Abstract
The silent information regulator protein (Sir2) and its homologs are NAD+-dependent deacetylase enzymes that play important roles in a variety of physiological processes. However, the functions of the Sir2 family in plants are poorly understood. Here, we report that Arabidopsis AtSRT2, a homolog of yeast Sir2, negatively regulates plant basal defense against the pathogen Pseudomonas syringae pv. tomato DC3000 (PstDC3000). In response to PstDC3000 infection, the expression of AtSRT2 was down-regulated in a salicylic acid (SA)-independent manner. In addition, knock-out of AtSRT2 (srt2) enhanced resistance against PstDC3000 and increased expression of pathogenesis-related gene 1 (PR1). Conversely, overexpression of AtSRT2 resulted in hypersusceptibility to PstDC3000 and impaired PR1 induction. Consistent with this phenotype, expression of PAD4, EDS5 and SID2, three essential genes in the SA biosynthesis pathway, were increased in the srt2 mutant and decreased in AtSRT2-overexpressing plants. Taken together, these results demonstrate that AtSRT2 is a negative regulator of basal defense, possibly by suppressing SA biosynthesis.
Keywords: AtSRT2, Basal defense, EDS5, PAD4, PstDC3000, SID2
Introduction
Silent information regulator 2 (Sir2) proteins, or sirtuins, are NAD+-dependent histone deacetylases (HDACs); NAD+ is required as a cofactor to deacetylate substrates (Blander and Guarente 2004, Dali-Youcef et al. 2007). Sir2 proteins contain sirtuin core domains, which are conserved from bacteria to humans (Brachmann et al. 1995, Frye 1999). Functional studies in yeast and mammalian cells have revealed that Sir2 proteins deacetylate both histone and non-histone substrates (Buck et al. 2004, Haigis and Guarente 2006, Sauve et al. 2006) and play important roles in numerous processes, including chromatin silencing, DNA repair, cell cycle, apoptosis and aging (Robyr et al. 2002, Blander and Guarente 2004, Yamamoto et al. 2007).
However, the functions of Sir2 proteins in plants are not fully understood. Sequence analysis has identified two Sir2 family genes in Arabidopsis (AtSRT1 and AtSRT2) and rice (OsSRT1 and OsSRT2) (Pandey et al. 2002). AtSRT1 and OsSRT1 belong to the same class of HDACs and showed a high sequence similarity (Pandey et al. 2002). Down-regulation of OsSRT1 by RNA interference (RNAi) enhances histone H3K9 acetylation on transposable elements and promoters of hypersensitive response (HR)-related genes (Huang et al. 2007). This increased H3K9 acetylation triggers HR-related gene expression and leads to hydrogen peroxide production, DNA fragmentation, cell death and lesions mimicking plant HR (Huang et al. 2007). Studies of OsSRT1 highlight the roles of plant Sir2 proteins in suppressing gene expression via histone H3 deacetylation. However, sequence analysis indicates that AtSRT2 and OsSRT1 are highly divergent, suggesting they may have different functions. The role of AtSRT2 is not clear, although a recent study has shown that mutation of AtSRT2 affects the Arabidopsis vernalization response (Bond et al. 2009).
Plants possess a complex network of defense strategies to deal with microbial pathogens. The small plant hormone molecule salicylic acid (SA) plays important roles in plant disease resistance. After detecting microbial pathogens, plants accumulate SA (Loake and Grant 2007, Vlot et al. 2008), which subsequently activates NPR1 (NON-EXPRESSER OF PR GENES 1) and results in defensive reaction including the expression of pathogen-related (PR) genes (Cao et al. 1997). Biosynthesis of SA in response to pathogens is believed to be controlled by PAD4 (PHYTOALEXIN DEFICIENT 4), EDS5 (ENHANCED DISEASE SUSCEPTIBILITY 5) and SID2 (SALICYLIC ACID INDUCTION DEFICIENT 2) (Shah 2003). PAD4 encodes a lipase-like protein that interacts with EDS1 (ENHANCED DISEASE SUSCEPTIBILITY 1) (Jirage et al. 1999, Feys et al. 2001). EDS5 is homologous to the bacterial multidrug and toxin extrusion transporter (MATE) protein and may be involved in transporting SA precursors (Nawrath et al. 2002, Shah 2003). SID2 encodes isochorismate synthase (ICS1), which controls pathogen-induced SA biosynthesis (Wildermuth et al. 2001). SA levels are significantly lower in pad4, eds5 and sid2 mutants compared with wild-type (WT) plants (Zhou et al. 1998, Nawrath and Metraux 1999, Wildermuth et al. 2001). As a result, pad4, eds5 and sid2 mutants are hypersusceptible to biotrophic pathogens and are deficient in PR1 induction (Rogers and Ausubel 1997, Zhou et al. 1998, Nawrath and Metraux 1999).
Transcription defense genes are tightly regulated because numerous transcription factors interact to fine-tune the defense response (Riechmann et al. 2000, Thilmony et al. 2006). In addition, eukaryotic DNA is wrapped around histone octamers. The resulting chromatin provides a higher level of regulation; chromatin configuration can be altered to allow or prevent transcription initiation (Nelissen et al. 2007). In both Arabidopsis and tobacco, SA-induced PR1 expression is associated with increased histone acetylation at the PR1 promoter (Butterbrodt et al. 2006, Mosher et al. 2006), indicating that histone acetylation regulates gene expression in the SA signaling pathway. Previous studies have shown that PAD4, EDS5 and SID2 are rapidly induced by pathogens (Jirage et al. 1999, Wildermuth et al. 2001, Nawrath et al. 2002). However, the mechanism by which transcription of PAD4, EDS5 and SID2 is regulated at the level of histone modification remains largely unclear.
