Abstract
This paper proposes a method to match diffusion tensor magnetic resonance images (DT-MRI) through the large deformation diffeomorphic metric mapping of tensor fields on the image volume, resulting in optimizing for geodesics on the space of diffeomorphisms connecting two diffusion tensor images. A coarse to fine multi-resolution and multi-kernel-width scheme is detailed, to reduce both ambiguities and computation load. This is illustrated by numerical experiments on DT-MRI brain and images.
1. Introduction
Diffusion Tensor Magnetic Resonance Imaging (DT-MRI) is a non-invasive technique that measures the diffusion of water molecules in biological tissues [16, 14, 20, 9]. Diffusion refers to the random translational movement of molecules (also called Brownian motion). The rate and direction of the movement is closely related to the structural anisotropy of the medium, hence it can be used to infer the tissue micro structure. Diffusion is described mathematically by a symmetric second order tensor. Orientation of the principal eigenvector of the diffusion tensor is known to align with fiber tracts [16, 20]. DT-MRI is rapidly gaining importance in neuroscience and other fields but tools for exploring and analyzing DT-MRI data are still not widely available.
Image registration is inevitable whenever images acquired from different subjects, at different times, or from different scanners need to be combined or compared for analysis or visualization. Alexander and Gee [1] adapted the multi-resolution elastic matching algorithm using tensor similarity measures for matching DT-MRIs. Tensor reorientation is not included in the energy function, but tensors are reoriented in each iteration according to the estimated displacement field. Ruiz-Alzola [19] proposed a unified framework for registration of medical volumetric multi-valued data by matching local areas with high structure. Local structure is detected by measuring “cornerness”. Then the Kriging estimator was used to interpolate the displacement field in the remaining areas. Tensor reorientation is performed after the registration. Guimond et al. [11] proposed a deformable registration method using tensor characteristics that are invariant to rigid transformations to avoid the complexity of tensor reorientation. The invariants used in their experiments are the tensor eigenvalues. Rohde et al. [18] proposed an intensity based registration method capable of performing affine and nonlinear registration of multi-channel images. The multi-channel similarity measure considered both the similarity between same-index image channels and the similarity between image channels with different index. But tensor reorientation is performed after affine registration and after the nonlinear registration is complete. Zhang et al [21] proposed a piecewise local affine registration algorithm. Leemans et al [12] proposed an affine multi-channel matching method for DT-MRIs based on mutual information. The Diffusion Weighted Images was used as the channels.
This paper proposes a method to match diffusion tensor magnetic resonance images through an extension of the Large Deformation Diffeomorphic Metric Mapping (LD-DMM) framework [5] to this type of dataset, resulting in optimizing for geodesics on the space of diffeomorphisms connecting two tensor fields. In [7], LDDMM framework was extended to matching vector fields. In the paper, we extends it further to the full tensor fields. Establishing this methodology continues our efforts on establishing the construction and computation of metric spaces for studying anatomical structures. Our method includes tensor reorientation in the optimization, hence the measure of difference is calculated correctly during the optimization. Convergence of the algorithm is guaranteed.
2. Formulation of the Problem
In our approach, anatomy is modeled as a deformable template. The images are defined on an open bounded set and are viewed as the orbit under the diffeomorphic transformations G acting on a template. In detail, let the background space Ω be a bounded domain in Rd and G a group of diffeomorphisms on Ω. Let the images be functions M: Ω → Rd×d that associate to each point x ∈ Ω the diffusion tensor, a symmetric second order tensor. G acts on the set I of all images.
Elements in G are defined through the solutions of the nonlinear Eulerian transport equation
| (1) |
For each t, vt is a vector field on Ω which belongs to some Hilbert space V. The meaning of this equation is that we have a path on the space of transformations ϕt, where t is the time. Under the action of this flow, a particle which starts at a point x(0), traces out a path x(t) = ϕt(x(0)), where ẋ(t) = vt(x(t)), vt is the speed at time t.
