Summary
Mammalian telomeres consist of tandem DNA repeats that bind protective protein factors collectively termed shelterins. Telomere disruption typically results in genome instability induced by telomere fusions. For reasons that are unclear, the mechanism of telomere fusion varies depending on the means of telomere disruption. Here, we investigate telomere fusions caused by overexpression of mutant telomerases that add mutated telomeric repeats, thereby compromising shelterin binding to telomeric termini. While all mutant telomeric sequences tested induced heterodicentric chromosome fusions in ATM-competent cells, only those mutant repeat sequences with significant self-complementarity induced ATM-independent sister chromatid and isodicentric chromosome fusions. Thus, once a telomere becomes dysfunctional, the terminal telomeric sequence itself determines the fate of that telomere. These results suggest that annealing of self-complementary DNA sequence engages an alternative telomere fusion pathway in human cells, and provide one explanation for the conspicuous lack of self-complementarity in the majority of known naturally-occurring eukaryotic telomeric sequences.
Introduction
Telomeres maintain genome stability by protecting the ends of linear chromosomes from recognition as DNA double-strand breaks (Blackburn, 2000). Telomeric DNA consists of a tract of duplex tandem repeats terminating with a single-stranded 3' overhang. In mammals, the telomeric repeat sequence 5'-TTAGGG-3' is bound by complexes of proteins collectively termed shelterins (Palm and de Lange, 2008). The ribonucleoprotein enzyme telomerase, which includes the core protein reverse transcriptase hTERT and the template-containing RNA hTER, maintains telomere length by replenishing TTAGGG repeats lost during cell division (Blackburn, 2000). Critical shortening of telomeres or experimental disruption of the protective shelterin complexes results in telomere deprotection, leading to a DNA damage response and telomere fusions (Palm and de Lange, 2008). These telomere fusions, which often occur through an ataxia-telangiectasia-mutated (ATM)-dependent mechanism, are an important source of genomic instability during carcinogenesis (Bailey and Murnane, 2006; Denchi and de Lange, 2007).
For reasons that are not fully understood, different types of telomere dysfunction ultimately result in different fusion patterns. For example, disruption of the shelterin component TRF2 predominately induces dicentric fusions, in which the chromatids of the two involved chromosomes are fused end-to-end (Smogorzewska et al., 2002), while disruption of the shelterin component POT1 also induces numerous sister chromatid fusions, in which the two chromatids of the same post-replicative chromosome fuse (Hockemeyer et al., 2006; Wu et al., 2006). Different mechanisms of telomeric attrition also result in distinct fusion patterns. For example, complete and acute loss of a single telomere often leads to sister chromatid fusions (Bailey and Murnane, 2006), while replicative shortening of telomeres frequently causes dicentric fusions (Zou et al., 2009). Collectively, these experiments have suggested a number of determinants of fusion outcomes, including differential disruption of the various shelterin components, cell cycle timing differences, and the varying availability of fusion partners.
Recent studies have highlighted a potential role of terminal DNA sequence in modulating mechanisms and outcomes of telomere fusion. In fission yeast, telomere attrition exposes homologous subtelomeric DNA sequences that mediate fusions through a single-strand annealing mechanism (Wang and Baumann, 2008). Analysis of telomere fusion junctions in human cells with critical telomere shortening has in many cases revealed microhomology involving subtelomeric terminal DNA sequences, and this microhomology has been proposed to induce fusions with complex rearrangements (Capper et al., 2007; Letsolo et al., 2009). Such findings suggest that the terminal DNA sequence itself may determine fusion outcomes, but the role of sequence has been difficult to separate from the other potential influences outlined above.
Here we directly examine the role of terminal DNA sequence in modulating telomere fusion outcomes in human cells by using overexpression of mutated hTer molecules (MT-hTers) containing alterations in the template sequence (Blackburn, 2005). After assembling with endogenous hTERT in the cell, MT-hTers direct addition of mutant telomeric repeats to the chromosome ends (Marusic et al., 1997; Stohr and Blackburn, 2008). Since the DNA-binding components of shelterin – TRF1, TRF2, and POT1 – are specific for wild-type telomeric repeat sequences, the mutant repeats are predicted to cause compromised shelterin binding at the telomeric termini (Broccoli et al., 1997; Loayza et al., 2004; Marusic et al., 1997). Thus, MT-hTers rapidly inhibit cancer cell proliferation and induce numerous telomere fusions (Guiducci et al., 2001; Stohr and Blackburn, 2008).
