Abstract
Versatile superresolution imaging methods, able to give dynamic information of endogenous molecules at high density, are still lacking in biological science. Here, superresolved images and diffusion maps of membrane proteins are obtained on living cells. The method consists of recording thousands of single-molecule trajectories that appear sequentially on a cell surface upon continuously labeling molecules of interest. It allows studying any molecules that can be labeled with fluorescent ligands including endogenous membrane proteins on living cells. This approach, named universal PAINT (uPAINT), generalizes the previously developed point-accumulation-for-imaging-in-nanoscale-topography (PAINT) method for dynamic imaging of arbitrary membrane biomolecules. We show here that the unprecedented large statistics obtained by uPAINT on single cells reveal local diffusion properties of specific proteins, either in distinct membrane compartments of adherent cells or in neuronal synapses.
Introduction
Over the last few years, superresolution optical microscopy techniques have revolutionized biomolecular imaging in cells (1). By pushing optical resolutions down to the scale of individual biomolecules, such techniques give us access to nanoscale molecular organizations. It is now crucial to develop versatile methods that can provide dynamic information about such organizations in living cells, while keeping the high-content information provided by superresolution imaging. In this context, wide-field illumination and camera-based methods are promising for recording fast dynamics on large cellular regions (2,3), compared to those based on highly localized illumination schemes (4). For instance, structured illumination microscopy has recently reached 11-Hz imaging rates at a resolution of ∼100 nm (2). In particular, single molecule imaging can reach very fast dynamics (5) and provide subdiffraction-pointing accuracies, which are only limited by the signal/noise ratio at which the isolated molecules are detected (6).
However, until recently, single molecule studies were restricted to only a few spatially isolated molecules sparsely labeled on living cells (7), despite improvements in image analysis to try to increase this number (8,9). To circumvent this limitation, a number of approaches have emerged that generate reconstructed images of single-molecule localizations at high density (10). The concept behind these approaches is in acquiring collections of images that contain distinct, sparsely located fluorescent entities while keeping the majority of the population in nonemissive states. By controlling their emission properties, a different subset of single emitters is thus imaged in each frame. Superresolved images are then reconstructed by gathering in all collected images, the localizations of the individual emitters determined with subdiffraction resolution. For this aim, blinking of fluorescent probes can be used (11,12), and by adjusting the local redox condition of the molecules, the blinking properties can be tuned on demand (13). In addition, stochastic optical reconstruction microscopy (14) and related methods (15,16) are based on the photoswitching of organic dyes between emissive and nonemissive states. This commonly requires oxygen removal and addition of fluorophore-specific oxidizing and reducing agents. Due to the imaging buffers needed to control the blinking or photoswitching properties of the fluorophores, these methods have been mainly applied to fixed-cells studies (17) or in vitro assays (18). More recently, reducing agents with thiol groups were used to perform dye molecule photoswitching at slightly basic pH. Images of immobile mRNA labeled with fluorescent oligomers could thus be obtained in the nucleus of living cells (19). On the other hand, photoactivatable localization microscopy (PALM) and related methods that use photoactivatable fluorescent proteins (20–22) are directly suitable for superresolution imaging on living cells (23,24). Using evanescent wave illumination, single-particle tracking can be obtained by sptPALM at high densities in the basal membrane of the cells (3). However, sptPALM is constrained in its versatility by the need for transfected fluorescent proteins, which prevents studying endogenous molecules. Furthermore, fusion of a fluorescent protein to a biomolecule of interest might alter its natural behavior (25). Fluorescent proteins also bear photophysical properties (22,26,27) that are not as optimal as some dye molecules (16,28). This currently restricts the trajectory numbers and lengths obtained by sptPALM on a single cell. In this context, none of these methods is readily applicable to study the dynamical properties of endogenous proteins at large densities on living cells.
Another single-molecule-based superresolution imaging approach, named points-accumulation-for-imaging-in-nanoscale-topography (PAINT), was introduced a few years ago (29). PAINT is based on targeting the surface of objects by fluorescent probes that diffuse in the solution and become fluorescent upon binding to the object. By using Nile red, a fluorescent molecule located in the hydrophobic environments of lipid bilayers, PAINT was applied to reveal supported bilayers topographies (29). However, due to the transient insertion (millisecond range) of the fluorescent molecules in the membrane, the dynamic information of molecules could not be obtained. Furthermore, because PAINT is not based on ligands targeting arbitrary biomolecules, it did not find, until now, many applications in biological systems. In order to study any membrane molecule on the surface of living cells, including endogenous proteins, we generalize here the principle of PAINT. Our method is based on continuously and stochastically labeling membrane biomolecules with fluorescent ligands in solution while imaging the samples with oblique illumination (30). This method, named universal PAINT (uPAINT), is a simple technique that provides superresolved images as well as long single-molecule trajectories (up to tens of seconds) at unprecedented high densities (up to 120 μm−2). Analysis of these trajectories produces high-content diffusion maps of the proteins at the surface of the cells. Different fluorophore/ligand combinations can be used depending on the label size, brightness, or photostability required for particular experiments. We demonstrate here the potential and versatility of uPAINT by using several fluorescent ligand systems targeting different membrane proteins. We show that the diffusion of a model transmembrane protein, TM-6His (31), is reduced at the fibroblast cell edge and that GPI-anchored proteins, displaying slow diffusion, accumulate in local domains of COS7 cells. Finally, we applied uPAINT to track endogenous AMPA glutamate receptors (AMPARs) on living neurons, revealing high receptor densities and reduced diffusion in synapses.
