatin decondensation, while ATM inhibition restored both transcription and associated chromatin decompaction at sites multiple kilobases from DSBs. Notably, neither DSB induction nor ATM caused a discernable change to locus area in the absence of transcriptional induction. This differential impact of ATM in transcriptionally active and inactive regions is a novel observation that may relate to the known role of ATM in preventing chromosomal translocations. Physiological DNA breaks can activate global transcriptional networks, as well as global chromatin relaxation, in an ATM-dependent manner (Bredemeyer et al., 2008; Ziv et al., 2006). With the aid of single cell analysis, we now describe a novel role for this kinase in modulating local chromatin architecture and transcriptional activity.
ATM dependent transcriptional silencing may prevent chromatin movement surrounding breaks, thus maintaining proximity of broken chromosomal termini. While future investigations are clearly required to reach such a conclusion, a correlation exists between transcriptional deregulation and the propensity for translocation (Mathas et al., 2009). Indeed, transcriptional activation by androgen receptor has recently been reported to promote translocations between one of its target genes and non-contiguous chromosomal sites (Lin et al., 2009) . Notably, ATM deficiency dramatically enhanced translocation incidence in this study.
ATM directed ubiquitin signals regulate transcriptional silencing at DSBs
We have observed an ATM kinase-mediated transcriptional silencing at DSBs that correlates with ubiquitylation of histone H2A (uH2A). Several independent pieces of data suggest a functional relationship between uH2A levels and DISC: (i) expression of an H2A allele that cannot be monoubiquitylated at lysine 119 partially rescues transcription (Figure 5A), (ii) inhibition of the known H2A E3 ubiquitin ligase RNF8 and the related RNF168 partially reverses silencing (Figure 6A,B), and (iii) depletion of the uH2A deubiquitylating enzyme USP16 prevents the reversal of silencing and diminution of uH2A at DSBs upon ATMi or cessation of damage (Figures 6,7).
uH2A accumulation at DSBs is dependent on the E3 ubiquitin ligases RNF8 and RNF168 . This mark has been primarily associated with K63Ub chains and recruitment of damage response proteins to chromatin. Our data, however, indicate multiple ubiquitin species exist at DSBs, with different functions and relative requirements on ATM for their maintenance (Figures 5E, S5), suggesting they, at least in part, represent unique signals. A complex ubiquitin landscape therefore appears to exist at sites of damage (Messick and Greenberg, 2009), with K63Ub being best correlated with DSB recognition, and uH2A more closely linked to DISC. This may be necessary to modulate parallel, yet distinct signaling pathways in the vicinity of DNA damage.
A potential role for damage induced silencing in heritable chromatin change
Cancer genomes demonstrate a greater degree of plasticity than normal somatic cells due to the inherent genomic instability present in most tumors. Cancer epigenomes also display alterations (Feinberg and Vogelstein, 1983), although mechanisms underlying epigenetic alterations are unclear. While DSB induced silencing was reversible following termination of nuclease action, it is possible that at some subset of DSBs, ubiquitin-mediated transcriptional silencing is converted to a long-term, heritable silencing through the formation of additional repressive modifications such as CpG island methylation.
The DUB USP16 was necessary for removal of uH2A at DSBs and reversal of silencing upon ATMi or IPTG treatments (Figures 6 and 7). In principle, USP16 deficiency could predispose genes within kilobases of a repaired DSB to stable and heritable silencing. Interestingly, biallelic mutations in UTX, a histone H3K27 demethylase, occur in 3% of a wide range of tumor samples (van Haaften et al., 2009). As with uH2A, H3K27 methylation is associated with stable gene silencing and has been reported to transiently accumulate at DSBs (O'Hagan et al., 2008).
DSBs are prevalent in all stages of malignancy as evidenced by elevated γH2AX and ATM dependent signaling events in human tumors and premalignant lesions (Bartkova et al., 2005; Gorgoulis et al., 2005). A general model of chromatin dynamics at DNA damage sites predicts nucleosomal clearing is necessary to facilitate DSB repair protein access to DNA damage. Following repair, nucleosome structure would be restored. This “Break-Access-Repair-Restore” model, coupled with the knowledge that numerous post-translational modifications transiently develop on chromatin flanking DSBs, creates a conceptual framework for the genesis of epigenetic change emanating from DNA repair (Green and Almouzni, 2002; Kastan et al., 1982).
Transcriptional silencing has also been observed at DNA structures resembling DSBs, such as telomeric heterochromatin and episomal viral genomes (Gottschling et al., 1990; Knipe and Cliffe, 2008). Thus, one wonders whether DSB induced silencing mechanisms have evolved to prevent transcription at a host of DNA structures in native and foreign genomes. Clearly, more work will be necessary prior to reaching any conclusions. Given the arsenal of small molecules that inhibit either DNA repair or chromatin modifying enzymes, and the reversible nature of chromatin modification, this area of investigation opens up new possibilities to explore connections between DSB repair and epigenetic alterations in a broad range of normal and pathophysiologic states.
