Abstract
More than 70% of eukaryotic proteins are composed of multiple domains. However, most studies of the search for DNA focus on individual protein domains and do not consider potential cross talk within a multidomain transcription factor. In this study, the molecular features of the DNA search mechanism were explored for two multidomain transcription factors: human Pax6 and Oct-1. Using a simple computational model, we compared a DNA search of multidomain proteins with a search of isolated domains. Furthermore, we studied how manipulating the binding affinity of a single domain to DNA can affect the overall DNA search of the multidomain protein. Tethering the two domains via a flexible linker increases their affinity to the DNA, resulting in a higher propensity for sliding along the DNA, which is more significant for the domain with the weaker DNA-binding affinity. In this case, the domain that binds DNA more tightly anchors the multidomain protein to the DNA and, via the linker, increases the local concentration of the weak DNA-binding domain (DBD). The tethered domains directly exchange between two parallel DNA molecules via a bridged intermediate, where intersegmental transfer is promoted by the weaker DBD. We found that, in general, the relative affinity of the two domains can significantly affect the cross talk between them and thus their overall capability to search DNA efficiently. The results we obtained by examining various multidomain DNA-binding proteins support the necessity of discrepancies between the DNA-binding affinities of the constituent domains.
Introduction
Multidomain proteins consist of two or more domains, which, according to the Structural Classification of Proteins database, are the evolutionary units of proteins. Multidomain proteins account for at least two-thirds of eukaryotic genomes (1,2). A domain can have an independent function or it can contribute to the function of a multidomain protein by cooperating with the other domains. For example, many proteins that are involved in regulating gene expression are multidomain and contain DNA-binding domains (DBDs) that recognize specific DNA sequences. Cross talk is often found between the various domains of multidomain transcription factors. In particular, cooperation between the DBDs of multidomain transcription factors has been reported to be crucial for efficient binding to the DNA promoter, and thus plays an essential role in cellular regulation (3–6). One might ask whether the two domains engage in cross talk while searching the DNA.
It has been known for about four decades that transcription factors can locate their specific binding sites on the DNA about two orders of magnitude faster than would be predicted for a bimolecular reaction guided by three-dimensional (3D) diffusion. On the basis of a study by Berg et al. (7), it is widely accepted that the target search process on DNA is facilitated by combining a 3D diffusion mechanism with other mechanisms, i.e., sliding, hopping, and intersegment transfer, performed in a lower-dimensional space (7–17). In the case of sliding, as was demonstrated by several studies (16,18–20), the protein linearly diffuses in a helical fashion along the phosphate-sugar rails (14,15,21). Hopping involves dissociation of the protein from the DNA followed by brief diffusion in free solution and reassociation a short distance farther along the DNA. Accordingly, hopping can be defined as linear diffusion along the DNA, where the protein is not restricted in terms of location to the major groove. In intersegment transfer, the protein directly transfers from one DNA fragment to another, presumably via a doubly bound intermediate (7,22).
The existence of intersegment transfer, which is known to accelerate the search for a specific target site on DNA (22–24), has been confirmed by a number of in vitro experiments in which the protein was treated globally, obscuring mechanistic details specific to protein regions and domains (19,25,26). An extensive computational study using a coarse-grained model (16) showed that homeodomain proteins jump between two DNA molecules through an intermediate in which the recognition helix of the protein is adsorbed to one DNA fragment while the disordered N-tail, which acts as a subdomain, is adsorbed to the other. Intersegment transfer is facilitated by the disordered tail, whose length and net charge, in combination with the charge distribution pattern along the tail, can promote direct transfer via the fly-casting mechanism (27) and consequently the number of jumping events. An elegant NMR study by the Clore group (28) demonstrated domain-specific kinetic data for translocation between high-affinity binding sites by the Oct-1 multidomain protein. The two DBDs of Oct-1 exchange between their cognate sites at significantly different rates, whereas the intersegment transfer of the whole multidomain occurs via a ternary intermediate in which the domains bridge two different DNA fragments simultaneously. Yet, not much is known about the molecular components that dictate the mechanism and efficiency of intersegment transfer by a multidomain DNA-binding protein.
In this study, we use coarse-grained molecular-dynamics (MD) simulations to characterize DNA search by two multidomain transcription factors: Pax6 and Oct-1. Both of these proteins consist of two helix-turn-helix DBDs connected by a flexible linker of 15 and 24 residues, respectively.
Pax proteins, which contain a conserved 128 amino acid DNA binding paired domain, play critical roles in mammalian development and oncogenesis (29–33). The two independent domains of Pax6 will be distinguished here on the basis of their position along the sequence. Accordingly, the domain located closer to the N-terminal will be referred to as Pax6N and that closer to the C-terminal will be referred to as Pax6C. The crystal structure of a complex containing the Drosophila paired domain with its DNA sequence provides a model for the docking of the N-domain (34), whereas the C-domain does not make any DNA contacts. Yet, for other paired domains, genetic and biochemical studies suggest that Pax6C has important functions and makes DNA contacts, and that both the Pax6C and Pax6N domains are required for efficient binding to promoters (3,4).
The Oct-1 multidomain belongs to the POU family, which contains transcription factors with similar structures: a 75 amino acid POU specific domain, a variable linker of 15–30 residues, and a 60 amino acid POU homeodomain. The specific domain and homeodomain of Oct1 will be referred to as OctS and OctHD, respectively. POU proteins serve as key transcription regulators during mammalian development and control many general cellular processes. The two tethered domains, which fold independently, directly interact with the DNA (5,6), and both are required to achieve high affinity and sequence-specific binding (35–37).
To understand how the individual domains cooperate in nonspecific DNA recognition and contribute to search efficiency and, in particular, intersegment transfer, we performed coarse-grained MD simulations in which nonspecific protein-DNA interactions were modeled solely by electrostatic interactions. We provide the microscopic mechanism and the molecular details of 1D diffusion and intersegment transfer by Pax6 and Oct-1 multidomains. We demonstrate the advantages a multidomain protein may have over a single-domain DNA-binding protein in searching DNA, and which biophysical properties the two domains must have to achieve facilitated search by tethering.
