Abstract
For the last couple of years, a method that permits the collection of precise positional information of transcriptional start sites (TSSs) together with digital information of the gene-expression levels in a high-throughput manner was established. We applied this novel method, ‘tss-seq’, to elucidate the transcriptome of tachyzoites of the Toxoplasma gondii, which resulted in the identification of 124 000 TSSs, and they were clustered into 10 000 transcription regions (TRs) with a statistics-based analysis. The TRs and annotated ORFs were paired, resulting in the identification of 30% of the TRs and 40% of the ORFs without their counterparts, which predicted undiscovered genes and stage-specific transcriptions, respectively. The massive data for TSSs make it possible to execute the first systematic analysis of the T. gondii core promoter structure, and the information showed that T. gondii utilized an initiator-like motif for their transcription in the major and novel motif, the downstream thymidine cluster, which was similar to the Y patch observed in plants. This encyclopaedic analysis also suggested that the TATA box, and the other well-known core promoter elements were hardly utilized.
Keywords: Toxoplasma, transcriptome, tss-seq, core promoter
1. Introduction
Toxoplasma gondii is an obligate apicomplexan parasite that can invade a wide variety of mammalian cells, including those of humans and many economically important domestic animals. Toxoplasmosis is a common opportunistic infection caused by T. gondii in immunocompromised individuals, such as AIDS patients, and congenital toxoplasmosis is also a serious protozoan disease that occurs in infants when mothers became infected with T. gondii for the first time during or just before pregnancy. Infection in livestock is another threat that causes a serious economic loss.1
Toxoplasma gondii is also one of the most intensively studied protozoans. Their life cycle is heteroxenous. That is, sexual proliferation occurs in members of Felidae as definitive hosts and asexual proliferation occurs in other worm-blooded intermediate hosts in two forms (acutely lytic tachyzoites and slowly proliferating bradyzoites in tissue cysts that cause chronic infection). To understand the molecular mechanism driving this stage conversion is a major issue in the field of Toxoplasma studies because chronic infection by bradyzoites makes it difficult for the immune system to exclude infected parasites. Furthermore, bradyzoites are also a causable element for inter-individual spreading of the parasites.
To give a molecular-based explanation for these unique biological natures, genomics and transcriptomics might be a promising approach. The genome of T. gondii has been sequenced, and around 8000 genes are predicted.2 A series of EST analyses has contributed to give reliability to the prediction.3,4 In addition, the identification of intron-spanning peptides provided by proteome analysis supports splice site annotation.5,6 The expression profiles have also been analysed by EST,3,4 microarray,7–9 and SAGE,10 and they have confirmed the existence of transcripts associated with the predicted ORFs. The result of a SAGE analysis, in particular, provides a substantial clue for the estimation of the 3′ terminal of each transcript.10 On the other hand, the majority of 5′ UTRs and translation start sites are estimated without any substantial evidence except for genes individually analysed. Therefore, experimental evidence for 5′ UTRs and fine-tuning of the Kozak rule in T. gondii11 are desired for a better gene annotation.