Using this system, we first demonstrate that all mutant telomeric sequences tested induce heterodicentric chromosome-type fusions at a similar frequency in cancer cells. In contrast, only those mutant sequences with significant self-complementarity also engage an ATM-independent fusion pathway, producing sister chromatid and isodicentric chromosome-type fusions. Thus, once a telomere becomes dysfunctional, the terminal telomeric sequence itself alters the fate of that telomere. These results suggest that annealing of self-complementary DNA sequence engages an alternative telomere fusion pathway in human cells, and provide one explanation for the lack of self-complementarity in the majority of known naturally-occurring eukaryotic telomeric sequences.
Results
MT-hTers with different template mutations have distinct effects on cancer cell proliferation and telomere fusion
We previously showed that MT-hTer-47A, which adds 5'-TTTGGG-3' telomeric repeats instead of wild-type 5'-TTAGGG-3' repeats, inhibits proliferation of LOX melanoma and UM-UC-3 bladder cancer cells and induces end-to-end telomere fusions through an ATM-dependent mechanism (Stohr and Blackburn, 2008). Inhibition of ATM by either shRNA-mediated depletion or the ATM-inhibiting compound KU-55933 rendered the cancer cells relatively resistant to the anti-proliferative effects of MT-hTer-47A and blocked MT-hTer-47A-induced telomere fusion (Stohr and Blackburn, 2008). Importantly, ATM inhibition did not block expression of MT-hTer-47A or incorporation of mutant 5'-TTTGGG-3' repeats at the telomeres. Instead, the ATM-depleted cancer cells grew robustly despite having long mutant telomeres (Stohr and Blackburn, 2008). Murine studies have similarly shown a direct role for ATM in the end-to-end fusion of telomeres rendered dysfunctional by loss of TRF2 (Denchi and de Lange, 2007).
In stark contrast to our prior results with MT-hTer-47A, MT-hTer-AU5, which is predicted to add 5'-TATATA-3' telomeric repeats, efficiently inhibited proliferation of LOX and UM-UC-3 cell lines independently of ATM status (Fig. 1A and B; Fig. S1A and B). In addition, unlike MT-hTer-47A, MT-hTer-AU5 induced the appearance of massively enlarged, multinucleate cells in both ATM-competent and ATM-depleted samples (Fig. 1C; Fig. S1C). Prior studies have suggested that these aberrant cells result from telomere fusion-induced genome instability and crisis (Guiducci et al., 2001; Stohr and Blackburn, 2008).
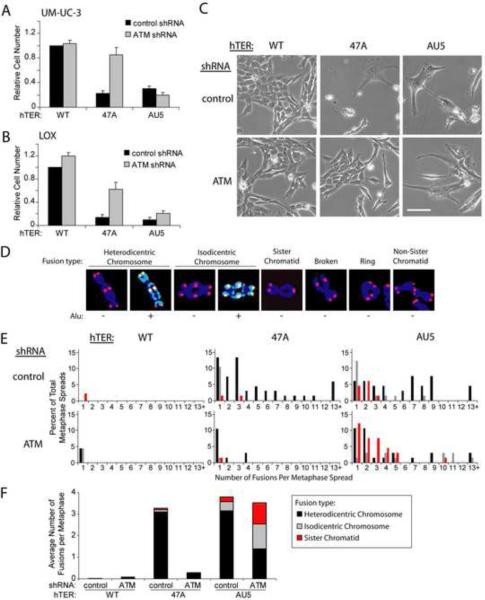
Figure 1.
Different MT-hTers have distinct effects on cancer cell proliferation and telomere fusion.
(A and B) UM-UC-3 bladder cancer (A) and LOX melanoma (B) cells were infected with lentivirus expressing the indicated shRNAs and hTERs. Cell number was determined 8 days after infection with hTER-expressing lentivirus. Cell counts were normalized to samples treated with the control shRNA and wild-type hTER. Values indicate the mean ± standard deviation of three independent hTER infections run in parallel (A) or three independent experiments (B). Rescue of proliferation of MT-hTer-47A-treated cells by prior ATM depletion was significant in both cell lines (p < 0.001 (A) and p = 0.01 (B) with the Student's t test).
(C) UM-UC-3 cells 7 days after infection with hTER-expressing lentivirus. Phase-contrast images were obtained at identical magnification; the scale bar is 100 μm.
(D) Different types of MT-hTer-induced telomere fusions in LOX cells. DAPI-stained chromosomes were labeled with probes for wild-type telomeric sequence (red) and Alu repeats (green).
