Abstract
SNAP-25B is a neuronal protein required for neurotransmitter (NT) release and is the target of Botulinum Toxins A and E. It has two SNARE domains that form a four-helix bundle when combined with syntaxin 1A and synaptobrevin. Formation of the three-protein complex requires both SNARE domains of SNAP-25B to align parallel, stretching out a central linker. The N-terminal of the linker has four cysteines within eight amino acids. Palmitoylation of these cysteines helps target SNAP-25B to the membrane; however, these cysteines are also an obvious target for oxidation, which has been shown to decrease SNARE complex formation and NT secretion. Because the linker is only slightly longer than the SNARE complex, formation of a disulfide bond between two cysteines might shorten it sufficiently to reduce secretion by limiting complex formation. To test this idea, we have carried out molecular dynamics simulations of the SNARE complex in the oxidized and reduced states. Indeed, marked conformational differences and a reduction of helical content in SNAP-25B upon oxidation are seen. Further differences are found for hydrophobic interactions at three locations, crucial for the helix-helix association. Removal of the linker induced different conformational changes than oxidation. The simulations suggest that oxidation of the cysteines leads to a dysfunctional SNARE complex, thus downregulating NT release during oxidative stress.
Introduction
SNAP-25B is a critical protein found in mature neurons where it is required for proper release of neurotransmitters. Its nearly identical isoform, SNAP-25A, is expressed early in development (1). The key role of SNAP-25B seems to be the formation of a protein complex leading to fusion of synaptic vesicles with the plasma membrane. The complex forms sequentially, with SNAP-25B first interacting with syntaxin 1A, anchored at the active zone of the cell membrane, and then with synaptobrevin within the synaptic vesicle membrane. The three proteins form a four-helix bundle with SNAP-25B contributing two of the four α-helices (helix 1 and helix 2). Each helix, known as a SNARE domain, is composed of ∼63 amino acids. The helices wind together in a parallel fashion, forming layers of highly conserved interacting residues (2). As a consequence, the two SNARE domains of SNAP-25B must stretch out a central linker region of 58 amino acids in order to align. The SNARE complex has been crystallized showing the coiled-coil nature of the three-protein complex (3), but no structure has been determined for the SNAP-25B linker.
The two SNARE domains of SNAP-25 are connected by a linker region, which has several unique features. First, removal of the linker slows down SNARE assembly (4) and decreases secretion from intact chromaffin cells (5). Secondly, the linker contains a conserved region of amino acids that aid in targeting SNAP-25 to the membrane (6). This membrane-binding domain is close to the N-terminal of the linker (Fig. 1 A) and includes a cysteine-rich domain that contains four cysteines within an eight-amino-acid section. Cysteines contribute to the targeting of SNAP-25 to the membrane through the palmitoylation of one or more of them (7,8). Binding to syntaxin may also help target SNAP-25 to membranes (9), but this is controversial (10).
Figure 1.
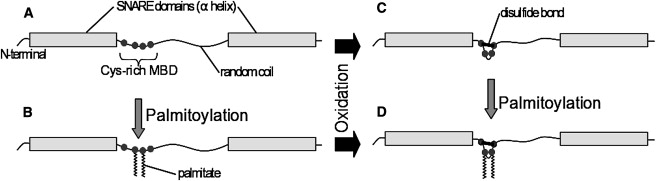
Putative SNAP-25 response to oxidative stress. (A) “Naked” SNAP-25 with its two SNARE domains and four cysteines (C85, C88, C90, and C92). (B) In vivo, SNAP-25 is posttranslationally palmitoylated, which adds 1–4 palmitic acids to cysteines within the weak membrane-binding domain (MBD); palmitoylation of the middle two cysteines is shown. (C) Oxidation of SNAP-25 could result in disulfide bond formation between any of the free cysteines (one of six possibilities is shown). (D) A specific example of a SNAP-25 with the inner two cysteines palmitoylated and the outer cysteines disulfide-linked.
It has generally been assumed that all four cysteines of SNAP-25 are palmitoylated and thus function solely in membrane targeting, but several other functions for these cysteines have been implied. Huang et al. (11) showed that the four cysteines of SNAP-25A can form an iron-sulfur cluster, although there is no data that such a cluster actually forms in vivo. Pallavi and Nagaraj (12) reported that peptides from the linkers of SNAP-25/23 can enhance membrane fusion depending on the extent of palmitoylation. Giniatullin et al. (13) showed that oxidation of SNAP-25 was the likely cause of the reduced secretion observed in neurons exposed to oxidative stress. Such stress is known to play a role in many neurodegenerative disorders (e.g., Alzheimer's and Parkinson's) (14,15). Although the site of oxidative modification was not identified, cysteines are the most susceptible residues and frequently are involved in redox control within cells through the formation of disulfide bonds (16).
Thus, the four cysteines of SNAP-25 may function as sites of palmitoylation, in membrane fusion, in heavy metal coordination, or in disulfide bond formation. These potential modifications need not be exclusive, because all three are reversible and one modification might have a physiological effect simply by preventing a different modification. Additionally, cysteine oxidation and partial palmitoylation could occur simultaneously (Fig. 1 D). Only two cysteines are required to form a single disulfide bond and palmitoylation of two cysteines would still provide a strong membrane anchor for the protein. The extent of palmitoylation appears to depend on cell type and environment. In HIT (insulin-secreting) cells, palmitoylation is not critical for function, because the replacement of all four cysteines still allowed significant secretion (8), but in PC12 cells the same modification prevented exocytosis (17).
The possibility that cysteine oxidation in SNAP-25 may have functional significance is intriguing, but brings two question to mind.
First, under what conditions might SNAP-25 be oxidized and how might oxidation appropriately alter the function of SNAP-25? The location of SNAP-25 at the synapse places it at a good location to sense oxidative stress from internal sources (mitochondria) and from the extracellular environment. Under such stresses, neuron survival would likely be enhanced if the energy-requiring steps of neurosecretion were temporarily reduced. Although the redox state of the intracellular environment is generally reduced, reactive oxygen species produced after intense neuronal activity (16,18,19) could oxidize the cysteines of SNAP-25. As previously noted by Giniatullin et al. (13), oxidation of SNAP-25 decreases formation of the SNARE complex and thus could be the direct cause of decreased secretion.