In control of transcription, it is widely accepted that transcription factors and cis-DNA elements play a key role. In the former case, it is reported in Apicomplexa that there are homologous genes of general transcription factors, such as the TATA box-binding protein or TATA association factors (TAFs).12 However, specific transcription factors (TFs) have not been identified, except for the ApiAP2 family, which was originally identified in plants.13–15 The cis-DNA elements can be categorized into transcription motifs and core promoter elements. As transcription motifs in T. gondii, short nucleotides consisting of WGAGACG upstream of SAG1, some GRA genes, and DHFR-TS16–18 and YGCATGCR in ribosomal protein have been reported.19 Specific sequences of 30 bp in NTPase and 20 bp in the BAG1 promoter are also identified as a responsible element in Compound 1 induction.20 Joint work with bioinformatics has also succeeded in finding several novel motifs.21 Although core promoters were originally considered to be invariant, it has been found that they have certain diversity in structure and function22 derived from motifs such as the TATA box, the initiator (INR), the upstream or downstream TFIIB recognition element (BREu or BREd), the downstream core promoter element (DPE), and the motif-10 element (MTE). Furthermore, it is suggested that diversity in the core promoter structure is involved in transcriptional regulation.22 However, in T. gondii core promoters, neither the TATA box nor other well-characterized motifs in mammals are observed in T. gondii promoters,17,18,23–26 with the exception of the initiator motif (INR). The INR motif is widely observed in mammalian cells, and YYANWYY is a consensus sequence that overlaps a transcriptional start site (TSS).27 Sequences found in upstream regions of SAG1, NTP1, and NTP3 ORFs correspond to the INR consensus, and it has also been demonstrated experimentally that the sequence is responsible for its transcription by reporter assay.24 However, the popularity of the motif in the T. gondii core promoter remains an open question, and novel motifs in their core promoter have not been discovered yet because a systematic analysis for core promoter structure, which is essential for these attempts, has not been executed. Here, the relative position of core promoter motifs is associated with each TSS in general;22,28,29 therefore, it is essential for systematic analysis to obtain sufficient information about reliable TSSs.
For the last couple of years, a method was established by combining the full-length cDNA method30 and the massively parallel sequencing technology. This makes it possible to collect precise positional information of TSSs together with digital information of the gene-expression levels in a high-throughput manner.31 In this study, we applied this method to elucidate the T. gondii transcriptome focusing on the estimation of transcription regions (TRs), their association for annotated ORFs, and the typical core promoter structure that could be helpful to accelerate an understanding of the nature of the parasites.
2. Materials and methods
2.1. Parasite culture and RNA preparation
The T. gondii RH strain was cultured and maintained in the monolayer of Vero cells. The infected cells were homogenized by passage through a 27-gauge needle, and cell debris was eliminated by a 5-µm pore size filter (Millex-SV). The supernatant was removed by centrifugation at 800 g. Parasites were suspended in Eagle's minimal essential medium, which resulted in 1 × 107 parasites per millilitre. ICR mice (CLEA Japan) were inoculated with 1 ml of the purified parasites per head by intraperitoneal injection. Three days after inoculation, proliferated parasites were harvested and purified by centrifugation and filtration as reported above. A total of 1 × 109 of purified parasites were suspended in 1 ml of PBS, and 20 ml of the TRI reagent (Sigma-Aldrich) was added. Total RNA was purified according to the manufacturer's instructions.
2.2. Oligo-capping and high-throughput sequencing
The detailed protocol for oligo-capping and sequencing has been described elsewhere.31 In brief, 200 µg of purified total RNA was dephosphorylated with Bacterial Alkaline Phosphatase, ligated with oligo-RNA (5′-AAUGAUACGGCGACCACCGAGAUCUACACUCUUUCCCUACACGACGCUCUUCCGAUCUGG-3′) using T4 RNA ligase, and cDNA was then synthesized from a random hexamer primer (5′-CAAGCAGAAGACGGCATACGANNNNNNC-3′) with Super Script II (Invitrogen). The cDNA was amplified by 15 cycles of PCR, and massive parallel sequencing was executed with Illumina GA sequencer (Illumina) to obtain tag with 36-nt length.
2.3. Data processing
The obtained sequences were mapped onto the T. gondii ME49 strain genomic sequencing (version 5.0 in toxoDB; http://toxodb.org/toxo/) with the sequence alignment program Eland after removing two nucleotides of margin nucleotides between the sequencing primer and transcripts. Unmapped or redundantly mapped tags were removed from the data set. Tags with more than two mismatches were also removed.
2.4. Characterization of TSSs
TSSs were clustered for further analysis. As a first step, the number of TSSs in a window of 20-nt size over the genome was counted, and the window was assigned as an active transcription window if it contained statistically significant counts that exceeded background distribution. According to the background distribution, it was assumed that each TSS distributed randomly over the genome, and its probability was defined as the window size per genome size. Because the probability was small enough, the Poisson distribution was applied as a model of background distribution. In practice, 0.66 tags in a 20-nt window were expected by the following calculation (there were 4 027 161 uniquely mapped valid tags). The genome size was offset from 61 769 183 into 60 930 771.6 bp on the basis of the ratio between uniquely mapped counts (4 027 161) and redundantly mapped counts (55 414). Then, (20/60 930 771.6 × 2) × 4 027 161 = 0.66. In this study, it was regarded as significant when there were more than 10 TSSs in each window. The transcription windows were merged when they overlapped, and they were referred to as the TR.
