Abstract
Autophagy is an evolutionarily conserved process to catabolize cytoplasmic proteins and organelles1, 2. During starvation, the target of rapamycin (TOR), a nutrient-responsive kinase, is inhibited, thereby inducing autophagy. In autophagy, double-membrane autophagosomes envelop and sequester intracellular components and then fuse with lysosomes to form autolysosomes which degrade their contents to regenerate nutrients. Current models of autophagy terminate with the degradation of autophagosome cargo in autolysosomes3-5, but the regulation of autophagy in response to nutrients and the subsequent fate of the autolysosome are poorly defined. Here we show that mTOR signaling is inhibited during autophagy initiation, but reactivated with prolonged starvation. mTOR reactivation is autophagy-dependent, and requires the degradation of autolysosomal products. Increased mTOR activity attenuates autophagy and generates proto-lysosomal tubules and vesicles that extrude from autolysosomes and ultimately mature into functional lysosomes, thereby restoring the full complement of lysosomes in the cell – a process we identify in multiple animal species. Thus, an evolutionarily-conserved cycle in autophagy governs nutrient sensing and lysosome homeostasis during starvation.
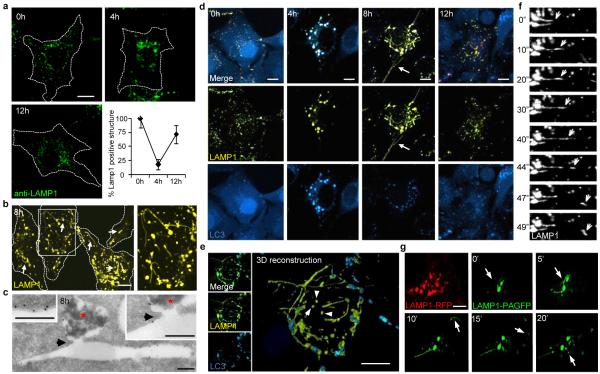
We observed that after nutrient deprivation of normal rat kidney (NRK) cells, multiple lysosomes fuse with each autophagosome such that after 4 hours, essentially all lysosomes were consumed into fewer and larger lysosomal-associated membrane protein 1 (LAMP1)-stained autolysosomes (Supplementary S1 as a movie; Fig. 1a). However, lysosome size and number had largely recovered after 12 hours of starvation, validated independently with cathepsin D as a lumenal marker (Fig. 1a and Supplementary S2, S3). We observed similar changes in vivo following starvation in the lysosomes of the fat body of Drosophila melanogaster expressing LAMP1-green fluorescent protein (GFP) (Supplementary S4), and also in cell lines derived from fish, amphibians, birds and other mammals (Supplementary S5), although the kinetics varied between different cell lines. Hence, a homeostatic cycle involving the consumption and restoration of lysosomes during starvation-induced autophagy appeared to be evolutionarily conserved.
Figure 1.
Lysosome homeostasis during starvation. (a) LAMP1+ lysosome size and quantity in NRK cells (dotted outline) starved for hours (h). Error bar show s.e.m (n=3). (b) Starved NRK cells expressing LAMP1-YFP show tubules (arrows). Box is expanded at right. (c) Immuno-TEM (gold marks LAMP1) of starved NRK cells. Continuity (arrow) illustrated between autolysosome (red star) and tubule. (d) LAMP1-YFP expressing NRK-LC3 cells as in (a). Arrows show reformation tubules. (e) 3-dimensional reconstruction of a cell from (d). (f) Time-lapse images (seconds) of starved LAMP1-YFP-expressing NRK cells. (g) Time-lapse images (minutes) of starved LAMP1-PAGFP or LAMP1-RFP-expressing NRK cells after photo-activation. Scale bars = 5 um.