Second, and this is the focus of this work, how might oxidation of SNAP-25 prevent complex formation? We hypothesize that because the linker is only slightly longer than the SNARE domain, formation of a disulfide bond between two cysteines could shorten the loop sufficiently to hinder or even block the proper alignment of the two SNARE domains in SNAP-25B and thus also the formation of the complete coiled-coil complex. To test this idea, we carried out multiple molecular dynamics (MD) simulations, which indeed showed several marked conformational changes of SNAP-25B induced by formation of a single disulfide bond.
Methods
In a first step, the length decreases due to the 10 different possibilities for disulfide bonds were estimated from short-term simulations of the involved peptide. The one with the largest effect was selected and studied in subsequent extended simulations of the whole solvated SNARE complex.
Ten different configurations of the stretch of 18 amino acids from the SNAP-25 linker, (G K F C (1) G L C (2) V C (3) P C (4) N K L K S S D), were constructed. The configurations differed in the number and position of disulfide bonds: none, C1–C3, C2–C4, C2–C3, C1–C4, C1–C2, C3–C4, C1–C4, and C2–C3, C1–C3, and C2–C4, and C1–C2 and C3–C4.
Each configuration was then relaxed and stretched through 41 iterations using proportional atom translations followed by energy minimization with the CHARMM22 force field (20). After each iteration, a 12.5 × 5 × 5 nm water box and 0.154 mol/L K+Cl− were added.
From each of the obtained 410 systems, an MD run was started using CHARMM version c35b1 (21) with the CHARMM22 force field, a time step of 2 fs, and periodic boundaries using a central cell of 12.5 × 5 × 5 nm. The N- and C-termini were restrained at their original separations with an energy penalty of 1000 kcal × mol−1 nm−1. Coordinates were stored every 2 ps. All bonds to hydrogen atoms were held rigid using the ShakeH algorithm as implemented in CHARMM. Langevin dynamics were used to maintain the pressure at 1 atm with a collision frequency of 20 ps−1, a pressure piston mass of 400 amu, and a piston bath temperature of 300 K. Each system was heated from 48 K to 298 K for 50 ps and equilibrated for another 50 ps. For each of the 410 trajectories, average distances between the N-terminus nitrogen and the C-terminus carbon and average energies were computed.
The SNARE complex system was constructed from the structure by Sutton et al. (3) (PDB code: 1SFC). It contains four α-helix fragments, where the C-terminal of helix 1 and the N-terminal of helix 2 do not extend as far as the other helices and the linker between them and other parts of the proteins have been removed by proteolysis. A more recent crystal structure of the SNARE complex, that includes the membrane-spanning region of syntaxin and synaptobrevin, shows helices extending continuously through the SNARE domain and membrane-spanning domain (22). Therefore, the short ends of helices 1 and 2 were extended by several coils to match the length of the other helices. All residues between helix 1 and 2 (C88-M127) were added as an unstructured linker.
After energy minimization, the complete complex was solvated in a dodecahedron box with a minimum distance of 1.5 nm between the proteins and the box boundary. Ions were added at a physiological concentration of 0.154 mol L−1 with the program GENION from the GROMACS suite (23). The total system size was ∼360,000 atoms. After a second energy-minimization, the solvent was equilibrated for 1 ns, with position restraints on the heavy atoms of the proteins using a force constant of k = 1000 kJ mol−1 nm−2.
Five sets of simulations were carried out, termed:
“Reduced” (without disulfide bond)
“Oxidized” (with disulfide bond)
“Cut-linker uncharged”
“Cut-linker charged” (with the linker cut between residues N107 and N108, using uncharged and charged termini for the linker, respectively)
“No-linker” (removed linker residues, N93–Q126; charged termini).
For the “reduced” simulations, the energy-minimized system was used. For the “oxidized” simulations, the sulfur atoms of the SNAP-25B residues C85 and C92 were pulled toward each other through gradually switching on the respective disulfide bond during a 1-ns simulation. For the “cut-linker charged” simulations, the peptide bond between N107 and N108 was removed, charged termini were added, and the system was simulated for 1 ns with position restraints on the protein heavy atoms. The same protocol was applied for the “cut-linker uncharged” simulations. Additionally, a simulation with removed linker (N93–Q126) was carried out (“no-linker”). All five systems were subsequently equilibrated without restraints for at least 100 ns.
To create five independent trajectories for each of the five models, four more simulations each were forked at 10, 20, 30, and 40 ns, respectively, with new velocities assigned. After 100 ns of the “oxidized” simulations, snapshots from the trajectory were taken, the disulfide bond was removed, and the system was energy-minimized and subsequently equilibrated with position restraints on the backbone atom for 1 ns. Subsequently, new simulations without position restraints were started (“removed disulfide”).
All simulations were carried out with GROMACS 4 (23), the OPLS all-atom force field (24), and the TIP4P (25) water model. Velocity rescaling (26) at T = 300 K with a coupling time constant of τT = 0.1 ps and a Parrinello-Rahman barostat (27) at p = 1 atm, a τp = 1 ps coupling time constant, and an 4.5 × 10−5 bar−1 isotropic compressibility was used. All bond lengths were constrained with the LINCS algorithm (28). Electrostatic interactions were calculated pairwise below 0.9 nm. Long-range electrostatic interactions were calculated by particle-mesh Ewald summation (29) with a grid spacing of 0.12 nm. Lennard-Jones interactions were calculated below 1.4 nm. Coordinates were recorded every 1 ps.
Root mean-square deviations (RMSD values) were calculated after fitting to the backbone of the helical residues present in the crystal structure.
To monitor the angles between different parts of the SNAP-25B helix, the orientation of each part was calculated for each frame by averaging the coordinates of the backbone atoms of every four successive residues, yielding positions close to the central axis of the helix. The eigenvector with the largest eigenvalue of the covariance matrix of these averaged coordinates was then used as orientation vector. For each frame, the angle between respective orientation vectors was recorded.
Secondary structure content was determined with the program DSSP (30).