For any M ∈ I and ϕ ∈ G, M can be decomposed as , where λ1, λ2 and λ3 are 3 eigenvalues, λ1 ≥ λ2 ≥ λ3, and e1, e2 and e3 are the corresponding eigenvectors of M. The action is defined as follows [2]:
| (2) |
where
| (3) |
| (4) |
| (5) |
Remark 1
This is the Gram-Schmidt ortho-normalization.
Here in Equation (2) to (5) everything depends on x and Dϕ denotes the Jacobian matrix of the function ϕ. In summary, the eigenvector deformation strategy assumes that the eigenvalues of the diffusion tensor do not change, the principal eigenvector changes according to the Jacobian, but preserve the length, i.e. e1 changes to Dϕ e1/||Dϕ e1||. The plane generated by the first and the second principal vectors e1 and e2 changes to the plane generated by Dϕ e1 and Dϕ e2. Although the exact mechanism for diffusion anisotropy is not well understood, it is clear that this anisotropy directly reflects the presence of spatially oriented structures in the tissue [6]. Assume the tissue microstructure doesn’t change during the deformation, then the shape of the diffusion tensor should not change, hence we assume the eigenvalues of the diffusion tensor do not change. The eigenvectors reflect the orientation of the tissue, so they will be affected by the deformation.
It can be shown that the action is well-defined, i.e., the choice of the eigenvectors of M doesn’t affect the value of ϕ.M. It can also be shown that the action defined above is a group action, i.e., ϕ.(ψ.M) = (ϕ○ψ).M, using the chain rule of differentiation and the fact that a Jacobian acts linearly.
Let the notation denote the composition . Then is the solution at time t of the equation with initial condition ys = x. The function is called the flow associated to v starting at s. It is defined on [0, 1] × Ω. In physical terms, is the motion of a particle which starts at x at time s. For s, r, t ∈ [0, 1], we have . This equation states that the movement from s to t is the same as the movements from s to r, and then from r to t.
Given two elements template Mtemp and target Mtarg, we would like to find an optimal matching ϕ in the space of diffeomorphisms between Mtemp and Mtarg which minimizes a distance function dist(ϕ.Mtemp, Mtarg) on the tensor fields. We use the Frobenius or Hilbert-Schmidt matrix norm,
| (6) |
| (7) |
where the notation A* denotes the conjugate transpose of A. The mapping ϕ is generated as the end-point ϕ = ϕ1 = ϕ01 of the flow of smooth time-dependent vector fields vt ∈ V via Equation (1).
Problem Statement
Estimation of the optimal diffeomorphic transformation connecting two images of tensor fields Mtemp and Mtarg is done via the following variational problem:
| (8) |
| (9) |
where ||·||V is an appropriate functional norm on the velocity field vt(·).
Remark 2
The Frobenius or Hilbert-Schmidt norm we have used to compare the tensor can be related to a noise model in which matrix coefficients are subject to additive independent Gaussian noise. From a metric point of view several authors [8, 15, 4] have recently introduced a specific affine invariant distance designed for the comparison of symmetric tensors. The squared distance between two tensors M1 and M2 is given by the sum of square of the logarithm of the eigenvalues of . Even if a direct use of this distance is not feasible here, a first order approximation of it, namely
| (10) |
(where I is the identity matrix) can be used with slight modification of the algorithm (will be introduced later) in our framework.
3. First order variation
To solve the variational problem (8), we need to compute the first order variation of the energy function with respect to the vector field v.
Definition
Let v and h be time-dependent vector fields, t ∈ [0, 1], such that for each t, vt, ht ∈ V and . For a function f(v), we define
| (11) |
Here ∂hfv is called the Gâteaux differential of f at v with increment h and ∇v f is called the Fréchet derivative of f [13].
