Abstract
From supercoiled DNA to the tight loops of DNA formed by some gene repressors, DNA in cells is often highly bent. Despite evidence that transcription by RNA polymerase (RNAP) is affected in systems where DNA is deformed significantly, the mechanistic details underlying the relationship between polymerase function and mechanically stressed DNA remain unclear. Seeking to gain additional insight into the regulatory consequences of highly bent DNA, we hypothesize that tightly looping DNA is alone sufficient to repress transcription. To test this hypothesis, we have developed an assay to quantify transcription elongation by bacteriophage T7 RNAP on small, circular DNA templates ∼100 bp in size. From these highly bent transcription templates, we observe that the elongation velocity and processivity can be repressed by at least two orders of magnitude. Further, we show that minicircle templates sustaining variable levels of twist yield only moderate differences in repression efficiency. We therefore conclude that the bending mechanics within the minicircle templates dominate the observed repression. Our results support a model in which RNAP function is highly dependent on the bending mechanics of DNA and are suggestive of a direct, regulatory role played by the template itself in regulatory systems where DNA is known to be highly bent.
Introduction
To comprehensively understand how gene expression is controlled through the regulation of transcription, it is necessary to discern the underlying biological mechanisms used to alter the behavior of RNA polymerase (RNAP) at the molecular level. Many general strategies for repressing RNAP function are now widely accepted, including competitive binding for the DNA template between a repressor protein and required components of the transcription machinery, steric blocking of an actively transcribing RNAP by a repressor bound to the template, and direct binding of a repressor to either RNAP itself or other required transcription factors (1). A notable feature of these regulatory strategies is that they typically involve, in addition to the RNAP and the DNA template, the necessary participation of a gene repressor. However, a body of evidence has emerged that is suggestive of another model for transcriptional repression whereby the DNA template mechanics directly modulate RNAP activity in the absence of auxiliary regulatory proteins (2–5). The DNA template in vivo is commonly subjected to nontrivial levels of bending deformation. Here, we seek here to quantify the influence of DNA bending on transcription. To illustrate the biological relevance of this work, we have identified two model systems that may include the application of mechanical stresses to the DNA template as a necessary feature of the molecular mechanism for RNAP regulation: gene repressors known to form tight loops of DNA and supercoiled DNA (Fig. 1).
Figure 1.
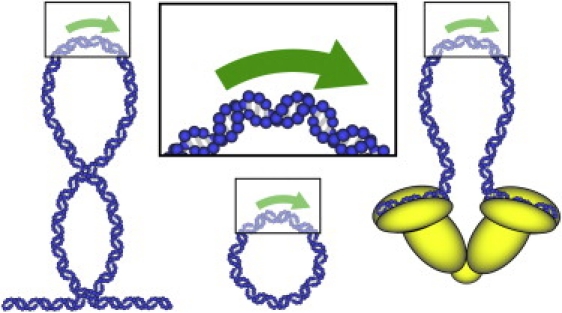
Mimicking biologically relevant cases where mechanically stressed templates may regulate transcription. DNA minicircles (bottom, center) were designed to sustain levels of bending stress that are comparable to what could be experienced locally by RNAP in DNA templates that are either (left) supercoiled or (right) bound by loop-forming repressors.
Many gene repressors bind to two closely spaced sites on DNA, forcing the DNA template to adopt a highly bent, looped structure sustaining significant mechanical stresses. Often, as with the lactose (lac) repressor in Escherichia coli (6,7) and the even-skipped (eve) repressor in Drosophila melanogaster (8), for example, the promoter sequence that is recognized and bound by RNAP is located within these repression loops. Curiously, unlike repression mechanisms involving competition between a repressor and RNAP for a single binding site, repressor-induced promoter looping leaves the RNAP binding site exposed and often not prevented on steric grounds from being bound by an RNAP. Additionally, although the binding of loop-forming repressors limits full-length RNA synthesis, several studies have shown that in some cases repressor binding does not preclude RNAP from carrying out early phases of transcription (9–13). Whereas it has been established that many loop-forming repressors depend on the formation of DNA loops to effectively regulate RNAP activity (11,14), it is unclear if DNA looping is itself causal to repression. Several repressor studies have posited that the mechanical state of the looped DNA is responsible for the induced repression (12,15). Yet, a direct observation of the ability of highly bent DNA templates to independently repress transcription has not been made, largely owing to the lack of an assay capable of quantifying the transcriptional competency of an RNAP from DNA templates sustaining well-defined levels of bending stress in the absence of DNA-binding proteins.
Previous attempts have been made to understand the relationship between torsional stress and RNAP activity by measuring the sensitivity of transcription to levels of DNA supercoiling (2–5,16). These studies have shown convincingly that the activity of RNAP depends strongly on the mechanical state (supercoiling) of DNA. However, the interpretation of these studies is inherently complicated by the tendency of supercoiling to act indiscriminately across relatively large regions of DNA and by the poorly defined bending and torsional stresses imparted onto the supercoiled DNA. Here, we have overcome these limitations by developing an assay that is capable measuring transcription activity from DNA templates sustaining well-defined levels of bending and torsion. We leverage the ability of small circular DNA to introduce tight bends into and maintain the torsional state of a transcription template. Importantly, the bending and twist sustained within minicircle template are defined only by the number of basepairs in the circular DNA and are applied in the absence of DNA-binding proteins. Furthermore, and in contrast to previous transcription assays using torsionally stressed, supercoiled plasmids, the degree of twist within the subpersistence length minicircle templates in our study cannot be relieved significantly by supercoiling (17). By using DNA minicircles containing an RNAP-specific promoter sequence in a transcription assay, any observed differences in RNAP activity must be directly attributed to the mechanical properties of the DNA template. To gain insight into the effects of highly bent DNA on transcription, we chose to prepare minicircle templates on the order of 100 bp in size to impart biologically relevant bending stresses onto the DNA that were comparable to those encountered at the apical loops of plectonemic supercoils (18) and in many known loop-forming repressor systems (19–21) (Fig. 1).