We hypothesized that a process of lysosome reformation might follow autolysosome establishment. In cells starved for 8 hrs and expressing Yellow-Fluorescent Protein-(YFP)-tagged LAMP1 (LAMP1-YFP) or stained for endogenous LAMP1, we observed tubular structures extending from autolysosomes (Fig. 1b and Supplementary S6a). The membrane limited nature of these tubules could be appreciated by transmission electron microscopy (TEM) and immuno-TEM which also revealed LAMP1 on the surface but little or no discernible lumenal contents from the autolysosomes (Fig. 1c and Supplementary S6b and S7). We next monitored NRK cells by live-cell imaging after serum deprivation following expression of LAMP1-YFP to illuminate lysosomes and autolysosomes and Cyan Fluorescent Protein (CFP)-tagged microtubule-associated light chain 3 (CFP-LC3) to distinguish autophagosomes and autolysosomes (note: lysosomes are LAMP1+, LC3− and autolysosomes are LAMP1+, LC3+). After 4 hours of starvation, most, if not all, lysosomes coalesced into enlarged autolysosomes (Fig. 1d). At 8 hours of starvation, we observed LAMP1-positive tubular structures extending from autolysosomes (Fig. 1d). At 12 hours after starvation, LC3 was dispersed and less punctate, indicating attenuated autophagy and the size and number of lysosomes, now devoid of LC3, had returned to pre-starvation levels (Fig. 1d). Similar LAMP1-positive tubules were observed in different cell types from various species in the animal kingdom (Supplementary S5b).
We next reconstructed Z-stacked confocal microscopic images of starving cells which confirmed that the LAMP1+, LC3− tubules emanated from autolysosomal membranes and appeared to give rise to LAMP1+, LC3− vesicles by direct budding (Fig. 1e). By time-lapse microscopy, we discerned that tubule extension is a continuous and highly dynamic process with the distal portions extruding free vesicles (Fig. 1f and Supplementary S8 as a movie). We also used a photoactivatable GFP-tagged LAMP1 (PAGFP-LAMP1) for a pulse-chase analysis of the autolysosomal membrane. After 4 hours of starvation, individual autolysosomes were laser-activated and followed by time-lapse microscopy. Within 20 minutes, we observed tubule formation followed by budding of LAMP1+ vesicles (Fig. 1g and Supplementary S9 as a movie). Vesicle formation was not inhibited by the protein synthesis inhibitor cycloheximide (Supplementary S10).
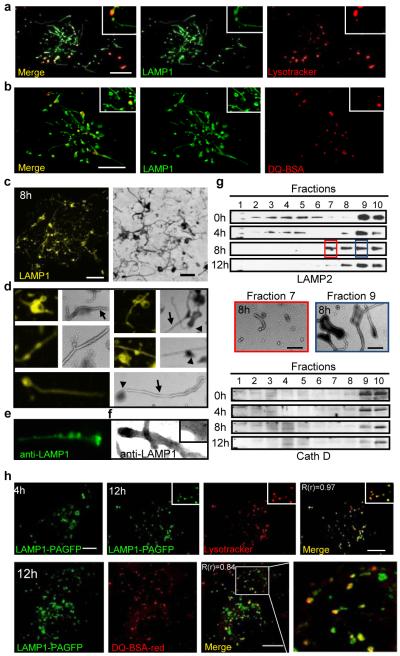
Testing for lysosomal properties, we found that the LAMP1-positive tubules were not stained by the acid-dependent dye Lysotracker despite strong staining of the autolysosomes (Fig. 2a). Furthermore, the lysosomal substrate DQ-BSA was degraded within the autolysosomes, but not the tubules (Fig. 2b). Thus, the newly formed tubules and vesicles lacked key biochemical characteristics of autolysosomes or lysosomes. We therefore preloaded starved cells with dextran, and found that it filled the autolysosomes but did not enter the tubules, confirming our observation from immuno-TEM that autolysosomal contents are withheld from the tubules (Supplementary S11).
Figure 2.