We construct the Hilbert space V using the theory of reproducing kernel Hilbert spaces (RKHS) [3]. The Hilbert space V is defined as follows [7]:
| (12) |
where K̃ is continuous, positive definite and has compact support in Ω. The inner product on V is defined as <v, v′>V =<u, u′>L2. V is a RKHS with reproducing kernel K(x, y) = ∫ΩK̃ (x, z)K̃(z, y)dz. To take advantage of the Fourier transform in the calculations related to the kernel, we define in our experiments, K̃(x, y) = η(x)K1(x, y)η(y)|Ω, where K1 is a Gaussian function and η(x) is a function which smoothes K1 near the boundary and has zero boundary conditions.
Let
| (13) |
| (14) |
where wt1, wt2 and wt3 are transformed eigenvectors of e01, e02 and e03 under the action of . Then
| (15) |
Here we denote Kx(y) = K(y, x) and K(α) the α-th column of K. See appendix for the details on calculations.
Remark 3
In our definition of the Hilbert space V, the element v is defined by means of L2 functions u. In our experiments, we find it is more efficient to work on u directly. We define
| (16) |
So, for hα(x) ∈ L2(Ω, Rd), we have
| (17) |
| (18) |
| (19) |
4. Numerical implementation
We extend the method used in [7] for the numerical implementation of the estimation of the optimal vector field v. Let Ω = [0, 1]2/[0, 1]3 be the 2D/3D background space. Ω is discretized into a uniform square/cube mesh. The time space is discretized as tn = nΔt, n = 0, 1, …, N and . Let denote the value of f at xj, in time step tn for kth iteration.
4.1. Gradient Descent Scheme Based Optimization
The variational optimization of the energy functional is performed in a steepest descent scheme
| (20) |
where (∇uE)t is computed using Equation (19). The derivatives are calculated via central differences with enforcement that Dxf (x) = 0 if the function f is a function of I0 and f(x) = 0. The integrations are calculated by Fourier transformation since they can be viewed as convolutions. We use the Gaussian kernel functions as K1 in our experiments. Other kernel functions can be used. The step size ε is decided by the golden section line search algorithm [17] in the direction of steepest descent. We use a second order semi-Lagrangian scheme [10] to integrate the velocity field v to generate the map ϕt0 and ϕt1.
4.2. Hierarchical Multi-resolution Multi-kernel- width Matching Strategy
A hierarchical multi-resolution matching strategy is used to reduce ambiguity issues and computation load. It is employed from coarse to fine, and results achieved on one resolution are considered as approximations for the next finer level. We generate the image pyramid by reducing the resolution from one level to the next by a factor of 2 using linear interpolation.
The width of the kernel significantly affects the deformation. When the width of the kernel is large, the deformation is smoother but the constraints on the deformation will also be stronger, hence the template may not have enough freedom to deform to the target, but it is also not easily trapped into the local minimum. When the width of the kernel is small, the template deforms more freely, hence the local matching will be better, but it may also be easily trapped into the local minimum. Hence, at the early stage, we should use bigger width to get a rough matching, then at later stage, use smaller width to refine the matching.
If we change the width of the kernel, the vector space V also changes. We need to find the relationship between two vector spaces with different kernel width. This is a difficult problem for general kernels, but with Gaussian kernels, we have a simple solution due to their special property. Recall that the vector space V is defined as follows:
| (21) |
where K̃(x, y) = η(x)K1(x, y) η(y)|Ω, K1 is a Gaussian function and η(x) is a function which smoothes K1 near the boundary and has zero boundary conditions.
Suppose V1 and V2 are two vector spaces with kernel size σ1 and σ2, σ1 > σ2 > 0. Then , and , where Z1 and Z2 are constants to make the integration of the kernel be 1.
| (22) |
| (23) |
| (24) |
| (25) |
with
| (26) |
| (27) |
Equation (27) provides the transition from u1 to u2 between the spaces V1 and V2.
4.3. The Matching Algorithm
At a given resolution and given kernel width:
1) Initialize the algorithm with k = 0, , j = 0, 1, …, N or any initial guess.
For each iteration k,
2) Calculate ϕtj0 and ϕtj1 using the second order semi-Lagrangian scheme [10].