In the present study, we characterized the functions of Arabidopsis deacetylase AtSRT2. We found that AtSRT2 was down-regulated by Pseudomonas syringae pv. tomato DC3000 (PstDC3000) infection. The protein encoded by AtSRT2 negatively regulates the plant basal defense and PR1 expression. Moreover, pathogen-induced expression of PAD4, EDS5 and SID2 was suppressed by AtSRT2, suggesting that AtSRT2 plays an important role in regulating SA synthesis.
Results
Nuclear localization of AtSRT2
Several HDACs are translocated to the nucleus to regulate gene expression (Hollender and Liu 2008), which is consistent with their functions in modifying chromatin. AtSRT2 has seven predicted splice variants (see Supplementary Fig. S1A); however, only the third transcript (AtSRT2-CDS3), which lacks the two C-terminal exons, has been characterized (Pandey et al. 2002). We amplified the seven putative transcripts of AtSRT2 by reverse transcription–PCR (RT–PCR), and found that AtSRT2-CDS3 was the predominant splice variant (data not shown).
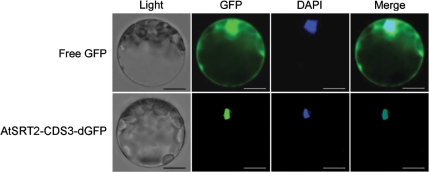
Sequence analysis has demonstrated that AtSRT2-CDS3 contains a typical nuclear localization signal (NLS) (Pandey et al. 2002). To determine the subcellular localization of AtSRT2-CDS3, we fused AtSRT2-CDS3 in-frame to the 5′ end of the green fluorescent protein (dGFP). The AtSRT2-CDS3-dGFP construct was introduced into Arabidopsis mesophyll protoplasts by polyethylene glycol (PEG)-mediated DNA transfection (Yoo et al. 2007). Green fluorescence was detected in the whole cell transformed with the GFP control (Fig. 1, upper panel), whereas the AtSRT2-CDS3–dGFP fusion protein was expressed exclusively in the nucleus (Fig. 1, lower panel), indicating that AtSRT2-CDS3 localizes to the nucleus.
Fig. 1.
Nuclear localization of AtSRT2-CDS3. Plasmids carrying green fluorescent protein (GFP control; upper panel) or AtSRT2-CDS3–GFP (bottom panel) were transformed into Arabidopsis mesophyll protoplasts. Fluorescent images were taken at 16 h after transfection. The nucleus was stained with 4′,6-diamidino-2-phenylindole (DAPI). Scale bar = 10 μm. The image is representative of experiments performed in triplicate.
Expression profile of AtSRT2
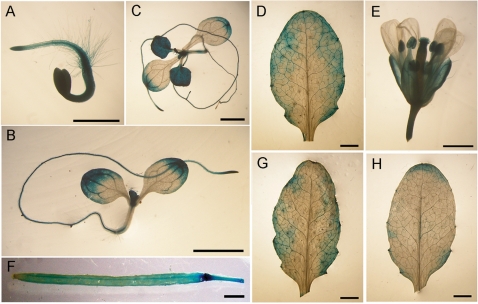
To determine the function of AtSRT2, we first evaluated its expression profile by fusing the AtSRT2 promoter to a β-glucuronidase (GUS) reporter gene. The resulting construct (pAtSRT2-GUS) was transformed into Arabidopsis. Four independent transgenic lines with a single insertion were obtained, and GUS activity was detected in different organs and development stages (Fig. 2A–F). In particular, AtSRT2 promoter activity was high in roots (Fig. 2A, 2B), leaves (Fig. 2B, D) and flowers (Fig. 2E).
Fig. 2.
Expression profile of AtSRT2. β-Glucuronidase (GUS) activity was detected by histochemistry in transgenic plants containing pAtSRT2-GUS. Typical GUS expression patterns are shown for (A) 3-day-old seedlings; (B) 6-day-old seedlings; (C) 12-day-old seedlings; (D) leaves from adult plants; (E) flowers; (F) siliques; (G) mock-treated leaves; and (H) leaves inoculated with PstDC3000. Scale bar = 1 mm. These images are representative of experiments performed in triplicate.
To assess whether the expression of AtSRT2 is responsive to pathogen infection, we compared GUS activity in transgenic plants before and after virulent PstDC3000 inoculation. GUS activity was reduced after pathogen inoculation but not after mock treatment (Fig. 2G, H), which suggests that PstDC3000 infection represses AtSRT2 expression.
Down-regulation of AtSRT2 by PstDC3000, inoculation
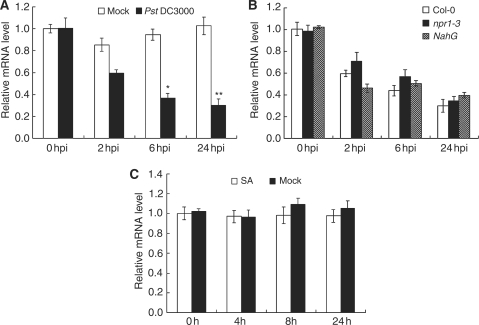
The down-regulation of AtSRT2 promoter activity by PstDC3000 infection (Fig. 2G, H) prompted us to evaluate the role of AtSRT2 in the plant basal defense. To gain more detailed insights into AtSRT2 expression upon PstDC3000 infection, we determined AtSRT2 mRNA levels by quantitative RT–PCR in PstDC3000-inoculated plants at different time points. As shown in Fig. 3A, pathogen infection markedly reduced AtSRT2 mRNA levels. Only about 30% of AtSRT2 transcripts remained at 24 h after pathogen inoculation, which is consistent with our promoter activity assay results (Fig. 2).