Results and Discussion
To test the hypothesis that the formation of a disulfide bond upon oxidation of two cysteines shortens the SNAP-25B linker sufficiently to hinder or slow down SNARE complex formation, we proceeded in two steps. First, we screened for the influence of all 10 possible disulfide bonds on the linker length with geometry optimizations. Second, to investigate the effect of the most restrictive disulfide bond on the SNARE complex, we modeled the complete complex with extended SNAP-25B helix ends and SNAP-25B linker, and compared simulations with (“oxidized”) and without disulfide bond (“reduced”). Additionally, simulations with the cut-linker and with removed-linker were carried out to investigate the effects of the linker on the SNARE complex and to separate these effects from the effect of the mechanical strain generated by the shortening of the linker.
Influence of different disulfide bonds on linker length
To estimate how much the linker shortens after formation of different disulfide bonds, we generated all six possible single-disulfide linkers and the three double-disulfide linkers and compared the length of each to the linker without disulfides. Using the N-terminal 18 amino acids of the linker for each of the 10 possibilities, molecular modeling was carried out for varying distance between the N- and C-terminal amino acids. Fig. 2 shows the energy of each system as a function of end-to-end distance imposed by constraints on the termini followed by energy minimization. As expected for elastic stretching, each curve can be fitted well by a semiharmonic function shifted by an offset. The offset change with respect to the offset of the reduced state was used to estimate the amount of shortening produced by the respective disulfide linkages (inset of Fig. 2 and Table S1 in the Supporting Material). The largest shortening (2.0 nm) was found for the C85–C92 single-disulfide and for the C85–C92 and C88–C90 double-disulfide. The single-disulfide bond 1–4 was therefore chosen for the MD simulations of the whole complex.
Figure 2.
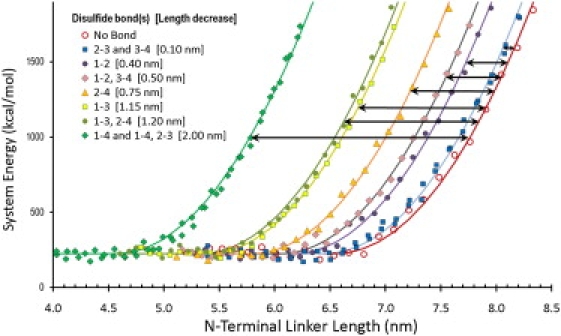
N-terminal linker system energy as a function of end-to-end distance for different combinations of disulfide bonds in SNAP-25B. Semiharmonic functions, , were used to fit the data with a common spring coefficient k and adjustable offsets Δx. (Brackets and solid arrows) Length decreases for the nine disulfide linkages with respect to the reduced state.
Effects of the disulfide bond on the complete SNARE complex
The disulfide bond was formed after pulling the respective sulfur atoms toward each other as described in Methods from an initial separation of 15 Å to 2.7 Å. To investigate the effects of the resulting linker shortening on the SNARE complex, five 140-ns “oxidized” simulations were started from the obtained model. For comparison, five 180-ns “reduced” simulations without disulfide bond were carried out. Fig. 3, B and C, shows ribbon representations of the “oxidized” and “reduced” starting structures. If our hypothesis is correct, one would expect that the SNARE complex starts to unfold in the “oxidized” simulations, while remaining stable in the “reduced” simulations. To further check whether conformational changes seen during the first 100 ns of the “reduced” simulations are reversible on short timescales, the disulfide bond in the oxidized structures was removed at 100 ns, and additional 100-ns simulations (“removed disulfide”) were started.
Figure 3.
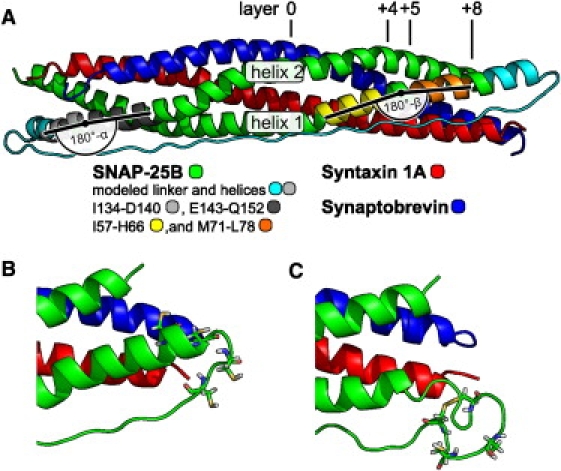
Ribbon representation of the model of the SNARE complex. (A) The complete model, (B) close up on cysteines for the model and (C) for the established disulfide bond, respectively. (Green, dark gray, orange, and yellow) Residues of SNAP-25B seen in the crystal. (Cyan and light gray) Modeled residues. (Red) Syntaxin. (Blue) Synaptobrevin. (Light gray, dark gray, yellow, and orange) Regions of the SNAP-25B helices that define the angles α and β, respectively. Location of the helix-helix interaction layers 0, +4, +5, and +8 are shown in part A.
Fig. 4 shows the mean RMSD of each residue for each of the three setups with respect to the modeled structure, calculated from the final 40 ns of each trajectory. As seen, the residues that compose helices in the crystal structure (“core” helices; dark shaded bars) show small RMSD values for all three proteins and for all setups. In contrast, the residues close to the termini exhibit larger RMSD values and also larger variances, reflecting their increased flexibility. The largest RMSD values are observed for the modeled SNAP-25B residues, including the newly established ends of the helices (light shaded bar) and the linker.
Figure 4.
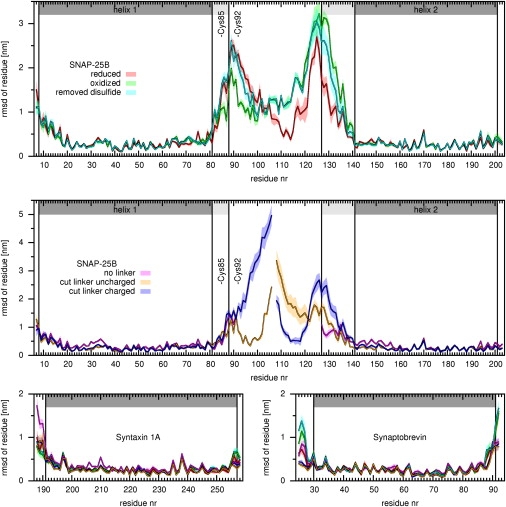
Structural deviation for the different setups discussed in the text (colors): Shown are mean RMSD values (bold lines) and standard errors (shaded area) of each residue, determined from all five simulations of each setup and averaged over the final 40 ns. (Medium- and light-shaded background) The helical part of the crystal structure and the modeled helix ends, respectively.