3) Calculate the new gradient ∇uk E using Equation (19).
4) Calculate the new velocity uk+1 = uk − ε∇ukE. Find the optimal step size ε using golden section line search algorithm [17] in the direction of steepest descent. If ε < TOL, stop the iteration, otherwise, return to step 2).
The multi-resolution matching algorithm is as follows: (1.) Rigid matching. (2.) Generate multi-resolution image pyramid by reducing the resolution from one level to the next by a factor of 2. (3.) Pick an initial kernel width, from the coarsest level to the finest level, do the following: i). Use the fixed resolution matching algorithm to find the optimal vector field u for that level. ii). Use linear interpolation to convert the optimal vector field u from one level to the next finer level. (4.) Reduce the kernel width, convert the optimal vector field u to the new space using Equation (27). (5.) Use the fixed resolution matching algorithm to find the optimal vector field u for the new kernel at the finest resolution. (6.) Calculate the corresponding v and ϕ01 and ϕ10.
5. Experimental Results and Discussions
The algorithm is implemented in C++ for 3D tensor field images. In our experiments, the time interval of the flow is discretized into 20 steps, with step size Δt = 0.05. We use c = 5. We first use 0.075 and then 0.025 as the standard deviation of the Gaussian kernel with the grid normalized to [0, 1]3.
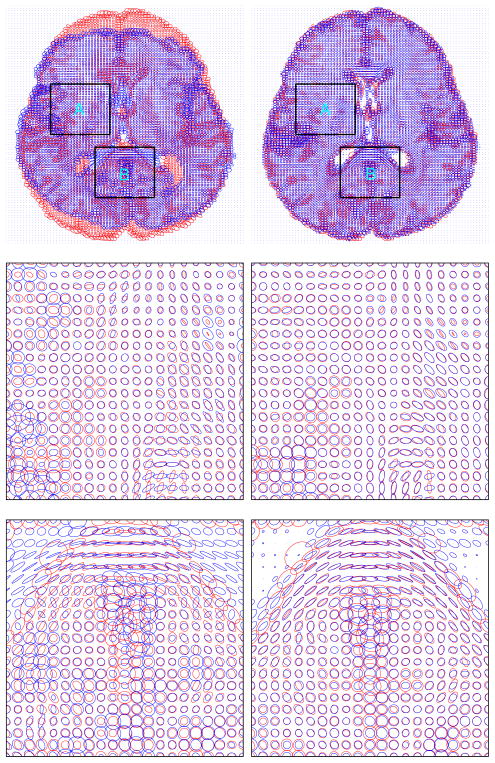
We have done experiments on various normal human brain diffusion tensor images. Figure 1 shows the tensor matching result for one pair of human diffusion tensor brain images in detail. We can see from the figures that the alignment of the diffusion tensor at each voxel improved a lot after the matching.
Figure 1.
3D tensor matching of two normal human brains. Left column shows the tensor distribution of slice 30 before matching; Right column shows the tensor distribution of slice 30 after matching. Template and target are superimposed with blue color the template and red color the target. First row shows the whole slice. Second row and third row shows the enlargement of region A and region B respectively.