The goal of this work is to understand the effects of bending stress on transcription in general terms. We chose to test our hypothesis using bacteriophage T7 RNA polymerase (T7 RNAP), a widely studied model RNAP that is remarkably resilient, as evidenced by its ability to transcribe through a variety of topological challenges in the transcription template (22). As a single-subunit RNAP displaying transcriptional activities common to other RNAPs, T7 RNAP has the advantage of not requiring additional transcription cofactors, the coordination of which may also be influenced by template mechanics. As the first step in addressing our broad hypothesis, we have focused our observations on the effects of mechanically stressed DNA on the elongation phase of transcription. Using highly bent circular DNA templates, we have measured the kinetics describing several characteristic behaviors of T7 RNAP, including elongation complex (EC) formation, EC stability, elongation velocity, and processivity. Here, we show that simply looping the template is sufficient to strongly repress elongation by T7 RNAP. To determine whether the observed repression was dominated by the bending or the torsional stresses sustained within the covalently closed, circular DNA templates, we have also characterized T7 RNAP elongation activity from DNA minicircles sustaining varying degrees of residual twist (torsional stress). From the three cases tested (highly untwisted, mildly untwisted, and overtwisted), we show that although the torsional stress sustained within the minicircle templates can influence the elongation velocity and processivity of T7 RNAP, these effects are relatively minor compared to the overall observed repression. We therefore conclude that the repression of T7 RNAP on minicircle templates is largely attributed to the bending of DNA.
Materials and Methods
Minicircle template synthesis
Sufficient quantities of minicircle templates were created by modifying the ligation-assisted minicircle accumulation technique previously described (23). Briefly, two double-stranded DNA sequences (each the length of the desired minicircle template) were designed such that the first half of one sequence is complementary to the second half of the other. Consequently, upon melting into single-stranded DNA, the DNA can reanneal as either double-stranded DNA or a doubly nicked minicircle. After ligation by a thermophilic DNA ligase, however, the closed minicircle is incapable of melting, and hence accumulates in solution. Both sequences were amplified by PCR and restricted with a blunt-end restriction enzyme. Taq DNA ligase (New England Biolabs, Beverly, MA) was then added to 12–15μg of each product, then thermocycled for 15 cycles as follows: melt at 95°C for 20 s, plunged to 4°C and held for 1 min, ligate at 65°C for 20 min. After digesting any remaining linear DNA using exonucleases I and III (New England Biolabs), the monomeric circular DNA was estimated by denaturing PAGE to comprise ∼99% of the exonuclease-resistant product (see Fig. S1 in the Supporting Material) and was used without further purification.
Transcription elongation assays
Minicircle template (0.5 nM) was incubated with 8.5 nM T7 RNAP (Fermentas, Hanover, MD) for 10 min in transcription buffer (40 mM Tris-acetate, 10 mM Mg-acetate, 0.05% v/v Tween-20, 10 mM DTT, pH 8.0), supplemented with 1 mM each of ATP, GTP, and CTP (Fermentas) and 20 nM 2′-o-methyl-RNA molecular beacons (Sigma Life Science, The Woodlands, TX); 5 mg/ml heparin (Sigma Life Science) was added to inactivate free T7 RNAP, and incubated for 5 min before the initiation of transcription with the addition of 1 mM UTP (Fermentas). The transcription reaction was terminated by heat inactivation of the polymerase (and destruction of secondary structure in the RNA transcript) at 80°C for 10 min. The solution was then slowly cooled at a rate of −0.1°C/s to allow molecular beacons to hybridize to RNA target sequences present.
Fluorescence was measured using an AMINCO-Bowman Series 2 luminescence spectrometer (Thermo Spectronic, Rochester, NY), equipped with an external photomultiplier tube module. All reaction solutions, 65 μl in volume, were excited with 535 nm light in a 50 μl Sub-Micro Quartz Fluorometer Cell (16.50F-Q-10/Z15, Starna Cells, Atascadero, CA), and the emission from the cuvette was first filtered with a longpass filter (OG570, Schott, Elmsford, NY). The fluorescence signal from molecular beacons hybridized to target RNA in solution was experimentally determined to scale as 0.4085 AU per nM of hybridized molecular beacon (see Fig. S3). The number of nucleotides transcribed was calculated from the fluorescence data by determining the concentration of hybridized beacon with the above calibration factor and multiplying the result by the number of nucleotides in the transcript to synthesize an additional beacon target sequence (i.e., the nucleotides transcribed in a single round of the minicircle template, either 100 bp, 106 bp, or 108 bp). This value was then normalized by the number of T7 RNAP experimentally determined to occupy the template after template and heparin incubations to account for the template-specific rates of EC formation and EC stability (described below).
A nonlinear least-squares fitting routine was implemented using MATLAB (The MathWorks, Natick, MA) to fit the time-dependent fluorescence data (N, the number of nucleotides transcribed per elongating T7 RNAP) to the solution to the first-order differential equation describing the reaction kinetics,
| (1) |
where the amplitude of the rising exponential (P) is equal to the transcription processivity (the number of nucleotides synthesized per T7 RNAP on average before dissociation) and kdiss is the dissociation rate of the enzyme from the template (in s−1). The elongation velocity, kcat, is equal to the maximal rate of transcription (the value of the first derivative of Eq. 1 at t = 0) and is calculated by multiplying P by kdiss.
Elongation complex formation assays
Concentrations and buffer conditions were identical to transcription elongation assays, with the following modifications: T7 RNAP was incubated with the template for varying periods of time before the addition of heparin. Heparin was added and, as above, the solution was incubated for 5 min before UTP addition. In all cases, transcription was allowed to proceed for 10 min before transcription was terminated by heat inactivation of T7 RNAP. The time-dependent occupancy data (Of, in percentage of template occupied after EC formation) were fit to the solution to the first-order differential equation describing the reaction kinetics,
| (2) |
where kfor is the rate of elongation complex formation.
Elongation complex stability assays
Experimental conditions were identical to transcription elongation assays, with the following modifications: T7 RNAP was incubated with the template for 10 min before the addition of heparin. Heparin was then added and the solution was incubated for varying periods of time before UTP addition. In all cases, transcription was allowed to proceed for 10 min before transcription was terminated by heat inactivation of T7 RNAP. The time-dependent occupancy data (Os, in percentage of template remaining occupied by stable ECs after heparin incubation) were fit to
| (3) |
where kdiss(stalled) is the dissociation rate of stalled ECs from the template (which is inversely related to EC stability).
Results
Minicircle transcription assay
To characterize the dependence of T7 RNAP activity on DNA template mechanics, we focused our observations on the elongation phase of transcription by using a well-established technique using heparin, a potent inhibitor of T7 RNAP binding to DNA. Briefly, we designed minicircle templates such that the first 20 bases could be transcribed in the presence of only ATP, CTP, and GTP (Fig. 2 A). Transcription assays were carried out by first incubating T7 RNAP with the minicircle template in the absence of UTP (Fig. 2 B), followed sequentially by the addition of heparin (Fig. 2 C) and UTP (Fig. 2 D). As a result, transcription was only allowed to occur from polymerases that had successfully transitioned into a stable elongation complex. Transcription activity was quantified by the fluorescence emission resulting from the hybridization of molecular beacons to a specific target sequence within the synthesized transcript (Fig. 2 E).