(E) LOX cell metaphase spreads were prepared 5 days after infection with hTER-expressing lentivirus. Between 45 and 67 metaphase spreads for each sample were scored for the presence of the different fusion types shown in (D). For clarity, only data for chromosome-type fusions and sister chromatid fusions are shown here. Data for other fusion types are included in Fig. S1E.
(F) Average number of different telomere fusion types per metaphase.
We compared the types and frequencies of telomere fusions induced by MT-hTer-47A and MT-hTer-AU5. Several different classes of chromosome fusions were observed by metaphase analysis (Fig. 1D). Heterodicentric chromosome-type fusions involve two nonhomologous chromosomes fused end-to-end at both chromatids and predominately result from fusion in G1 of the cell cycle (Smogorzewska et al., 2002). In contrast, sister chromatid fusions occur in S or G2 phase following DNA replication. Isodicentric chromosome-type fusions consist of identical or homologous chromosomes attached at both chromatids in a “mirror-image” orientation and likely predominately result from segregation without breakage and subsequent replication of sister chromatid fusions (Fig. 1D and S1D) (Martin et al., 2005). To better distinguish heterodicentric and isodicentric chromosome-type fusions, we designed a probe to Alu repeats, which gives a banding pattern that is generally unique to each chromosome. Other fusion types observed included “broken” chromosome-type fusions, ring fusions, and non-sister chromatid-type fusions (Fig. 1D).
As observed previously (Stohr and Blackburn, 2008), MT-hTer-47A induced a significant number of heterodicentric chromosome fusions through an ATM-dependent mechanism (Fig. 1E, black bars in histograms; Fig. S1E). The marked drop in MT-hTer-47A-induced fusions caused by ATM depletion was not a consequence of cell cycle alterations, as ATM depletion had only a modest impact on cell cycle parameters (Fig. S1F). Strikingly, while MT-hTer-AU5 similarly induced heterodicentric fusions, it also induced a significant number of sister chromatid and isodicentric chromosome-type fusions (Fig. 1E, red and gray bars respectively) in both ATM-competent and ATM-depleted LOX cells. As a result, the total number of fusions in MT-hTer-AU5-treated cells changed very little with ATM depletion (Fig. 1F), likely explaining why cell proliferation was not substantially rescued in the ATM-depleted cells. We conclude that MT-hTer-AU5, unlike MT-hTer-47A, induces sister chromatid and isodicentric chromosome fusions through an ATM-independent pathway.
Self-complementary MT-hTers induce ATM-independent telomere fusions and inhibit proliferation of ATM-depleted cancer cells
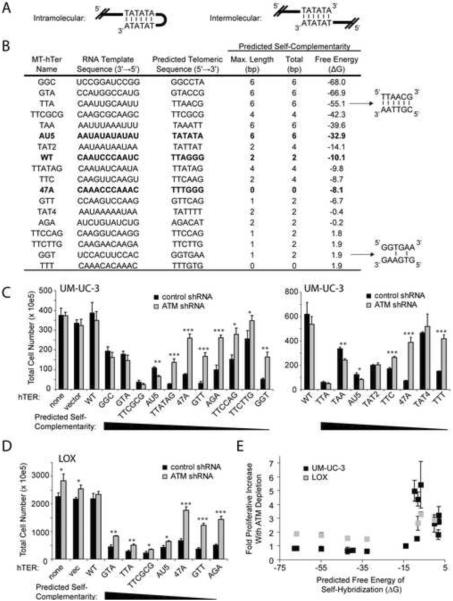
We noted that MT-hTer-AU5, but not MT-hTer-47A, is predicted to add terminal mutant telomeric repeat sequences with perfect self-complementarity, thereby potentially mediating intramolecular or intermolecular annealing (Fig. 2A). To test whether this characteristic explained the unexpected behavior of MT-hTer-AU5, we designed additional MT-hTers predicted to add tandem telomeric repeats with a wide range of sequences and self-complementarity (Fig. 2B). These MT-hTer template sequences differ from wild-type at 2 or more of the 6 positions in the repeat unit (Table S1), so all of the predicted telomeric repeats would be expected to disrupt shelterin binding (Broccoli et al., 1997; Loayza et al., 2004; Marusic et al., 1997). Indeed, in vitro binding assays with a representative subset of mutant telomeric repeat sequences demonstrated that all had significantly reduced affinity for TRF2 and POT1 (Fig. S2), although two of the tested non-self-complementary mutant sequences had measurable residual affinity for POT1.
Figure 2.
Proliferation of ATM-depleted cancer cells is efficiently blocked by self-complementary MT-hTers
(A) Potential intramolecular and intermolecular annealing of self-complementary MT-hTer-AU5 mutant telomeric repeats.