Materials and Methods
Studied proteins
We selected two multidomain proteins, the Pax6 paired domain (PDB code: 6PAX (38)) and Oct-1 (PDB code: 1OCT (39)), for examination in our study of DNA search by multidomain DNA-binding proteins, and used their resolved structures to parameterize the native topology-based Hamiltonians. We also studied three additional variants of Pax6 in which the binding affinity of the Pax6C domain was modified by changing the charge of its seven positively charged residues (R and K residues) to neutral (Pax6°) or by increasing the three positively charged residues in the recognition helix (residues R122, R125, and K131) from the wild-type (WT) value of +1 (Pax6+1) to +1.5 (Pax6+1.5) or +2 (Pax6+2). In the case of WT Oct-1+1, the crystal structure of the human multidomain with a 14 bp DNA was used. The Oct-1+2 variant was generated by increasing the charge of three positively charged residues in Oct-1S (residues R20, R49, and K62) from +1 to +2. Only one of these residues is located in the recognition helix (residue 49). Isolated domains of Pax6 and Oct-1 proteins were studied by eliminating the linkers that connect the Pax6 domains (residues 61–76) and the Oct-1 domains (residues 76–99).
Simulation model
The molecular and dynamic nature of a protein search of DNA was studied using a reduced model that allows sampling of long-timescale processes such as sliding, hopping, 3D diffusion, and intersegment transfer. The DNA was modeled as having three beads per nucleotide, representing phosphate, sugar, and base. A negative point charge was assigned to beads representing the DNA phosphate groups. In the simulations, a 100 bp B-DNA molecule was used to study protein dynamics on a single dsDNA molecule, and two 100 bp B-DNA molecules separated by 60 Å were applied to investigate intersegment transfer.
The protein was represented by a single bead for each residue located at the Cα of that residue. Beads representing charged amino acids (K, R, D, and E) were charged in the model. In contrast to the DNA, which remained in place and rigid throughout the simulations, the protein remained flexible and could undergo folding and unfolding events. We simulated the protein with a native topology-based model corresponding to a perfectly funneled energy landscape where native protein interactions were attractive and all other interactions were repulsive (40). In addition to the native interactions, electrostatic interactions between all charged residues of the protein and the phosphate bead of the DNA were included and were modeled by the Debye-Huckel potential, which accounts for the ionic strength of a solute immersed in aqueous solution (15). The dynamics of each protein were studied at various salt concentrations in the range of 0.01–0.2 M, using a dielectric constant of 40 at the temperature at which the protein was completely folded. We emphasize that the information obtained from the crystal structure was used only to model the protein, and the interface between the protein and DNA was modeled solely by electrostatic and repulsive interactions. Accordingly, our model does not include any bias toward the specific binding mode. More details regarding the simulation and structural classification of protein sliding, hopping, 3D diffusion, and intersegment transfer can be found elsewhere (15,16,41).
Electrostatic contribution to protein-DNA binding energy
Electrostatic interactions play a key role in protein-DNA binding and are relevant to any DNA sequence. Consequently, they dictate nonspecific interactions between proteins and DNA. The electrostatic energy of various molecular systems (e.g., the salt dependence of ligand-DNA binding (42–44)) was successfully estimated using the nonlinear Poisson-Boltzmann equation. In these calculations, the molecular surface was chosen as the boundary between the solute low dielectric and the solvent dielectric. Dong et al. (45) previously demonstrated that the electrostatic interaction energy is sensitive to the choice of dielectric boundary, such that the sign of the energy can be altered from positive to negative when the choice of dielectric boundary is changed from the molecular surface to the van der Waals (vdW) surface (45,46). It has also been found that calculated salt effects are insensitive to the details of the charge distribution (43) and the choice of the dielectric boundary (45,46). Thus, experimental salt effects cannot be used to discriminate between the two choices of the dielectric boundary. However, better agreement between the calculated and experimental effects of charge mutations is obtained when the calculation utilizes vdW-based estimates of the dielectric boundary (45,46).
The contribution of electrostatic interactions between a protein and a DNA to the binding free energy, ΔGelbinding, was calculated as ΔGelbinding = ΔGelcomplex − (ΔGelprot + ΔGelDNA), where ΔGelprot, ΔGelDNA, and ΔGelcomplex are the electrostatic energies of the protein, DNA, and protein-DNA complex, respectively, that result from charging up the solute molecule estimated by solving the Poisson-Boltzmann equation. A negative value of ΔGelbinding indicates that electrostatic interactions are stabilizing for protein-DNA binding. The electrostatic binding energies were evaluated using the Adaptive Poisson-Boltzmann Solver (APBS) software package (47). In all electrostatic calculations, the parameters of charges and bind radii were adopted from the AMBER force field and Bondi radii. The temperature was set to 298.15 K, and the solute and solvent dielectric constants were 4 and 80, respectively. The solvent was modeled with an ionic strength of 150 mM. The dielectric boundary was set to the vdW surface. The calculation began with a coarse grid with a 2.0 Å spacing and then moved to a finer grid with a 1.2 Å spacing, both centered at the geometric center of the solute molecule. The dimensions of all grids were 97 Å×97 Å×97 Å.
Using the scheme described above, we estimated the electrostatic contribution to the binding affinity of each domain of multidomain DNA-binding proteins whose structures in complex with DNA were resolved by either x-ray or NMR. For multidomain DNA binding proteins with more than two domains, data are shown for the two domains that have the most diverse DNA binding affinities (see Fig. 6).
Figure 6.

Electrostatic binding free energy for the N-domain versus the C-domain for several DNA-binding multidomain proteins. The energy was calculated using the nonlinear Poisson-Boltzmann model for each domain with its specific DNA taken from the PDB (the corresponding PDB ID is indicated in the plot). All presented proteins, except for 1a1l and 1aay, are composed of two domains (N- and C-domains). The structures of the proteins are shown in Fig. S7. These results suggest that individual fragments in many multidomain proteins bind DNA with different affinities.