Figure 2.
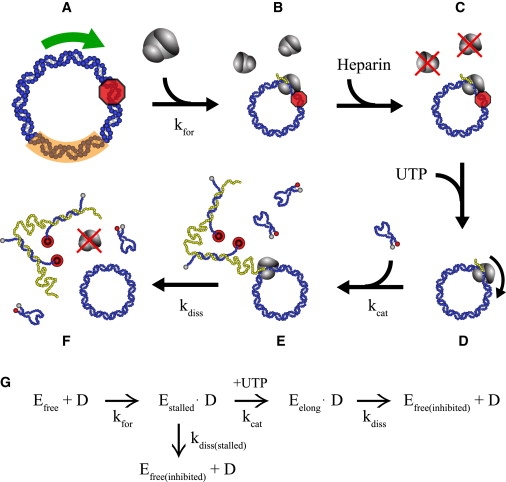
Minicircle transcription assays. (A) Looped DNA topologies containing a highly bent promoter (arrow) are recreated with DNA minicircles, eliminating any influence on transcription owing to either the presence of a repressor protein or variations in bending and torsional stresses within supercoiled DNA. The minicircle template contains a T7 RNAP promoter sequence, a site at which T7 RNAP ECs will stall in the absence of UTP (shaded octagon), and a target sequence (shaded arc) from which the corresponding RNA transcript will hybridize to a designed molecular beacon sequence. (B) T7 RNAP (Efree) is incubated with the DNA template without UTP, and forms stalled ECs (Estalled) at some rate of complex formation (kfor). (C) Heparin addition inactivates free T7 RNAP (Efree(inhibited)). (D) UTP addition initiates transcription from active ECs (Eelong), which synthesize RNA at the elongation velocity (kcat). (E) Molecular beacons hybridize to target transcript sequence. (F) Elongating T7 RNAP detach from the template at some rate of dissociation (kdiss). (G) A simplified kinetic equation governing T7 RNAP behavior in the assay.
Molecular beacons offer a highly sensitive measure of transcription that allows low concentrations of template to be used in our assay. Given the significant challenge to produce large quantities of highly stressed, circular templates that are energetically unfavorable to form, the use of low template concentrations is particularly advantageous. Molecular beacons also make it possible for us to account for the possibility that the target sequence may be transcribed multiple times by a single T7 RNAP making multiple rounds on the circular template. In this case, the number of molecular beacons that hybridize to the transcript is equal to the number of times the target sequence appears in the transcript. The resulting fluorescence accumulates over time, owing to the high stability of the hybridized transcript-beacon complex, and serves to report the number of complete rounds made by an elongating T7 RNAP transcribing a given minicircle template (Fig. 2 F).
The elongation-specific transcription assay we have developed makes it possible to observe several T7 RNAP behaviors of interest. The fluorescence intensity at any point in time is directly related to both the number of transcribing polymerases and the average elongation velocity from a given transcription template. Because all elongating polymerases are prohibited from rebinding the template in the presence of heparin, the accumulation of fluorescence over time is governed by the kinetics for a single step, irreversible reaction. The signal therefore increases exponentially in time to saturation. These time-dependent fluorescence data are then fit to a single exponential function, where the maximal slope is proportional to the average elongation velocity (kcat) (Fig. 2 G), the fitted rate constant is equal to the polymerase dissociation rate from the template (kdiss) (Fig. 2 G), and the amplitude is linearly related to average processivity of the enzyme (the number of nucleotides transcribed before dissociation).
To quantify the effects of template bending on transcription by T7 RNAP, we designed circular templates 100 bp, 106 bp, and 108 bp in length (Fig. 3 A) and modified the ligation-assisted minicircle accumulation technique (23) to produce homogeneous solutions of minicircle templates, as confirmed by cryo-electron microscopy (Fig. 3 B) and gel electrophoresis (Fig. S1). All three species experience varying levels of torsional stress, which results from forming closed, circular templates from linear DNA that is not an integral number of helical turns in length. To qualitatively determine the relative levels of torsional stress within the minicircles, each species is incubated with Bal-31 nuclease, which is known to specifically digest untwisted circular DNA (24). We observe that the 100 bp minicircle is quickly digested (Fig. 3 C), indicating that it is the most highly untwisted template (denoted as +UT), whereas the 106 bp minicircle is resistant to Bal-31, demonstrating that it either sustains no residual twist or is overtwisted (OT). The 108 bp minicircle is sensitive to Bal-31, but less so than +UT, and so we conclude that it is only mildly untwisted (UT). Our interpretation of the Bal-31 data with respect to the torsional state of each minicircle is confirmed by the cyclization of a 104 bp linear DNA (sharing sequence homology with the three minicircle species tested). The resulting 104 bp circular DNA is distributed between two topoisomers, one overtwisted and one untwisted (Fig. S2). Because two topoisomers are expected to occur when the linear DNA is a semi-integer number of helix turns in length, we draw from this observation that the 106 bp minicircle is likely to be overtwisted, whereas the 108 bp minicircle is untwisted but is actually closer to being torsionally relaxed than either the 106 bp or 100 bp minicircle (see Supporting Material for further discussion).
Figure 3.
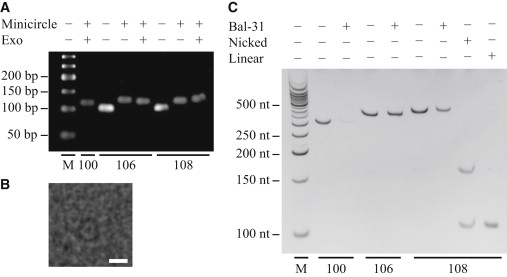
Minicircle transcription templates. (A) Native agarose gel electrophoresis of DNA (3% agarose, 0.5× TBE, 10 V/cm) of the specified length (100 bp, 106 bp, 108 bp), either as linear DNA before cyclization into minicircle templates or immediately after the cyclization procedure. Minicircle products were resistant to a prolonged incubation with exonucleases I and III (Exo), confirming the presence of a circular DNA species. (B) Cryo-EM image of 100 bp minicircle confirms that the cyclization procedure generates circular monomers (white scale bar = 10 nm). (C) Denaturing PAGE of minicircle templates (10% polyacrylamide, 7 M urea, 1× TBE, 30 V/cm) reveals that the 100 bp minicircle is highly untwisted (and hence quickly digested by Bal-31), the 106 bp minicircle is overtwisted (resistant to Bal-31), and the 108 bp minicircle is modestly untwisted (slowly digested). Minicircle templates were incubated with 0.015 U/μL for 15 min at 30°C. Singly nicked and linearized 108 bp minicircles provide references for linear and circular single-stranded DNA species under denaturing conditions.