Fig. 3.
AtSRT2 expression is repressed by pathogen infection. (A) Four-week-old WT Arabidopsis plants (Col-0) were infiltrated with 10 mM MgCl2 (open bars) or PstDC3000 (filled bars; OD600 = 0.2 in 10 mM MgCl2). The infiltrated leaves were collected at the indicated time for quantitative RT–PCR analysis. (B) Four-week-old WT (Col-0), npr1-3 and NahG Arabidopsis plants were infiltrated with PstDC3000 (OD600 = 0.2 in 10 mM MgCl2). The infiltrated leaves were collected at the indicated time for quantitative RT–PCR analysis. (C) Two-week-old WT (Col-0) seedlings grown on MS medium were untreated (open bars) or treated with 0.5 mM salicylic acid (SA; filled bars). Seedlings were collected at the indicated time for quantitative RT–PCR analysis. UBQ10 was used as an internal control. Data represent the mean ± SD from four independent experiments. The statistical significance of the difference was confirmed by Student’s t-test, *P < 0.05; **P < 0.01.
The SA-mediated signaling pathway regulated by NPR1 is one of the most important pathways in plant defense (Durrant and Dong 2004, Loake and Grant 2007). To assess the roles of SA and NPR1 in the pathogen-induced down-regulation of AtSRT2, we determined AtSRT2 mRNA levels in the npr1-3 mutant and SA-deficient NahG transgenic plants. As shown in Fig. 3B, AtSRT2 expression was still inhibited by PstDC3000 infection in npr1-3 mutants and NahG plants, indicating that AtSRT2 expression is not dependent on SA or NPR1. Consistent with this result, we also found that the AtSRT2 mRNA levels were not affected by exogenous SA treatment in WT plants (Fig. 3C).
Disruption of AtSRT2 enhances plant basal defense and PR1 expression
To characterize the functions of AtSRT2 in vivo, we obtained a homozygous T-DNA insertion line (SALK_149295) for AtSRT2 from the Arabidopsis Biological Resource Center (ABRC). The precise insertion position was determined by PCR with primers specific to AtSRT2 and the T-DNA sequence, followed by sequencing of the PCR product. We found that SALK_149295 carries a T-DNA insertion in the second exon of AtSRT2 (see Supplementary Fig. S1A). AtSRT2 mRNA was not detected in the srt2 mutant by RT–PCR (see Supplementary Fig. S1B).
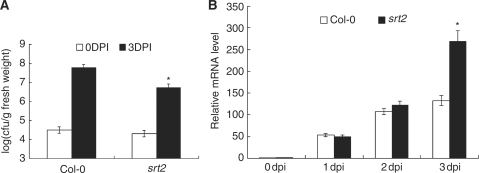
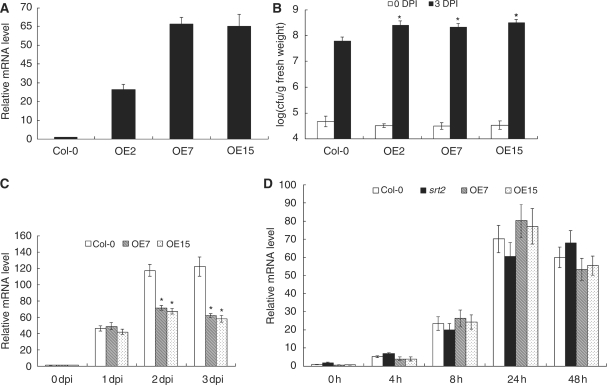
The down-regulation of AtSRT2 by pathogen infection prompted us to investigate the potential role of AtSRT2 in regulating the plant basal defense. After inoculating the srt2 mutant and WT plants with PstDC3000, we compared bacterial growth rates. As shown in Fig. 4A, at 0 day post-inoculation (dpi), srt2 and WT plants contained the same amount of PstDC3000, indicating equal initial bacterial doses. However, at 3 dpi, the bacterial pathogen accumulated in the srt2 mutant was 10-fold lower than that of WT plants in three independent experiments, suggesting that the srt2 mutation enhanced the plant basal defense. PR genes have been widely used as markers of the plant basal defense system (Durrant and Dong 2004). We determined PR1 mRNA expression in PstDC3000-inoculated srt2 and WT plants by quantitative RT–PCR and Northern blot. As shown in Fig. 4B and Supplementary Fig. S2, PstDC3000 treatment induced PR1 expression strongly in both WT and mutant plants, and PR1 transcripts levels were higher in srt2 mutants at 3 dpi compared with the WT.
Fig. 4.