Results and Discussion
Cooperation between domains increases affinity to DNA, and tethering affects the propensity to slide, hop, and 3D diffuse
In this study, we investigated the roles played by various domain combinations in searches of nonspecific DNA sequences by quantifying the molecular characteristics of sliding, hopping, and 3D diffusion undertaken by Pax6 and Oct-1 multidomain transcription factors. For each multidomain protein, the search of a 100 bp B-DNA molecule by the tethered and isolated domains of Pax6C and Oct-1S was simulated at a wide range of salt concentrations using a simple computational model (15,16) in which protein-DNA interactions were represented solely by electrostatic forces. To investigate the effect of the relative affinity with which the two domains bind DNA, we studied the interactions with DNA of the WT domains and of variants in which the affinity of the weaker DBDs (Pax6C and Oct-1S domains) was modulated by increasing the strength of its electrostatic interface with DNA. The WT proteins will be designated Pax6+1 and Oct-1+1 (since the positive charges at the interface are +1 under physiological conditions). The variants in which the Pax6C and Oct-1S domains have a higher affinity for DNA (while the Pax6N and Oct-1HD remain unchanged) will be designated Pax6+1.5, Pax6+2, and Oct-1+2 (since the positive charges of the R and K residues at the protein-DNA interface were increased to 1.5 or 2, respectively). As a control simulation, for Pax6 we also studied a variant without positive charges on the C-domain (designated as Pax6°).
To characterize the configuration of the multidomain proteins during sliding (simulated at a low salt concentration), we compared the distances between all of the protein residues and their closest DNA atoms during sliding with the equivalent distances in the crystal structure of the specific complex. These distances were calculated for the connected and isolated domains of each protein (Fig. 1). A strong similarity between the nonspecific binding modes of the connected and isolated domains is seen, indicating that tethering the two domains via a linker does not perturb nonspecific binding. Furthermore, a reasonable degree of similarity is found between the specific complex and the nonspecific association of the multidomain protein with DNA. Similar observations were previously demonstrated by NMR measurements for HoxD9 (14,19) and by MD studies (15,16) for other DNA-binding proteins, and are a consequence of the sliding being performed while the protein is situated in the major groove and performing a helical motion along the DNA backbone. Yet, some differences between the specific and nonspecific binding modes are clearly expected due to their different affinities, dynamics, and hydration natures.
Figure 1.

Comparison of protein-DNA interfaces for specific and nonspecific binding. The average distance of each residue of the multidomain Pax6 (a) and Oct-1 (b) transcription factors and their isolated constituent domains from the closest DNA atom when they slide along DNA at a salt concentration of 0.01 M is shown. The tethered and isolated domains are shown in gray and blue, respectively. The corresponding distances of the specific complexes of Pax6 (PDB ID 6PAX) and Oct-1 (PDB ID 1OCT) are shown in black. The correlation coefficient between the distances of the tethered domains and those of the crystal structure are R = 0.25 and 0.55 for the connected N- and C-domains, respectively. The correlation coefficients between the distances of the specific (S) domain and homeodomain (HD) of Oct-1 found during sliding and in the crystal structures are 0.29 and 0.71, respectively. A pictorial representation of snapshots of sliding of Pax6 (c) and Oct-1 (d) along DNA illustrates the variation in dXY and dZ distances between the two tethered domains.
Overall, the nonspecific binding of the multidomain protein is similar to that seen in the crystal structures; however, the two tethered domains that comprise either Pax6 or Oct-1 deviate in terms of their binding mode to DNA. The Pax6N and Oct-1HD domains are closer to the DNA than their tethered Pax6C and Oct-1S component domains, indicating that the latter domains are more detached from the DNA in the nonspecific search because they have a lower affinity for the DNA (Fig. 1). The two tethered domains slide along DNA, and the flexible linker allows variations in their separation distance along the DNA, dZ, and in the XY plane, dXY (Fig. 1 and Fig. S1 in the Supporting Material).
The propensity to search DNA using a sliding mechanism diminishes as salt concentration rises, because the electrostatic attraction between the protein and the DNA weakens. Instead, the protein searches the DNA via the hopping and 3D diffusion mechanisms (Fig. 2 and Fig. S2). Clearly, the dynamics of domains with different nonspecific binding affinities to the DNA will be affected differently by increased salt concentration. One might expect the interplay between sliding, hopping, and 3D search to be different for the two tethered domains that comprise the multidomain proteins because they have different affinities to DNA and will respond differently upon a change in salt concentration. Indeed, the connected and isolated Pax6N and Oct-1HD domains perform more sliding than the corresponding Pax6C and Oct-1S domains, which weakly interact with DNA. In the isolated Pax6N and Oct-1HD domains, the hopping search mode is highly populated at moderate salt concentrations, but Pax6C and Oct-1S hop only at low salt concentrations because they mostly perform 3D diffusion at moderately low salt concentrations (Fig. 2 and Fig. S2).
Figure 2.
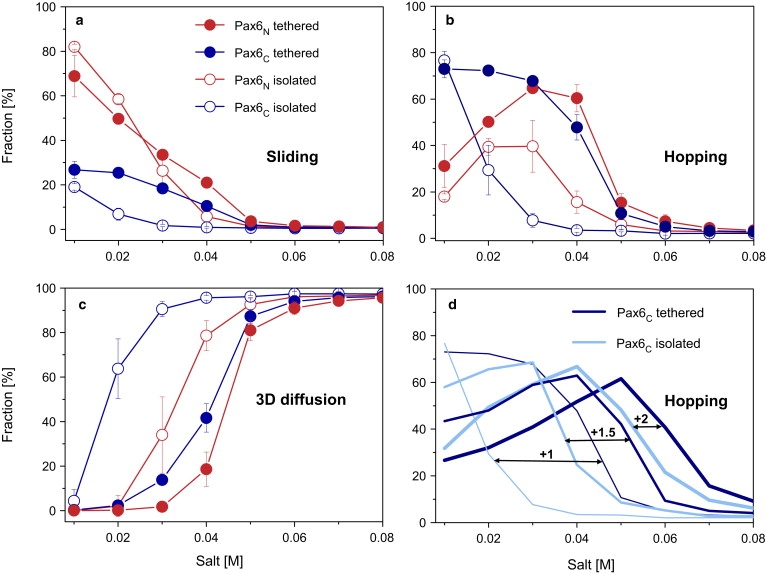
Effect of tethering the domains on the interplay between DNA search mechanisms by Pax6 variants at different salt concentrations. (a–c) DNA search via (a) sliding, (b) hopping, and (c) 3D diffusion was investigated for tethered (solid circles) and isolated (open circles) Pax domains. The N- and C-domains of Pax6 (Pax6N and Pax6C) are colored red and blue, respectively. (d) The effect of the DNA affinity of each Pax6C domain on its hopping propensity compared with the corresponding isolated variants. Isolated and tethered Pax6C domains are indicated by light blue and dark blue, respectively. The affinity strength (modeled by increasing the positive charges of Pax6C residues from +1 to +2) is designated by the thickness of the line.