Determining template occupancy
To accurately characterize the elongation behavior using the molecular beacon assay, it is necessary to first know [Estalled] (Fig. 2 G) at the time that UTP is added to resume transcription elongation. To determine [Estalled] for a particular set of incubation conditions before UTP addition, we modified our assay to measure the percentage of templates that remained occupied by functional, stalled T7 RNAP as a function of time for the two incubation steps used in the elongation-specific assay. Driven by the kinetics governing EC formation, the template occupancy during the RNAP incubation (Fig. 2, B and C) increases as a function of time. Conversely, the template occupancy during the heparin incubation (Fig. 2, C and D) decreases over time according to the dissociation kinetics of stalled T7 RNAP from the template, a characteristic reflective of EC stability.
To quantitatively determine the time-dependence of EC formation, we define a pseudo first-order rate constant, kfor (Fig. 2 G), to describe the overall kinetics of promoter binding, open complex formation, and transcription initiation (25). kfor can be determined by varying the time T7 RNAP is incubated with template DNA before the addition of heparin (Fig. 2, B and C) and measuring the concentration of hybridized beacon resulting from otherwise identical transcription conditions. The resulting signal is therefore directly proportional to the number of ECs formed at the time of heparin addition. A single exponential fit to the fluorescence intensity as a function of the RNAP incubation time with DNA templates yields an apparent rate constant, kfor, describing the formation of stalled T7 RNAP ECs (see Supporting Material).
We hypothesized that tightly bending DNA would be highly repressive to the pre-elongation phases of transcription. We also thought that progressively higher levels of untwist in the template (such as in UT and +UT) would relieve the torsional load acting against the polymerase during these early phases, and hence increase kfor as the template became more untwisted. Surprisingly, we observe that kfor is reduced for the highly untwisted (+UT, 2.0 × 10−3 s−1) and overtwisted (OT, 2.4 × 10−3 s−1) minicircles, compared to the mildly untwisted UT (4.8 × 10−3 s−1) (Fig. 4). To relate the value of kfor measured from minicircle templates to unrepressed T7 RNAP behavior, we measured the kinetics of EC formation using a 2536 bp circular plasmid that was purified from E. coli (and therefore supercoiled at native density). From the supercoiled plasmid, we observe that the rate of EC formation (kfor ∼1.3 × 10−3 s−1) is slower than for any of the minicircle templates tested. In attempting to interpret these data, it is important to note that we must limit our conclusions to the cumulative effects of mechanical stress on all pre-elongation phases because our assay is specifically designed to monitor transcription elongation. More targeted studies would be required to address the effects of mechanical stress on any particular phase of transcription before elongation.
Figure 4.
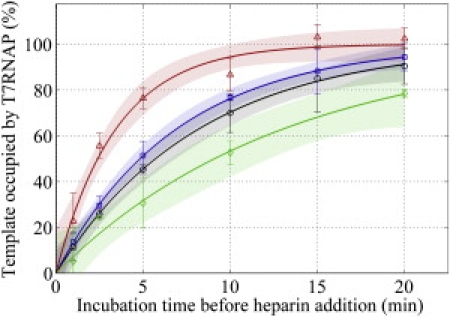
Monitoring the formation of stable T7 RNAP ECs. Transcription from templates allowed to incubate with T7 RNAP for a defined period of time is proportional to the number of T7 RNAPs successfully loaded onto the templates and awaiting the addition of UTP. The rates at which ECs are forming on OT (blue squares) and +UT (black circles) do not seem to be significantly different, but ECs form more quickly onto UT (red triangles). EC formation occurs more quickly on all three minicircle templates than on a supercoiled, 2536 bp plasmid (green diamonds).
Defining the fraction of templates occupied by competent, stalled EC complexes before allowing elongation to proceed (Fig. 2 D) requires, in addition to knowing the rate of EC formation, knowledge of the dissociation rate from the template during the heparin incubation. The procedure to determine the dissociation rate of stalled ECs from the template is analogous to measuring kfor. However, instead of varying the time for the RNAP incubation (Fig. 2, B and C), the heparin incubation time is varied (Fig. 2, C and D). Consequently, the concentration of hybridized beacon is directly proportional in this case to the number of ECs remaining bound after a defined incubation of stalled ECs with heparin. The single exponential fit to the fluorescence data yields a rate constant, kdiss(stalled) (Fig. 2 G), that describes the dissociation rate of stalled ECs from the template during heparin incubation, and is reflective of EC stability (see Supporting Material).
We hypothesized that stalled ECs would dissociate from the template faster when challenged by increasing levels of template stress. Therefore, we expected that tightly bending DNA would decrease EC stability and that EC stability would increase as the template became more untwisted. We do observe that kdiss(stalled) appears to decrease as the torsional state of the template progresses from overtwisted (OT, 3.6 × 10−3 s−1), to mildly untwisted (UT, 3.1 × 10−3 s−1), to highly untwisted (+UT, 2.5 × 10−3 s−1). Interestingly, and in contrast to our expectations, stalled ECs dissociate more quickly from the supercoiled, 2536 bp plasmid (kdiss(stalled) ∼5.2 × 10−3 s−1) than from any of the minicircle templates (Fig. 5).
Figure 5.
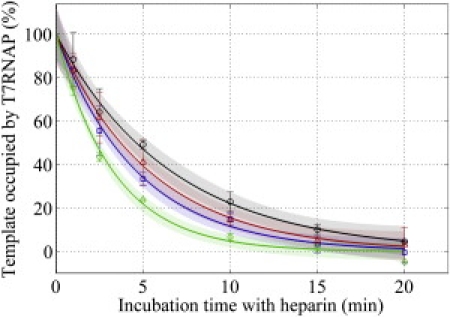
Quantifying EC stability. ECs stalled at the UTP stall site were incubated with heparin for varying periods of time before UTP addition. The fluorescence signal resulting from transcription under otherwise identical conditions is proportional to the number of stalled ECs remaining associated with the template. Stalled ECs dissociate most quickly from (and hence are least stable on) the supercoiled 2536 bp plasmid (green diamonds), followed sequentially by OT (blue squares), UT (red triangles), and +UT (black circles).