The srt2 mutant is more resistant to pathogen infection. (A) WT (Col-0) plants and srt2 mutants were infiltrated with a suspension of PstDC3000 (OD600 = 0.0001 in 10 mM MgCl2). Bacterial growth was determined at 0 dpi (open bars) or 3 dpi (filled bars). Each data point consisted of at least six samples. Error bars indicate the SD. The statistical significance of the difference was confirmed by Student’s t-test, *P < 0.05. (B) Pathogen-induced PR1 expression. WT (Col-0) plats and srt2 mutants were infiltrated with a suspension of PstDC3000 (OD600 = 0.0001 in 10 mM MgCl2). Total RNA was extracted at the time indicated for quantitative RT–PCR analysis. UBQ10 was used as internal control. Data represent the mean ± SD from three independent experiments. The statistical significance of the difference was confirmed by Student’s t-test, *P < 0.05.
Overexpression of AtSRT2-CDS3 compromises plant basal defense and PR1 expression
To characterize further the function of AtSRT2 in the basal defense system, we generated transgenic Arabidopsis plants that overexpress AtSRT2-CDS3. The AtSRT2-CDS3 full-length cDNA was cloned behind the cauliflower mosaic virus (CaMV) 35S promoter, and this construct was transformed into Arabidopsis plants. Three independent transgenic lines (OE2, OE7 and OE15) were chosen for further analysis. Our quantitative RT–PCR results revealed constitutively elevated expression of AtSRT2 in all the three transgenic plants, while the expression level of AtSRT2 in OE2 was lower than that in OE7 and OE15 (Fig. 5A).
Fig. 5.
Overexpression of AtSRT2-CDS3 attenuates the plant defense response. (A) AtSRT2 expression of 4-week-old WT (Col-0) and AtSRT2-CDS3-overexpressing Arabidopsis plants was determined by quantitative RT–PCR. UBQ10 was used as an internal control. Data represent the mean ± SD from two independent experiments. (B) WT (Col-0) and AtSRT2-CDS3-overexpressing plants were infiltrated with a suspension of PstDC3000 (OD600 = 0.0001 in 10 mM MgCl2). Samples were taken at 0 dpi (open bars) or 3 dpi (filled bars) to determine bacterial growth. Each data point consisted of at least six samples. Error bars indicate the SD. The statistical significance of the difference was confirmed by Student’s t-test, *P < 0.05. (C) Pathogen-induced PR1 expression. WT (Col-0) and AtSRT2-CDS3-overexpressing plants (OE7 and OE15) were treated with a suspension of PstDC3000 (OD600 = 0.0001 in 10 mM MgCl2). Inoculated leaves were collected for quantitative RT–PCR analysis. UBQ10 was used as internal control. Data represent the mean ± SD from three independent experiments. The statistical significance of the difference was confirmed by Student’s t-test, *P < 0.05. (D) Two-week-old WT (Col-0), srt2 and AtSRT2-CDS3-overexpressing (OE7 and OE15) Arabidopsis plants were treated with 0.5 mM SA to induce PR1 expression. Total RNA was extracted at different time points for quantitative RT–PCR analysis. UBQ10 was used as internal control. Data represent the mean ± SD from two independent experiments.
After PstDC3000 inoculation of plants, more bacterial pathogen was detected in the overexpressing transgenic lines compared with WT plants at 3 dpi in three independent experiments (Fig. 5B), indicating that overexpression of AtSRT2-CDS3 made plants more susceptible to PstDC3000 infection. In addition, OE2 was susceptible to PstDC3000 at a similar level to that of OE7 and OE15, suggesting that pathogen susceptibility in overexpressing plants might be independent of the expression level of AtSRT2. Consistent with these findings, PR1 transcripts were reduced in the AtSRT2-overexpressing lines compared with WT plants (Fig. 5C and Supplementary Fig. S2). These results are consistent with our findings in the srt2 mutant. Thus, analysis of both loss-of-function AtSRT2 mutants and gain-of-function AtSRT2-CDS3-overexpressing plants indicates that AtSRT2 functions as a negative regulator in plant basal defense.
Besides pathogen inoculation, we also analyzed SA-induced PR1 expression in WT, srt2 and AtSRT2-CDS3-overexpressing plants to determine the mechanism by which AtSRT2 regulates the SA signaling pathway. As shown in Fig. 5D, we did not see a significant difference in PR1 transcript levels among WT, srt2 mutant and overexpression plants, suggesting that AtSRT2 does not influence downstream gene expression in the presence of SA.
AtSRT2 negatively regulates EDS5, PAD4 and SID2 expression
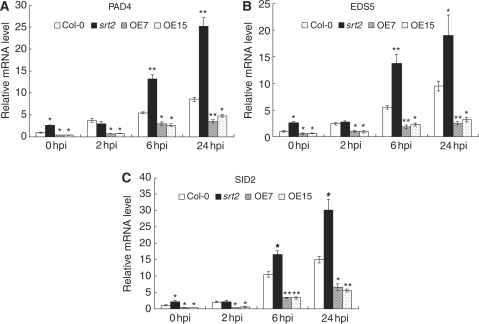
Biosynthesis of SA, which requires a series of enzymes, is an essential step in the plant defense against biotrophic pathogens (Shah 2003, Durrant and Dong 2004). We found that AtSRT2 repressed pathogen-induced PR1 expression but had little effect on SA-induced PR1 expression (Figs. 4B, 5C, D), suggesting that AtSRT2 is involved downstream of pathogen recognition but upstream of SA signaling. To better understand the role of AtSRT2 in SA biosynthesis in the plant basal defense system, we analyzed the expression of SA biosynthesis-related enzymes PAD4, EDS5 and SID2 under various conditions. As shown in Fig. 6, expression of PAD4, EDS5 and SID2 at 0 dpi was higher in the srt2 mutant but lower in AtSRT2-CDS3-overexpressing lines compared with the WT, suggesting that these three genes are repressed by AtSRT2 even in the absence of pathogens. Treatment with PstDC3000 increased expression of PAD4, EDS5 and SID2, which is consistent with results of previous studies (Jirage et al. 1999, Wildermuth et al. 2001, Nawrath et al. 2002). Furthermore, pathogen-induced expression of all the three genes was significantly higher in srt2 plants but lower in AtSRT2-CDS3-overexpressing plants compared with the WT (Fig. 6). Taken together, these results indicate that AtSRT2 negatively regulates both basal and pathogen-induced expression of SA biosynthesis-related genes, possibly a determinant for its role in suppressing plant basal defense.