Experimental Procedures
Cells
Reporter cells were cultured in DMEM (GIBCO) containing 10% Tet-system approved FBS (Clontech), 1% Penicillin/Streptomycin, 200 mg/ml G418, and 100 mg/ml hygromycin B. 293T cells were used to produce FokI WT and D450A containing lentivirus.
Immunofluorescence
IF was performed as previously described (Sobhian et al., 2007). A complete list of antibodies and dilutions used can be found in Supplementary Materials.
Image capture and analysis
Following immunostaining, 8-bit gray scale images of approximately 30-100 cells per condition were captured with a QImaging RETIGA-SRV camera connected to a Nikon Eclipse 80i microscope driven by ImagePro 6.2 software. All images for a given experiment were captured on the same day with the same exposure times, calculated to avoid saturation. Image analyses were performed using ImageJ software from the NIH. For locus area analyses, the freehand selection tool was used to analyze mCherryFokI (WT or D450A) areas in representative cells selected based on YFP-MS2 intensity. For intensity analyses, transfected cells were chosen at random and the elliptical tool was used to select a region of interest for which the mean fluorescence intensity was measured. For each cell, a separate region within the nucleus was measured for background subtraction. Some images were enhanced using a nearest neighbors deconvolution algorithm (ImagePro 6.2 software). The same deconvolution settings were used for each image within a given experiment. All quantification was performed on unprocessed images. Grayscale images were overlayed and pseudocolored with Adobe Photoshop CS4 software.
Transfections
Transient transfections were performed with LipoD293 (Signagen), and siRNA transfections with Lipofectamine RNAiMax (Invitrogen) according to manufacturer's protocols. Analyses were performed 12-48 hours after transient transfection of plasmids, 48-72 hours after siRNA transfection, and 36-72 hours after lentivirus transduction. A complete list of siRNA sequences used can be found in Supplementary Materials.
Chromatin Immunoprecipitation
ChIP analyses were carried out as described in Supplemental Materials. Briefly, cells were crosslinked 10′ at RT with 1% formaldehyde. Sample were precleared, then immunoprecipitated with 10 μg antibody overnight. Antibody information and primer sequences can be found in Supplementary Materials.
Southern Blot
Southern blot analyses were performed with a DIG labeling and detection kit (Roche). Oligonucleotides indicated by the colored lines in Figure 1H were Dig labeled and Southern analyses carried out per manufacturer's instructions.
Statistical analyses
Graphs were created and statistical analyses performed using Prism software (GraphPad). For comparative analyses in which one data series was normalized to 1, a one-sample t-test was performed on the other sample with a hypothetical mean set to 1. For all other comparisons, a Student's unpaired two-tailed t-test was performed without assuming equal variances.
Supplementary Material
Acknowledgements
We thank Genentech for providing linkage specific antibodies; E. Brown, B. Johnson, G. Blobel, M. Lampson, and members of the Greenberg lab for critical discussion. RAG gratefully acknowledges funding from: 1R01CA138835-01 from the NCI, An ACS Research Scholar Grant, K08 awards 1K08CA106597-01 and 3K08CA106597-04S1 from the NCI, the Sidney Kimmel Foundation Scholar Award, and funds from the Abramson Family Cancer Research Institute, and from the Penn Genomes Frontiers Institute. NMS was supported by training grant 1-T32-CA-115299-01A2.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anindya R, Aygun O, Svejstrup JQ. Damage-induced ubiquitylation of human RNA polymerase II by the ubiquitin ligase Nedd4, but not Cockayne syndrome proteins or BRCA1. Mol Cell. 2007;28:386–397. doi: 10.1016/j.molcel.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- Bredemeyer AL, Helmink BA, Innes CL, Calderon B, McGinnis LM, Mahowald GK, Gapud EJ, Walker LM, Collins JB, Weaver BK, et al. DNA double-strand breaks activate a multi-functional genetic program in developing lymphocytes. Nature. 2008;456:819–823. doi: 10.1038/nature07392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregman DB, Halaban R, van Gool AJ, Henning KA, Friedberg EC, Warren SL. UV-induced ubiquitination of RNA polymerase II: a novel modification deficient in Cockayne syndrome cells. Proc Natl Acad Sci U S A. 1996;93:11586–11590. doi: 10.1073/pnas.93.21.11586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper EM, Cutcliffe C, Kristiansen TZ, Pandey A, Pickart CM, Cohen RE. K63-specific deubiquitination by two JAMM/MPN+ complexes: BRISC-associated Brcc36 and proteasomal Poh1. Embo J. 2009;28:621–631. doi: 10.1038/emboj.2009.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Yao J, Larson DR, Causse SZ, Bosanac L, de Turris V, Ruda VM, Lionnet T, Zenklusen D, Guglielmi B, et al. Imaging transcription in living cells. Annu Rev Biophys. 2009;38:173–196. doi: 10.1146/annurev.biophys.050708.133728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, Jeggo PA. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Jr., Kastrinakis NG, Levy B, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- Green CM, Almouzni G. When repair meets chromatin. First in series on chromatin dynamics. EMBO Rep. 2002;3:28–33. doi: 10.1093/embo-reports/kvf005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, et al. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo HY, Zhai L, Yang C, Nie S, Erdjument-Bromage H, Tempst P, Chang C, Wang H. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature. 2007;449:1068–1072. doi: 10.1038/nature06256. [DOI] [PubMed] [Google Scholar]