Materials and Methods
Cell culture
Immortalized mouse embryonic fibroblasts and COS 7 cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco, Invitrogen, Carlsbad, CA) with 10% fetal bovine serum. Transient transfection of plasmids encoding EGFP, GFP-GPI and TM-6His were performed using Fugene 6 (Roche, Basel, Switzerland). On the day of the experiments, cells were detached with trypsin/EDTA (0.05% for 2 min), the trypsin inactivated with 10% fetal bovine serum DMEM, the cells suspended in serum free condition in Ringer (in mM: 140 NaCl, 5 KCl, 2 CaCl2, 2 MgCl2, 10 HEPES, 11 Glucose, pH 7.4), and incubated for 30 min before plating on collagen I (Roche) glass surface. Experiments were performed 3 h after cell plating. Cultures of hippocampal neurons were prepared from E18 Sprague-Dawley rats after the method previously described (32). Cells were plated at a density of 100–200 × 103 cells/mL on poly-lysine precoated coverslips. To localize excitatory postsynapses, neurons were transfected with Homer-1C::GFP at 8 days in vitro using Effecten (Qiagen, Valencia, CA) and processed two weeks later. Cultures were maintained in serum-free neurobasal medium (Invitrogen, Carlsbad, CA) and kept at 37°C in 7.4% CO2 for 10–15 days in vitro.
TrisNTA-AT647N synthesis
TrisNTA, synthesized as previously described (33), was coupled on solid phase to the N-terminus of the proline linker peptide, PPPPCA. After deprotection and cleavage from the resin, the cysteine residue was labeled with AT647N-maleimide (Atto-Tec, Siegen-Weidenau, Germany) via standard thiol chemistry. Products were purified via reversed-phase C18 HPLC. The trisNTA-AT647N was loaded with nickel and its concentration was determined via the absorption of the fluorophore as described (34).
Anti-GluR2 antibody preparation
The murine anti-GluR2 hybridoma cell lines were prepared by standard procedures, using purified GluR2 receptor in detergent solution as the antigen. From the initial panel of hybridomas, cell culture supernatants were screened for production of antibodies recognizing tertiary epitopes, as previously described (35). The binding of promising candidates to the GluR2 receptor was then verified by fluorescence-detection size-exclusion chromatography (36), using recombinant GluR2 receptor expressed in baculovirus-infected insect cells and solubilized in n-dodecyl-β-D-maltoside containing buffer (37).
Labeling procedure of antibodies
Labeling of the anti-GFP (Roche, Strasbourg, France) antibodies and anti-GluR2 with ATTO647N-NHS-ester (Atto-Tec) or Cy5 (Amersham Biosciences, Uppsala, Sweden) was performed by using modified versions of the manufacturers' procedures. Briefly, 100 μg of antibodies were dialyzed for 6 h in phosphate-buffered saline medium then incubated with 2 μL of ATTO647N-NHS-ester in the presence of NaHCO3 (0.1 M) for 3 h at room temperature. Separation of labeled antibodies from unbounded dyes was performed in size-exclusion columns (Sephadex G25; Pharmacia, New Market, NJ). The resulting fluorophore-antibody labeling ratio was determined by spectrofluorometric measurement. On average, anti-GFP was labeled with 2–3 dye molecules and anti-GLuR2 with 0.8 dye molecules.
Detection and trajectory analysis of single fluorescent ligand molecules
A standard single molecule epifluorescence microscope was used for detecting individual fluorophores (28). In brief, an HeNe laser was used for excitation of samples mounted on an inverted microscope (model No. IX71; Olympus America, Melville, NY) equipped with a high 100× objective (NA = 1.4) and a charge-coupled device camera (Cascade 128; Roper Scientific, Princeton Instruments, Trenton, NJ). In each image of the recorded sequence, the single fluorescent ligands appear as diffraction-limited bright spots. For image analysis, single dye identification is performed by a custom-made program (MATLAB, The MathWorks, Natick, MA) (25). The program obtains the positions and positioning errors for each fluorescent signal (spots) by fitting a two-dimensional Gaussian to the intensity (I) patterns of the spot (28,38,39), as
where n is the image number, An is the amplitude of the Gaussian (given as a grayscale value in the images), and σn is related to the full width at half-maximum of the pattern by FWHM = 2 (2 ln 2)1/2σn. Given the point-spread of our setup, FWHM = 360 ± 40 nm. The parameters xn and yn give the central positions of the spot. The errors in xn and yn, which represent the positioning accuracy, depend on the signal/noise-ratio (6). They are well below the optical resolution limit, typically 50 nm in this work. Importantly, in order to take into account the molecule movement contribution during the 50-ms integration time, this positioning accuracy was measured from the value found at time zero in the mean-square displacement (MSD) analysis of single molecule trajectories (25).
Once the fluorescent spots are detected in each image, the two-dimensional trajectories of single molecules in the plane of focus are constructed by correlation analysis between consecutive images using a Vogel algorithm (25,40). This method connects simultaneously several pairs of spots detected in two successive images by minimizing the deviation of the mean diffusion constant of the connected spots with respect to a given diffusion constant. Because labeling occurs while imaging, only molecules that were present in trajectories that lasted more than several points (five) were kept for further analysis. This allowed us to reject false single-molecule detection that could occur due to noise of the detectors, or any unbound molecules, in order to obtain trajectories long enough for further analysis.