Fig. 6.
AtSRT2 negatively regulates EDS5, PAD4 and SID2 expression. Four-week-old WT (Col-0), srt2 and AtSRT2-CDS3-overexpressing (OE7 and OE15) Arabidopsis plants were inoculated with PstDC3000 (OD600 = 0.2 in 10 mM MgCl2). Total RNA was extracted at the indicated time points for quantitative RT–PCR analysis of PAD4, EDS5 and SID2. UBQ10 was used as an internal control. Data represent the mean ± SD from three independent experiments. The statistical significance of the difference was confirmed by Student’s t-test, *P < 0.05, **P < 0.01.
Discussion
Histone modification, especially acetylation, is essential for transcriptional regulation. In general, histone hyperacetylation is associated with gene activation, whereas histone deacetylation by HDACs leads to gene repression (Hebbes et al. 1988, Hollender and Liu 2008). Plant genomes contain a large number of HDACs (Pandey et al. 2002), making it important, but challenging, to determine the function of each specific HDAC.
Our findings also demonstrated that AtSRT2 functions as a negative regulator of the plant basal defense. First, we generated transgenic Arabidopsis plants that stably expressed the GUS gene under the control of the AtSRT2 promoter. GUS staining was observed in roots (Fig. 2A, B), leaves (Fig. 2B, D) and flowers (Fig. 2E). AtSRT2 was found to affect the expression of FLC and the vernalization response of Arabidopsis (Bond et al. 2009).Our GUS staining result suggested that AtSRT2 may have effects on plant growth and development. GUS staining also revealed that AtSRT2 promoter activity was reduced upon PstDC3000 inoculation (Fig. 2G, H), indicating that AtSRT2 may be involved in the PstDC3000-induced defense response. Quantitative RT–PCR analysis confirmed that AtSRT2 expression was repressed by PstDC3000 infection (Fig. 3A, B) in an NPR1- and SA-independent manner (Fig. 3B, C).
Secondly, knock-out of AtSRT2 enhanced resistance against PstDC3000 infection and increased PR1 expression (Fig. 4 and Supplementary Fig. S2), suggesting a negative regulatory role for AtSRT2 in the pathogen-induced defense response. This conclusion was supported by findings in AtSRT2-CDS3-overexpressing plants; both independent homozygous transgenic lines were more susceptible to PstDC3000 infection (Fig. 5B) and attenuated PstDC3000-induced PR1 expression (Fig. 5C and Supplementary Fig. S2).
Thirdly, pathogen-induced expression of PAD4, EDS5 and SID2, three key regulators of SA biosynthesis, was increased in the srt2 mutant but markedly reduced in AtSRT2-CDS3- overexpressing lines compared with WT plants (Fig. 6). The AtSRT2 attenuation of SA biosynthesis-related genes indicates suppression of SA-mediated signaling. Further, exogenous SA treatment did not affect AtSRT2 expression (Fig. 3C), and exogenous SA-induced PR1 expression was unaffected by the srt2 mutation or overexpression (Fig. 5D). We also studied the potential function of AtSRT2 in response to an avirulent strain of PstDC3000. We measured the ionic conductivity of the released electrolyte after PstDC3000 (AvrRpt2) infection but did not observe any significant difference among WT, srt2 and AtSRT2-CDS3-overexpressing plants (data not shown).
In the present study, we characterized the function of AtSRT2, which is a member of the second HDAC subfamily in Arabidopsis. AtSRT2 has seven predicted splice variants (Pandey et al. 2002); we observed that the third transcript (AtSRT2-CDS3) was the predominant splice variant (data not shown). In addition, we showed that the AtSRT2-CDS3–dGFP fusion protein was located exclusively in the nucleus (Fig. 1), suggesting a role for AtSRT2-CDS3 in regulating gene expression.
SA is essential in plant disease resistance. In response to hemi-biotrophic pathogens such as PstD3000, plants accumulate SA and rapidly activate SA signaling (Nimchuk et al. 2003, Akira et al. 2006). However, SA itself can be harmful to the growth, reproduction and survival of plants, especially at high doses (Heil and Baldwin 2002). In Arabidopsis, constitutive overproduction of SA results in a strongly dwarfed phenotype and decreased seed production (Mauch et al. 2001). Thus negative regulation of SA biosynthesis-related genes is needed to avoid SA toxicity (Heil and Baldwin 2002). Expression of PAD4, EDS5 and SID2 was enhanced in the srt2 mutant but reduced in the AtSRT2-CDS3-overexpressing lines; therefore, we propose that AtSRT2 inhibits SA accumulation by suppressing SA biosynthesis-related genes. The antagonistic effects of AtSRT2 on SA synthesis may prevent an effective response to pathogen infections (Figs. 4A, 5B); therefore, negative regulation of AtSRT2 expression occurs as early as 2 h after pathogen inoculation (Fig. 3A). However, the mechanism by which AtSRT2 is regulated at this early stage of the defense response requires further investigation.