Proto-lysosome formation and maturation. (a) - (c) Starved LAMP1-YFP-NRK cells stained with Lysotracker or DQ-BSA. (c, right) TEM of purified lysosomes. Magnification (4x) (d), immunofluorescence (e) or immuno-EM (f) using LAMP1 antibody of (c). (g) Density gradient fractions of NRK cells starved for hours (h), blotted for LAMP2 or Cathepsin D. 8h fractions 7 and 9 analyzed by TEM (center). Fraction 10 = gradient top. (h) Starved LAMP1-PAGFP NRK cells were photo-activated (4h) and imaged at 12 hours with either Lysotracker (upper panels) or DQ-BSA (lower panel) with enlargements (boxes) and Pearson’s coefficients (R(r)). Scale bars = 5 um.
We next used Optiprep (60% w/v iodixanol in H2O) gradients to purify the autolysosome fraction from starved NRK cells which yielded an extensive tangle of autolysosomes and tubules whose morphology by TEM resembled tubules observed in vivo (Fig. 2c-f). Sequestration of the autolysosomal contents from the generally unfilled tubules was clearly evident (Fig. 2d). Using detailed gradient fractionation, we found that prior to starvation (time = 0 hr), LAMP2 staining, lysosomal cathepsins D and B, and acid phosphatase activity were chiefly detected in mature lysosome fractions at the top of the gradient (fractions 9 and 10) (Fig. 2g and supplementary S12). After 4 hours of starvation, when autophagosomes fuse to lysosomes, LC3 becomes transiently coincident with LAMP2 in Fraction 9, biochemically confirming the evanescence of autolysosomes (Fig. 2g and supplementary S12). Simultaneously, LAMP2 in fraction 10 is reduced reflecting lysosome consumption during autophagy. At 8 hours, as tubules and budding vesicles appear, two denser fractions (7 and 8) become LAMP2+, but not LC3+ and lack cathepsins or acid phosphatase (Fig. 2g and supplementary S12). In fraction 7, TEM revealed mainly short tubules and vesicles, while fraction 9 contained autolysosomes extending tubules (Fig. 2g). These data reinforce the concept that reformation tubules and tubule-derived vesicles represent recycling of the lysosomal membrane components, but not autolysosomal contents.
Co-localization analyses showed that LAMP1+ tubules and vesicles were not acidic at 8 hrs of starvation (Fig. 2a). However, by 12 hours, the LAMP1+ vesicles became acidic (Supplementary S13), possibly maturing into functional lysosomes. We therefore visually tracked vesicles derived from PAGFP-LAMP1-labelled autolysosomes. Cells were starved for 4 hours, PAGFP-LAMP1 was activated, and starvation was continued for 8 additional hours, during which we found that tubule-derived vesicles (smaller than autolysosomes) became acidic (Fig. 2h, upper panels, 12h). Moreover, these vesicles also acquired the capacity to cleave DQ-BSA (Fig. 2h, lower panels). Thus, the autolysosome-derived vesicles apparently mature into functionally active lysosomes and are henceforth referred to as proto-lysosomes.
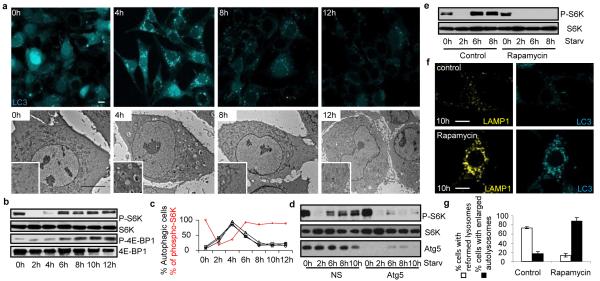
Autophagy is generally held to be a constitutive process that is strongly induced during starvation1,6,7. In agreement with recent literature8,9, we observed that the number of autophagic cells is transient during starvation and peaks after 4 hours but steadily decreases to almost none after 12 hours (Fig. 3a, c and Supplementary S14). This attenuation of autophagy correlated with the initiation of autophagic lysosome reformation (ALR), hinting at a common coordinating mechanism. mTOR is a key governor of cell growth and metabolism that is rapidly inhibited during starvation which triggers autophagy6,10. We found that phosphorylation of the mTOR substrates S6 Kinase (S6K) and 4E-BP1 were inhibited after 2 hours of starvation, but remarkably restored by 6 hours and thereafter indicating that mTOR signaling is evidently re-activated despite ongoing starvation (Fig. 3b, c).