2.5. Comparison among TRs and annotated ORFs
Subsequently, associations between the assigned TRs and ORFs were examined. For that purpose, 7923 annotated ORFs on the chromosome were selected from a database, ToxoDB version 5.0. The relative distance between the peak TSS, a TSS with the most abundant tag in a TR, and the ORFs was evaluated. In this study, a score of 1 was given if the peak TSS was located from 0 to 1000 bp relative to 5′ of the ORF. If the peak TSS was located from 1000 to 5000 bp, the score was linearly decreased from 1 to 0. In addition, if the peak TSS was located inside of the exons and introns, the score was linearly redacted from 1 at 5′ end to 0 at the 3′ end. The maximum score for both the TR to ORFs and each ORF to the TRs was examined and assigned as a pair when the scores exceeded 0.5. A start codon for ENO2 was referred to the previous report describing its promoter structure.26
2.6. Core promoter analysis
To analyse the T. gondii core promoter structure, representative TSSs and their surrounding sequences were examined. TSSs with the most tags in each TR were selected as the representative TSSs when they had more than 10 tags. A test data set named core ± 100 was constructed to consist of –100 to +100 regions from the TSSs. The initial severance was executed by counting the numbers of adenine, thymine, guanine, and cytosine at each position in the core ± 100. For the initiator motif analysis, a sub-data set consisting of seven nucleotides from −2 to +5 in the core ± 100 was constructed, and the number of unique sequences was counted and then sorted according to their occurrence. The 30 most frequently occurring sequences were selected, and the position frequency matrixes (PFMs) for the T. gondii-specific initiator motif (tgINRT and tgINRA) were constructed according to the sequence. To find a specific pattern in the thymidine-rich region from +3 to +14, short sequences consisting of six nucleotides were selected, and their occurrence was examined. Finally, the PFM for the downstream thymidine cluster (DTC) was constructed in the same manner as reported above.
Specific localization of the motifs was evaluated by scanning for each motif in the core ± 100. PFMs for the mammalian TATA box and INR32 were also examined. These PFMs were transformed into a position weight matrix (PWM) according to the function.33 For all probable nucleotide combinations, e.g. 16 384 variations for seven nucleotides of the INRs, their similarity score to each motif was evaluated using the PWMs, and the highest 1% sequences, e.g. 163 unique sequences out of the 16 384, were selected as positive. In addition, the occurrence frequency and location of these positive sequences in the core ± 100 were examined. To investigate the correlations between core elements (tgINRT, tgINRA, and DTC at +3, +9, and +19) and promoter activity, tag counts belonging to each promoter were tallied and the Mann–Whitney test was executed.
The calculations reported above were executed mainly with the programming language Ruby.
3. Results and discussion
3.1. Statistics for obtained tags
A de novo analytical method using a next-generation sequencer, tss-seq, provides a short-tag sequence with one-nucleotide resolution and massively high coverage; therefore, it is possible to collect digital information of the TSS position with transcriptional quantity.31 In this study, we analysed the transcriptome of T. gondii for the first time in the Apicomplexa parasite with this method and obtained 6 801 945 tags consisting of 1 336 060 unique sequences from the T. gondii RH strain tachyzoites propagated in ICR mice (Table 1). To define the position of each tag in a genome, the tags were mapped onto the genome sequence of the T. gondii ME49 strain because the full genome sequence for the T. gondii RH strain is not currently available and that of the T. gondii ME49 is the most authorized sequence among the T. gondii strains. This inevitable discordance among specimens and the reference sequence causes SNPs; therefore, tags with equal to or fewer than two mismatches for the T. gondii ME49 genome were accepted. Then, 4 082 575 tags consisting of 450 852 unique sequences were assigned as valid (Table 1). Of these tags, 4 027 161 tags consisting of 440 690 unique sequences were uniquely mapped on the genome (Table 1). A total of 55 414 tags consisting of 10 162 unique sequences were redundantly mapped, and these tags were excluded as exceptions to reduce the complexity (Table 1). The remaining tags were tallied according to the mapped position, which resulted in the acquisition of 124 217 TSSs (Table 1). These data are available on a Website, Full-Toxoplasma (http://fullmal.hgc.jp/), with graphic interface.