Tethering the component domains to each other via a linker increases the affinity of the resulting multidomain protein (Pax6 or Oct-1) to DNA (48), as shown by the greater propensity of the tethered component domains to engage in sliding compared to their isolated counterparts (Fig. 2 and Fig. S2). Although both the isolated and tethered domains of Pax6 can slide along DNA, tethering increases the propensity of the PaxC domain to slide (Fig. 2 a) and, more importantly, to hop (Fig. 2 b). The higher propensity of the tethered Pax6C domain to slide and hop comes at the expense of the 3D diffusion mechanism favored by isolated Pax6C (Fig. 2 c). Pax6C exhibits increased affinity to DNA because it is tethered to Pax6N, which has a stronger DNA binding affinity. The Pax6N thus anchors the Pax6C domain closer to the DNA and increases its local concentration around the DNA. Indeed, a higher salt concentration is needed to dissociate tethered compared to isolated domains from the DNA (Fig. 2 c). To examine the ability of tethering to enhance binding affinity, we studied the search mechanism used by Pax6 variants in which the Pax6C domain has a higher affinity to DNA than is found in the WT. We found that the higher the affinity of the Pax6C domain to the DNA, the smaller the increase in hopping that is achieved by tethering it to the Pax6N domain (Fig. 2 d). Tethering was observed to yield very similar effects for the Oct-1 multidomain protein (Fig. S2).
The value of the 1D diffusion coefficient, D1, of multidomain variants linearly diffusing along the DNA (by sliding or hopping) increases with increasing salt concentration due to an increase in hopping events. Hopping is a faster dynamic than helical sliding along the DNA backbone because it is not constrained to the major groove (15,16) (Fig. 3). The isolated domains of Pax6 diffuse much faster than the corresponding tethered domains. Isolated Pax6C diffuses faster than isolated Pax6N because of its lower affinity to DNA and its relatively high propensity to hop, even at a low salt concentration. Tethering the Pax6N and Pax6C domains to each other not only produces a multidomain protein that diffuses more slowly than either of its component domains, it also abolishes the discrepancy between the D1 values of the component domains. To examine the effect of the DNA affinity of one domain on the linear diffusion coefficient of the domain tethered to it, we tethered the Pax6N domain to various Pax6C domains with different DNA affinities and measured its D1. Fig. 3 b shows that the diffusion rate of tethered Pax6N domains becomes slower as the DNA affinity of Pax6C increases. Similar behavior is observed for the diffusion of Oct-1 domains (Fig. S3). This observation, which illustrates that the effect of coupling between the two domains depends on their relative affinities, can be explained by an increase in the slower search mode (sliding) at the expense of the faster search mode (hopping) for overcharged Pax6C, which decelerates the Pax6N domain tethered to it.
Figure 3.

Characteristics of linear diffusion along the DNA. (a) The linear diffusion coefficient D1 as a function of salt concentration calculated for the sliding and hopping dynamics of WT Pax6+1. Tethered (solid circles) and isolated (open circles) of Pax6N+1 and Pax6C+1 are shown in red and blue, respectively. (b) The D1 of variants of Pax6N connected to Pax6C with various affinities to DNA. For comparative purposes, the D1 of the isolated WT Pax6N+1 domain is shown as well.
Cooperation between tethered domains facilitates intersegment transfer via a monkey-bar mechanism
We previously demonstrated that the existence of a disordered tail in homeodomain proteins can significantly enhance intersegment transfer between two parallel DNA molecules (16). The tail acts as an additional subdomain that interacts with DNA and hence increases the overall protein affinity to the DNA and promotes intersegmental transfer via a fly-casting mechanism (27). To determine whether the presence of two tethered DBDs facilitates intersegment transfer, as it does for homeodomains, or inhibits it due to tighter binding to the DNA, we studied the dynamics of the Pax6 and Oct-1 multidomain proteins in the presence of two parallel 100 bp DNA molecules separated by 60 Å. Free-energy surfaces as a function of the locations of the center of mass of the recognition helices of each of the N- and C-domains comprising either Pax6 (Fig. 4) or Oct-1 (Fig. S4) illustrate the mechanism of intersegment transfer. The energy landscape for multidomain jumps is composed of four minima. Two minima correspond to cases in which both tethered domains are concurrently bound to either DNA I or DNA II. The other two minima correspond to cases in which each domain interacts with a different DNA molecule. This suggests that the transfer from one DNA to the other proceeds through a bridged intermediate in which the domains are simultaneously attached to two different DNA molecules (Fig. 4). This mechanism resembles the motion of children as they swing along monkey bars (16). In similarity to the way a child transfers one hand at a time when swinging from bar to bar, the multidomain proteins cross from one DNA to another by transferring first one domain and then, after a certain lag time, the other domain. Experimental evidence for a ternary intermediate (i.e., a multidomain protein plus two DNA fragments) in intersegment transfer was provided by a recent NMR study by Doucleff and Clore (28) on the intermolecular translocation of Oct-1. In addition, a flexible, positively charged tail can also serve as a domain that stabilizes the bridged intermediate, as was proposed by NMR measurements of HoxD9 homeodomain (20) and demonstrated by a computational study of several homeodomain proteins (16).
Figure 4.

Intersegment transfer mechanism of Pax6. (a) The free-energy surface is projected along the locations of the center of mass of the recognition helix of WT Pax6N and Pax6C at a salt concentration of 0.05 M. It suggests a bridged intermediate for an intersegment transfer mechanism (the centers of the DNA molecules are placed at [30, 0, z] and [−30, 0, z]). (b) Pictorial representation of four frames from the Pax6 trajectory corresponding to the locations indicated in panel a. Snapshots 1 and 4 correspond to diffusion on DNA I and DNA II, respectively. Snapshots 2 and 3 correspond to intersegment transfer events.