Figure 3.
mTOR reactivation inhibits autophagy and initiates lysosome reformation. (a) TEM (bottom, scale bar = 5 um) or fluorescence (top, scale bar, 10 um) of NRK-LC3 cells starved for hours (h) with enlargements (boxes). (b) and (d) immunoblots of (a) with indicated antibodies after transfection (d) with non-specific (NS) or atg5 RNAi. (c) Autophagy (black lines = independent experiments) and phospho-S6K (densitometry of b, red line). (e)-(g) Two hour starved LAMP1-YFP-NRK-LC3 cells treated with 100 nM rapamycin for another 4 (6 h) or 6 (8 h) hours analyzed by blotting (e) as in (b) or microscopy (f, scale bar, 5 um) with quantitation (g). Error bars show s.e.m (n=3).
mTOR is activated by growth factors and nutrients, such as carbohydrates, amino acids and ATP10, and the chief role of autophagy during starvation is to provide nutrients by degrading cellular contents1. We therefore tested whether intracellular nutrients generated by autophagy could stimulate mTOR signaling, and thereby provide a negative feedback signal to down-regulate autophagy and trigger ALR. Indeed, knock-down of atg5 or atg7 markedly inhibited the recovery of S6K phosphorylation (Fig. 3d and Supplementary S15). We also found that mTOR reactivation apparently triggers ALR because adding the mTOR inhibitor rapamycin at 2 hrs of starvation also blocked S6K phosphorylation and ALR, resulting in enlarged autolysosomes persisting even after 10 hours of starvation (Fig. 3e-g). Note that at the time of rapamycin addition, mTOR activity was already inhibited by starvation, so rapamycin was blocking mTOR reactivation. We also found, as shown previously11, that knocking down mTOR increases autophagy but totally inhibits ALR leaving giant autolysosomes which persist even after 12 hours of starvation (Supplementary S16). mTOR was recently reported to localize to endosomes12, so we examined its subcellular localization during starvation and found that a small fraction of mTOR colocalizes with LC3 after autolysosome formation suggesting that it may directly regulate ALR (Supplementary S17).
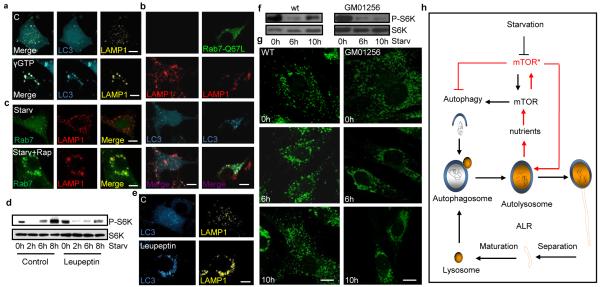
Additional studies provided further insights into the molecular requirements for ALR. First, we found proto-lysosomes were closely associated with and decorate microtubules (Supplementary S18a). Also, disruption of microtubules with nocodazole arrested ALR and induced giant persisting autolysosomes (Supplementary S18b). Thus, microtubules are required for ALR, possibly providing a scaffold for the extrusion of reformation tubules. Second, we found that the small GTPase Rab7 plays a key regulatory role. Treating cells with GTPγS, a non-hydrolyzable analogue of GTP, completely inhibited ALR leaving only enlarged autolysosomes (Fig. 4a). Rab7 has been shown to reside on autolysosomes and mediate autophagosome-lysosome fusion3. We detected Rab7 in lysosomal fractions but not in proto-lysosome and tubule fractions after 8 or 12 hrs of starvation (Supplementary S19). We conjectured that Rab7 must dissociate from tubules before reformation can proceed. We therefore overexpressed constitutively active Rab7, which is permanently membrane-associated, and observed that it abrogates ALR, resulting in enlarged and long-lasting autolysosomes (Fig. 4b). Interestingly, inhibition of ALR by rapamycin blocks Rab7 disassociation from the distended autolysosomes it produced, indicating that mTOR potentially regulates ALR through Rab7 (Fig. 4c).