Furthermore, assuming three-dimensional isotropic diffusion for fluorescent ligands, freely diffusing in solution, the diffusion coefficient of these ligands D is well known and relates the ligand hydrodynamic diameter d and the fluid viscosity η by the Stokes-Einstein relation,
where kB is the Boltzmann constant and T is the temperature. For ligand hydrodynamic sizes of d ∼ 5–10 nm, D is ∼40–80 μm2/s and the average time they spend in the two-dimensional light sheet of Δx2≈2-μm thickness is 2Δx2/2D, resulting in times <100 ms. This indicates that unbound fluorescent ligands diffusing in solution do not statistically appear in more than two consecutive images. Such unwanted events were then rejected from the analysis by omitting the first two molecular detections in each trajectory.
Mean-square displacement analysis
For a trajectory of N data points (coordinates x(t), y(t) at times t = 0 to N∗ Δt with Δt = 50 ms), the MSD for time intervals τ = nΔt is calculated using the formulae
For the time interval τ = nΔt, the MSD and its error bar are thus calculated on N-n points for which the individual accuracy is 50 nm. For each MSD, we calculated the instantaneous diffusion coefficient, D, from linear fits of the first 3–5 points (corresponding to 150–250 ms) of the MSD using MSD(τ) = 〈r2〉 (τ) = 4Dτ.
Data visualization
A specific software (SuperRes) was developed to have a comprehensive three-dimensional representation and quantification of the molecules localizations and trajectories obtained by uPAINT. As hundreds of thousands of data points may be provided in a single acquisition, this visualization software was developed in C++ with OpenGL support.
Three different representations/quantifications are available:
First, superresolved images are produced from two-dimensional density maps of the individual molecules localizations obtained from Gaussian fitting with pixel sizes <50 nm. A two-dimensional Gaussian filter of width equal to the pointing accuracy (50 nm) is then applied on each pixel of the two-dimensional density maps to obtain the superresolved images.
Second, the trajectories of the molecules are displayed on a two-dimensional image, with a color-code relating the time the trajectory was detected in the acquisition sequence. Different colors on the same location indicate that different molecules visited the location at different times.
Third, two-dimensional maps of the molecule mobilities are produced by displaying, in a color-coded manner, each image pixel (whose size is chosen by the user, typically 200 nm) and the median of the step lengths (corresponding to the distance between two consecutive points of a trajectory) found in this pixel. Blue pixels correspond to smallest step lengths (thus regions where molecules display low diffusion behaviors) and red to the largest ones.
Results and Discussion
The 633-nm line of a HeNe laser was used to excite either individual ATTO-647N molecules (AT647N), an organic dye with remarkable photostability properties (4) or individual Cy5 molecules. Fluorescent images were obtained with an integration time of 50 ms with up to three stacks of 5000 consecutive charge-coupled device frames. In these imaging conditions, the pointing accuracy of each detected individual fluorophore is 50 nm. Individual fluorescent beads adsorbed on the glass coverslips were used as immobile reference objects to correct long-term mechanical instabilities of the microscope. Oblique illumination of the sample was performed by translating the excitation beam with respect to the axis of the objective (30). This allowed us to image individual fluorescent ligands that are bound to the cell surface without illuminating the molecules in the solution above, decreasing photobleaching and background noise (Fig. 1). The illumination beam angle on the specimen was deduced from the measurement of the beam angle at the output of the microscope objective, taking into account the refractive index of the recording medium as well as the transverse spatial extension of the beam. The angle was chosen to obtain an illumination thickness of Δx ∼2 μm in the center of the field of view such that unbound fluorescent molecules freely diffusing in solution do not statistically appear in more than two consecutive images. Such unwanted events were rejected from the analysis performed with homemade software by omitting the first two molecular detections in each trajectory (see Materials and Methods). Moreover, only resulting trajectories that lasted more than five points were kept for analysis.
Figure 1.
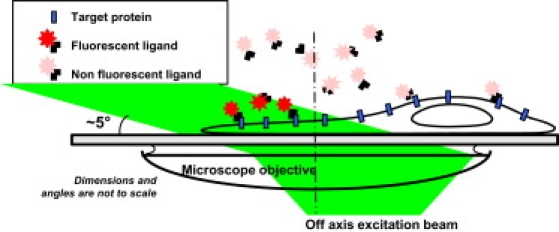
Schematics of the experimental setup. A low concentration of fluorescent ligands is introduced in the extracellular medium such that a constant rate of membrane molecules is being labeled during the imaging sequence. Oblique illumination of the sample is used to excite predominantly fluorescent ligands that have bound to the cell surface while not illuminating the molecules in the above solution.