(B) Self-complementarity of tested MT-hTers. “Maximum length” indicates the longest stretch of uninterrupted self-complementarity within each 6-base-pair repeat, while “Total” indicates the maximum total number of self-complementary base pairs within each 6-base-pair repeat. Maximum self-complementarity was determined by considering all possible alignments of tandem telomeric repeats. The last column shows the predicted free energy of self-hybridization for 8 single-stranded tandem repeats of the indicated telomeric sequences, determined using DINAMelt (Markham and Zuker, 2005).
(C and D) UM-UC-3 (C) and LOX (D) cells were infected with lentivirus expressing the indicated shRNAs and hTERs. Cell counts were obtained 9 days after infection with the hTER-expressing virus. Values indicate the mean ± standard deviation of three independent hTER infections run in parallel. Student's t test was used to identify significant differences between matched control and ATM-depleted samples (* = p <0.05, ** = p < 0.005, *** = p < 0.0005).
(E) Correlation between predicted free energy of self-hybridization and proliferative rescue by ATM depletion. All MT-hTers from panels (C) and (D) are included except for MT-hTer-TAT4 and MT-hTer-TTCTTG, which were excluded from the graph due to their minimal effect on proliferation of UM-UC-3 control cells. For UM-UC-3 infections with MT-hTer-47A and MT-hTer-AU5, results from the two graphs in (C) were averaged and included as single points in (E). Values indicate the mean ± standard deviation of three independent hTER infections run in parallel.
As expected based on previous studies (Kim et al., 2001; Rivera and Blackburn, 2004), different MT-hTers had widely different effects on proliferation of ATM-competent cancer cells (Fig. 2C and D, black bars). One potential explanation for this variation is the differing efficiencies of MT-hTers in adding mutant repeats onto wild-type telomeres (Rivera and Blackburn, 2004). Strikingly, despite this variability, the ability of ATM depletion to rescue cell growth was perfectly correlated with the self-complementarity of each MT-hTer-specified sequence across the entire group of template sequences tested (Fig. 2C–E). Specifically, prior ATM depletion had little to no effect on the proliferation of cells treated with MT-hTers with strong self-complementarity, while rescuing the proliferation of cells treated with MT-hTers lacking significant self-complementarity. Given that all of the mutant template sequences were designed essentially randomly with respect to predicted sequence-specific shelterin binding affinity, this perfect correlation argues very strongly against inherent shelterin binding affinity as the relevant variable (see Discussion). Rather, we conclude that the degree of predicted self-complementarity of a given MT-hTer-specified sequence is the major determinant of whether ATM depletion will rescue proliferation.
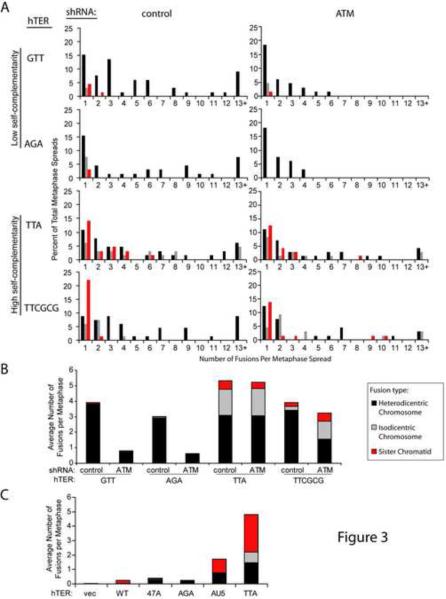
Metaphase analysis was performed with a representative subset of the MT-hTers. The strongly self-complementary MT-hTer-TTA and MT-hTer-TTCGCG induced significant numbers of sister chromatid and isodicentric chromosome fusions in both ATM-competent and ATM-depleted LOX cells (Fig. 3A and B; Fig. S3A), similar to MT-hTer-AU5. In contrast, MT-hTer-GTT and MT-hTer-AGA, which each lack significant self-complementarity, primarily induced ATM-dependent heterodicentric chromosome fusions (Fig. 3A and B; Fig. S3A), similar to MT-hTer-47A. We conclude that induction of sister chromatid and isodicentric chromosome fusions is a general property of self-complementary MT-hTers, and that these fusions at least partly explain the ability of these MT-hTers to block proliferation of ATM-depleted cancer cells. Notably, while our data suggest that self-complementary MT-hTers may also induce ATM-independent heterodicentric chromosome-type fusions (Fig. 1 and 3), such an interpretation is complicated by several caveats, including further rearrangements of isodicentric fusions that can mimic heterodicentric fusions (Fig. S3B and C). Thus, we focus on the sister chromatid and isodicentric chromosome fusions in the discussion below.