Table 1.
Global statistics
| Number of tags | |
|---|---|
| Total obtained tags | 6 801 945 |
| Unique obtained tags | 1 336 060 |
| Total valid tags with 0–2 mismatches | 4 082 575 |
| Uniquely mapped tags | 4 027 161 |
| Without mismatches | 2 590 719 |
| With 1 mismatch | 972 811 |
| With 2 mismatches | 463 631 |
| Redundantly mapped tags | 55 414 |
| Unique valid tags with 0–2 mismatches | 450 852 |
| Uniquely mapped tags | 440 690 |
| Redundantly mapped tags | 10 162 |
| TSSs | 124 217 |
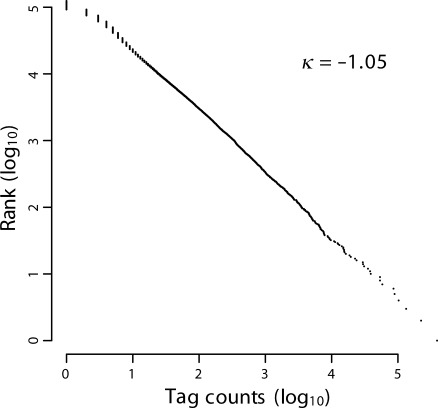
To characterize the distribution of the TSSs, we plotted the logarithmic converted tag counts on a horizontal axis and the logarithmic converted rank of each TSS on a vertical axis (Fig. 1). As a result, we found a linear correlation similar to the Power Law distribution. The same trend was observed in transcriptome analysis with microarrays.34 Therefore, our observation suggested that the power law distribution was involved not only in the gene-expression level but also in the selection of TSSs. The power law distribution was observed in various situations, for example, on an airline route map, metabolic networks, and protein–protein interaction.35–37 However, the question remains open as to whether there is a general theory for determining the distribution and whether there is any biological meaning.
Figure 1.
Statistical profile of TSSs in T. gondii. Counts of each unique tag were converted to a logarithm and plotted on a horizontal axis. The rank of the tag according to its counts was converted to a logarithm and plotted on a vertical axis; 440 609 tags consisting of 124 217 TSSs were examined.
3.2. Characterization of the TR
The advantages of the tss-seq method also play a key role in detailed analysis of TRs, which are difficult to conduct using existing exhaustive methods, such as the microarray. In this study, TRs were assigned by clustering TSSs, and, more specifically, we adopted a null hypothesis in which tags were distributed without control; therefore, an area with accumulated tags could be under the control of transcription machinery if the number of tags in the area were statistically higher than expected. The Poisson distribution expresses the probability of events if these events occurred independently of others with a known average rate. Furthermore, it can be applied to systems with a large number of possible events with low frequency. In particular, the events corresponded to the mapping of the massive number of obtained tags, and its frequency was quite low because a tag position must be selected from a very large genome. Therefore, the situation met the requirement for the Poisson distribution. With this method, it was possible to demonstrate the cut-off value supported by statistics, which then made it possible to compare multiple data sets even if the size of each system was considerably different. It was expected that this advantage would become more critical when TSS databases became expanded.