Comparing the “oxidized” and “reduced” states in the region of the two cysteines C85 and C92, the RMSD values are smaller for the “oxidized” than for the “reduced” simulations. In contrast, the RMSD values of residues 106–124 and 126–132 are significantly larger for the “oxidized” state. Remarkably, these differences are seen at distances from the disulfide bond as large as 4.8 up to 11.2 nm. When comparing the “removed disulfide” simulations with those of the two other setups, it can be seen that the RMSD values for the residues in the central part of the linker (residues 98–127) are more similar to those of the “oxidized” than to those of the “reduced” simulations. In contrast, the RMSD values for residues 127–140 fall between the values for the “oxidized” and “reduced” simulations.
Overall, the RMSD values reflect a marked influence of the disulfide bond on the linker conformation and in particular on the conformation of the newly established helix ends. In contrast, no influence on the “core” helices is seen. Further, the RMSD values calculated for the “removed disulfide” simulations show that parts of the structure start to approach the conformation of the “reduced” simulations.
Fig. 5, A–C, shows the final SNARE complex conformations obtained from the five trajectories of the “reduced”, “oxidized”, and “removed disulfide” simulations. The most pronounced difference between the conformations of the “oxidized” and “reduced” simulations is a kink of the partially unfolded extended helix end of helix 2, residues I134–D140 (light shaded), Fig. 5 B, which is not seen in Fig. 5 A. Notably, the same part of helix 2 is straight at the end of two of the “removed disulfide” simulations (Fig. 5 C). In addition, the C-terminal end of helix 1 becomes bent for three of five “oxidized” structures compared to the conformations obtained for the “reduced” structures.
Figure 5.
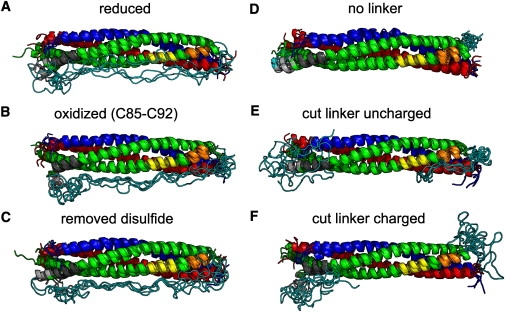
Ribbon representation of the final frames of the simulations. For each setup, the conformations at the end of the five trajectories are overlaid. (Colors as in Fig. 3.)
To analyze these drastic structural changes, Fig. 6, A and B, shows the angle α between two parts of helix 2, I134–D140 and E143–Q152 (light and dark shading in Fig. 3), during the simulation, and its frequency distribution. The angle fluctuates around a mean value of 30° for all “reduced” simulations, whereas it increases to >80° for the five “oxidized” simulations. Remarkably, in one of the “removed disulfide” simulations, α reverts to <30° within 60 ns. Additionally, in the “removed disulfide” simulation that started with a low value of α, the helix remained stretched. The three other trajectories did not yet revert to low α values within the 100–130 ns simulation time, although one of them tends toward the stretched conformation. This result shows that reverting pathways exist.
Figure 6.
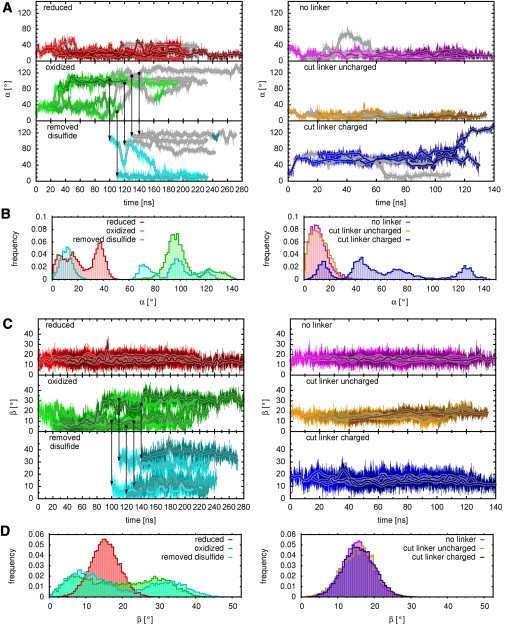
Kinking of the SNAP-25B helices: (A) Angle α between parts of helix 2, I134–D140 and E143–Q152, as a function of time. (Shaded lines) Parts of the trajectories in which the fraction of residues from I134–D140 in an α-helical secondary structure is <0.5. (B) Histogram of the angle α from the final 40 ns of each simulation. (C) Angle β between parts of helix 1, L57–H66 and M71–L78, as a function of simulation time and (D) a histogram of the angle β from the final 40 ns of each simulation. (Solid vertical lines) Points in time where the disulfide bond was removed from snapshots of the “oxidized” simulations and the “removed disulfide” simulations were started.
Closer structural inspection of the trajectories reveals that, overall, the α-helical content of the helix extension decreases during the simulations compared to α-helical content of the initial model. Those parts of the trajectories, for which the fraction of the residues I134–D140 that are in an α-helical secondary structure is <0.5, are shown as gray lines in Fig. 6 A. In the “reduced” simulations, the trajectories alternate between states where the fraction is either < or >0.5. In contrast, the fraction is <0.5 at the end of four of five “oxidized” simulations. In two cases, the helix first kinked and then unfolded, whereas in two other cases the helix first unfolded and then α increased. This finding suggests that the equilibrium conformation of these residues is much closer to random coil in the “oxidized” state than it is for the “reduced” state.