Figure 4.
Autophagic lysosome reformation. (a) LAMP1/LC3-NRK cells were starved for 4 hours and treated with GTPγS for 6 hours starvation. (b) Rab7Q67L-GFP transfected LC3-CFP/Lamp1-RFP-NRK cells starved for 10 hours. (c) /Rab7-CFP-NRK cells starved for 2 hrs and treated with 100 nM rapamycin for 8 hours starvation. (d) LAMP1-YFP/ NRK-LC3 cells starved for hours (h) with 1ug/ml leupeptin and blotted with indicated antibodies or (e) imaged. (f) Fibroblast cells from Scheie syndrome (GM01256) patients or controls starved and blotted with indicated antibodies or (g) imaged. Lysosome size classified by IPP software. (h) Provisional model of the autophagic lysosome reformation (ALR) cycle. Scale bars = 5 um.
We also observed that inhibition of autolysosomal protein degradation by the lysosomal inhibitors leupeptin or E64/pepstatin A during starvation abolished mTOR reactivation (Fig. 4e and Supplementary S20). Inhibition of lysosomal proteases also abrogated autophagy attenuation and ALR resulting in enlarged and long-lasting autolysosomes (Fig. 4e). Significantly, Scheie syndrome (GM01256), Fabry disease (GM00636), and Aspartylglucosaminuria (GM02056) fibroblasts derived from lysosome storage disease (LSD) patients show impaired mTOR reactivation and defective lysosome reformation (Fig. 4f, g and Supplementary S21, S22). The LAMP1 structures in leupeptin-treated or LSD cells are slightly elongated compared to those treated with rapamycin, possibly due to undegraded material accumulating in the autolysosome. Although we cannot exclude the possibility that altered pH or other defects contribute to aberrant ALR in LSD cells, these observations are consistent with the conclusion that lysosomal degradative capability is required for ALR.
Previous work has documented that autophagy leads to fusion of lysosomes and autophagosomes to form autolysosomes, but whether this restores a nutrient balance and how this affects lysosome homeostasis is not understood. We found that a large percentage of lysosomes are subsumed into autolysosomes at the peak of autophagy. However, we also observed that autophagy was eventually consummated and the normal complement of lysosomes was restored by an unknown mechanism. We now report the discovery of a negative feedback mechanism that reverses autophagy and restores lysosome homeostasis that we term autophagic lysosome formation (ALR). The degradation of macromolecules and release of intracellular substituents following autophagy appears to trigger mTOR reactivation, which inhibits autophagy and stimulates the recycling of proto-lysosomal membrane components via tubules and then vesicles that mature in to new lysosomes (Fig. 4h). Genetic or chemical inhibition of lysosome function arrests ALR despite starvation. This feedback mechanism tightly couples nutritional status to the induction and cessation of autophagy. Thus, autophagy is self-regulated so that nutrient replenishment prevents excess autophagy, which could lead to autophagic cell death13-15. As part of this cycle of lysosome consumption and restitution, ALR allows the cell to reuse a critical component required for further autophagy, the lysosomal membrane and associated proteins, in a time of scarce resources.