It is important to note that because the minicircle templates were designed such that an EC would stall at the +20 position from the transcription start site, a second T7 RNAP is capable of binding the exposed promoter and initiating transcription behind a stalled EC (26). Consequently, there may be a concern that the actual elongation behavior could be overestimated in our assay (arising from multiple RNAPs when only one is assumed to be active). We consider this possibility to be unlikely for two reasons: the second polymerase will remain in an unstable initiation complex (26) and initiation complexes are unlikely to survive a prolonged incubation with heparin (27). Indeed, the EC stability data (Fig. 5) support our contention that each template is unlikely to be occupied by more than one polymerase because >50% of the templates are unoccupied after the 5-min heparin incubation used in our elongation assays.
To determine the percentage of the minicircle templates occupied by functional, stalled ECs before initiating transcription elongation with the addition of UTP, we calculate the occupancy arising from EC formation (given the time the template was incubated with T7 RNAP before heparin addition) and the occupancy owing to stalled EC stability (given the time over which stalled ECs were incubated with heparin before adding UTP). The percentage of template occupied after both incubation steps is then equal to the product of the template occupancies after each incubation step. All transcription elongation data are normalized by this calculated occupancy for the chosen regimen of incubation conditions to account for the number of templates from which productive elongation actually occurs.
Repression of transcription elongation
Three minicircle templates were used in our assay with the expectation that the varying amounts of residual twist sustained within each species would reveal the contribution to template-induced repression that arises from the torsional state of the DNA minicircles. That is, if the transcription activity from the three templates does not differ significantly, then torsional stress can effectively be ruled out as a causal factor to any observed repression. We first characterized transcription elongation from the mildly untwisted minicircle (UT), fitting the observed time-dependent fluorescence data to a single exponential to calculate kcat (the maximal slope) and processivity (the amplitude of the fitted exponential function). From this fit, we determined that T7 RNAP had a catalytic rate (kcat) of 4.1 nt/s and an overall processivity of 6708 nt on mildly untwisted minicircle templates (Fig. 6, red triangles). We next considered the case where untwist was relieved (OT, Fig. 6, blue squares), and observed that kcat was not affected (4.2 nt/s), but the processivity was more than doubled (14,440 nt) compared to UT. By contrast, when we used template minicircles that are more untwisted than UT (as in +UT), the processivity did not significantly change (6773 nt), but kcat increased nearly fourfold (15.6 nt/s) (Fig. 6, black circles). Although these data support a clear relationship between T7 RNAP behavior and template twist, transcription from these minicircle templates remains in all cases significantly reduced compared to wild-type T7 RNAP activity typically reported in the literature (kcat ∼230 nt/s (28), processivity >30,000 nt (29)).
Figure 6.
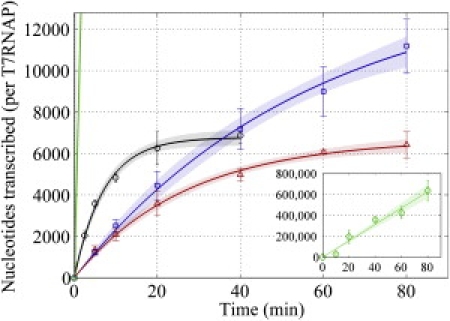
T7 RNAP elongation behavior from highly bent templates. Transcription from the overtwisted template (OT, blue squares) is roughly four times slower than from the most untwisted template (+UT, black circles), but is also more processive. The moderately untwisted template (UT, red triangles), by comparison, shows an elongation velocity comparable to OT, but the processivity of +UT. Both the elongation velocity and processivity measured from each minicircle is significantly repressed compared to a negatively supercoiled, 2536 bp circular plasmid (green diamonds, inset).
As a positive control for our assay, we compared the transcription elongation data from the minicircle templates to that from a supercoiled, 2536 bp circular template and observed a kcat of ∼130 nt/s (Fig. 6, inset). Although the exact mechanical stresses encountered by a transcribing T7 RNAP on the supercoiled template is not entirely defined due to the potential variations in bending and twisting in supercoiled templates (see Discussion), these data verify that our molecular beacon assay is faithfully reporting quantitative information regarding transcription elongation. Relative to transcription from the large plasmid, we observe that transcription from highly bent templates reduced kcat by ∼10–30 times and depressed processivity by >40–90 times (Table 1), depending on the minicircle template. It should be noted, however, that T7 RNAP is so processive in elongation on the 2536 bp circular plasmid that the time-dependent fluorescence data are best fit to a linear function, suggesting that the characteristic time constant for the expected exponential function is significantly >80 min. After 80 min, the final fluorescence signal corresponds to an average transcript size in excess of 625,000 nt in length, and it is therefore expected that the average processivity for T7 RNAP on the large plasmid is well above this value. Therefore, because the average number of nucleotides transcribed by T7 RNAP before dissociation from a larger plasmid has not been measured directly, our stated value for the change in polymerase processivity during transcription of highly bent minicircle templates provides only a lower bound for the actual template-induced repression.
Table 1.
Transcription activity is sensitive to mechanical state of the template
| Species | Twist | Size (bp) | kfor (×10−3 s−1) | kdiss(stalled) (×10−3 s−1) | kcat (nt × s−1) | kdiss (×10−3 s−1) | Processivity (×103 nt) |
|---|---|---|---|---|---|---|---|
| +UT | − | 100 | 2.0 (1.3, 4.1) | 2.5 (1.8, 4.2) | 15.6 (12.6, 19.8) | 2.3 (2.0, 2.8) | 6.8 (6.4, 7.2) |
| UT | − | 108 | 4.8 (3.4, 8.0) | 3.1 (2.3, 4.9) | 4.1 (3.2, 5.4) | 0.6 (0.5, 0.8) | 6.7 (6.2, 7.2) |
| OT | + | 106 | 2.4 (1.8, 3.6) | 3.6 (2.8, 5.0) | 4.2 (2.4, 8.4) | 0.3 (0.2, 0.5) | 14.4 (11.2, 17.7) |
| Control | − | 2536 | ∼1.3 | ∼5.1 | ∼130 | n/a∗ | >625 |
Mean values for kfor, kdiss(stalled), kcat, kdiss, and processivity are determined from the best-fit exponential function fitted to data. The 95% confidence intervals from the fits are shown in parentheses.
These data were best fit to a linear function and therefore are not well described by an exponential constant.