In the “removed disulfide” simulation in which the helix was kinked but not unfolded, the angle decreased. Further, in the simulation, which started with unkinked and helical residues I134–D140, these residues remained helical and the angle remained small. In contrast, only slightly decreasing angles are observed for the simulations that started with a large angle and unfolded residues I134–D140. A short refolding of the extended helix is only observed in one of these cases.
The kinking at the opposite C-terminal part of helix 1 was characterized by the angle β between L57–H66 and M71–L78 (colored yellow and orange in Fig. 3). As above, Fig. 6, C and D, shows β and its frequency distribution. For the “reduced” trajectories, β fluctuates around a mean value of 15°; however, in the “oxidized” simulations β increases to 30° in two of the trajectories. Further, in the “removed disulfide” simulations, which start with a large value of β, the angle does not decrease to the values of the “reduced” state.
To examine whether these drastic structural differences between oxidized and reduced SNAP-25B also affect the central hydrophobic layers that are crucial for the association of the four SNARE helices (31), RMSD values for the model structure for the residues of each layer were calculated after appropriate Cα fitting. Fig. 7 A shows the mean RMSD values for each of the 16 interacting layers −7 through +8 and each setup, averaged over the final 40 ns of the respective trajectories. Error bars were estimated from the observed RMSD standard deviation. The RMSD values of the layers are small compared, e.g., to the linker region, but also compared to the modeled helices (see Fig. 4), which implies that the layers are quite rigid. Further, the RMSD values for the different setups are similar for most of the layers. However, marked differences in RMSD values between the “oxidized” and “reduced” states are seen in layers +4, +5, and +8, where the RMSD values for the “oxidized” state are larger.
Figure 7.
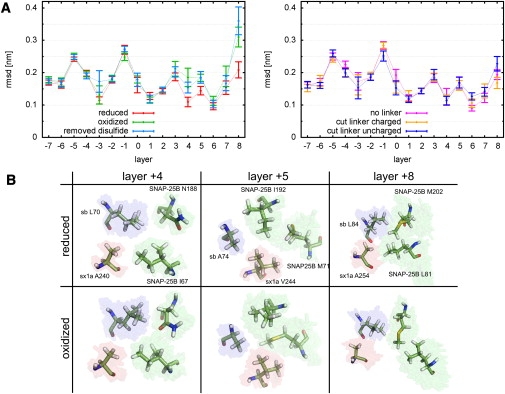
Structural deviations of the residues which contribute to the hydrophobic layers for the six simulations discussed in the text. (A) Shown are the means and the standard errors for the RMSD values calculated from the final 40 ns of each simulation. The RMSD of each layer was calculated after fitting the positions of the Cα-atoms to those of the modeled structure. (B) (Stick representation) For “reduced” and “oxidized” setups SNAP-25B, syntaxin 1A (sx1a), and synaptobrevin (sb) residues contributing to layers +4, +5, and +8. (Solid sticks) One conformation at the end of a trajectory. (Colored region) An ensemble from the final 40 ns of all trajectories.
In particular, the largest difference between RMSD values for these two states is observed for layer +8, with residue L81 of SNAP-25B located only four residues away from C85. Remarkably, the conformational differences expand further into the SNARE complex (layer +4 and +5). The RMSD values of the “removed” disulfide setup are close to those of the setup with disulfide bond; thus, the conformations of the layers did not change drastically after the removal of the disulfide bond during the relatively short simulations.
Fig. 7 B shows the conformations of the residues contributing to layers +4, +5, and +8 during the final 40 ns of each trajectory of the “reduced” and “oxidized” state. The residues of layer +4 show increased flexibility in the “oxidized” state. In layer +5, the residues also show more conformational diversity in the “oxidized” ensemble than in the “reduced” ensemble. In particular, the methyl group of M71 also occupies the center of the layer in the “oxidized” simulations. The residues of layer +8 show drastic conformational differences between the “reduced” and “oxidized” simulations. L81 of SNAP-25B moves out of the center of the layer and loses contact with the residues L84 of synaptobrevin and A254 of syntaxin 1A. M202 of SNAP-25B also moves out of the center, maintaining contact with L81. In summary, oxidation of C85 and C92 is found to induce significant rearrangements in these layers.
Role of the SNAP-25B linker
To further investigate the role of the SNAP-25B linker, simulations with the linker (N93–Q126) removed (“no-linker”) and with the linker cut (“cut-linker”) between residues N107 and N108 were performed. Both setups remove the mechanical stress exerted by the linker, whereas only the latter setup retains most nonbonded interactions between linker and helices. Comparison of these simulations with the previous simulations should therefore allow us to distinguish between effects caused by the mechanical strain of the linker and those caused by purely nonbonded interactions.
Two sets of simulations were performed: one set of simulations where the termini of the linker were charged (“cut-linker charged”) and a second set where the termini were left uncharged (“cut-linker uncharged”). The first set mimics SNAP-25B cut chemically by a protease, whereas the second set is intended to remove the mechanical strain only, without further chemical or electrostatic modifications.
To characterize the structural differences among the three setups (i.e., “no-linker”, “cut-linker uncharged”, and “cut-linker charged”), Fig. 4 shows the RMSD of the residues with respect to the initial model of the complex. The RMSD values of the residues in the two “cut-linker” setups show significant differences in the loop region, which suggests that the charge of the termini strongly influences the conformation of the loop. Similar to the setups with the complete linker, the RMSD values of the “core” helices (dark shaded bars) in the simulations with cut linker and without linker are small. Furthermore, the RMSD values calculated for residues 25–35 and 142–156 from the “no linker” trajectories are significantly larger than those of the “cut-linker” trajectories.
The ribbon representations of the obtained “no-linker” conformations (see Fig. 5 D) show that the modeled helix end (I134–D140, light shading) is still α-helical and the helix is bent to a lesser extent than in the “reduced” setup (Fig. 5 A). The cut-linker with uncharged termini (Fig. 5 E) behaves very differently from the cut linker with charged termini (Fig. 5 F). Whereas the part of the cut-linker carrying the N-terminal interacts with the helices when charged, and the modeled helix-end (I134–D140) is kinked and unfolds in three out of five simulations, it is very flexible when uncharged and interacts only weakly with the helices. In addition, the SNAP-25 helix remains unkinked. In contrast, the C-terminal of the linker is less flexible in the simulations with the uncharged termini. For the uncharged linker, helix 2 of SNAP-25 is bent to a lesser extent as compared to the structures of the “reduced” setup, implying that the mechanical stress caused by the linker suffices to bend the helix.