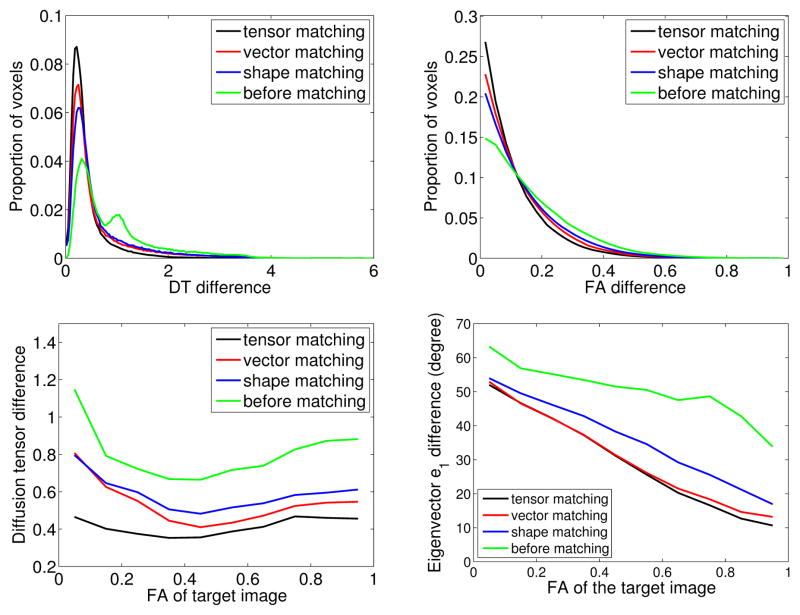
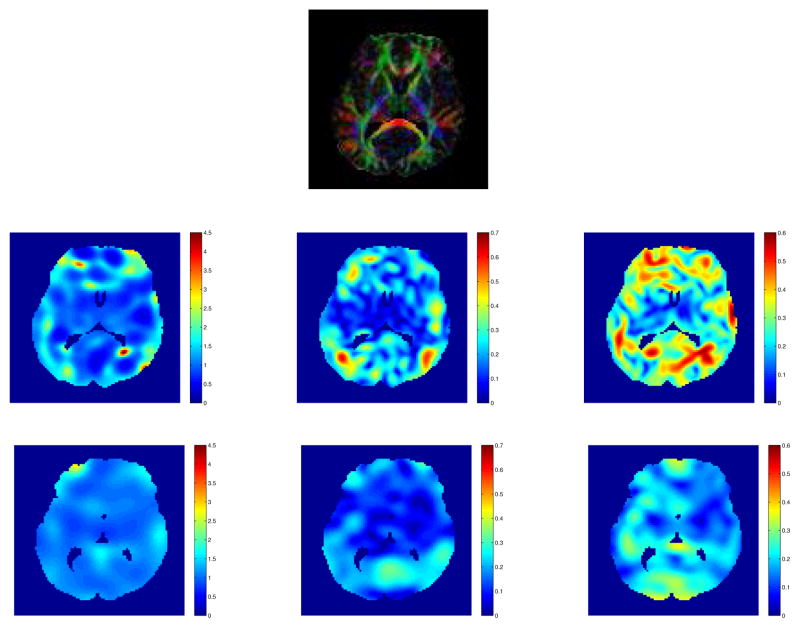
Figure 2 shows the comparison of several matching methods on one pair of normal human brain diffusion tensor images. The shape matching method refers to matching the geometrical shape of the two brains. The vector matching method refers to matching the principal eigenvector of the diffusion tensor at each voxel. All the methods are implemented under the LDDMM framework. It is clear that tensor matching is better than vector matching and vector matching is better than shape matching. This result is as expected since the tensor matching method uses the most information from the data and the shape matching method the least information among the three methods. Figure 3 shows the comparison of the deformations result from LD-DMM vector matching and tensor matching schemes. For the vector matching scheme, the deformation in areas with low fractional anisotropy (FA) values is very large since the principal directions of the tensor in those areas are not clear and kind of random. Forcing matching them causes unnecessary deformations. For the tensor matching scheme, the deformation in areas with low FA values is not forced. From Figure 2 and Figure 3, we can see that tensormatching scheme improves the matching quality in areas with low FA values. In the areas with high FA values, the two schemes produce similar results.
Figure 2.
Comparison of the deformed template and the target for different LDDMM matching schemes. Top left panel shows the histogram of the tensor difference (Frobenius norm) at each voxel. Top right panel shows the histogram of the FA difference at each voxel. Bottom left panel shows the mean diffusion tensor difference (Frobenius norm) between corresponding voxels as function of FA value. Bottom right panel shows the mean difference between corresponding principal eigenvectors as function of FA value.
Figure 3.