METHODS SUMMARY
NRK cells were cultured and maintained in DMEM medium with or without (starvation condition) 10% FBS and 2 mM L-glutamine. Cells were transfected using Amaxa nucleofection™ using solution T and program X-001, using 200 pmol RNAi or total 2 ug DNA of expression plasmids prepared by standard molecular biology methods. Transfected cells were re-plated in LabTek chamber coverglasses (NUNC) grown overnight at 37°C with 5% CO2 in a PeCon open chamber (PeCon, Erbach, Germany) for microscopy. Stains for lysosomal markers and Lysotracker were carried out using the manufacturers’ recommended procedures. Images were digitally acquired by a Leica TCS SP5 confocal microsystem and analyzed using instrument software and by the IMARIS package (Bitplane, Switzerland). For immuno-EM, cell were fixed in 4% formaldehyde in 0.1 M phosphate buffer (pH 7.4), and then in 4% formaldehyde. Ultrathin cryosectioning and immunogold labeling were previously described16. Lysosome isolation kit is from Sigma-Aldrich. Cell fractionation and lysosome isolation were performed according to manufacturer’s instructions. Protein extraction procedures, gel electrophoresis, and immunoblotting were detailed previously15, 17.
Supplementary Material
S1. Movie showing that multiple lysosomes fuse with GFP-LC3-labeled autophagic vesicles. GFP-LC3-expressing NRK cells grown in chambers were induced to form autophagic vesicles by starvation. After induction, cells were labeled with the acidotropic pH-sensitive dye LysoTracker Red for five minutes. Excess dye was then washed out. Dual-channel live cell imaging revealed fusion events between individual GFP-LC3-positive autophagosomes and multiple low-pH lysosomes. Lysosome fusion causes the degradation of GFP-LC3. Arrow indicates autophagosome (green) multiple lysosomes (red) fusing to it.
S8. Movie of tubules extending from autolysosomal membranes (arrow) and small Lamp1-positive vesicles pinching off from the tips of tubules. 8 hours after serum starvation, LAMP1-YFP-expressing NRK cells were imaged by time lapse microscopy. Time (seconds) is indicated in the top right of each panel.
S9. Movie of tubules breaking away from autolysosomes and condensing into Lamp1-positive vesicles. NRK cells transfected with Lamp1 (photoactivatable) PAGFP/Lamp1-RFP (3:1) were starved for 6 hours. LAMP1-PAGFP was photo-activated by a 405 laser, and Lamp1-GFP structures were followed by time-lapse photography. Time (minutes) is indicated in the top left of each panel.
Acknowledgments
We are grateful to NIAID imaging core facility and Olympus China for providing technical support, O. Schwartz, Juraj Kabat, Lily Koo, Meggan Czapiga (BIF, NIAID, NIH), Qi Dong (Olympus China) for assistance with confocal microscopy and imaging processing, K. Nagashima and M. J. de la Cruz (NCI) for TEM analyses, J. Lippincott-Schwartz, Harris Bernstein, and J. Bonifacino for helpful discussions, and D. Yamamoto, G. Davis, and H. Kramer for constructs and fly strains. We thank M. v. Peski and R. Scriwanek for assistance with the preparation of the EM figures. This research was supported by the Division of Intramural Research of the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services and NIH Grant GM079431 to EB, 973 program 2010CB833704, NSF grant 20091300700, and Tsinghua University grant 20091081391 to YL. J.K. is the recipient of VICI grant 918.56.611 of the Netherlands Organisation for Scientific research (NWO).
Appendix
Online Methods
Reagents and Antibodies
Lysotracker-red, dextran-red (MW 10000), DQ-BSA-red, Hoechst and DQ-ovalbumin-green were from Invitrogen (Carlsbad, CA). Leupeptin is from Millipore (Billerica, MA). Cycloheximide and anti-LAMP1 antibody was from Sigma-Aldrich (St. Louis, MO). Anti-P-S6K, Anti-S6K, anti-S6K, anti-P-4E-BP, anti-4E-BP, anti-atg7, anti-LC3 were from Cell Signaling Technology (Danvers, MA). anti-AP-3 was from Developmental Studies Hybridoma Bank, Anti-Cath B was from Santa Cruz Biotechnology, anti-Cath D is from BD Bioscience. Western blots were performed as previously described14.