These data unequivocally demonstrate that bending stress represses transcription of an otherwise competent transcription template, and suggest that torsional stress also plays a role in influencing the kinetics of elongation by T7 RNAP. The modulation of kcat due to untwisting the DNA can be put into context when one considers that the promoter must be untwisted by T7 RNAP during the isomerization phase of transcription, where the polymerase accesses the nucleotide bases (30). As a direct consequence of this step, the remaining DNA in a circular template is necessarily overtwisted. Therefore, by requiring T7 RNAP to transcribe templates that already sustain some degree of untwist, the overtwist generated by the polymerase during open complex formation can be partially alleviated. We therefore interpret the restorative effects of DNA untwist on kcat as reflecting the relief of the torsional load encountered by T7 RNAP as it transcribes the circular DNA. It remains unclear why overtwisted templates increase the processivity of T7 RNAP, as does the molecular mechanism driving this interplay between elongation velocity and processivity as a function of template twist.
Discussion
We have sought to directly address our hypothesis that tight looping of the DNA template alone is sufficient to repress transcription. Toward this end, we have developed a novel transcription assay capable of quantifying RNAP activity from circular DNA templates sustaining fixed amounts of bending and torsional stresses. From these minicircle templates, we observe that the elongation velocity and processivity of T7 RNAP are highly repressed compared to wild-type levels. Our data suggest that overtwisting a highly bent template enhances processivity whereas untwisting increases elongation velocity. We have also observed differences, albeit to a lesser extent, in the rates of EC formation and stability. We believe the results presented in this study constitute a significant advance in our understanding of the molecular mechanisms underlying template-induced repression of RNAP activity. To frame our results in the context of previous observations, we wish to highlight two particular examples where DNA bending mechanics may play a significant role in transcriptional modulation: DNA supercoiling and loop-forming gene repressors.
It has long been known that RNAP activity is dependent on the mechanical state of the DNA template. Since the initial observation that transcriptional activity was enhanced from DNA templates that were negatively supercoiled (2), many other studies have attempted to understand the influence of the mechanical state of DNA on transcription (3–5,16). Important first steps have been made toward elucidating the structure-function relationship between template mechanics and RNAP activity, exemplified by the work of Gopal et al. (16) where the thumb subdomain of T7 RNAP was implicated in stabilizing the EC on supercoiled DNA. However, the use of supercoiled DNA in transcription studies presents significant and unavoidable complications in interpreting the data on a mechanistic level, particularly with respect to understanding the molecular interactions between RNAP and highly bent and twisted templates. Specifically, supercoiled DNA displays two problematic characteristics: the torsional stress within the DNA can vary dynamically at the expense of bending energy (31) and the steady-state average magnitudes of bending and torsional stresses are exclusively governed by the geometrical parameters defining the DNA tertiary (superhelical) structure (32). Previous transcription studies using supercoiled DNA do not have a detailed description of the tertiary structure of supercoiled DNA, and as a direct consequence, are not capable of defining the bending and torsional stresses sustained by the DNA template at the site of RNAP activity. It is therefore not possible in these cases to accurately determine the dependence of transcription on the twist and bending sustained within the DNA template. Consequently, conclusions from these studies are largely restricted to the qualitative demonstration that negative torsional stress is often sufficient to enhance transcription.
The dependence of RNAP activity on DNA bending has been investigated in a number of studies using intrinsically curved DNA sequences inserted within the transcription template (33–37). These studies have shown that intrinsic (stress-free) curvature within the DNA template can modulate transcription, but they do not address the effects of stresses applied to the template (a case that is more more biologically relevant to understanding the role of tightly looped DNA induced by DNA-binding proteins or DNA supercoiling). Perhaps the most direct attempt to address the effect of forcibly bending DNA on transcription was carried out by TenHarmsel and Biggin (15). In this study, a 245-bp circular DNA template was designed for in vitro transcription by Drosophila RNA polymerase II (RNAP II) in an attempt to demonstrate that the loop formed by the eve repressor was causal to transcriptional repression in this system. The authors reported that overall transcription was repressed from their circular templates, and attributed the decline in RNAP activity to the inhibition of binding of the general transcription factor TFIID.
As a positive control, TenHarmsel and Biggin (15) used the TATA-binding protein (TBP), a subunit of the multiprotein TFIID complex that was capable of binding to the circular template. However, whereas TBP was able to support basal transcription on linear templates, RNAP activity remained highly repressed on the circular templates. This result showed that template bending was capable of interfering with some other step(s) in the kinetic pathway of transcription. It remains unclear if transcription from circular templates using TFIID shares the unidentified rate-limiting step when TBP is used. In the context of our observation of highly bent DNA directly repressing an otherwise competent RNAP, it is possible that the observed repression of RNAP II activity originates largely from the mechanical stress sustained within the template itself, where TFIID binding inhibition represents only a minor contribution to the overall repression. The results reported by TenHarmsel and Biggin (15) provide strong evidence of the regulatory role played by the mechanical state of DNA. Yet this important study also highlights the need to use more detailed, reductionistic approaches using precisely characterized DNA templates to determine the underlying molecular interactions between RNAP and mechanically stressed DNA to identify the mechanistic basis of template-induced repression.
Taken together, the observation that DNA bending can repress transcription in both eukaryotic RNAP II (shown by TenHarmsel and Biggin (15)) and an evolutionarily unrelated bacteriophage RNAP (this study) suggest that DNA bending may be a common mechanism capable of regulating gene expression among evolutionarily distant RNAPs. We contend that the existence of DNA loop-forming repressors in species ranging from λ phage and E. coli to Drosophila and yeast (19–21) is suggestive of some shared regulatory feature in these systems. Because the presence of a tightly looped DNA template is a common variable in these otherwise unrelated systems, we consider it possible that the mechanics of the template itself may serve a direct role in repression by loop-forming repressors.
If DNA bending indeed serves as a ubiquitous regulatory mechanism, then previous studies of regulatory proteins that form tightly bent loops within the DNA template should be consistent with this overarching framework. As one illustrative example, let us consider the case of the well-studied lactose repressor LacI. It has been generally accepted that loop formation serves to effectively enhance repressor binding (20,38–40). In the context of a model for repressor function where RNAP activity is simply dependent on the presence or absence of a bound repressor, the loop-dependent increase in repressor binding kinetics seems to explain how loop formation represses gene expression. However, this model is challenged to some extent by the observation that the in vivo repression of E. coli RNAP actually increases as LacI is required to form smaller loops of DNA (6,14). This functional relationship between repression efficiency and loop size does not seem to be explained through considerations of repressor binding kinetics, which should decrease as the length of DNA (and hence bending and twisting flexibility) decreases (40). We propose that the dependence of RNAP activity on loop size could be explained if the model for LacI function were revised to include, as a component of the induced repression, the mechanical stress within the loop. In this revised model, although the rate of loop formation by LacI decreases as the repressor loops become smaller and more energetically costly to bend, once tight loops are formed, RNAP is more efficiently repressed by the significant bending stresses sustained within the loop. Indeed, our data support this model, albeit with an unrelated RNAP.