- Kastan MB, Gowans BJ, Lieberman MW. Methylation of deoxycytidine incorporated by excision-repair synthesis of DNA. Cell. 1982;30:509–516. doi: 10.1016/0092-8674(82)90248-3. [DOI] [PubMed] [Google Scholar]
- Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;316:1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- Kim ST, Lim DS, Canman CE, Kastan MB. Substrate specificities and identification of putative substrates of ATM kinase family members [In Process Citation] J Biol Chem. 1999;274:37538–37543. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- Knipe DM, Cliffe A. Chromatin control of herpes simplex virus lytic and latent infection. Nature reviews. 2008;6:211–221. doi: 10.1038/nrmicro1794. [DOI] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science (New York, NY. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak M, Crouch EE, Orlov M, Montano C, Gorski SA, Nussenzweig A, Misteli T, Phair RD, Casellas R. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature. 2007;447:730–734. doi: 10.1038/nature05842. [DOI] [PubMed] [Google Scholar]
- Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol. 2008;9:759–769. doi: 10.1038/nrm2514. [DOI] [PubMed] [Google Scholar]
- Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, Zhang J, Rose DW, Fu XD, Glass CK, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- Mathas S, Kreher S, Meaburn KJ, Johrens K, Lamprecht B, Assaf C, Sterry W, Kadin ME, Daibata M, Joos S, et al. Gene deregulation and spatial genome reorganization near breakpoints prior to formation of translocations in anaplastic large cell lymphoma. Proc Natl Acad Sci U S A. 2009;106:5831–5836. doi: 10.1073/pnas.0900912106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Messick TE, Greenberg RA. The ubiquitin landscape at DNA double-strand breaks. J Cell Biol. 2009;187:319–326. doi: 10.1083/jcb.200908074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller WG, Walker D, Hager GL, McNally JG. Large-scale chromatin decondensation and recondensation regulated by transcription from a natural promoter. J Cell Biol. 2001;154:33–48. doi: 10.1083/jcb.200011069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton K, Matsumoto ML, Wertz IE, Kirkpatrick DS, Lill JR, Tan J, Dugger D, Gordon N, Sidhu SS, Fellouse FA, et al. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 2008;134:668–678. doi: 10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]
- O'Brien T, Hardin S, Greenleaf A, Lis JT. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature. 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- O'Hagan HM, Mohammad HP, Baylin SB. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet. 2008;4:e1000155. doi: 10.1371/journal.pgen.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Shao G, Lilli DR, Patterson-Fortin J, Coleman KA, Morrissey DE, Greenberg RA. The Rap80-BRCC36 de-ubiquitinating enzyme complex antagonizes RNF8-Ubc13-dependent ubiquitination events at DNA double strand breaks. Proc Natl Acad Sci U S A. 2009;106:3166–3171. doi: 10.1073/pnas.0807485106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff R, Arbel-Eden A, Pilch D, Ira G, Bonner WM, Petrini JH, Haber JE, Lichten M. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol. 2004;14:1703–1711. doi: 10.1016/j.cub.2004.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, Ried T, Danuser G, Misteli T. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol. 2007;9:675–682. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E, Misteli T. Activation of the cellular DNA damage response in the absence of DNA lesions. Science (New York, NY. 2008;320:1507–1510. doi: 10.1126/science.1159051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–434. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, Edkins S, Hardy C, O'Meara S, Teague J, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Elledge SJ. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci U S A. 2007;104:20759–20763. doi: 10.1073/pnas.0710061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- Waugh DS, Sauer RT. Single amino acid substitutions uncouple the DNA binding and strand scission activities of Fok I endonuclease. Proc Natl Acad Sci U S A. 1993;90:9596–9600. doi: 10.1073/pnas.90.20.9596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Transcriptional regulation by histone ubiquitination and deubiquitination. Genes Dev. 2003;17:2733–2740. doi: 10.1101/gad.1156403. [DOI] [PubMed] [Google Scholar]
- Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, Lukas J, Bekker-Jensen S, Bartek J, Shiloh Y. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol. 2006;8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.