Cell Culture and Transfection
NRK cells were obtained from American Type Culture Condition (ATCC) and cultured in DMEM (Life Technologies) medium supplemented with 10% FBS (5% CO2). Cells were transfected via Amaxa nucleofection™ using solution T and program X-001, using 200 pmol RNAi or total 2 ug DNA. Cells were then cultured in growth medium for further analysis. For two rounds of transfection, cells were transfected with 200 pmol RNAi, and 72 hours after transfection, cells were transfected again with 100 pmol RNAi and up to 2 ug DNA. Starvation medium is DMEM medium without the addition of serum and glutamine. HELA, C166, MRC-5, SH-SY5Y, MEF, OP9, Vero, HuH7, DF-1, ZF4,A6, HepG2, OMK, A549, H4-II-E-C3 are from ATCC and cultured according to the repository instructions.
Constructs
The human spinster construct was kindly provided by Dr. D. Yamamoto, Waseda University, Japan. LAMP1 and LC3 constructs with CFP, RFP, YFP, GFP or PAGFP tags were provided by Dr. J. Lippincott-Schwartz, NICHD, NIH. Rab7-GTP, and Rab7 were provided by Dr. J. Bonifacino, NICHD, NIH. RNAi were synthesized from invitrogen. mTOR, CAGCAGCTGGTACATGACAAGTACTTT, atg5, GGCATTATCCAATTGGCCTACTGTT
Live Cell Imaging
Transfected cells were re-plated in Lab Tek Chambered coverglass (NUNC) the night before imaging, and cells were maintained at 37°C with 5% CO2 in a PeCon open chamber (PeCon, Erbach, Germany). Images were acquired by a Leica sp5 confocal microscope. 3D models were constructed by collecting images by Z-stack scanning at 0.5 um intervals, and images were collapsed to construct 3D models by IMRIS (Bitplane, Zurich, Switzerland). For fly experiments, 3rd instar larvae either expressing tub-LAMP1-GFP (control), or trans-heterozygous spinster mutant larvae expressing LAMP1-GFP (tub-LAMP1-GFP; spinster10403/spinsterK09905) were starved in moist petri dishes lacking food at 25°C. The fat body was dissected and stained with Hoechst, and samples were imaged immediately without fixation on a Zeiss AxiovisionZ.1 microscope with fluorescence. Lysosome number was measured using Zeiss Automeasure software. Lysosomes were quantified in at least 7 different larvae per time-point, in fat cells from one fat body per larva and 2 fields per fat body. Each field contained 3-4 cells. The number of lysosomes per timepoint was normalized to the average number of lysosomes in non-starved (0h) larvae. Error bars indicate standard deviation.
Cell fractionation
Lysosome isolation kit is from Sigma-Aldrich. Cell fractionation and lysosome isolation were performed according to manufacture’s manual.
Staining
Cells were washed with Phosphate Buffered Saline (PBS), fixed in 2% paraformaldehyde for 10 min, and permeabilized in 0.2% Triton X-100 for 5 min. They were blocked with 10% FBS in PBS for 30 min, stained with 10 ug/ml of rabbit anti-LAMP1 (Sigma-Aldrich) in blocking buffer for 1 hour, and washed with PBS three times. Cells were then stained with fluorescein isothiocyanate conjugated (FITC)-anti rabbit secondary antibody (Becton-Dickinson, San Jose, CA) in PBS for 1 hour and washed with PBS three times.
Electron Microscopy
Cells were fixed in 3% glutaraldehyde in 0.1 M MOPS buffer (pH 7.0) for 8 hrs at room temperature, then 3% glutaraldehyde/1% paraformaldehyde in 0.1 M MOPS buffer (pH 7.0) for 16 hours at 4°C. They were then post-fixed in 1% osmium tetroxide for 1 hour, and embedded in Spurr’s resin, sectioned, doubly stained with uranyl acetate and lead citrate, and analyzed using a Zeiss EM 10 transmission electron microscope.