It has also been suggested that loop-forming repressors inhibit open complex formation by stabilizing the torsional inflexibility of the looped DNA (12). Applying this model to our transcription system, it would be expected that transcriptional repression would be largely relieved as templates sustain increasing levels of untwist. Although we do observe that torsional stress is capable of directly modulating elongation velocity and processivity (Fig. 6), these effects are modest in comparison to the apparent influence of bending on the activity of T7 RNAP. Additionally, although we cannot speak to open complex formation specifically using our molecular beacon-based assay, we note that the rate of EC formation by T7 RNAP on minicircle templates seems to be greater than on the larger, supercoiled plasmid (Fig. 4). Because the rate of EC formation in our assay necessarily includes all steps before elongation (promoter recognition and binding, open complex formation, initiation, and abortive transcription), it is not possible to precisely define the underlying mechanism explaining the observed difference in the rates of EC formation.
In summary, we have developed a transcription assay that unequivocally shows that tight looping of an otherwise competent transcription template can repress transcription elongation by T7 RNAP. Conclusions from this work build on long-standing efforts to understand the relationship between supercoiled DNA and RNAP function. Our data suggest that transcriptional repression by gene repressors known to significantly deform the DNA template may involve, in part, a direct role played by the mechanically stressed DNA template. We therefore propose a model of gene regulation in which targeted genes can be specifically regulated at the DNA level through local mechanical stresses within the template. Given the important role that the mechanical and topological properties of DNA inherently play in all known transcription systems, it will now be of critical importance to apply our assay to more complex, multisubunit RNAP systems to determine if template-induced repression serves as an evolutionarily conserved mechanism common to otherwise unrelated regulatory systems. Given the observed interplay between the torsional state of the DNA template and the elongation velocity and processivity, more directed studies must now be carried out with templates sustaining additional states of torsional stress to precisely characterize the effects of this particular mechanical feature on transcription elongation by T7 RNAP.
Acknowledgments
The authors thank N. Perkins, T. Lillian, and D. Wiener (University of Michigan) for helpful discussions. Cryo-EM images were courtesy of A. Stasiak and D. Demurtas (University of Lausanne).
This work was supported by the National Science Foundation and National Institutes of Health (E.M.), the National Science Foundation Graduate Research Fellowship (T.A.L.), and the National Defense Science and Engineering Graduate Fellowship (T.A.L.).
Supporting Material
References
- 1.Rojo F. Mechanisms of transcriptional repression. Curr. Opin. Microbiol. 2001;4:145–151. doi: 10.1016/s1369-5274(00)00180-6. [DOI] [PubMed] [Google Scholar]
- 2.Wang J.C. Interactions between twisted DNAs and enzymes: the effects of superhelical turns. J. Mol. Biol. 1974;87:797–816. doi: 10.1016/0022-2836(74)90085-0. [DOI] [PubMed] [Google Scholar]
- 3.Mirkin S.M., Bogdanova E.S., Larionov O.A. DNA supercoiling and transcription in Escherichia coli: influence of RNA polymerase mutations. Mol. Gen. Genet. 1979;177:169–175. doi: 10.1007/BF00267267. [DOI] [PubMed] [Google Scholar]
- 4.Revyakin A., Ebright R.H., Strick T.R. Promoter unwinding and promoter clearance by RNA polymerase: detection by single-molecule DNA nanomanipulation. Proc. Natl. Acad. Sci. USA. 2004;101:4776–4780. doi: 10.1073/pnas.0307241101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qureshi M., Eydmann T., Dixon R. Torsional constraints on the formation of open promoter complexes on DNA minicircles carrying sigma 54-dependent promoters. Biochemistry. 1997;36:12303–12316. doi: 10.1021/bi9701179. [DOI] [PubMed] [Google Scholar]
- 6.Muller J., Oehler S., Muller-Hill B. Repression of lac promoter as a function of distance, phase and quality of an auxiliary lac operator. J. Mol. Biol. 1996;257:21–29. doi: 10.1006/jmbi.1996.0143. [DOI] [PubMed] [Google Scholar]
- 7.Kramer H., Niemoller M., Muller-Hill B. lac repressor forms loops with linear DNA carrying two suitably spaced lac operators. EMBO J. 1987;6:1481–1491. doi: 10.1002/j.1460-2075.1987.tb02390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.TenHarmsel A., Austin R.J., Biggin M.D. Cooperative binding at a distance by even-skipped protein correlates with repression and suggests a mechanism of silencing. Mol. Cell. Biol. 1993;13:2742–2752. doi: 10.1128/mcb.13.5.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuhnke G., Theres C., Ehring R. RNA polymerase and gal repressor bind simultaneously and with DNA bending to the control region of the Escherichia coli galactose operon. EMBO J. 1989;8:1247–1255. doi: 10.1002/j.1460-2075.1989.tb03498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Straney S.B., Crothers D.M. Lac repressor is a transient gene-activating protein. Cell. 1987;51:699–707. doi: 10.1016/0092-8674(87)90093-6. [DOI] [PubMed] [Google Scholar]
- 11.Choy H.E., Adhya S. Control of gal transcription through DNA looping: inhibition of the initial transcribing complex. Proc. Natl. Acad. Sci. USA. 1992;89:11264–11268. doi: 10.1073/pnas.89.23.11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choy H.E., Park S.W., Adhya S. Transcription regulation by inflexibility of promoter DNA in a looped complex. Proc. Natl. Acad. Sci. USA. 1995;92:7327–7331. doi: 10.1073/pnas.92.16.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J., Goldfarb A. lac repressor acts by modifying the initial transcribing complex so that it cannot leave the promoter. Cell. 1991;66:793–798. doi: 10.1016/0092-8674(91)90122-f. [DOI] [PubMed] [Google Scholar]