Comparison of the deformations result from LDDMM vector matching and tensor matching schemes. Top panel shows the FA weighted color-coded orientation map of the target. Second row shows the deformations from vector matching, third row shows the deformation from tensor matching. First column shows the the determinant of the Jacobian matrix.. Second column shows the rotation part of the Jacobian matrix, the rotation angle in degree. Third column shows the normalized difference of the eigenvalues of the Jacobian matrix. Tensor matching scheme improves the matching quality in areas with low FA values.
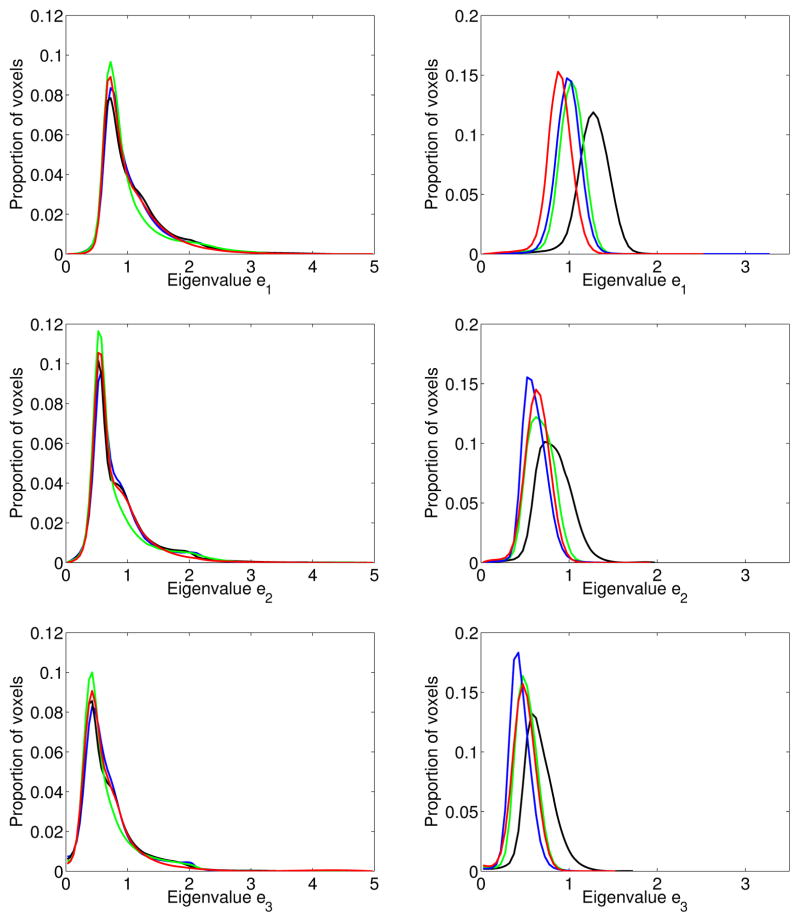
In our tensor matching method, we assume the eigenvalues of the diffusion tensor doesn’t change. This is based on the assumption that the tissue micro-structure doesn’t change during the deformation and different subjects have similar tissue types. Left column of Figure 4 shows the histogram of the 3 eigenvalues of different normal brain diffusion tensor images. From the figure we can see that the distributions for different subjects are very similar. They are peaked at the similar values and have similar ranges. This shows our assumption is reasonable. However, due to changes of conditions, sometimes the eigenvalues did change for different scans. Right column of Figure 4 shows the histogram of the 3 eigenvalues of different normal canine heart diffusion tensor images. The distribution are quite different. This figure shows sometimes it is necessary to normalize the eigenvalues of the tensors before matching. We will investigate that problem in the future.
Figure 4.
Left column shows the histogram of 3 tensor eigenvalues for several normal human brain DT-MRI datasets, right column shows the histogram of 3 tensor eigenvalues for several normal canine heart DT-MRI datasets. Same color means the same dataset. This figure shows sometimes it necessary to normalize the eigenvalues of the tensors before matching.