Here, we assigned 10 508 TRs by clustering 124 217 TSSs according to the method described above (Table 2); however, this number could change depending on the parameters, in particular, the cut-off value for significance under the Poisson distribution. In this study, we applied the cut-off value of 10 in each window, where a probability for the existence of more than 10 tags in the window became 0.24 × 10−8 according to the Poisson distribution if mapping occurred randomly. Furthermore, the number of windows was twice the genome size; therefore, the expected false positive was determined to be under 0.32. This value was quite stringent; however, it could prevent overestimation for associations between TRs and annotated ORFs. As a result, we assigned 10 508 TRs, which was in good agreement with 7923 of the annotated number of T. gondii genes if alternative promoters38 were utilized in T. gondii as well. In addition, the number was higher than the 898 genes detected by SAGE10 in the T. gondii RH strain tachyzoites even under such stringent conditions. Obviously, it would be possible to apply other cut-off values. For example, 0.41 × 10−9, derived from the P-value of 0.05 denominated by 120 M nucleotides according to the Bonferroni method, could be applicable. This was quite safe, but it caused too many false negatives, especially for the purposes of this study. On the other hand, we might have lost the positive TRs under the parameters in this study, in which case a milder cut-off value could be applied to detect subtle TRs; therefore, the cut-off value should be selected depending on the purpose of each study.
Table 2.
Positional comparison between TSS cluster and gene
| Number of items | |
|---|---|
| TSS cluster | 10 508 |
| With assigned ORF | 7447 |
| Without assigned ORF | 3061 |
| ORF | 7923 |
| With assigned TSS cluster | 4363 |
| Without assigned TSS cluster | 3560 |
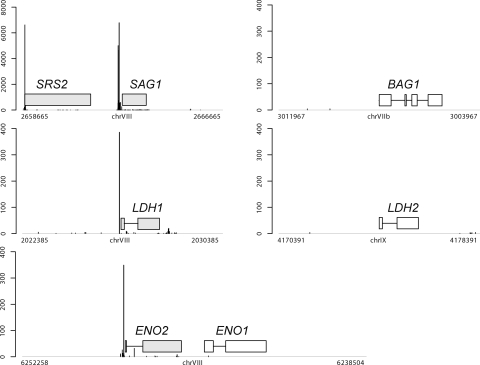
The correspondence between the resulting cluster and known transcription profile might be an important concern. For this reason, we checked the expression profile for representative tachyzoite- and bradyzoite-specific genes and confirmed that our observation was in good agreement with previous findings. For example, the tags for four representative tachyzoite-specific genes (SAG1, SRS2, LDH1, and ENO2) were successfully observed (Fig. 2). In detail, SAG1 had eight TRs with a total of 33 773 tags, SRS2 had four TRs with a total of 1297 tags, LDH1 had two TRs with a total of 439 tags, and ENO2 had three TRs with a total of 589 tags (Supplementary Tables S1 and S2), in agreement with other reports,39–44 even though, for ENO2, there was a difference of ∼30 nucleotides in position between the peak TSSs observed in this study and in a previous report.26 This minor discordance might be caused by differences in the subjected strains and culture conditions in each case. On the other hand, no clusters were assigned around three representative bradyzoite-specific genes, BAG1, LDH2, and ENO1 (Fig. 2, Supplementary Tables S1 and S2).
Figure 2.
Distribution of tag sequences mapped on the genome of T. gondii. The coding regions for representative tachyzoite-specific genes, SAG1, SRS2, LDH1, and ENO2, are shown by grey boxes. Those for bradyzoite-specific genes, BAG1, LDH2, and ENO1, are shown by white boxes. The length of the horizontal lines represents the number of observed tags, and the position represents the TSS.
Association of the assigned 10 508 TRs to the 7923 annotated ORFs was examined. If a TR was located within 3 kb upstream of an ORF or within the anterior half of an ORF, we determined that the TRs were proximal to the ORFs unless they were regarded as orphan TRs; as a result, 7447 TRs had proximal ORFs, but other 3061 TRs were orphan (Table 2 and Supplementary Table S1). In the former case, it was suggested that the TRs were responsible for these ORFs in transcription. In the latter case, it was probable that they were undiscovered genes or non-coding RNA. On the other hand, there were 3560 ORFs out of 7923 that did not have proximal TRs in their upstream region (Table 2 and Supplementary Table S2); then, it was suggested that these ORFs were incorrectly predicted or transcribed in the other stages or strains.