Immuno-electron microscopy
NRK LAMP-1-YFP cells were starved for 8 hours and then fixed for immuno-electron microscopy by adding freshly-prepared 4% formaldehyde in 0.1 M phosphate buffer (pH 7.4) to an equal volume of culture medium. After 10 min, the fixative was replaced for post-fixation in 4% formaldehyde at 4°C overnight. Ultrathin cryosectioning and immunogold labeling were performed as previously described 16. The LAMP-1-YFP construct was detected by using polyclonal rabbit anti-LAMP-1/LGP120 M3 at a dilution of 1:650. This antibody is a kind gift of Dr. I Mellman (Genentech, San Francisco). The antibody was marked by protein A-10 nm gold particles (Cell Microscopy Center Utrecht, The Netherlands).
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
References
- 1.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 2.Xie Z, Klionsky D. J. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 3.Jager S, et al. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117:4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 4.Jahreiss L, Menzies FM, Rubinsztein DC. The itinerary of autophagosomes: from peripheral formation to kiss-and-run fusion with lysosomes. Traffic. 2008;9:574–587. doi: 10.1111/j.1600-0854.2008.00701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 6.Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 7.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 8.Massey AC, Follenzi A, Kiffin R, Zhang C, Cuervo AM. Early cellular changes after blockage of chaperone-mediated autophagy. Autophagy. 2008;4:442–456. doi: 10.4161/auto.5654. [DOI] [PubMed] [Google Scholar]
- 9.Mizushima N, Kuma A. Autophagosomes in GFP-LC3 Transgenic Mice. Methods Mol Biol. 2008;445:119–124. doi: 10.1007/978-1-59745-157-4_7. [DOI] [PubMed] [Google Scholar]
- 10.Dennis PB, et al. Mammalian TOR: a homeostatic ATP sensor. Science. 2001;294:1102–1105. doi: 10.1126/science.1063518. [DOI] [PubMed] [Google Scholar]
- 11.Ravikumar B, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 12.Sancak Y, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu L, Lenardo MJ, Baehrecke EH. Autophagy and caspases: a new cell death program. Cell Cycle. 2004;3:1124–1126. [PubMed] [Google Scholar]
- 14.Yu L, Strandberg L, Lenardo MJ. The selectivity of autophagy and its role in cell death and survival. Autophagy. 2008;4:567–573. doi: 10.4161/auto.5902. [DOI] [PubMed] [Google Scholar]
- 15.Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S, Baehrecke EH, Lenardo MJ. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- 16.Slot JW, Geuze HJ. Cryosectioning and immunolabeling. Nat Protoc. 2007;2:2480–2491. doi: 10.1038/nprot.2007.365. [DOI] [PubMed] [Google Scholar]
- 17.Bidère N, et al. Casein kinase 1alpha governs antigen-receptor-induced NF-kappaB activation and human lymphoma cell survival. Nature. 2009;458:92–96. doi: 10.1038/nature07613. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S1. Movie showing that multiple lysosomes fuse with GFP-LC3-labeled autophagic vesicles. GFP-LC3-expressing NRK cells grown in chambers were induced to form autophagic vesicles by starvation. After induction, cells were labeled with the acidotropic pH-sensitive dye LysoTracker Red for five minutes. Excess dye was then washed out. Dual-channel live cell imaging revealed fusion events between individual GFP-LC3-positive autophagosomes and multiple low-pH lysosomes. Lysosome fusion causes the degradation of GFP-LC3. Arrow indicates autophagosome (green) multiple lysosomes (red) fusing to it.
S8. Movie of tubules extending from autolysosomal membranes (arrow) and small Lamp1-positive vesicles pinching off from the tips of tubules. 8 hours after serum starvation, LAMP1-YFP-expressing NRK cells were imaged by time lapse microscopy. Time (seconds) is indicated in the top right of each panel.
S9. Movie of tubules breaking away from autolysosomes and condensing into Lamp1-positive vesicles. NRK cells transfected with Lamp1 (photoactivatable) PAGFP/Lamp1-RFP (3:1) were starved for 6 hours. LAMP1-PAGFP was photo-activated by a 405 laser, and Lamp1-GFP structures were followed by time-lapse photography. Time (minutes) is indicated in the top left of each panel.