UPAINT is first applied to study the dynamic organization of a transmembrane protein (TM-6His) expressed in fibroblasts. The TM-6His protein consists of the transmembrane domain of the PDGF receptor fused to a six-histidine (6His) tag on its extracellular side. For labeling, we used the nanometer-sized ligand Ni2+ tris N-nitrilotriacetic acid (trisNTA), which has nanomolar affinity for His-tagged proteins (33,34) conjugated with one AT647N molecule. Fibroblasts, cotransfected with TM-6His and cytosolic GFP to identify transfected cells (Fig. 2 A), were spread on glass coverslips. The ligand was added to the extracellular solution at a final concentration of ∼15 nM immediately before recordings to prevent receptor labeling saturation. Fig. 2 B is a superresolved image constructed from >50,000 single fluorescent spots detected on a 75-μm2 cell area during 12-min experimental recordings. This also provides a two-dimensional spatial map of the molecular densities. Inhomogeneous TM-6His localization is readily observed with higher densities found at the cell edges. After trajectory reconstruction, >2000 single-molecule trajectories are obtained on the same area (Fig. 2 C). From 19,191 trajectories, an average trajectory density of 45 ± 8 μm−2 (mean ± SE, n = 4 cells, Fig. 2 G) is found in 12 min, reaching up to 120 μm−2 in some subcellular areas. This corresponds to a local rate of 10 trajectories per μm2/min. As a control, we performed similar experiments on cells transfected with cytosolic GFP only. In this case, a low density of trajectories is found on the cell surface (<5 μm−2 trajectories, Fig. 2 G, and Fig. S1 in the Supporting Material). Such trajectories correspond to trisNTA fluorescent ligands, which have bound nonspecifically to the cell membranes. The number of trajectories obtained here are orders-of-magnitude larger than those obtained by methods using consecutive labeling steps (41) and significantly larger than those obtained by sptPALM (3).
Figure 2.
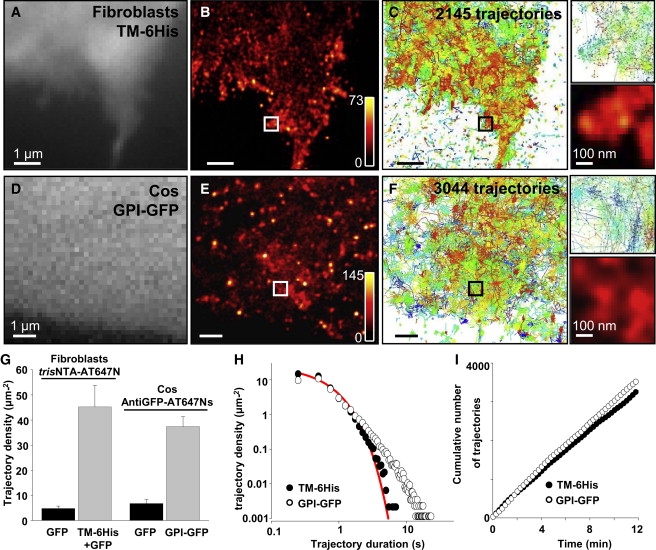
uPAINT imaging with two different ligand/receptor systems. (A) Wide-field fluorescence image of a fibroblast expressing TM-6His and GFP. (B) Superresolved image of TM-6His labeled with trisNTA-AT647N obtained by uPAINT and (C) corresponding trajectories. (D–F) Same as panels A–C, but for GPI-GFP labeled with anti-GFP-AT647Ns on a COS 7 cell. (G) Surface density of the trajectories recorded on TM-6His/GFP or GPI-GFP transfected cells (19,191 and 31,936 trajectories, respectively), and on GFP-transfected control cells for the two protein/ligand complexes (n = 4–5 cells in each condition). (H) Distributions of the trajectory lengths measured with trisNTA-AT647N and anti-GFP-ATTO-647Ns. (Solid line) Fit with an exponential curve. (I) Cumulative number of trajectories as a function of time for the two protein-ligand complexes.
The versatility provided by uPAINT is also demonstrated by imaging and tracking GPI-anchored GFP (GPI-GFP) expressed at the surface of COS 7 cells. In these experiments, the fluorescent ligand was a monoclonal anti-GFP IgG antibody carrying either Cy5 or AT647N molecules. The use of Cy5 dyes, although successful, resulted in shorter trajectories than with AT647N because of the decreased photostabilities of Cy5 when linked to antibodies (42) (not shown). AT647N, molecules were thus preferred (Fig. 2, D–F). Unlike standard single-molecule methods, single ligands carrying multiple fluorophores can be used with uPAINT while ensuring that the biomolecules of interest are studied at the single-molecule level. Indeed, real-time visualization of individual ligands binding to their target molecules ensures that the majority of diffraction-limited fluorescent-spots found in trajectories stems from individual labeled molecules. This feature can be used to increase the trajectory lengths by coupling multiple dye molecules per ligand. We coupled ∼2–3 AT647N molecules per anti-GFP antibodies and reduced the excitation intensity accordingly (∼0.5 kW/cm2 compared to ∼1 kW/cm2 for trisNTA-AT647N imaging). Fig. 2 H shows the normalized histograms of the trajectory lengths obtained for trisNTA-AT647N and anti-GFP-AT647Ns.
In the first case, which corresponds to ligands coupled to one AT647N only, 13% of the trajectories lasted >1 s. In the second case where multiple fluorophores per ligand are used, this proportion increased significantly and reached 22% (n = 19,191 trajectories, four cells; and 31,936 trajectories, five cells, respectively). In both cases, these proportions are significantly larger than obtained by sptPALM, where only 3% of the trajectories last >750 ms (3). Interestingly, when ligands coupled to individual fluorophores are used, the trajectory length distribution is reasonably well adjusted by an exponential curve (solid line in Fig. 2 H). This indicates that the trajectories are terminated by stochastic events. The trajectory lengths are, in principle, a function of two factors: the photobleaching and the dissociation rate of the fluorescent ligands. In our case, the dissociation rate, which is low (1.7 10−4 s−1 for trisNTA-AT647N/TM-6His, see Fig. S2), did not play a significant role on the timescale of the trajectories; thus, photobleaching was the limiting stochastic process. We also noticed that, for anti-GFPs carrying multiple AT647N dyes, the trajectory length distribution cannot be adjusted by a monoexponential curve (Fig. 2 H). The heterogeneity in the dye number carried by each anti-GFP can account for this effect. The number of trajectories obtained during a given time by uPAINT is a function of the ligand affinity (especially through the association rate constant), the density of molecules at the cell surface, and the concentration of fluorescent ligand in solution. Under our experimental conditions, we found that the rate of detected molecules was constant during the 12-min acquisition time (Fig. 2 I). Given the low trisNTA-AT647N/TM-6His dissociation rate compared to the timescale of the experiments, this indicates that TM-6His labeling was not saturating. A constant labeling rate was also found for GPI-GFP at the same experimental conditions (Fig. 2 I).