3.3. Cis-elements in core promoter regions
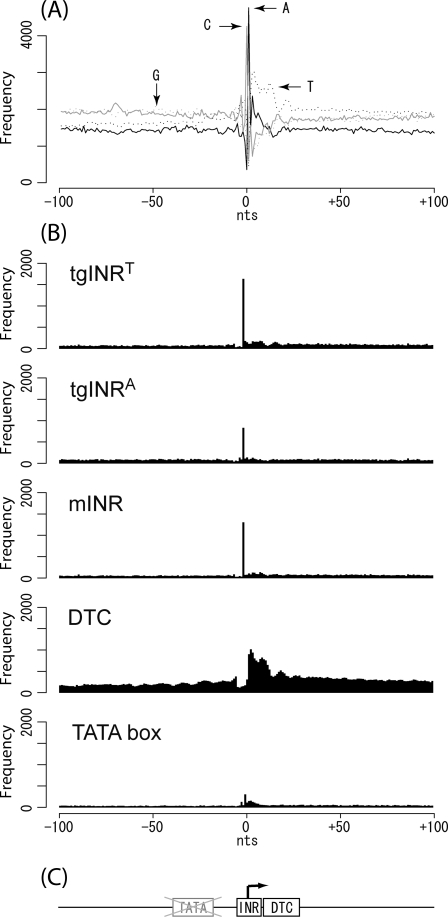
A typical promoter is composed of regulatory domains and a core promoter. Identification of regulatory domains is essential for a better understanding of the transcriptome; however, since comparative transcriptome analysis among various conditions is desirable for a reliable estimation, we focused on another factor, namely, the core promoter in this study. For that purpose, the peak TSSs with more than 10 tags (6898 out of 10 508) were selected, a data set consisting of −100 to +100 nucleotides around the TSSs, a core ± 100, was constructed, and the distribution of each nucleotide was surveyed, then a significant localization of cytosine at −1, adenine at +1 (i.e. TSS), and thymidine from +3 to +14 was observed (Fig. 3A). Because the C + A localization on a TSS corresponded to the underlined nucleotides of the initiator consensus, YYANWYY, a sub-data set consisting of a sequence with seven nucleotides from −2 to +5 from the core ± 100 was constructed and examined to determine whether it was identical to the initiator consensus by sorting the seven nucleotide sequences according to their frequency (Table 3). As a result, TCACTTT, TCATTTT, and CCACTTT were the most popular sequences (88, 63, and 60 times out of 6898, respectively), which was consistent with the initiator consensus. Even though every C + A was conserved at least within the top 30 sequences (Table 3), the following four nucleotides were divided into two patterns. The major pattern (25 out of 30 listed in Table 3) was T-rich, and the minor one (5 out of 30 listed in Table 3) was A-rich. PFMs were constructed from both the T-rich and A-rich sequences (Table 4) and labelled as the tgINRT and tgINRA, respectively. According to the significant thymidine localization, every six nucleotides on from +3 to +14 in the core ± 100 were examined, and poly thymidines were observed in high frequency (Table 5); thus, their PFM was constructed (Table 6) and labelled as the DTC. Their positional specificity and frequency in the core promoters of the tgINRs (tgINRT and tgINRA), mammalian initiator (mINR), and DTC were examined together with the TATA box motif32 by scanning the core ± 100 data set with their PWMs. Then, the obvious localization of the tgINRs on the TSS and following DTC was confirmed (Fig. 3B). The mINR was also observed, but its frequency was less than that of tgINRT. On the other hand, the TATA box was not observed at around −30, its expected location45 even though there was a weak peak around +1 that presumably appeared due to biased nucleotide utilization (Fig. 3A) or a certain similarity among INR consensus and TATA box.
Figure 3.