Figure 3.
Self-complementary MT-hTers induce a distinct pattern of telomere fusion.
(A) LOX cell metaphase spreads were prepared 5 days after infection with hTER-expressing lentivirus. Between 64 and 71 metaphase spreads for each sample were scored for the presence of the different fusion types shown in Fig. 1D. For clarity, only data for chromosome-type fusions and sister chromatid fusions are shown here. Data for other fusion types are included in Fig. S3A.
(B) Average number of different telomere fusion types per metaphase in LOX cells.
(C) MRC-5-TERT cell metaphase spreads were prepared 5 days after infection with hTER-expressing lentivirus. Between 25 and 35 metaphase spreads were scored for the presence of the different fusion types shown in Fig. 1D. For clarity, only data for chromosome-type fusions and sister chromatid fusions are shown. The average number of different telomere fusion types per metaphase spread is shown, while the full distribution of these fusions is presented in Fig. S3D.
We tested a representative subset of the MT-hTers in the primary human fibroblast cell line MRC-5 expressing exogenous wild-type hTERT (MRC-5-TERT). As in cancer cells, only MT-hTers with a high degree of self-complementarity induced significant numbers of sister chromatid and isodicentric chromosome fusions (Fig. 3C and S3D), indicating that our findings extend to a checkpoint-proficient setting. Intriguingly, while all MT-hTers tested induced similar numbers of fusions in ATM-competent cancer cells (Fig. 1F and 3B), self-complementary MT-hTers induced a much higher number of telomere fusions in MRC-5-TERT cells compared to MT-hTers lacking self-complementarity (Fig. 3C and S3D).
MT-hTers induce fusions by adding the expected terminal mutant repeats, not by promoting telomere loss
We tested whether self-complementary MT-hTers were indeed adding the expected mutant repeats to the terminal telomeres. Previous studies have used Southern blot analysis to show that MT-hTer-47A adds the expected TTTGGG repeats to the telomere ends (Marusic et al., 1997; Stohr and Blackburn, 2008). However, MT-hTers with significant self-complementarity are not amenable to such analysis, since the required probes would form stable secondary structures. We therefore developed a new strategy using restriction enzyme analysis, which confirmed that all three self-complementary MT-hTers tested for incorporation (TTA, GTA, and GGC) indeed directed incorporation of the expected mutant sequences at the telomeric ends (Fig. S4A).
We examined whether any of the different fusion types we observed were induced by telomere shortening or loss. We analyzed telomere spot signal intensities (reflecting the length of wild-type telomeric repeat tracts) in metaphase spreads from ATM-competent and ATM-depleted LOX cells expressing MT-hTer-47A or MT-hTer-AU5. For all fusion types, the average intensity of any fused telomere was approximately equal to the sum of the intensities of two adjacent unfused telomeres (Fig. S4B). This result indicates that, on average, the bulk length of telomeres that undergo fusion matches the bulk length of the original telomere population. Taken together, these data indicate that MT-hTers induce telomere fusions by adding terminal mutant repeats, not by promoting telomere shortening.
Discussion
Here, we have used MT-hTers to directly address the ability of terminal DNA sequence to modulate dysfunctional telomere fusion in human cells. We found that while all MT-hTers tested induce heterodicentric chromosome-type fusions at a similar frequency in ATM-competent cancer cells, only MT-hTers which direct the addition of self-complementary telomeric repeats induce ATM-independent sister chromatid and isodicentric chromosome-type fusions. Thus, telomeric sequence itself alters the fate of telomeres that have become dysfunctional.
A likely explanation for the unique fusion profile of self-complementary telomeric sequences is their ability to promote intra- or intermolecular strand annealing, which could generate sister chromatid fusions through two distinct mechanisms (Fig. S4C). These mechanisms have been implicated in sister chromatid fusions at non-telomeric locations in organisms ranging from yeast to mammals (Lobachev et al., 2002; Okuno et al., 2004; Tanaka et al., 2007; Tanaka and Yao, 2009; VanHulle et al., 2007). Furthermore, both pathways can mediate fusion of critically short yeast telomeres in certain contexts (Maringele and Lydall, 2004; Wang and Baumann, 2008). Our data now provide evidence for the activity of such a pathway at dysfunctional human telomeres.