The effect of domain cooperation and salt concentration on the number of intersegment transfer events was explored for Pax6 and Oct-1 (Fig. 5 and Fig. S5). In our simulation model, we found that a significant population of intersegment transfer events occurs at salt concentrations of 0.04–0.06 M for domains that bind DNA with low affinity and 0.06–0.1 M for domains that bind DNA with high affinity. The existence of an optimal salt concentration for this mechanism stems from the need to balance the strength of the electrostatic attraction between the protein and the DNA, which must be strong enough to allow helical sliding along the DNA but not so strong as to prevent 3D diffusion.
Figure 5.

Quantitative characteristics of intersegment transfer. The number of intersegment transfer events by Pax6N (red) and Pax6C (blue) variants as a function of salt concentration is shown. (a) The number of intersegmental transfer events of isolated (open circles) and tethered (solid circles) WT Pax6 (solid circles). (b) The number of intersegmental transfer events performed by tethered Pax6 variants (WT Pax6+1 (thin line), Pax6+1.5 (medium line), and Pax6+2 (thick line)). The higher the affinity of the C-domain to DNA, the greater is the number of intersegment transfer events performed by the N-domain in the connected variants. The number of DNA positions that were probed by sliding of tethered and isolated WT Pax6 (c) as well as by the variant Pax6+2 (d) of two DNA molecules.
In Pax6, tethered Pax6C and Pax6N domains perform more intersegment transfer events than their isolated counterparts (Fig. 5 a). The increase in the number of jumps is significantly greater for the tethered Pax6C domain than for the tethered Pax6N domain. In Oct-1, tethering the two domains significantly increases the number of jumps of Oct-1S, but it barely affects Oct-1HD compared to the corresponding isolated domains (Fig. S5). This is supported by an extensive study of Oct-1 dynamics on cognate DNA high-affinity binding sites that demonstrated that the average Oct-1S exchange rate is ∼1.5 times faster than the average Oct-1HD exchange rate at physiological salt concentrations (28). For a DBD whose affinity to DNA is weak, tethering improves its ability to perform searches via intersegmental transfer.
Since the different affinities of the domains when they are linked to each other results in a different tendency to jump, we next examined how the variants in which the affinity of one DBD is modulated scan DNA via jumping. Enhancing the affinity of Pax6C to the DNA reduces the number of intersegmental transfer events it performs, and causes a parallel increase in the number of jump events performed by the complementary Pax6N (Fig. 5 b). For example, tethered WT Pax6C and Pax6N perform ∼55 and ∼30 jumps, respectively, per simulation at an optimal salt concentration, whereas tethered C-overcharged Pax6C+2 and Pax6N+2 perform ∼10 and 80 jumps, respectively. These results suggest that the dissociation of a domain from one DNA molecule and its transfer to an adjacent DNA molecule while the other domain remains attached to the first DNA is more favorable when the other domain has a higher affinity to DNA and acts as a strong anchor. In the case of WT Pax6 and Oct-1 multidomain proteins, the domains with a higher DNA affinity (i.e., the Pax6N and Oct-1HD domains) act as anchors, whereas the lower DNA affinity domains (i.e., Pax6C and Oct-1S) scan binding sequences on different DNA fragments. When the DNA affinity of the domain with the lower DNA affinity is increased, the reverse picture is obtained and Pax6C and Oct-1S act as anchors.
Search efficiency is improved by domain cooperation and intersegment transfer
To understand the effect of domain cooperation on search efficiency, we calculated the number of DNA positions that are probed using sliding by Pax6 and Oct-1 multidomain variants as a function of salt concentration, following the procedure described by Givaty and Levy (15). For isolated Pax6N (or Oct-1HD) on a single DNA molecule, optimal search efficiency is achieved at moderate salt concentrations (at lower salt concentrations the protein will mostly slide, and at higher concentrations it will mostly diffuse in the bulk), as was previously found for single-domain proteins (7,11,16). By contrast, Pax6C and Oct-1S, which possess a low affinity to DNA, have a narrow range of salt concentrations in which they search the DNA efficiently (Fig. 5 c and Fig. S6). These DBDs show a low search efficiency at moderate salt concentrations because under such conditions their tendency to slide is negligible (Fig. 2 and Fig. S2). Fig. 5 c suggests that cooperation between Pax6N and Pax6C in the tethered multidomain Pax6 improves search efficiency, as reflected by the increase in the number of DNA sites that are probed using sliding at a larger range of salt concentrations compared to search by isolated domains. Cooperation between the Oct-1S and Oct-1N domains in the tethered Oct-1 compensates for the narrow salt concentration range in which Oct-1S is able to search efficiently, and shifts the efficient search range of the multidomain to moderate salt concentrations (Fig. S6).
Recent theoretical (22,23) and computational (16) studies showed that intersegment transfer notably improves search efficiency by DNA-binding proteins. The high search efficiency in a salt concentration range of 0.03–0.06 M is linked to the larger number of intersegment transfer events seen for tethered Pax6 domains in this range (Fig. 5). We propose that the cooperation achieved by tethering two domains with moderate DNA affinities to each other will result in a better search efficiency than can be achieved with each isolated domain. Brachiating the protein into individual domains with different DNA affinities supports intersegmental transfer and therefore efficient search. When the affinity of one of the tethered domains to DNA is too high, the search efficiency of both domains may be reduced.
Individual domains that comprise multidomain proteins differ in their binding affinities to DNA
Our results suggest that the different affinities to DNA observed for the domains that comprise Pax6 and Oct-1 have a significant effect on search efficiency. This observation raises the question of whether the phenomenon of DBDs having different affinities to DNA is common to other multidomain DNA-binding proteins. To address this question, we calculated the nonspecific binding affinity of various DBD components of multidomain DNA-binding proteins to DNA by estimating the electrostatic contribution to the binding energy of the protein to its specific DNA sequence. We performed this calculation for all multidomain DNA-binding proteins for which solved structures in complex with DNA are available.