Core promoter structure in T. gondii. (A) Distribution of the nucleotide in the core ± 100. The core promoter area ±100 nucleotides around the TSS is shown on a horizontal axis, and the observed frequency is plotted on a vertical axis. The black line, dotted black line, grey line, and dotted grey line represent the occurrence frequency for adenine, thymine, cytosine, and guanine, respectively. (B) Distribution of motifs in the core ± 100. Their PWM and threshold applied for evaluation are described in the text. (C) Model of the core promoter structure in T. gondii. The two boxes represent the tgINRs consensus sequence and DTC. An arrowhead represents a TSS. The region where the TATA box should be is shown by a crossed-out grey box.
Table 3.
Frequently observed sequences at TSSs
| Sequence | Freq. | Sequence | Freq. |
|---|---|---|---|
| TCACTTTa | 88 | TCACAAAb | 26 |
| TCATTTTa | 63 | CCAGTTCa | 25 |
| CCACTTTa | 60 | ACACTTTa | 25 |
| GCATTTTa | 40 | ACATTTTa | 25 |
| TCAGTTTa | 39 | CCAGAAAb | 24 |
| TCACTTCa | 34 | CCATTTCa | 23 |
| CCAGTTTa | 33 | TCATTCTa | 22 |
| GCAGAAAb | 32 | CCACTTCa | 22 |
| TCATTTCa | 32 | TCAGTTCa | 21 |
| TCAGAAAb | 31 | ACATTTCa | 20 |
| GCATTTCa | 27 | ACAGAAAb | 20 |
| CCATTTTa | 27 | TCAATTTa | 19 |
| TCACATTa | 26 | GCAGTTTa | 19 |
| GCACTTTa | 26 | CCATTCTa | 19 |
| ACAGTTTa | 26 | GCATTCTa | 18 |
aSequences subjected to the frequency matrix tgINRT consensus.
bSequences subjected to the frequency matrix tgINRA consensus.
Table 4.
PFM for the tgINRT and tgINRA
| First | Second | Third | Fourth | Fifth | Sixth | Seventh | |
|---|---|---|---|---|---|---|---|
| tgINRT | |||||||
| A | 0.12 | 0.00 | 1.00 | 0.02 | 0.03 | 0.00 | 0.00 |
| T | 0.44 | 0.00 | 0.00 | 0.41 | 0.97 | 0.92 | 0.74 |
| G | 0.17 | 0.00 | 0.00 | 0.21 | 0.00 | 0.00 | 0.00 |
| C | 0.27 | 1.00 | 0.00 | 0.36 | 0.00 | 0.08 | 0.26 |
| tgINRA | |||||||
| A | 0.15 | 0.00 | 1.00 | 0.00 | 1.00 | 1.00 | 1.00 |
| T | 0.43 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| G | 0.24 | 0.00 | 0.00 | 0.80 | 0.00 | 0.00 | 0.00 |
| C | 0.18 | 1.00 | 0.00 | 0.20 | 0.00 | 0.00 | 0.00 |
Table 5.
Frequently observed sequences from +3 to +14
| Rank | Sequence | Freq. | Rank | Sequence | Freq. |
|---|---|---|---|---|---|
| 1 | TTTTCT | 510 | 16 | TGTTTT | 220 |
| 2 | TTTTTC | 483 | 17 | TTTCTG | 212 |
| 3 | TTTTTT | 455 | 18 | TTTTCG | 207 |
| 4 | TTTCTT | 425 | 19 | TCTTTC | 204 |
| 5 | TTCTTT | 360 | 20 | TTTTTG | 201 |
| 6 | TCTTTT | 324 | 21 | TTTTGT | 197 |
| 7 | TTTCTC | 320 | 22 | TTGTTT | 183 |
| 8 | TTTTCC | 319 | 23 | TTCCTT | 183 |
| 9 | CTTTTC | 290 | 24 | TTTGTT | 174 |
| 10 | TTCTTC | 282 | 25 | TCTCTT | 169 |
| 11 | TTTCCT | 256 | 26 | CTTTCT | 168 |
| 12 | GTTTTT | 240 | 27 | TTTCGT | 164 |
| 13 | TTCTCT | 236 | 28 | ATTTTT | 162 |
| 14 | CTTTTT | 232 | 29 | GTTTTC | 162 |
| 15 | TCTTCT | 225 | 30 | TTTTCA | 161 |
| 44 | AAAAAA | 123 | 71 | AAAAAG | 98 |
| 61 | AAGAAA | 103 | 79 | AAAGAA | 93 |
| 67 | AAAAGA | 100 | 89 | GAAAAA | 87 |
| 69 | AGAAAA | 99 | 109 | AAAACA | 80 |
A-rich hexamers are selected and shown under the dotted line.