The large number of trajectories obtained by uPAINT allows statistical analysis to be performed not only on individual cells but also between subcellular regions of a given cell. First, we built a histogram of the diffusion constants of GPI-GFP molecules from 24,100 trajectories. This high-content histogram from only five cells revealed the presence of two distinct mobile populations as well as molecules having a diffusion constant <10−3 μm2/s and representing <3.5% of the population (Fig. 3 A). Interestingly, the high statistics obtained by uPAINT allowed us to depict the contribution of ligands which have bound nonspecifically to the cell membrane or to the glass. For instance, Fig. 3 A shows that trajectories with diffusion constants <10−3 μm2/s mainly correspond to nonspecific binding events and not ligands bound to GPI-GFP molecules. To reveal information about diffusion heterogeneities in specific subcellular regions, we produced superresolved images (Fig. 3 B) and two-dimensional spatial maps of the displacement steps of the molecules during 50 ms (Fig. 3 C). From such images, a correlation diagram between the molecule surface density and the displacement steps can be obtained on each subcellular region with high statistics (Fig. 3 D). Such analysis unambiguously indicates that GPI-GFP, which is known to be enriched in lipid microdomains (43), is concentrated and displays slower diffusion in micrometer domains, which are stable on the minute timescale (Fig. S3, A–C).
Figure 3.
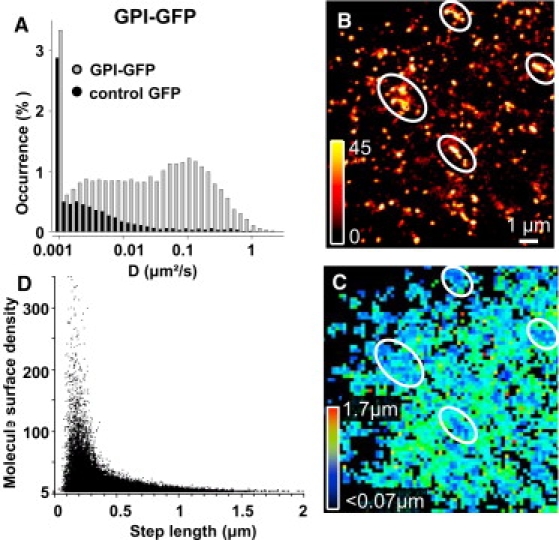
Statistical analysis of GPI-GFP spatio-temporal localization on COS 7 cells. (A) Distribution of instantaneous diffusion constants for GPI-GFP on COS 7 cells. Control corresponds to cells expressing GFP alone. (B) Four microdomains presenting a high surface density of GPI-GFP are highlighted on a superresolved image. (C) Corresponding color-coded two-dimensional map of the mean GPI-GFP step lengths during 50 ms within 200 × 200 nm2 pixels. (D) Two-dimensional plot of the molecule densities as a function of mean step lengths measured within 200 × 200 nm2 pixels (n = 487,112 steps, five cells) revealing reduced movements in the high density regions.
A similar analysis was performed for TM-6His molecules in fibroblasts (Fig. 4). Similar to GPI-GFP, the high content histogram of TM-6His diffusion constants indicates that the majority of TM-6His are mobile (Fig. 4 A), but with a much larger proportion of molecules in the fast mobile population than for GPI-GFP. Two-dimensional spatial maps of the molecular densities (Fig. 4 B) and displacement steps (Fig. 4 C) were also produced. Strikingly, Fig. 4 C indicates that TM-6His diffusion is notably reduced at the fibroblast cell edges (∼1 μm) (Fig. S3, D–F). This effect is more evident in Fig. 4 D where the histograms of displacements steps are constructed as a function of subcellular localization. This observation can be attributed to specific protein/lipid compositions and membrane curvatures that can affect the membrane topology and molecular crowding near the cell edges (44).
Figure 4.
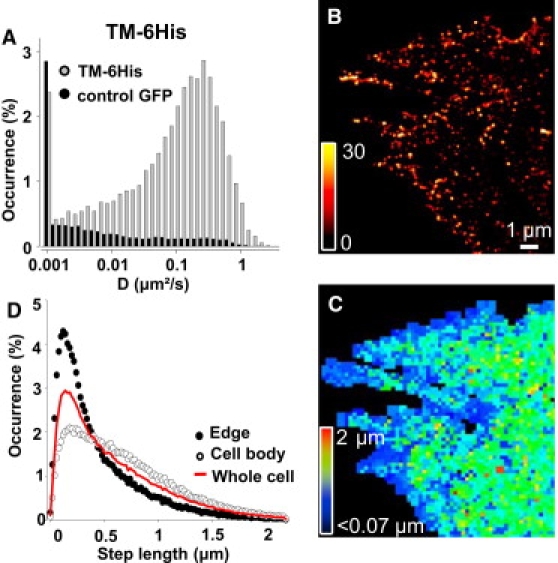
Statistical analysis of TM-6His spatio-temporal localization on fibroblasts. (A) Distributions of instantaneous diffusion constants for TM-6His on fibroblasts. (B) Superresolved image and (C) corresponding color-coded two-dimensional map of the mean TM-6His step lengths during 50 ms within 200 × 200 nm2 pixels, revealing reduced movements at the cell edges. (D) Step-length distributions for TM-6His (n = 214,089 steps, n = 4) during 50 ms without localization information, at the cell edges and on the cell body.