We calculated the electrostatic free-energy change on binding of several characterized domains to their specific DNA, ΔGelbinding, using the nonlinear Poisson-Boltzmann equation and the APBS software package (47). The dielectric boundary was set to the vdW surface. The electrostatic binding energy of the N-terminal domain to the specific DNA-binding site, in comparison with that of the C-terminal domain, reveals that most individual fragments in multidomain proteins bind with significantly different affinities to promoter sites (Fig. 6). For example, Oct-1HD has a lower electrostatic free energy than Oct-1S (−11.2 vs. 0.8 kcal/mol) and Pax6N has a lower electrostatic free energy than Pax6C (−6.4 vs. −1.9 kcal/mol) on binding with their specific DNA sites, in agreement with their nonspecific affinities discussed above. The variety in attraction to DNA seen for individual domains in multidomain DNA-binding proteins may suggest that there is a general biological relevance for specific or nonspecific DNA binding by individual fragments with different affinities.
Conclusions
Many proteins involved in the regulation of gene expression contain several DBDs, each of which is crucial for specific recognition. These domains very often lack any interface and can cooperate only via a flexible linker. The degree of cooperation between domains and their role in nonspecific DNA searches is not well understood. Understanding the molecular ingredients that allow proteins to search DNA efficiently via sliding, hopping, and intersegment transfer is essential for understanding the cellular network at the molecular level. It was recently shown that protein-DNA electrostatic forces, which can be significantly modulated by adjusting the ionic strength, are pivotal factors in dictating search efficiency. As such, homologous domains with different nonspecific affinities to DNA will search DNA differently. Furthermore, decoration of the globular domain with a disordered tail can change the timescale of the search.
In this work we performed a computational study of the molecular mechanisms of DNA search employed by two multidomain transcription factors, Pax6 and Oct-1. Our motivation was to formulate the biophysical criteria for achieving facilitated DNA search by a multidomain protein compared with isolated domains. We show that the domains that comprise either Pax6 or Oct-1 proteins engage in nonspecific binding to DNA with significantly different affinities. Our results are supported by experimental evidence, such as the finding that isolated Oct-1HD binds to the human H2B histone promoter 150 times more tightly than isolated Oct-1S (49). Tethering the two domains that constitute either Pax-6 or Oct-1 strengthens the affinity of the resulting multidomain protein to the DNA (48) by increasing the population of the hopping search mode at the expense of 3D diffusion. Of more importance, the lower the affinity exhibited by the isolated domain to the DNA, the greater will be the effect of tethering to enhance its affinity to the DNA. Accordingly, tethering a DBD with weak affinity to DNA to a DBD with a higher affinity increases its local concentration around the DNA and its propensity to linearly diffuse along the DNA. The DBD with a high DNA affinity serves as an anchor to the DNA and improves the sliding performance of the DBD with the weaker affinity, but at the same time the tethering results in slower linear diffusion along the DNA than is exhibited by the isolated domains.
A multidomain protein searches two parallel DNA molecules with which it interacts nonspecifically by exchanging between them via a bridged intermediate in which each domain is adsorbed onto a different DNA molecule, suggestive of a monkey-bar mechanism (16). Intersegment transfer is facilitated by the existence of two domains, which act as two DNA-binding motifs and whose electrostatic potential can significantly affect the number of jumping events. As expected from the different affinity of the tethered DBD compared to its isolated counterpart, the intersegment transfer exchange rate of each domain in Pax6 and Oct-1 is also different. Our quantitative evaluation of jumping events performed by Pax6 and Oct-1 variants suggests that the direct transfer of a domain from one DNA molecule to an adjacent one is more favorable when the complementary domain has a higher affinity to DNA and acts as a strong anchor. In the case of WT Pax6 and Oct-1, the Pax6N and Oct-1HD domains act as anchors, whereas Pax6HD and Oct-1S act as explorers that scan potential DNA-binding sites. Experimental investigations of the exchange rate of lac repressor (26) and glucocorticoid receptor (25) have demonstrated the existence of different rates for specific dimeric subunits on nonsymmetric DNA-binding sites. NMR data for Oct-1 provide the exchange rates of each domain, which demonstrate that Oct-1S jumps ∼1.5 times faster than Oct-1HD. Thus, brachiating the protein into two domains can be advantageous for brachiating proteins that are using the monkey-bar mechanism to jump between two DNA molecules (50).
Different kinetic parameters for tethered domains at particular promoters may result in differential transcription activation or repression by the multidomain cofactors, as was shown for single-basepair mutations in Oct-1HD (51) and Oct-1S (52) hemisites, and for missense mutations within the paired domains of the Pax genes (3,53,54). In our simulations, mutations that strengthen or weaken domain-DNA affinity significantly influenced the interplay among sliding, hopping, and intersegment transfer, and affected search efficiency. On the basis of these results, we propose that cooperation between two domains with moderate affinities to DNA will yield a better search efficiency compared to that achieved by each isolated domain.
Our study indicates that cross talk between two domains is mediated by the flexible linker even when there is no physical interface between the two domains. This cross talk can lead to a significant improvement in the search efficiency that strongly depends on the relative affinities to DNA of the two domains. To examine the prevalence of this phenomenon, we examined the electrostatic contribution made by the individual domains of multidomain DNA-binding proteins to DNA-binding affinity. We found that, indeed, tethered domains tend to have different DNA-binding affinities, which may suggest that both specific and nonspecific DNA binding has biological relevance. Together, our results of enhanced DNA search for multidomains with polarized DNA-binding affinity and the observation that the individual domains of multidomain DNA-binding proteins have different affinities to DNA suggest that there is an evolutional drift toward the tethering of domains with very different affinities to DNA.
Acknowledgments
We thank Nathan Baker, David Gohara, and Yong Huang for their help with the APBS software.
This work was supported by the Kimmelman Center for Macromolecular Assemblies and the Minerva Foundation with funding from the Federal German Ministry for Education and Research. Y.L. holds the Lillian and George Lyttle Career Development Chair.