6. Conclusion
In conclusion, we have presented in this paper the large deformation diffeomorphic metric mapping of tensor fields. The optimal mapping is the endpoint of a geodesic path on the manifold of diffeomorphisms connecting two tensor fields. Finding the optimal mapping and the geodesic path is formulated as a variational problem over a vector field. A gradient descent based multi-resolution multi-kernel-width algorithm is implemented. We did the experiments on various 3D normal brain diffusion tensor magnetic resonance images. This method gives good results when the assumption that the eigenvalues of the diffusion tensor do not change during transformation holds. We expect this method be a useful tool for analysis diffusion tensor magnetic resonance images and other images which have similar properties.
Acknowledgments
This work was supported in part by NIH grant P41-RR15241, NIH grant P50 MH71616, NIH 1U24RR0211382-01, NIH grant 1-R01-HL70894 and the Falk Medical Trust.
A. Calculation of First Order Variation
To solve the variational problem (8), we need to compute the first order variation of the energy function with respect to the vector field v.
Let
| (28) |
| (29) |
where wt1, wt2 and wt3 are transformed eigenvectors of e01, e02 and e03 under the action of . Let v and h be time-dependent vector fields, t ∈ [0, 1], such that for each t, vt, ht ∈ V and .
| (30) |
| (31) |
Equation (31) defines the function f. Let g = id + εf, then g−1 = id − εf + o(ε) and Dg = id + εDf.
| (32) |
where and are transformed eigenvectors of wt1, wt2 and wt3 under the action of g. It is easy to show that
| (33) |
| (34) |
then
| (35) |
| (36) |
| (37) |
Lemma A.1
Let v and h in χ(Ω), then for x ∈ Ω,
This lemma is proved in [5], but stated in a different way. Using this lemma, we can prove
| (38) |
| (39) |
Let
| (40) |
We have
| (41) |
| (42) |
| (43) |
| (44) |
Suppose K is the self-reproducing kernel of V, then K is a d × d matrix. We denote Kx(y) = K(y, x). Let K(α) be the α-th column of K. We have
| (45) |
| (46) |
then
| (47) |
So
| (48) |
Contributor Information
Yan Cao, Email: yan@cis.jhu.edu, Center for Imaging Science, Johns Hopkins University.
Michael I. Miller, Email: mim@cis.jhu.edu, Center for Imaging Science, Johns Hopkins University
Susumu Mori, Email: susumu@mri.jhu.edu, Department of Radiology and Kennedy Krieger Institute, Johns Hopkins University, School of Medicine.
Raimond L. Winslow, Email: rwinslow@bme.jhu.edu, Center for Cardiovascular Bioinformatics & Modeling and the Whitaker Biomedical Engineering Institute, Johns Hopkins University
Laurent Younes, Email: younes@cis.jhu.edu, Center for Imaging Science, Johns Hopkins University.
References
- 1.Alexander DC, Gee JC. Elastic matching of diffusion tensor images. Computer Vision and Image Understanding. 2000;77:233–250. [Google Scholar]
- 2.Alexander DC, Pierpaoli C, Basser P, Gee JC. Spatial transformations of diffusion tensor magnetic resonance images. IEEE TMI. 2001;20(11):1131–1139. doi: 10.1109/42.963816. [DOI] [PubMed] [Google Scholar]
- 3.Aronszajn N. Theory of reproducing kernels. Trans Amer Math Soc. 1950 May;68(3):337–404. [Google Scholar]
- 4.Batchelor PG, Moakher M, Atkinson D, Calamante F, Connelly A. A rigorous framework for diffusion tensor calculus. Magnetic Resonance in Medicine. 2005;53:221–225. doi: 10.1002/mrm.20334. [DOI] [PubMed] [Google Scholar]
- 5.Beg MF, Miller MI, Trouvé A, Younes L. Computing metrics via geodesics on flows of diffeomorphisms. IJCV. 2005;61(2):139–157. [Google Scholar]