The addition of self-complementary mutant telomeric repeats could potentially influence fusion mechanisms through other telomeric effects as well. First, terminal hairpins might hinder the formation of t-loops, which are proposed to offer protection against telomere fusion (Palm and de Lange, 2008). Secondly, hairpins are known to interfere with the progression of replication forks (Mirkin and Mirkin, 2007), which could conceivably encourage sister chromatid fusions. Third, annealing of self-complementary sequence at the telomeric terminus might exacerbate loss of protective single-stranded binding proteins such as POT1 or the newly identified CTC1-STN1-TEN1 complex, thereby altering fusion pathways (Denchi and de Lange, 2007; Miyake et al., 2009; Surovtseva et al., 2009). While some of the predicted mutant sequences lacking self-complementarity do show residual POT1 binding affinity, the lack of correlation between this residual binding affinity and telomere fusion outcomes argues against differential POT1 binding as an explanation for the unique behavior of self-complementary mutant repeats. Finally, we cannot entirely exclude the possibility that self-complementary MT-hTers add a different number of mutant telomeric repeats than non-self-complementary MT-hTers, which could potentially influence downstream fusion pathways. However, the fact that both classes of MT-hTers induce a remarkably similar number of heterodicentric chromosome-type fusions in ATM-competent LOX cells (Fig. 1F and 3B) suggests that they induce a comparable overall level of telomere dysfunction.
The vast majority of known naturally occurring telomeric sequences exhibit clear strand bias, with the strand terminating in a 3' end rich in guanosine and lacking in cytosine (Palm and de Lange, 2008). The 5'-TTAGGG-3' sequence common to all vertebrates, and many other phylogenetic groups, is a good example of this pattern. Explanations for this strand bias include a preference of telomerase for C-rich templates (Forstemann et al., 2003), evolutionary constraints imposed by the binding specificities of multiple protective proteins (Teixeira and Gilson, 2005), and a role for G-quartets in protecting the single-stranded 3' telomeric end (Teixeira and Gilson, 2005).
Our findings support an additional, non-mutually-exclusive explanation for the observed strand base composition bias of telomeric repeats: relegating guanosine and cytosine residues to opposite strands effectively limits self-complementarity of the tandem telomeric repeats. Our results suggest that by reducing self-complementarity, strand bias may protect against genome instability caused by sister chromatid fusions. Notably, we found that checkpoint-proficient primary cells are particularly susceptible to induction of telomere fusions by self-complementary terminal sequence (Fig. 3C and S3D). Given the rapid kinetics with which self-complementary sequence can form intramolecular secondary structures such as hairpins, even very transient deprotection of the telomeric ends, such as that which occurs during DNA replication (Verdun et al., 2005), might lead to genome instability if self-complementary sequence were available. The marked lack of self-complementarity found in the naturally-occurring eukaryotic telomere sequences listed in the Telomerase Database is consistent with selection against self-complementary sequences (Podlevsky et al., 2008).
Experimental procedures
Cell lines
LOX melanoma cells were maintained in RPMI 1640, and UM-UC-3 bladder cancer cells and MRC-5-TERT fetal lung fibroblasts were maintained in DMEM. Medium was supplemented with 10% fetal bovine serum, and cells were grown at 37°C in 5% CO2.
Plasmids and lentivirus
Construction of shRNA- and hTER-expressing lentivectors was described previously (Li et al., 2004; Stohr and Blackburn, 2008; Xu and Blackburn, 2004; Zufferey et al., 1997). The shRNA target sequences are as follows: 5'-GTTCTACAACGTAACGAGGTT-3' (untargeted control) and 5'-GCAACATACTACTCAAAGA-3' (ATM). Lentivirus was prepared as described (Xu and Blackburn, 2004). Lentiviral titers were obtained by counting green fluorescent protein (GFP)-positive foci as described (Stohr and Blackburn, 2008). Each lentiviral infection was performed with ~75–150 transducing units per cell, such that >95% of cells were infected as measured by GFP expression.
Lentiviral infections
Cells were infected with shRNA-expressing lentivirus at day −2, followed by infection with hTER-expressing lentivirus at day 0. Cells were passaged as needed to maintain logarithmic growth. To evaluate proliferation, cells were harvested and stained with Trypan blue, and viable cells were counted using a hemocytometer.
Metaphase fluorescence in situ hybridization
Telomeric fluorescence in situ hybridization and imaging was performed as described previously (Xu and Blackburn, 2007). The probes were TMR-OO-5'-(CCCTAA)3-3' (telomeric sequence) and AlexaFluor488-OO-5'-TGTAATCCCAGCACTTTG-3' (Alu repeat sequence). Fusions were scored in a blinded fashion.