Finally, we applied uPAINT to study endogenous proteins on living cells (Fig. 5). For demonstration, we choose AMPA glutamate receptors (AMPARs), which are well studied on live cultured neurons (7,28,45). For labeling, anti-GluR2 antibodies that bind with high affinity to three-dimensional epitopes on the native receptor were used. Similar to anti-GFP, anti-GluR2s were labeled with multiple AT647Ns to obtain long trajectories of GluR2 containing AMPARs. To differentiate synaptic versus extrasynaptic receptors, synapses were identified by the excitatory postsynaptic protein Homer1C::GFP (7,46) (Fig. 5 B). Experiments were conducted at 35°C. High-density tracking and superresolved images indicate that AMPARs accumulate in synaptic zones, due to the good accessibility of fluorescent ligands in the restricted subcellular environment of synaptic clefts (25) (Fig. 5, C and D). This feature is more evident when zooming on dendritic spines (Fig. 5, G and H). Indeed, on a single spine, the largest AMPAR accumulation and immobilization colocalizes with the postsynaptic density visualized by Homer-1C::GFP. The high-content histogram of the diffusion constants obtained from 19,412 trajectories in 11 imaging fields reveals one population of immobile receptors and two main populations of mobile receptors with median diffusions of 0.1 μm2/s and 6.3 × 10−3 μm2/s (Fig. 5 E). This is in accordance with previous low-density single-molecule studies (7,32). The two-dimensional diffusion map (Fig. 5, F and I) and correlation diagram between the molecule surface density and the displacement steps (inset of Fig. 5 E) indicate that the slowly mobile AMPARs are mainly located in regions of high AMPAR accumulation, which includes synaptic sites and their vicinities (28,46).
Figure 5.
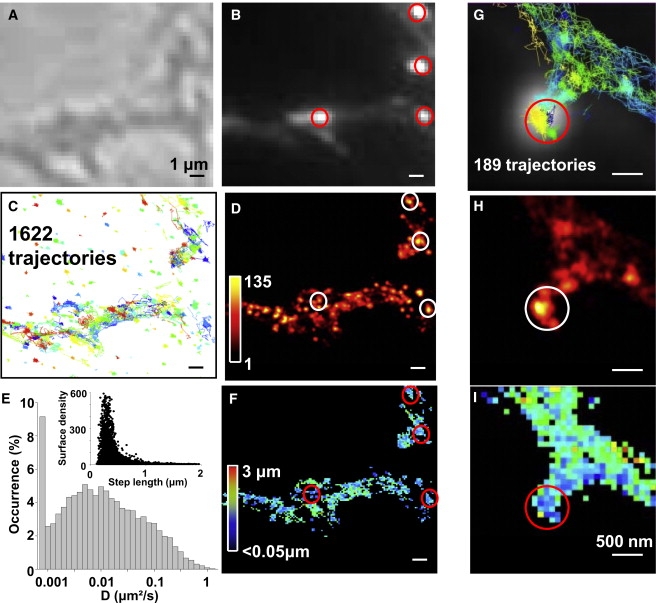
uPAINT on endogenous AMPA receptors in live neurons. (A) Wide-field Nomarski and (B) fluorescence image of live neurons transfected with Homer1C::GFP to reveal postsynaptic sites (red outlines). (C) Trajectories (n = 1622) of GluR2 containing AMPARs labeled with antiGluR2-AT647Ns. (D) Superresolved image corresponding to panels A–C. (E) Distributions of instantaneous diffusion constants (19,412 trajectories, 11 fields of view). (Inset) Two-dimensional plot of the molecule densities as a function of mean step lengths measured within 100 × 100 nm2 pixels revealing reduced movements in the high density regions. (F) Color-coded two-dimensional-map of the mean AMPARs step lengths during 50 ms within 100 × 100 nm2 pixels (n = 28,446 steps) corresponding to panels A–D. (G–I) Zoom on a dendritic spine, scale bar 500 nm. (G) Trajectories (n = 189) superimposed with fluorescent image of Homer1C::GFP. Corresponding superresolved image (H) and diffusion maps (I), as in panels E and F.
Conclusions
By providing a large level of dynamic information concerning the diffusion and localization properties of individual membrane molecules with subwavelength accuracies, uPAINT bridges the gap between single-molecule detection and ensemble live cell imaging. The method can be implemented on a standard single-molecule microscope operating with oblique illumination. In particular, uPAINT allows recording unprecedented densities of long single-molecule trajectories on a single cell. In addition, it has the unique advantage to be suitable for endogenous molecule studies in living cells. Interestingly, this method can be easily applied to multicolor labeling—opening the route for colocalization and molecular interaction studies at the nanometer scale, on living cells, with high-content information collected.