Numerous HDACs suppress gene expression by reducing histone acetylation (Hollender and Liu 2008). OsSRT1, an SIR2-related protein in rice, was found to deacetylate histone H3K9 and repress HR-related genes (Huang et al. 2007). Our results indicate that AtSRT2 negatively regulates the plant basal defense, presumably by down-regulating PAD4, EDS5 and SID2 expression. Sequence similarity among AtSRT2 and other SIR2 family members suggests that AtSRT2 may negatively regulate PAD4, EDS5 and SID2 by histone deacetylation of their promoters.
The understanding of plant defense regulation is still limited. In particular, the balance between activation and deactivation of defense-related genes to fine-tune the plant basal defense response remains largely unclear. Our results demonstrate that AtSRT2 attenuates the plant basal defense by reducing SA biosynthesis-related gene expression, providing insights into deactivation of SA signaling in the plant basal defense.
Materials and Methods
Plant materials
Seeds of Arabidopsis thaliana ecotype Columbia (Col-0) were surface-sterilized with 10% NaClO for 15 min and then washed five times with sterile water. Sterile seeds were suspended in 0.12% agarose and plated on Murashige and Skoog (MS) medium (Murashige and Skoog 1962) plus 3% sucrose. Plants were stratified in the dark for 48 h at 4°C and then grown in a controlled growth chamber with a relatively short photoperiod (10 h light at 22°C/14 h dark at 20°C) with approximately 75% relative humidity. After 2 weeks, seedlings were potted in soil.
Isolation of the T-DNA insertion mutant
Seeds of WT Arabidopsis and the srt2 mutant (SALK_149295) were obtained from the ABRC. The homozygous mutant was isolated according to the Salk protocol (http://signal.salk.edu/tdnaprimers.2.html). Plants homozygous for the T-DNA insertion were confirmed by PCR amplification using primers corresponding to the sequences flanking the T-DNA insertion and gene-specific primers. Primer sequences are shown in Supplementary Table S1.
Overexpression
The sequence of AtSRT2-CDS3 was amplified from the cDNA of WT (Col-0) plants using a high-fidelity DNA polymerase, KOD-plus (Toyobo, Osaka, Japan). Forward and reverse primer sequences are shown in Supplementary Table S1. The PCR product was inserted into the NcoI restriction sites of vector pRTL2-dGFP (a derivative of pRTL2). The coding sequence of AtSRT2 was fused in-frame to the N-terminus of the first GFP-coding sequence and driven by the CaMV 35S promoter. The resulting pRTL2-AtSRT2-dGFP construct was also used in the cellular localization assay. A restriction fragment containing AtSRT2 was released from pRTL2-AtSRT2-dGFP using HindIII and ligated into the binary vector pCAMBIA 1301 (http://www.cambia.org). The binary plasmid pCAMBIA1301-AtSRT2 was transformed into Agrobacterium tumefaciens strain GV3101 (pMP90).
Arabidopsis transformation was performed with the floral dip method (Clough and Bent 1998). To screen for transformants, seeds were grown on MS medium plates containing 40 μg ml−1 hygromycin B (Roche Diagnostics, Mannheim, Germany). Resistant plants were transferred to soil for further analysis.
Chemical treatment
Two-week-old seedlings grown on MS medium were transferred to fresh MS solution containing 0.5 mM SA (Sigma, USA). Samples were collected at different time points.
Northern blot
Total RNA was isolated from treated plants with TRIzol reagent (Invitrogen, USA) according to the manufacturer’s instructions. Approximately 5 μg of total RNA from each sample was separated on a 1.2% formaldehyde agarose gel (Mao et al. 2007). After transferring the separated RNA to Hybond-N membranes (Amersham Biosciences, UK), the membranes were hybridized with digoxigenin (DIG)-labeled probes. Immunodetection was performed according to the manufacturer’s instructions (Roche).
Pathogen inoculation
Pseudomonas syringae pv. tomato DC3000 strain was propagated at 28°C on King’s B medium containing rifampicin (50 μg ml−1). For disease testing, at least six 4-week-old plants were infiltrated with 10 mM MgCl2 (mock treatment) or a bacterial suspension of PstDC3000 (OD600 = 0.0001 in 10 mM MgCl2). After 3 d, leaves were harvested, homogenized in 10 mM MgCl2 and then serially diluted and spread on King’s B medium containing rifampicin (50 μg ml−1). Plates were incubated at 28°C for 2 d, and the colony number was then determined. Data analyses were performed using the computer program Sigma Plot Version 10.0 software and were considered significantly different at the 0.05 level. To determine expression of AtSRT2, PAD4, EDS5 and SID2, a bacterial suspension of PstDC3000 (OD600 = 0.2 in 10 mM MgCl2) was used.