The kink angle α of helix 2 (Fig. 6, A and B) calculated from the “cut-linker uncharged” and “no-linker” simulations remains at values at ∼10° during the simulation, whereas the five “cut-linker charged” simulations are heterogeneous.
Obviously, and despite its thermodynamic stability, the SNARE complex is very sensitive to these small changes. In particular, as the “cut-linker” simulations show, the outcome of an experiment that includes cutting of the linker likely depends on the chemical details.
The bending angle of helix 1 β for the “no-linker”, “cut-linker uncharged”, and “cut-linker charged” simulations (Fig. 6, C and D) fluctuates around a mean value of 15°, similar to the “reduced” trajectories, which suggests that the influence of the reduced linker on the bending of helix 1 is small.
The RMSD values of the hydrophobic layers (Fig. 7 A) from the three setups without linker or with a cut linker are similar to the RMSD values calculated for the “reduced” setup, except for layers –4, –3, and +6. In layers –4 and –3, the “no-linker” simulations show a larger RMSD value than for the “reduced” simulations. Additionally, layer –3 for the “cut-linker charged” simulations and layer +6 for the “cut-linker uncharged” simulations show slightly larger RMSD values as compared to the “reduced” simulations. This finding suggests that the different states, cut- and removed linker, lead to conformational changes in some of the layers but the influence is not as pronounced as it is in the oxidized state.
Helicity of SNAP-25B
For the residues of the SNAP-25B helix “cores” (with the first and last two residues omitted), the fraction of residues contributing to an α-helix was recorded during the simulations (see Fig. S1 A in the Supporting Material). The histograms over the final 40 ns of each trajectory show that the helical content for the “oxidized” state is markedly smaller than for the “reduced” state. The distribution for the “removed disulfide” simulations is similar to that for the “reduced” simulations. Further, the simulations with a cut linker show large frequencies for large helical contents, whereas the distribution is flatter for the simulations with the removed linker.
These results suggest that the disulfide bond causes a partial unfolding of the helices. After its removal, the helices do not refold during the relatively short simulation times. The fact that the distribution is shifted to larger helical content for the cut-linker simulations as compared to the “reduced” setup suggests that the secondary structure is mainly perturbed by the mechanical stress induced by the linker rather than by interactions with the complex. However, the fact that complete removal of the linker further enhances the frequencies of states with lower helical content suggests that these interactions further stabilize the secondary structure.
To identify the origin of the differences in the overall helicity of SNAP-25B, Fig. S1 B analyzes which SNAP-25B residues contribute to the loss of helical content. Shown is the frequency of α-helical secondary structure, calculated for each SNAP-25B residue from the final 40 ns of each trajectory. For the “no-linker” simulations, helicity is lower in three out of five cases around residues 53 and 155. None of the other data sets shows a loss of helicity at these residues, suggesting that the linker interacts specifically at these locations.
Conclusions
We studied how the formation of a disulfide bond in the SNAP-25B linker region affects the conformation of the SNARE complex. The SNARE complex is a coiled-coil of proteins critical for membrane fusion, and in neurons is composed of three proteins: syntaxin 1A, synaptobrevin, and SNAP-25B. In SNAP-25B, there are four cysteines within a region of eight amino acids of the linker. Formation of a single disulfide bond (oxidation) between C85 and C92 shortens the linker by 2.0 nm (Fig. 2).
To that aim, the complete SNARE complex was simulated in explicit solvent with both reduced and oxidized SNAP-25B linker. Marked conformational changes after oxidation were seen, as compared to both the “reduced” state as well as the oxidized and subsequently reduced (“removed disulfide”) state. Not surprisingly, formation of the disulfide bond was found to reduce the flexibility of the linker in the region of the disulfide bond (residues 85–96), but unexpectedly conformational changes were also seen at the far end of the linker (residues 126–132), which is 10-nm apart. Removal of the linker was seen to destabilize the SNARE complex, as testified by an RMSD increase in several regions of helix 1 and 2 for the “no-linker” setup (e.g., close to residue 155 of Fig. 4) and by a decrease in helix frequency in some of these regions as compared to the “reduced” setup (Fig. S1 B). However, when the individual residues at each layer of the complex were examined, as in Fig. 7 A, there is no marked difference between “reduced-“ and “no-linker” at the corresponding layers –7 and –6, whereas marked differences are found nearby at layers –4 and –3. Because the data in Fig. 7 A were obtained by fitting the coordinates of each layer separately, the increase in RMSD seen in Fig. 4 reflects a general bending or rotation of the whole complex after removal of the linker. Although our simulations, taken alone, do not establish functional significance of this bending, removal of the linker is known to have profound effects in vivo (5,32).
Closer examination of both ends of the linker showed a strong bend at the end of helix 2 and, somewhat weaker, within helix 1. These two bends, characterized by angles α and β (Fig. 3 A), show quite different behavior after formation of the disulfide bond (Fig. 6). The β-kink is particularly interesting because it resides within helix 1 and forms part of the SNARE complex.
With respect to oxidation of SNAP-25, a key difference between the two SNAP-25 homologs is the relocation of the third cysteine of SNAP-25B at C88 upstream to C84 (Table 1) in SNAP-25A. The significance of this change in the two isoforms is not known, but we would predict that such a change would make SNAP-25A more sensitive than SNAP-25B to oxidative stress and iron exposure. This prediction is based on three different possible molecular mechanisms, any of which would lower stability of the SNARE complex:
-
1.
The formation of a C84-C92 disulfide bond (in SNAP-25A) would shorten the linker by one more amino acid (∼2.4 nm instead of 2.0 nm), and thus, would more strongly prevent complete zippering of the SNARE complex.
-
2.
The formation of a disulfide bond at C84 would more likely unwind the C-terminal of helix 1, leading to greater disruption of layer +8 and/or larger changes in angle β.
-
3.