Acknowledgments
We thank O. Thoumine, A. Frouin, and C. Breillat for technical help, O. Rossier, B. Lounis, and C. Leduc for helpful discussions, and A.N.G. Parra-Vasquez for critical reading of the manuscript.
This research was funded by Centre National de la Recherche Scientifique (CNRS), the Région Aquitaine, and the Agence Nationale pour la Recherche. E.H. thanks the CNRS for financial support. The design and synthesis of the trisNTA-fluorophores by R.T. was supported by the German Research Foundation and the Federal Ministry of Education and Research (BMBF) program Nanobiotechnology (grant Nos. 0312031 and 0312034 to R.T.). The preparation and selection of the 14B11 GluR2 antibodies was supported by the National Institutes of Health (A.I.S., M.P.R., and E.G.). E.G. is an investigator with the Howard Hughes Medical Institute.
Footnotes
Michael P. Rosconi's present address is Regeneron Pharmaceuticals, Protein Chemistry Sciences, 777 Old Saw Mill River Road, Tarrytown, NY 10591.
Supporting Material
References
- 1.Hell S.W. Far-field optical nanoscopy. Science. 2007;316:1153–1158. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- 2.Kner P., Chhun B.B., Gustafsson M.G. Super-resolution video microscopy of live cells by structured illumination. Nat. Methods. 2009;6:339–342. doi: 10.1038/nmeth.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manley S., Gillette J.M., Lippincott-Schwartz J. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat. Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- 4.Westphal V., Rizzoli S.O., Hell S.W. Video-rate far-field optical nanoscopy dissects synaptic vesicle movement. Science. 2008;320:246–249. doi: 10.1126/science.1154228. [DOI] [PubMed] [Google Scholar]
- 5.Fujiwara T., Ritchie K., Kusumi A. Phospholipids undergo hop diffusion in compartmentalized cell membrane. J. Cell Biol. 2002;157:1071–1081. doi: 10.1083/jcb.200202050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bobroff N. Position measurements with a resolution and noise-limited instrument. Rev. Sci. Instrum. 1986;57:1152–1157. [Google Scholar]
- 7.Heine M., Groc L., Choquet D. Surface mobility of postsynaptic AMPARs tunes synaptic transmission. Science. 2008;320:201–205. doi: 10.1126/science.1152089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sergé A., Bertaux N., Marguet D. Dynamic multiple-target tracing to probe spatiotemporal cartography of cell membranes. Nat. Methods. 2008;5:687–694. doi: 10.1038/nmeth.1233. [DOI] [PubMed] [Google Scholar]
- 9.Jaqaman K., Loerke D., Danuser G. Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods. 2008;5:695–702. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang B., Bates M., Zhuang X. Super-resolution fluorescence microscopy. Annu. Rev. Biochem. 2009;78:993–1016. doi: 10.1146/annurev.biochem.77.061906.092014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lidke K.A., Rieger B., Heintzmann R. Superresolution by localization of quantum dots using blinking statistics. Opt. Express. 2005;13:7052–7062. doi: 10.1364/opex.13.007052. [DOI] [PubMed] [Google Scholar]
- 12.Steinhauer C., Forthmann C., Tinnefeld P. Superresolution microscopy on the basis of engineered dark states. J. Am. Chem. Soc. 2008;130:16840–16841. doi: 10.1021/ja806590m. [DOI] [PubMed] [Google Scholar]
- 13.Vogelsang J., Cordes T., Tinnefeld P. Controlling the fluorescence of ordinary oxazine dyes for single-molecule switching and superresolution microscopy. Proc. Natl. Acad. Sci. USA. 2009;106:8107–8112. doi: 10.1073/pnas.0811875106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rust M.J., Bates M., Zhuang X.W. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat. Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heilemann M., van de Linde S., Sauer M. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew. Chem. Int. Ed. Engl. 2008;47:6172–6176. doi: 10.1002/anie.200802376. [DOI] [PubMed] [Google Scholar]
- 16.Bossi M., Fölling J., Hell S.W. Multicolor far-field fluorescence nanoscopy through isolated detection of distinct molecular species. Nano Lett. 2008;8:2463–2468. doi: 10.1021/nl801471d. [DOI] [PubMed] [Google Scholar]
- 17.van de Linde S., Sauer M., Heilemann M. Subdiffraction-resolution fluorescence imaging of proteins in the mitochondrial inner membrane with photoswitchable fluorophores. J. Struct. Biol. 2008;164:250–254. doi: 10.1016/j.jsb.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 18.Endesfelder U., van de Linde S., Heilemann M. Subdiffraction-resolution fluorescence microscopy of myosin-actin motility. ChemPhysChem. 2010;11:836–840. doi: 10.1002/cphc.200900944. [DOI] [PubMed] [Google Scholar]
- 19.Heilemann M., van de Linde S., Sauer M. Super-resolution imaging with small organic fluorophores. Angew. Chem. Int. Ed. Engl. 2009;48:6903–6908. doi: 10.1002/anie.200902073. [DOI] [PubMed] [Google Scholar]