Supporting Material
References
- 1.Teichmann S.A., Park J., Chothia C. Structural assignments to the Mycoplasma genitalium proteins show extensive gene duplications and domain rearrangements. Proc. Natl. Acad. Sci. USA. 1998;95:14658–14663. doi: 10.1073/pnas.95.25.14658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apic G., Gough J., Teichmann S.A. Domain combinations in archaeal, eubacterial and eukaryotic proteomes. J. Mol. Biol. 2001;310:311–325. doi: 10.1006/jmbi.2001.4776. [DOI] [PubMed] [Google Scholar]
- 3.Azuma N., Nishina S., Yamada M. PAX6 missense mutation in isolated foveal hypoplasia. Nat. Genet. 1996;13:141–142. doi: 10.1038/ng0696-141. [DOI] [PubMed] [Google Scholar]
- 4.Epstein J.A., Glaser T., Maas R.L. Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes Dev. 1994;8:2022–2034. doi: 10.1101/gad.8.17.2022. [DOI] [PubMed] [Google Scholar]
- 5.Aurora R., Herr W. Segments of the POU domain influence one another's DNA-binding specificity. Mol. Cell. Biol. 1992;12:455–467. doi: 10.1128/mcb.12.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verrijzer C.P., Alkema M.J., van der Vliet P.C. The DNA binding specificity of the bipartite POU domain and its subdomains. EMBO J. 1992;11:4993–5003. doi: 10.1002/j.1460-2075.1992.tb05606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berg O.G., Winter R.B., von Hippel P.H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 8.Zhou H.X., Szabo A. Enhancement of association rates by nonspecific binding to DNA and cell membranes. Phys. Rev. Lett. 2004;93:178101. doi: 10.1103/PhysRevLett.93.178101. [DOI] [PubMed] [Google Scholar]
- 9.Halford S.E. An end to 40 years of mistakes in DNA-protein association kinetics? Biochem. Soc. Trans. 2009;37:343–348. doi: 10.1042/BST0370343. [DOI] [PubMed] [Google Scholar]
- 10.Mirny L., Slutsky M. How a protein searches for its site on DNA: the mechanism of facilitated diffusion. J. Phys. A: Math. Theor. 2009;42:434013. [Google Scholar]
- 11.Halford S.E., Marko J.F. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klenin K.V., Merlitz H., Langowski J., Wu C.X. Facilitated diffusion of DNA-binding proteins. Phys. Rev. Lett. 2006;96:018104. doi: 10.1103/PhysRevLett.96.018104. [DOI] [PubMed] [Google Scholar]
- 13.Blainey P.C., van Oijen A.M., Xie X.S. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc. Natl. Acad. Sci. USA. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwahara J., Clore G.M. Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature. 2006;440:1227–1230. doi: 10.1038/nature04673. [DOI] [PubMed] [Google Scholar]
- 15.Givaty O., Levy Y. Protein sliding along DNA: dynamics and structural characterization. J. Mol. Biol. 2009;385:1087–1097. doi: 10.1016/j.jmb.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 16.Vuzman D., Azia A., Levy Y. Searching DNA via a “monkey bar” mechanism: the significance of disordered tails. J. Mol. Biol. 2010;396:674–684. doi: 10.1016/j.jmb.2009.11.056. [DOI] [PubMed] [Google Scholar]
- 17.Slutsky M., Mirny L.A. Kinetics of protein-DNA interaction: facilitated target location in sequence-dependent potential. Biophys. J. 2004;87:4021–4035. doi: 10.1529/biophysj.104.050765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tafvizi A., Huang F., van Oijen A.M. Tumor suppressor p53 slides on DNA with low friction and high stability. Biophys. J. 2008;95:L01–L03. doi: 10.1529/biophysj.108.134122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwahara J., Zweckstetter M., Clore G.M. NMR structural and kinetic characterization of a homeodomain diffusing and hopping on nonspecific DNA. Proc. Natl. Acad. Sci. USA. 2006;103:15062–15067. doi: 10.1073/pnas.0605868103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iwahara J., Clore G.M. Direct observation of enhanced translocation of a homeodomain between DNA cognate sites by NMR exchange spectroscopy. J. Am. Chem. Soc. 2006;128:404–405. doi: 10.1021/ja056786o. [DOI] [PubMed] [Google Scholar]
- 21.Blainey P.C., Luo G., Xie X.S. Nonspecifically bound proteins spin while diffusing along DNA. Nat. Struct. Mol. Biol. 2009;16:1224–1229. doi: 10.1038/nsmb.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu T., Shklovskii B.I. How a protein searches for its specific site on DNA: the role of intersegment transfer. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2007;76:051909. doi: 10.1103/PhysRevE.76.051909. [DOI] [PubMed] [Google Scholar]
- 23.Sheinman M., Kafri Y. The effects of intersegmental transfers on target location by proteins. Phys. Biol. 2009;6:016003. doi: 10.1088/1478-3975/6/1/016003. [DOI] [PubMed] [Google Scholar]
- 24.Loverdo C., Bénichou O., Desbiolles P. Quantifying hopping and jumping in facilitated diffusion of DNA-binding proteins. Phys. Rev. Lett. 2009;102:188101. doi: 10.1103/PhysRevLett.102.188101. [DOI] [PubMed] [Google Scholar]
- 25.Lieberman B.A., Nordeen S.K. DNA intersegment transfer, how steroid receptors search for a target site. J. Biol. Chem. 1997;272:1061–1068. doi: 10.1074/jbc.272.2.1061. [DOI] [PubMed] [Google Scholar]
- 26.Fried M.G., Crothers D.M. Kinetics and mechanism in the reaction of gene regulatory proteins with DNA. J. Mol. Biol. 1984;172:263–282. doi: 10.1016/s0022-2836(84)80026-1. [DOI] [PubMed] [Google Scholar]
- 27.Trizac E., Levy Y., Wolynes P.G. Capillarity theory for the fly-casting mechanism. Proc. Natl. Acad. Sci. USA. 2010;107:2746–2750. doi: 10.1073/pnas.0914727107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doucleff M., Clore G.M. Global jumping and domain-specific intersegment transfer between DNA cognate sites of the multidomain transcription factor Oct-1. Proc. Natl. Acad. Sci. USA. 2008;105:13871–13876. doi: 10.1073/pnas.0805050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mansouri A., Hallonet M., Gruss P. Pax genes and their roles in cell differentiation and development. Curr. Opin. Cell Biol. 1996;8:851–857. doi: 10.1016/s0955-0674(96)80087-1. [DOI] [PubMed] [Google Scholar]