The new cysteine configuration in SNAP-25A allows it to bind iron (11), which would also shorten the linker and suppress complex formation.
If the two isoforms have similar sensitivity to oxidative stress, a reasonable scenario is also that the alternate splicing endows SNAP-25A with iron sensitivity, which would render the complex sensitive to both oxidation and iron exposure and thus allow the synapse to shut down under either environmental stress. Such a response may have developmental consequences, because SNAP-25A is expressed early in development (1).
A possible explanation for the alternate splicing between SNAP-25A and SNAP-25B is also that the two different sequences may alter the effectiveness of cellular enzymes to modify each protein. For example, the extent of palmitoylation of each cysteine in each isoform will depend on its effectivity as a target for the relevant palmitoylation enzyme. The stability of palmitoylation will depend on its substrate efficiency for any depalmitoylation enzyme (33). Thus, rearrangements of cysteines could change enzyme binding and thereby explain the apparent inconsistencies reported on the stability of palmitoylation of SNAP-25. Specifically, Lane and Liu (32) reported that in PC12 cells, palmitoylation of SNAP-25 (primarily SNAP-25A) rapidly turned over with a half-life of 3 h, whereas Kang and associates (34,35) observed no turnover in primary neuronal cultures (primarily SNAP-25B).
A report on SNAP-23 (36) is consistent with the interpretation that cellular palmitoylation of SNAP-25 is cysteine-dependent and not simply a nonspecific modification of all cysteines. SNAP-23 is a cellular homolog of the neuronal SNAP-25, and has yet another arrangement of cysteines in its linker (Table 1). Vogel and Roche (36) report that in transiently transfected HeLa cells, the extent of SNAP-23 palmitoylation was only 4% of that of SNAP-25. If the palmitoyl transferase nonspecifically palmitoylated every cysteine in each linker, one would expect SNAP-23 to be palmitoylated by 25% more than SNAP-25, because it has one more cysteine than SNAP-25. Instead, the change in sequence drastically reduced palmitoylation of SNAP-23 (or enhanced depalmitoylation). These data support a model consistent with Fig. 1, in which only certain cysteines are specifically palmitoylated/depalmitoylated, potentially leaving other cysteines as targets for oxidation.
Table 1.
Amino-acid sequence of the cysteine-rich regions of several SNAP-25B homologs
| Protein |
Amino-acid sequence |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 84 | 85 | 88 | 90 | 92 | ||||||||||||||
| SNAP-25B | D | L | G | K | F | C | G | L | C | V | C | P | C | N | K | L | K | S |
| SNAP-25A | D | L | G | K | C | C | G | L | F | I | C | P | C | N | K | L | K | S |
| SNAP-23A | E | L | N | K | C | C | G | L | C | V | C | P | C | N | R | T | K | N |
| SNAP-23B | E | L | N | K | C | C | G | L | C | V | C | P | C | N | S | I | T | N |
Numbering corresponds to both SNAP-25s. For SNAP-23, the first cysteine is at 79.
Unpalmitoylated cysteines, although susceptible to oxidation, would normally not be oxidized due to the reducing environment maintained in healthy cells; however, during times of high-energy output, reactive oxygen species (ROS) produced by the mitochondria could oxidize newly synthesized SNAP-25 (16,18,19). Another possibility for cellular oxidation of SNAP-25 is by external ROS that cross the cell membrane. Because SNAP-25 binds membranes (even without palmitoylation), it might be oxidized by such entering ROS. This would also be true after partial palmitoylation of SNAP-25 by a membrane-bound palmitoyl transferase, because the palmitoylation of even one cysteine would bury the nearby free cysteines deeper in the membrane where they become isolated from the reducing environment of the cell and exposed to external oxidative stress. Finally, it is possible that a disulfide bond-containing SNAP-25 is a better target for palmitoylation by the relevant palmitoyl transferase and that it is only after oxidation, palmitoylation, and reduction that the SNAP-25 becomes available for its normal role in exocytosis. All these scenarios are, of course, speculative, but testable.
In our simulations, formation of the disulfide bond also altered helix-helix interactions at layers +4, +5, and +8. This change was noted as an increase in the RMSD of residues in these layers (Fig. 7 A) and a shift in the averaged location of the residues in helix 1 of SNAP-25 (Fig. 7 B). Sørensen et al. (31) investigated the effects of mutations in the hydrophobic layers on exocytosis. Mutations in the layer closest to the cysteines (+8) slowed down secretion, and even more so with an additional mutation in layer +7. Further, mutations in layer +4 and +5 also drastically decreased secretion. Therefore, our observations of changes in layers +4, +5, and +8 in the oxidized SNAP-25B are consistent with the hypothesis that an oxidation-induced conformational change may be a chemomechanical regulator of SNARE complex formation, leading to downregulation of NT release during oxidative stress.
Acknowledgments
The authors thank David D. Busath for valuable discussions and guidance throughout this project and Jon Willes and S. Jason Bluth for their significant time and effort in preliminary modeling studies. We thank R. Jahn for helpful discussions and for reading the manuscript.
Contributor Information
Helmut Grubmüller, Email: hgrubmu@gwdg.de.
Dixon J. Woodbury, Email: dixon_woodbury@byu.edu.