Supplementary Material
Acknowledgements
We thank Sveta Makovets for key experimental insights; Peter Lansdorp (BC Cancer Research Centre) for advice on Alu probe design; Tanya Williams, Jue Lin, Tetsuya Matsuguchi, Imke Listerman, and Beth Cimini for critical review of the manuscript; and Kyle Lapham for assistance with data analysis. This work was supported by the Bernard Osher Foundation, NIH Breast Cancer SPORE CA58205, and NIH R01 CA096840 to E.H.B.; and American Cancer Society Postdoctoral Fellowship PF-05-111-01 and NIH K08 CA134552 to B.A.S. Special thanks to Elizabeth Warrens Claypool and Robert and Joanne Kidd for their financial support of this research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bailey SM, Murnane JP. Telomeres, chromosome instability and cancer. Nucleic Acids Res. 2006;34:2408–2417. doi: 10.1093/nar/gkl303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH. Telomere states and cell fates. Nature. 2000;408:53–56. doi: 10.1038/35040500. [DOI] [PubMed] [Google Scholar]
- Blackburn EH. Telomerase and Cancer: Kirk A. Landon--AACR prize for basic cancer research lecture. Mol Cancer Res. 2005;3:477–482. doi: 10.1158/1541-7786.MCR-05-0147. [DOI] [PubMed] [Google Scholar]
- Broccoli D, Smogorzewska A, Chong L, de Lange T. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat Genet. 1997;17:231–235. doi: 10.1038/ng1097-231. [DOI] [PubMed] [Google Scholar]
- Capper R, Britt-Compton B, Tankimanova M, Rowson J, Letsolo B, Man S, Haughton M, Baird DM. The nature of telomere fusion and a definition of the critical telomere length in human cells. Genes Dev. 2007;21:2495–2508. doi: 10.1101/gad.439107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- Forstemann K, Zaug AJ, Cech TR, Lingner J. Yeast telomerase is specialized for C/A-rich RNA templates. Nucleic Acids Res. 2003;31:1646–1655. doi: 10.1093/nar/gkg261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiducci C, Cerone MA, Bacchetti S. Expression of mutant telomerase in immortal telomerase-negative human cells results in cell cycle deregulation, nuclear and chromosomal abnormalities and rapid loss of viability. Oncogene. 2001;20:714–725. doi: 10.1038/sj.onc.1204145. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Daniels JP, Takai H, de Lange T. Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell. 2006;126:63–77. doi: 10.1016/j.cell.2006.04.044. [DOI] [PubMed] [Google Scholar]
- Kim MM, Rivera MA, Botchkina IL, Shalaby R, Thor AD, Blackburn EH. A low threshold level of expression of mutant-template telomerase RNA inhibits human tumor cell proliferation. Proc Natl Acad Sci U S A. 2001;98:7982–7987. doi: 10.1073/pnas.131211098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letsolo BT, Rowson J, Baird DM. Fusion of short telomeres in human cells is characterized by extensive deletion and microhomology, and can result in complex rearrangements. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Rosenberg JE, Donjacour AA, Botchkina IL, Hom YK, Cunha GR, Blackburn EH. Rapid inhibition of cancer cell growth induced by lentiviral delivery and expression of mutant-template telomerase RNA and anti-telomerase short-interfering RNA. Cancer Res. 2004;64:4833–4840. doi: 10.1158/0008-5472.CAN-04-0953. [DOI] [PubMed] [Google Scholar]
- Loayza D, Parsons H, Donigian J, Hoke K, de Lange T. DNA binding features of human POT1: a nonamer 5'-TAGGGTTAG-3' minimal binding site, sequence specificity, and internal binding to multimeric sites. J Biol Chem. 2004;279:13241–13248. doi: 10.1074/jbc.M312309200. [DOI] [PubMed] [Google Scholar]
- Lobachev KS, Gordenin DA, Resnick MA. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell. 2002;108:183–193. doi: 10.1016/s0092-8674(02)00614-1. [DOI] [PubMed] [Google Scholar]
- Maringele L, Lydall D. Telomerase- and recombination-independent immortalization of budding yeast. Genes Dev. 2004;18:2663–2675. doi: 10.1101/gad.316504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham NR, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33:W577–581. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, Genesca A, Latre L, Jaco I, Taccioli GE, Egozcue J, Blasco MA, Iliakis G, Tusell L. Postreplicative joining of DNA double-strand breaks causes genomic instability in DNA-PKcs-deficient mouse embryonic fibroblasts. Cancer Res. 2005;65:10223–10232. doi: 10.1158/0008-5472.CAN-05-0932. [DOI] [PubMed] [Google Scholar]