- 30.Dahl E., Koseki H., Balling R. Pax genes and organogenesis. Bioessays. 1997;19:755–765. doi: 10.1002/bies.950190905. [DOI] [PubMed] [Google Scholar]
- 31.Strachan T., Read A.P. PAX genes. Curr. Opin. Genet. Dev. 1994;4:427–438. doi: 10.1016/0959-437x(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 32.Stuart E.T., Kioussi C., Gruss P. Mammalian Pax genes. Annu. Rev. Genet. 1994;28:219–236. doi: 10.1146/annurev.ge.28.120194.001251. [DOI] [PubMed] [Google Scholar]
- 33.Noll M. Evolution and role of Pax genes. Curr. Opin. Genet. Dev. 1993;3:595–605. doi: 10.1016/0959-437x(93)90095-7. [DOI] [PubMed] [Google Scholar]
- 34.Xu W.G., Rould M.A., Pabo C.O. Crystal structure of a paired domain-DNA complex at 2.5 Å resolution reveals structural basis for Pax developmental mutations. Cell. 1995;80:639–650. doi: 10.1016/0092-8674(95)90518-9. [DOI] [PubMed] [Google Scholar]
- 35.Sturm R.A., Herr W. The POU domain is a bipartite DNA-binding structure. Nature. 1988;336:601–604. doi: 10.1038/336601a0. [DOI] [PubMed] [Google Scholar]
- 36.Ingraham H.A., Flynn S.E., Rosenfeld M.G. The POU-specific domain of Pit-1 is essential for sequence-specific, high affinity DNA binding and DNA-dependent Pit-1-Pit-1 interactions. Cell. 1990;61:1021–1033. doi: 10.1016/0092-8674(90)90067-o. [DOI] [PubMed] [Google Scholar]
- 37.Verrijzer C.P., Kal A.J., van der Vliet P.C. The oct-1 homeo domain contacts only part of the octamer sequence and full oct-1 DNA-binding activity requires the POU-specific domain. Genes Dev. 1990;4:1964–1974. doi: 10.1101/gad.4.11.1964. [DOI] [PubMed] [Google Scholar]
- 38.Xu H.E., Rould M.A., Pabo C.O. Crystal structure of the human Pax6 paired domain-DNA complex reveals specific roles for the linker region and carboxy-terminal subdomain in DNA binding. Genes Dev. 1999;13:1263–1275. doi: 10.1101/gad.13.10.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klemm J.D., Rould M.A., Pabo C.O. Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell. 1994;77:21–32. doi: 10.1016/0092-8674(94)90231-3. [DOI] [PubMed] [Google Scholar]
- 40.Levy Y., Onuchic J.N., Wolynes P.G. Fly-casting in protein-DNA binding: frustration between protein folding and electrostatics facilitates target recognition. J. Am. Chem. Soc. 2007;129:738–739. doi: 10.1021/ja065531n. [DOI] [PubMed] [Google Scholar]
- 41.Marcovitz A., Levy Y. Arc-repressor dimerization on DNA: folding rate enhancement by colocalization. Biophys. J. 2009;96:4212–4220. doi: 10.1016/j.bpj.2009.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Misra V.K., Hecht J.L., Honig B. Electrostatic contributions to the binding free energy of the lambdacI repressor to DNA. Biophys. J. 1998;75:2262–2273. doi: 10.1016/S0006-3495(98)77671-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zacharias M., Luty B.A., McCammon J.A. Poisson-Boltzmann analysis of the lambda repressor-operator interaction. Biophys. J. 1992;63:1280–1285. doi: 10.1016/S0006-3495(92)81723-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fogolari F., Elcock A.H., McCammon J.A. Electrostatic effects in homeodomain-DNA interactions. J. Mol. Biol. 1997;267:368–381. doi: 10.1006/jmbi.1996.0842. [DOI] [PubMed] [Google Scholar]
- 45.Dong F., Vijayakumar M., Zhou H.X. Comparison of calculation and experiment implicates significant electrostatic contributions to the binding stability of barnase and barstar. Biophys. J. 2003;85:49–60. doi: 10.1016/S0006-3495(03)74453-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qin S.B., Zhou H.X. Do electrostatic interactions destabilize protein-nucleic acid binding? Biopolymers. 2007;86:112–118. doi: 10.1002/bip.20708. [DOI] [PubMed] [Google Scholar]
- 47.Baker N.A., Sept D., McCammon J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou H.X. The affinity-enhancing roles of flexible linkers in two-domain DNA-binding proteins. Biochemistry. 2001;40:15069–15073. doi: 10.1021/bi015795g. [DOI] [PubMed] [Google Scholar]
- 49.Klemm J.D., Pabo C.O. Oct-1 POU domain-DNA interactions: cooperative binding of isolated subdomains and effects of covalent linkage. Genes Dev. 1996;10:27–36. doi: 10.1101/gad.10.1.27. [DOI] [PubMed] [Google Scholar]
- 50.Levin M.D., Shimizu T.S., Bray D. Binding and diffusion of CheR molecules within a cluster of membrane receptors. Biophys. J. 2002;82:1809–1817. doi: 10.1016/S0006-3495(02)75531-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu L.R., Du Z.W., Zhang J.W. T to C substitution at -175 or -173 of the γ-globin promoter affects GATA-1 and Oct-1 binding in vitro differently but can independently reproduce the hereditary persistence of fetal hemoglobin phenotype in transgenic mice. J. Biol. Chem. 2005;280:7452–7459. doi: 10.1074/jbc.M411407200. [DOI] [PubMed] [Google Scholar]
- 52.Di Rocco G., Gavalas A., Zappavigna V. The recruitment of SOX/OCT complexes and the differential activity of HOXA1 and HOXB1 modulate the Hoxb1 auto-regulatory enhancer function. J. Biol. Chem. 2001;276:20506–20515. doi: 10.1074/jbc.M011175200. [DOI] [PubMed] [Google Scholar]
- 53.Baldwin C.T., Hoth C.F., Milunsky A. Mutations in PAX3 that cause Waardenburg syndrome type I: ten new mutations and review of the literature. Am. J. Med. Genet. 1995;58:115–122. doi: 10.1002/ajmg.1320580205. [DOI] [PubMed] [Google Scholar]
- 54.Prosser J., van Heyningen V. PAX6 mutations reviewed. Hum. Mutat. 1998;11:93–108. doi: 10.1002/(SICI)1098-1004(1998)11:2<93::AID-HUMU1>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