- 14.Oehler S., Amouyal M., Muller-Hill B. Quality and position of the three lac operators of E. coli define efficiency of repression. EMBO J. 1994;13:3348–3355. doi: 10.1002/j.1460-2075.1994.tb06637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.TenHarmsel A., Biggin M.D. Bending DNA can repress a eukaryotic basal promoter and inhibit TFIID binding. Mol. Cell. Biol. 1995;15:5492–5498. doi: 10.1128/mcb.15.10.5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gopal V., Brieba L.G., Sousa R. Characterization of structural features important for T7 RNAP elongation complex stability reveals competing complex conformations and a role for the non-template strand in RNA displacement. J. Mol. Biol. 1999;290:411–431. doi: 10.1006/jmbi.1999.2836. [DOI] [PubMed] [Google Scholar]
- 17.Le Bret M. Catastrophic variation of twist and writhing of circular DNAs with constraint? Biopolymers. 1979;18:1709–1725. doi: 10.1002/bip.1979.360180710. [DOI] [PubMed] [Google Scholar]
- 18.Boles T.C., White J.H., Cozzarelli N.R. Structure of plectonemically supercoiled DNA. J. Mol. Biol. 1990;213:931–951. doi: 10.1016/S0022-2836(05)80272-4. [DOI] [PubMed] [Google Scholar]
- 19.Matthews K.S. DNA looping. Microbiol. Rev. 1992;56:123–136. doi: 10.1128/mr.56.1.123-136.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schleif R. DNA looping. Annu. Rev. Biochem. 1992;61:199–223. doi: 10.1146/annurev.bi.61.070192.001215. [DOI] [PubMed] [Google Scholar]
- 21.Adhya S. Multipartite genetic control elements: communication by DNA loop. Annu. Rev. Genet. 1989;23:227–250. doi: 10.1146/annurev.ge.23.120189.001303. [DOI] [PubMed] [Google Scholar]
- 22.Rong M., Durbin R.K., McAllister W.T. Template strand switching by T7 RNA polymerase. J. Biol. Chem. 1998;273:10253–10260. doi: 10.1074/jbc.273.17.10253. [DOI] [PubMed] [Google Scholar]
- 23.Du Q., Kotlyar A., Vologodskii A. Kinking the double helix by bending deformation. Nucleic Acids Res. 2008;36:1120–1128. doi: 10.1093/nar/gkm1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gray H.B., Jr., Ostrander D.A., Robberson D.L. Extracellular nucleases of Pseudomonas BAL 31. I. Characterization of single strand-specific deoxyriboendonuclease and double-strand deoxyriboexonuclease activities. Nucleic Acids Res. 1975;2:1459–1492. doi: 10.1093/nar/2.9.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skinner G.M., Baumann C.G., Hoggett J.G. Promoter binding, initiation, and elongation by bacteriophage T7 RNA polymerase. A single-molecule view of the transcription cycle. J. Biol. Chem. 2004;279:3239–3244. doi: 10.1074/jbc.M310471200. [DOI] [PubMed] [Google Scholar]
- 26.Zhou Y., Martin C.T. Observed instability of T7 RNA polymerase elongation complexes can be dominated by collision-induced “bumping”. J. Biol. Chem. 2006;281:24441–24448. doi: 10.1074/jbc.M604369200. [DOI] [PubMed] [Google Scholar]
- 27.Mentesana P.E., Chin-Bow S.T., McAllister W.T. Characterization of halted T7 RNA polymerase elongation complexes reveals multiple factors that contribute to stability. J. Mol. Biol. 2000;302:1049–1062. doi: 10.1006/jmbi.2000.4114. [DOI] [PubMed] [Google Scholar]
- 28.Golomb M., Chamberlin M. Characterization of T7-specific ribonucleic acid polymerase. IV. Resolution of the major in vitro transcripts by gel electrophoresis. J. Biol. Chem. 1974;249:2858–2863. [PubMed] [Google Scholar]
- 29.McAllister W.T., Morris C., Studier F.W. Utilization of bacteriophage T7 late promoters in recombinant plasmids during infection. J. Mol. Biol. 1981;153:527–544. doi: 10.1016/0022-2836(81)90406-x. [DOI] [PubMed] [Google Scholar]
- 30.Cheetham G.M., Jeruzalmi D., Steitz T.A. Structural basis for initiation of transcription from an RNA polymerase-promoter complex. Nature. 1999;399:80–83. doi: 10.1038/19999. [DOI] [PubMed] [Google Scholar]
- 31.Fuller F.B. The writhing number of a space curve. Proc. Natl. Acad. Sci. USA. 1971;68:815–819. doi: 10.1073/pnas.68.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glaubiger D., Hearst J.E. Effect of superhelical structure on the secondary structure of DNA rings. Biopolymers. 1967;5:691–696. doi: 10.1002/bip.1967.360050803. [DOI] [PubMed] [Google Scholar]
- 33.Bracco L., Kotlarz D., Buc H. Synthetic curved DNA sequences can act as transcriptional activators in Escherichia coli. EMBO J. 1989;8:4289–4296. doi: 10.1002/j.1460-2075.1989.tb08615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gartenberg M.R., Crothers D.M. Synthetic DNA bending sequences increase the rate of in vitro transcription initiation at the Escherichia coli lac promoter. J. Mol. Biol. 1991;219:217–230. doi: 10.1016/0022-2836(91)90563-l. [DOI] [PubMed] [Google Scholar]
- 35.Rojo F., Salas M. A DNA curvature can substitute phage phi 29 regulatory protein p4 when acting as a transcriptional repressor. EMBO J. 1991;10:3429–3438. doi: 10.1002/j.1460-2075.1991.tb04907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohyama T., Nagumo M., Sakuma S. Alteration of the curved helical structure located in the upstream region of the beta-lactamase promoter of plasmid pUC19 and its effect on transcription. Nucleic Acids Res. 1992;20:1617–1622. doi: 10.1093/nar/20.7.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohyama T. Bent DNA in the human adenovirus type 2 E1A enhancer is an architectural element for transcription stimulation. J. Biol. Chem. 1996;271:27823–27828. [PubMed] [Google Scholar]
- 38.Muller-Hill B. The function of auxiliary operators. Mol. Microbiol. 1998;29:13–18. doi: 10.1046/j.1365-2958.1998.00870.x. [DOI] [PubMed] [Google Scholar]
- 39.Saiz L., Vilar J.M. DNA looping: the consequences and its control. Curr. Opin. Struct. Biol. 2006;16:344–350. doi: 10.1016/j.sbi.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 40.Mossing M.C., Record M.T., Jr. Upstream operators enhance repression of the lac promoter. Science. 1986;233:889–892. doi: 10.1126/science.3090685. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