- Marusic L, Anton M, Tidy A, Wang P, Villeponteau B, Bacchetti S. Reprogramming of telomerase by expression of mutant telomerase RNA template in human cells leads to altered telomeres that correlate with reduced cell viability. Mol Cell Biol. 1997;17:6394–6401. doi: 10.1128/mcb.17.11.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin EV, Mirkin SM. Replication fork stalling at natural impediments. Microbiol Mol Biol Rev. 2007;71:13–35. doi: 10.1128/MMBR.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake Y, Nakamura M, Nabetani A, Shimamura S, Tamura M, Yonehara S, Saito M, Ishikawa F. RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol Cell. 2009;36:193–206. doi: 10.1016/j.molcel.2009.08.009. [DOI] [PubMed] [Google Scholar]
- Okuno Y, Hahn PJ, Gilbert DM. Structure of a palindromic amplicon junction implicates microhomology-mediated end joining as a mechanism of sister chromatid fusion during gene amplification. Nucleic Acids Res. 2004;32:749–756. doi: 10.1093/nar/gkh244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- Podlevsky JD, Bley CJ, Omana RV, Qi X, Chen JJ. The telomerase database. Nucleic Acids Res. 2008;36:D339–343. doi: 10.1093/nar/gkm700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera MA, Blackburn EH. Processive utilization of the human telomerase template: lack of a requirement for template switching. J Biol Chem. 2004;279:53770–53781. doi: 10.1074/jbc.M407768200. [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, Karlseder J, Holtgreve-Grez H, Jauch A, de Lange T. DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr Biol. 2002;12:1635–1644. doi: 10.1016/s0960-9822(02)01179-x. [DOI] [PubMed] [Google Scholar]
- Stohr BA, Blackburn EH. ATM mediates cytotoxicity of a mutant telomerase RNA in human cancer cells. Cancer Res. 2008;68:5309–5317. doi: 10.1158/0008-5472.CAN-08-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtseva YV, Churikov D, Boltz KA, Song X, Lamb JC, Warrington R, Leehy K, Heacock M, Price CM, Shippen DE. Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol Cell. 2009;36:207–218. doi: 10.1016/j.molcel.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Cao Y, Bergstrom DA, Kooperberg C, Tapscott SJ, Yao MC. Intrastrand annealing leads to the formation of a large DNA palindrome and determines the boundaries of genomic amplification in human cancer. Mol Cell Biol. 2007;27:1993–2002. doi: 10.1128/MCB.01313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Yao MC. Palindromic gene amplification--an evolutionarily conserved role for DNA inverted repeats in the genome. Nat Rev Cancer. 2009;9:216–224. doi: 10.1038/nrc2591. [DOI] [PubMed] [Google Scholar]
- Teixeira MT, Gilson E. Telomere maintenance, function and evolution: the yeast paradigm. Chromosome Res. 2005;13:535–548. doi: 10.1007/s10577-005-0999-0. [DOI] [PubMed] [Google Scholar]
- VanHulle K, Lemoine FJ, Narayanan V, Downing B, Hull K, McCullough C, Bellinger M, Lobachev K, Petes TD, Malkova A. Inverted DNA repeats channel repair of distant double-strand breaks into chromatid fusions and chromosomal rearrangements. Mol Cell Biol. 2007;27:2601–2614. doi: 10.1128/MCB.01740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdun RE, Crabbe L, Haggblom C, Karlseder J. Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol Cell. 2005;20:551–561. doi: 10.1016/j.molcel.2005.09.024. [DOI] [PubMed] [Google Scholar]
- Wang X, Baumann P. Chromosome fusions following telomere loss are mediated by single-strand annealing. Mol Cell. 2008;31:463–473. doi: 10.1016/j.molcel.2008.05.028. [DOI] [PubMed] [Google Scholar]
- Wu L, Multani AS, He H, Cosme-Blanco W, Deng Y, Deng JM, Bachilo O, Pathak S, Tahara H, Bailey SM, et al. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006;126:49–62. doi: 10.1016/j.cell.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Xu L, Blackburn EH. Human Rif1 protein binds aberrant telomeres and aligns along anaphase midzone microtubules. J Cell Biol. 2004;167:819–830. doi: 10.1083/jcb.200408181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Blackburn EH. Human cancer cells harbor T-stumps, a distinct class of extremely short telomeres. Mol Cell. 2007;28:315–327. doi: 10.1016/j.molcel.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Misri S, Shay JW, Pandita TK, Wright WE. Altered states of telomere deprotection and the two-stage mechanism of replicative aging. Mol Cell Biol. 2009;29:2390–2397. doi: 10.1128/MCB.01569-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.