Quantitative RT–PCR
Total RNA was extracted with Trizol Reagent (Invitrogen) and treated with RNase-free DNase I (TAKARA Biotechnology, Dalian, China). First-strand cDNA was synthesized with SuperScript III Reverse Transcriptase (Invitrogen) and then diluted for use as template for quantitative RT–PCR. Primer sequences are shown in Supplementary Table S1. PCR was carried out using SYBR Green Real-time PCR Master Mix (Toyobo, Japan) on an Opticon 2 continuous fluorescence detection system (CFD-3220, MJ Research, USA). The specific mRNA abundance relative to constitutively expressed UBQ10 was calculated using the 2−ΔΔCt method (Livak and Schmittgen 2001, Ferreira et al. 2006).
Histochemical GUS detection
To generate the pAtSRT2-GUS construct, a 1.2 kb fragment upstream of the AtSRT2 gene was amplified by PCR from genomic DNA. After sequence analysis, the promoter fragment was cloned into pCAMBIA1300-221 (Chu et al. 2007). Four independent transgenic lines, each containing a single T-DNA insertion, were tested for GUS activity. Tissues were incubated overnight in GUS staining buffer [2 mM X-gluc, 0.1 M sodium phosphate buffer (pH 7.0), 0.1% Triton X-100, 1 mM potassium ferricyanide, 1 mM potassium ferrocyanide, 10 mM EDTA] at 37°C in the dark. Samples were destained with 75% ethanol solution and examined by a light microscope (Olympus SZX-ILLD2-200, Olympus Corporation, Tokyo, Japan).
Cellular localization assay
The pRTL2-AtSRT2-dGFP plasmid was introduced into Arabidopsis mesophyll protoplasts; pRTL2-dGFP was used as a control with the DNA–PEG–calcium method as described previously (Yoo et al. 2007). After transfection, protoplasts were maintained for 16 h at room temperature in the dark. GFP was detected by fluorescence microscopy (Type 020-525.021, Leica Microsystems Ltd., Germany) and photographed with a KX Series Imaging System (Model KX32E, Apogee Instruments Inc., Logan, UT, USA).
Supplementary data
Supplementary data are available at PCP online.
Funding
This work was supported by grants from 973 [contract No. 2006CB101905, 2007CB10920204 programs to Y.L.]
Supplementary Material
Acknowledgments
We thank the ABRC for srt2 seeds, Dr. Biao Ding for pRTL2:dGFP (dimeric GFP), and Dr. Qi Xie for pCambia 1300-221. The virulent pathogen PstDC3000 was kindly provided by Dr. Jian-Min Zhou.
Glossary
Abbreviations
- CaMV
cauliflower mosaic virus
- DIG
digoxigenin
- EDS1
enhanced disease susceptibility 1
- EDS5
enhanced disease susceptibility 5
- GFP
green fluorescent protein
- HDAC
histone deacetylase
- HR
hypersensitive response
- GUS
β-glucuronidase
- MS
Murashige and Skoog
- NahG
salicylate hydroxylase
- NPR1
non-expresser of PR genes 1
- PAD4
phytoalexin deficient 4
- PEG
polyethylene glycol
- PR
pathogenesis-related
- PstDC3000
Pseudomonas syringae pv. tomato DC3000
- RT–PCR
reverse transcription–PCE
- SA
salicylic acid
- SID2
salicylic acid induction deficient 2
- Sir2
silent information regulator 2
- WT
wild type.
Footnotes
Nucleotide sequence data for the genes described in this study have been deposited in the GenBank/EMBL data libraries with the following accession numbers: AtSRT2 (At5g09230); AtSRT1 (At5g55760); PAD4 (At3g52430); EDS5 (At4g39030); EDS1 (At3g48090); SID2 (At1g74710); NPR1 (At1g64280); PR1 (At2g14610).
References
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- Bond DM, Dennis ES, Pogson BJ, Finnegan EJ. Histone acetylation, VERNALIZATION INSENSITIVE 3, FLOWERING LOCUS C, and the vernalization response. Mol. Plant. 2009;2:724–737. doi: 10.1093/mp/ssp021. [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Sherman JM, Devine SE, Cameron EE, Pillus L, Boeke JD. The Sir2 gene family, conserved from bacteria to humans, functions in silencing, cell-cycle progression, and chromosome stability. Genes Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- Buck SW, Gallo CM, Smith JS. Diversity in the Sir2 family of protein deacetylases. J. Leukoc. Biol. 2004;75:939–950. doi: 10.1189/jlb.0903424. [DOI] [PubMed] [Google Scholar]
- Butterbrodt T, Thurow C, Gatz C. Chromatin immunoprecipitation analysis of the tobacco PR-1a- and the truncated CaMV 35S promoter reveals differences in salicylic acid-dependent TGA factor binding and histone acetylation. Plant Mol. Biol. 2006;61:665–674. doi: 10.1007/s11103-006-0039-2. [DOI] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong XN. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell. 1997;88:57–63. doi: 10.1016/s0092-8674(00)81858-9. [DOI] [PubMed] [Google Scholar]
- Chu Z, Chen H, Zhang Y, Zhang Z, Zheng N, Yin B, et al. Knockout of the AtCESA2 gene affects microtubule orientation and causes abnormal cell expansion in Arabidopsis. Plant Physiol. 2007;143:213–224. doi: 10.1104/pp.106.088393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Dali-Youcef N, Lagouge M, Froelich S, Koehl C, Schoonjans K, Auwerx J. Sirtuins: the ‘magnificent seven’, function, metabolism and longevity. Ann. Med. 2007;39:335–345. doi: 10.1080/07853890701408194. [DOI] [PubMed] [Google Scholar]