Supporting Material
References
- 1.Bark C., Bellinger F.P., Wilson M.C. Developmentally regulated switch in alternatively spliced SNAP-25 isoforms alters facilitation of synaptic transmission. J. Neurosci. 2004;24:8796–8805. doi: 10.1523/JNEUROSCI.1940-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fasshauer D., Sutton R.B., Jahn R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl. Acad. Sci. USA. 1998;95:15781–15786. doi: 10.1073/pnas.95.26.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sutton R.B., Fasshauer D., Brunger A.T. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 4.Misura K.M., Gonzalez L.C., Jr., Weis W.I. Crystal structure and biophysical properties of a complex between the N-terminal SNARE region of SNAP25 and syntaxin 1a. J. Biol. Chem. 2001;276:41301–41309. doi: 10.1074/jbc.M106853200. [DOI] [PubMed] [Google Scholar]
- 5.Wang L., Bittner M.A., Holz R.W. The structural and functional implications of linked SNARE motifs in SNAP25. Mol. Biol. Cell. 2008;19:3944–3955. doi: 10.1091/mbc.E08-04-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gonzalo S., Greentree W.K., Linder M.E. SNAP-25 is targeted to the plasma membrane through a novel membrane-binding domain. J. Biol. Chem. 1999;274:21313–21318. doi: 10.1074/jbc.274.30.21313. [DOI] [PubMed] [Google Scholar]
- 7.Veit M., Söllner T.H., Rothman J.E. Multiple palmitoylation of synaptotagmin and the t-SNARE SNAP-25. FEBS Lett. 1996;385:119–123. doi: 10.1016/0014-5793(96)00362-6. [DOI] [PubMed] [Google Scholar]
- 8.Gonelle-Gispert C., Molinete M., Sadoul K. Membrane localization and biological activity of SNAP-25 cysteine mutants in insulin-secreting cells. J. Cell Sci. 2000;113:3197–3205. doi: 10.1242/jcs.113.18.3197. [DOI] [PubMed] [Google Scholar]
- 9.Vogel K., Cabaniols J.P., Roche P.A. Targeting of SNAP-25 to membranes is mediated by its association with the target SNARE syntaxin. J. Biol. Chem. 2000;275:2959–2965. doi: 10.1074/jbc.275.4.2959. [DOI] [PubMed] [Google Scholar]
- 10.Loranger S.S., Linder M.E. SNAP-25 traffics to the plasma membrane by a syntaxin-independent mechanism. J. Biol. Chem. 2002;277:34303–34309. doi: 10.1074/jbc.M202125200. [DOI] [PubMed] [Google Scholar]
- 11.Huang Q., Hong X., Hao Q. SNAP-25 is also an iron-sulfur protein. FEBS Lett. 2008;582:1431–1436. doi: 10.1016/j.febslet.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pallavi B., Nagaraj R. Palmitoylated peptides from the cysteine-rich domain of SNAP-23 cause membrane fusion depending on peptide length, position of cysteines, and extent of palmitoylation. J. Biol. Chem. 2003;278:12737–12744. doi: 10.1074/jbc.M208598200. [DOI] [PubMed] [Google Scholar]
- 13.Giniatullin A.R., Darios F., Giniatullin R. SNAP25 is a pre-synaptic target for the depressant action of reactive oxygen species on transmitter release. J. Neurochem. 2006;98:1789–1797. doi: 10.1111/j.1471-4159.2006.03997.x. [DOI] [PubMed] [Google Scholar]
- 14.Butterfield D.A., Perluigi M., Sultana R. Oxidative stress in Alzheimer's disease brain: new insights from redox proteomics. Eur. J. Pharmacol. 2006;545:39–50. doi: 10.1016/j.ejphar.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 15.Facheris M., Beretta S., Ferrarese C. Peripheral markers of oxidative stress and excitotoxicity in neurodegenerative disorders: tools for diagnosis and therapy? J. Alzheimers Dis. 2004;6:177–184. doi: 10.3233/jad-2004-6210. [DOI] [PubMed] [Google Scholar]
- 16.Janssen-Heininger Y.M., Mossman B.T., van der Vliet A. Redox-based regulation of signal transduction: principles, pitfalls, and promises. Free Radic. Biol. Med. 2008;45:1–17. doi: 10.1016/j.freeradbiomed.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Washbourne P., Cansino V., Wilson M.C. Cysteine residues of SNAP-25 are required for SNARE disassembly and exocytosis, but not for membrane targeting. Biochem. J. 2001;357:625–634. doi: 10.1042/0264-6021:3570625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finkel T. Oxygen radicals and signaling. Curr. Opin. Cell Biol. 1998;10:248–253. doi: 10.1016/s0955-0674(98)80147-6. [DOI] [PubMed] [Google Scholar]
- 19.Giniatullin A.R., Grishin S.N., Giniatullin R.A. Reactive oxygen species contribute to the presynaptic action of extracellular ATP at the frog neuromuscular junction. J. Physiol. 2005;565:229–242. doi: 10.1113/jphysiol.2005.084186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brooks B., Bruccoleri R., Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 21.Brooks B.R., Brooks C.L., 3rd, Karplus M. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stein A., Weber G., Jahn R. Helical extension of the neuronal SNARE complex into the membrane. Nature. 2009;460:525–528. doi: 10.1038/nature08156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hess B., Kutzner C., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 24.Kaminski G., Friesner R., Jorgensen W. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J. Phys. Chem. B. 2001;105:6474–6487. [Google Scholar]
- 25.Jorgensen W., Chandrasekhar J., Klein M. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 26.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 27.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 28.Hess B., Bekker H., Fraaije J. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 29.Essmann U., Perera L., Pedersen L. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 30.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 31.Sørensen J.B., Wiederhold K., Fasshauer D. Sequential N- to C-terminal SNARE complex assembly drives priming and fusion of secretory vesicles. EMBO J. 2006;25:955–966. doi: 10.1038/sj.emboj.7601003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lane S.R., Liu Y. Characterization of the palmitoylation domain of SNAP-25. J. Neurochem. 1997;69:1864–1869. doi: 10.1046/j.1471-4159.1997.69051864.x. [DOI] [PubMed] [Google Scholar]
- 33.Prescott G.R., Gorleku O.A., Chamberlain L.H. Palmitoylation of the synaptic vesicle fusion machinery. J. Neurochem. 2009;110:1135–1149. doi: 10.1111/j.1471-4159.2009.06205.x. [DOI] [PubMed] [Google Scholar]
- 34.Kang R., Swayze R., El-Husseini A. Presynaptic trafficking of synaptotagmin I is regulated by protein palmitoylation. J. Biol. Chem. 2004;279:50524–50536. doi: 10.1074/jbc.M404981200. [DOI] [PubMed] [Google Scholar]
- 35.Kang R., Wan J., El-Husseini A. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature. 2008;456:904–909. doi: 10.1038/nature07605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vogel K., Roche P.A. SNAP-23 and SNAP-25 are palmitoylated in vivo. Biochem. Biophys. Res. Commun. 1999;258:407–410. doi: 10.1006/bbrc.1999.0652. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