- 6.Bihan DL, van Zijl P. From the diffusion coefficient to the diffusion tensor. NMR in Biomedicine. 2002;15:431–434. doi: 10.1002/nbm.798. [DOI] [PubMed] [Google Scholar]
- 7.Cao Y, Miller MI, Winslow RL, Younes L. Large deformation diffeomorphic metric mapping of vector fields. IEEE TMI. 2005;24(9):1216–1230. doi: 10.1109/tmi.2005.853923. [DOI] [PubMed] [Google Scholar]
- 8.Corouge I, Fletcher PT, Joshi S, Gilmore JH, Gerig G. Fiber tract-oriented statistics for quantitative diffusion tensor mri analysis. MICCAI. 2005 doi: 10.1007/11566465_17. [DOI] [PubMed] [Google Scholar]
- 9.Dou J, Tseng WYI, Reese TG, Wedeen VJ. Combined diffusion and strain mri reveals structure and function of human myocardial laminar sheets in vivo. Magnetic Resonance in Medicine. 2003;50:107–113. doi: 10.1002/mrm.10482. [DOI] [PubMed] [Google Scholar]
- 10.Durran DR. Numerical Methods for Wave Equations in Geophysical Fluid Dynamics. Springer; 1999. [Google Scholar]
- 11.Guimond A, Guttmann CRG, Warfield SK, Westin C-F. Deformable registration of dt-mri data based on transformation invariant tensor characteristics. Proceedings of 2002 IEEE International Symposium on Biomedical Imaging; July 2002. [Google Scholar]
- 12.Leemans A, Sijbers J, Backer SD, Vandervliet E, Parizel PM. Affine coregistration of diffusion tensor magnetic resonance images using mutual information. Advanced Concepts for Intelligent Vision Systems: 7th International Conference, ACIVS 2005, LNCS 3708; September 2005.pp. 523–530. [Google Scholar]
- 13.Luenberger DG. Optimization by Vector Space Methods. John Wiley & Sons, Inc; 1969. [Google Scholar]
- 14.Mori S, Barker PB. Diffusion magnetic resonance imaging: Its principle and applications. The Anatomical Record (New Anat) 1999;257:102–109. doi: 10.1002/(SICI)1097-0185(19990615)257:3<102::AID-AR7>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 15.Pennec X, Fillard P, Ayache N. A riemannian framework for tensor computing. IJCV. 2005;66:41–66. doi: 10.1007/11566489_116. [DOI] [PubMed] [Google Scholar]
- 16.Pierpaoli C, Jezzard P, Basser PJ, Barnett A, Chiro GD. Diffusion tensor mr imaging of the human brain. Radiology. 1996;201(3):637–648. doi: 10.1148/radiology.201.3.8939209. [DOI] [PubMed] [Google Scholar]
- 17.Press WH, Teukolsky SA, Vetterling WT, Flannery BP. Numerical Recipes in C, The Art of Scientific Computing. 2. Cambridge University Press; 1992. [Google Scholar]
- 18.Rohde GK, Pajevic S, Pierpaoli C. Multi-channel registration of diffusion tensor images using directional information. Proceedings of 2004 IEEE International Symposium on Biomedical Imaging: From Nano to Macro; April 2004. [Google Scholar]
- 19.Ruiz-Alzola J, Westin CF, Warfield SK, Alberola C, Maier S, Kikinis R. Nonrigid registration of 3d tensor medical data. Medical Image Analysis. 2002;6:143–161. doi: 10.1016/s1361-8415(02)00055-5. [DOI] [PubMed] [Google Scholar]
- 20.Scollan DF, Holmes A, Winslow RL, Forder J. Histological validation of myocardial microstructure obtained from diffusion tensor magnetic resonance imaging. American Journal of Physiology (Heart and Circulatory Physiology) 1998;275:2308–2318. doi: 10.1152/ajpheart.1998.275.6.H2308. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H, Yushkevich PA, Gee JC. Towards diffusion profile image registration. Proceedings of the IEEE International Symposium on Biomedical Imaging (ISBI); 2004. [Google Scholar]