- Durrant WE, Dong X. Systemic acquired resistance. Annu. Rev. Phytopathol. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- Ferreira ID, Rosario VE, Cravo PV. Real-time quantitative PCR with SYBR Green I detection for estimating copy numbers of nine drug resistance candidate genes in Plasmodium falciparum. Malaria J. 2006;5:6. doi: 10.1186/1475-2875-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJ, Moisan LJ, Newman MA, Parker JE. Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 2001;20:5400–5411. doi: 10.1093/emboj/20.19.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Guarente LP. Mammalian sirtuins—emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- Hebbes TR, Thorne AW, Cranerobinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil M, Baldwin IT. Fitness costs of induced resistance: emerging experimental support for a slippery concept. Trends Plant Sci. 2002;7:61–67. doi: 10.1016/s1360-1385(01)02186-0. [DOI] [PubMed] [Google Scholar]
- Hollender C, Liu Z. Histone deacetylase genes in Arabidopsis development. J Integr. Plant Biol. 2008;50:875–885. doi: 10.1111/j.1744-7909.2008.00704.x. [DOI] [PubMed] [Google Scholar]
- Huang L, Sun Q, Qin F, Li C, Zhao Y, Zhou DX. Down-regulation of a SILENT INFORMATION REGULATOR2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiol. 2007;144:1508–1519. doi: 10.1104/pp.107.099473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage D, Tootle TL, Reuber TL, Frost LN, Feys BJ, Parker JE, et al. Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl Acad. Sci. USA. 1999;96:13583–13588. doi: 10.1073/pnas.96.23.13583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Loake G, Grant M. Salicylic acid in plant defence—the players and protagonists. Curr. Opin. Plant Biol. 2007;10:466–472. doi: 10.1016/j.pbi.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Mao P, Duan M, Wei C, Li Y. WRKY62 transcription factor acts downstream of cytosolic NPR1 and negatively regulates jasmonate-responsive gene expression. Plant Cell Physiol. 2007;48:833–842. doi: 10.1093/pcp/pcm058. [DOI] [PubMed] [Google Scholar]
- Mauch F, Mauch-Mani B, Gaille C, Kull B, Haas D, Reimmann C. Manipulation of salicylate content in Arabidopsis thaliana by the expression of an engineered bacterial salicylate synthase. Plant J. 2001;25:67–77. doi: 10.1046/j.1365-313x.2001.00940.x. [DOI] [PubMed] [Google Scholar]
- Mosher RA, Durrant WE, Wang D, Song JQ, Dong XN. A comprehensive structure–function analysis of Arabidopsis SNI1 defines essential regions and transcriptional repressor activity. Plant Cell. 2006;18:1750–1765. doi: 10.1105/tpc.105.039677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962;15:473–477. [Google Scholar]
- Nawrath C, Heck S, Parinthawong N, Metraux JP. EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell. 2002;14:275–286. doi: 10.1105/tpc.010376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C, Metraux JP. Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell. 1999;11:1393–1404. doi: 10.1105/tpc.11.8.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen H, Boccardi TM, Himanen K, Van Lijsebettens M. Impact of core histone modifications on transcriptional regulation and plant growth. Crit. Rev. Plant Sci. 2007;26:243–263. [Google Scholar]
- Nimchuk Z, Eulgem T, Holt BE, Dangl JL. Recognition and response in the plant immune system. Annu. Rev. Genet. 2003;37:579–609. doi: 10.1146/annurev.genet.37.110801.142628. [DOI] [PubMed] [Google Scholar]
- Pandey R, Muller A, Napoli CA, Selinger DA, Pikaard CS, Richards EJ, et al. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 2002;30:5036–5055. doi: 10.1093/nar/gkf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang CZ, Keddie J, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- Robyr D, Suka Y, Xenarios I, Kurdistani SK, Wang A, Suka N, et al. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell. 2002;109:437–446. doi: 10.1016/s0092-8674(02)00746-8. [DOI] [PubMed] [Google Scholar]
- Rogers EE, Ausubel FM. Arabidopsis enhanced disease susceptibility mutants exhibit enhanced susceptibility to several bacterial pathogens and alterations in PR-1 gene expression. Plant Cell. 1997;9:305–316. doi: 10.1105/tpc.9.3.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauve AA, Wolberger C, Schramm VL, Boeke JD. The biochemistry of sirtuins. Annu. Rev. Biochem. 2006;75:435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- Shah J. The salicylic acid loop in plant defense. Curr. Opin. Plant Biol. 2003;6:365–371. doi: 10.1016/s1369-5266(03)00058-x. [DOI] [PubMed] [Google Scholar]
- Thilmony R, Underwood W, He SY. Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157: H7. Plant J. 2006;46:34–53. doi: 10.1111/j.1365-313X.2006.02725.x. [DOI] [PubMed] [Google Scholar]
- Vlot AC, Klessig DF, Park SW. Systemic acquired resistance: the elusive signal(s) Curr. Opin. Plant Biol. 2008;11:436–442. doi: 10.1016/j.pbi.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature. 2001;414:562–565. doi: 10.1038/35107108. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Schoonjans K, Auwerx J. Sirtuin functions in health and disease. Mol. Endocrinol. 2007;21:1745–1755. doi: 10.1210/me.2007-0079. [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- Zhou N, Tootle TL, Tsui F, Klessig DF, Glazebrook J. PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell. 1998;10:1021–1030. doi: 10.1105/tpc.10.6.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.