- 20.Betzig E., Patterson G.H., Hess H.F. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 21.Hess S.T., Girirajan T.P., Mason M.D. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys. J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Egner A., Geisler C., Hell S.W. Fluorescence nanoscopy in whole cells by asynchronous localization of photoswitching emitters. Biophys. J. 2007;93:3285–3290. doi: 10.1529/biophysj.107.112201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hess S.T., Gould T.J., Zimmerberg J. Dynamic clustered distribution of hemagglutinin resolved at 40 nm in living cell membranes discriminates between raft theories. Proc. Natl. Acad. Sci. USA. 2007;104:17370–17375. doi: 10.1073/pnas.0708066104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shroff H., Galbraith C.G., Betzig E. Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat. Methods. 2008;5:417–423. doi: 10.1038/nmeth.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Groc L., Lafourcade M., Cognet L. Surface trafficking of neurotransmitter receptor: comparison between single-molecule/quantum dot strategies. J. Neurosci. 2007;27:12433–12437. doi: 10.1523/JNEUROSCI.3349-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harms G.S., Cognet L., Schmidt T. Autofluorescent proteins in single-molecule research: applications to live cell imaging microscopy. Biophys. J. 2001;80:2396–2408. doi: 10.1016/S0006-3495(01)76209-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Habuchi S., Ando R., Hofkens J. Reversible single-molecule photoswitching in the GFP-like fluorescent protein Dronpa. Proc. Natl. Acad. Sci. USA. 2005;102:9511–9516. doi: 10.1073/pnas.0500489102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tardin C., Cognet L., Choquet D. Direct imaging of lateral movements of AMPA receptors inside synapses. EMBO J. 2003;22:4656–4665. doi: 10.1093/emboj/cdg463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharonov A., Hochstrasser R.M. Wide-field subdiffraction imaging by accumulated binding of diffusing probes. Proc. Natl. Acad. Sci. USA. 2006;103:18911–18916. doi: 10.1073/pnas.0609643104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharonov A., Bandichhor R., Hochstrasser R.M. Lipid diffusion from single molecules of a labeled protein undergoing dynamic association with giant unilamellar vesicles and supported bilayers. Langmuir. 2008;24:844–850. doi: 10.1021/la702600w. [DOI] [PubMed] [Google Scholar]
- 31.Chen I., Howarth M., Ting A.Y. Site-specific labeling of cell surface proteins with biophysical probes using biotin ligase. Nat. Methods. 2005;2:99–104. doi: 10.1038/nmeth735. [DOI] [PubMed] [Google Scholar]
- 32.Groc L., Heine M., Choquet D. Differential activity-dependent regulation of the lateral mobilities of AMPA and NMDA receptors. Nat. Neurosci. 2004;7:695–696. doi: 10.1038/nn1270. [DOI] [PubMed] [Google Scholar]
- 33.Lata S., Gavutis M., Piehler J. Specific and stable fluorescence labeling of histidine-tagged proteins for dissecting multi-protein complex formation. J. Am. Chem. Soc. 2006;128:2365–2372. doi: 10.1021/ja0563105. [DOI] [PubMed] [Google Scholar]
- 34.van der Does C., Presenti C., Tampé R. Kinetics of the ATP hydrolysis cycle of the nucleotide-binding domain of Mdl1 studied by a novel site-specific labeling technique. J. Biol. Chem. 2006;281:5694–5701. doi: 10.1074/jbc.M511730200. [DOI] [PubMed] [Google Scholar]
- 35.Shaffer P.L., Goehring A., Gouaux E. Structure and mechanism of a Na+-independent amino acid transporter. Science. 2009;325:1010–1014. doi: 10.1126/science.1176088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawate T., Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 2006;14:673–681. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 37.Sobolevsky A.I., Rosconi M.P., Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt T., Schütz G.J., Schindler H. Imaging of single molecule diffusion. Proc. Natl. Acad. Sci. USA. 1996;93:2926–2929. doi: 10.1073/pnas.93.7.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheezum M.K., Walker W.F., Guilford W.H. Quantitative comparison of algorithms for tracking single fluorescent particles. Biophys. J. 2001;81:2378–2388. doi: 10.1016/S0006-3495(01)75884-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmidt T., Schuetz G.J., Schindler H. Characterization of photophysics and mobility of single molecules in a fluid lipid membrane. J. Phys. Chem. 1995;99:17662–17668. [Google Scholar]
- 41.Guignet E.G., Segura J.-M., Vogel H. Repetitive reversible labeling of proteins at polyhistidine sequences for single-molecule imaging in live cells. ChemPhysChem. 2007;8:1221–1227. doi: 10.1002/cphc.200700065. [DOI] [PubMed] [Google Scholar]
- 42.Gruber H.J., Hahn C.D., Knaus H.G. Anomalous fluorescence enhancement of Cy3 and Cy3.5 versus anomalous fluorescence loss of Cy5 and Cy7 upon covalent linking to IgG and noncovalent binding to avidin. Bioconjug. Chem. 2000;11:696–704. doi: 10.1021/bc000015m. [DOI] [PubMed] [Google Scholar]
- 43.Jacobson K., Mouritsen O.G., Anderson R.G.W. Lipid rafts: at a crossroad between cell biology and physics. Nat. Cell Biol. 2007;9:7–14. doi: 10.1038/ncb0107-7. [DOI] [PubMed] [Google Scholar]
- 44.McMahon H.T., Gallop J.L. Membrane curvature and mechanisms of dynamic cell membrane remodeling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 45.Petrini E.M., Lu J.Y., Choquet D. Endocytic trafficking and recycling maintain a pool of mobile surface AMPA receptors required for synaptic potentiation. Neuron. 2009;63:92–105. doi: 10.1016/j.neuron.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sugiyama Y., Kawabata I., Okabe S. Determination of absolute protein numbers in single synapses by a GFP-based calibration technique. Nat. Methods. 2005;2:677–684. doi: 10.1038/nmeth783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