Table 6.
PFM for the DTC
| First | Second | Third | Fourth | Fifth | Sixth | |
|---|---|---|---|---|---|---|
| A | 0.02 | 0.00 | 0.00 | 0.00 | 0.00 | 0.02 |
| T | 0.84 | 0.85 | 0.84 | 0.75 | 0.68 | 0.63 |
| G | 0.05 | 0.03 | 0.02 | 0.02 | 0.05 | 0.08 |
| C | 0.09 | 0.12 | 0.14 | 0.22 | 0.27 | 0.27 |
Subsequently, we examined the involvement of the motifs (tgINRs and DTC) in transcriptional activities. For the tgINRs, 1628 tgINRT-positive, 826 tgINRA-positive, and 4444 INR-negative promoters were subjected for the Mann–Whitney test according to their rank of tag count. As a result, it was implied that there were significant increase in tag count in promoters with both tgINRs rather than without the motifs (Table 7). This observation was consistent with an in vitro experiment with a reporter assay for NTP3.24 Besides it was also implied that the tgINRT positives tended to have more tags than the tgINRA positives (Table 7). For the DTC, they distributed over 20 nucleotides then we selected three representatives (at +3, +9, and +19) according to their peak positions and subjected for the Mann–Whitney test. As a result, it was implied that there were significant increases in tag count for DTC-positive promoters in all three cases rather than without it (Table 7). On the contrary, significant diversity was not observed according to their position among the three (Table 7). In this study, we analysed a massive number of core promoters; therefore, it was possible to detect statistical differences even though the differences were small enough as shown by medians in same cases (Table 7). Therefore, further experimental analysis would be desirable to conclude the functional involvement of the motifs into transcriptional activity.
Table 7.
The Mann–Whitney test for core promoter motifs and transcription activities
| Object 1 |
Object 2 |
P-value | ||
|---|---|---|---|---|
| Motif | Median | Motif | Median | |
| tgINRT | 32 | INR negative | 22 | 0* |
| tgINRA | 27 | INR negative | 22 | 8.02 × 10−8 |
| tgINRT | 32 | tgINRA | 27 | 3.11 × 10−4 |
| DTC at +3 | 29 | DTC negative at +3 | 24 | 2.42 × 10−5 |
| DTC at +9 | 27 | DTC negative at +9 | 24 | 9.78 × 10−4 |
| DTC at +19 | 29 | DTC negative at +19 | 24 | 8.50 × 10−3 |
| DTC at +3 | 29 | DTC at +9 | 27 | 8.22 × 10−1 |
| DTC at +3 | 29 | DTC at +19 | 29 | 9.83 × 10−1 |
| DTC at +9 | 27 | DTC at +19 | 29 | 8.72 × 10−1 |
*z-value was enough large (11.083) to calculate the exact P-value.
According to the definition of Butler and Kadonaga,45 a core promoter is the minimal stretch of contiguous DNA sequences that is sufficient to direct the accurate initiation of transcription by the RNA polymerase II machinery, and, in particular, its physical length is about ±40 bp around the TSS.45 The TATA box, initiator (INR), upstream or downstream TFIIB recognition element (BREu or BREd), DPE, and MTE are commonly found in the core promoters as motifs.28,29,45,46 Additionally, the pyrimidine patch (Y patch) has been reported in the plant core promoter.47,48 In this study, we found two motifs, tgINRs and DTC, in the T. gondii core promoter region (Fig. 3, Tables 4 and 6), and the combination of tgINRs and DTC was suggested as a typical core promoter structure (Fig. 3C). The INR element encompasses the TSS and is identified in a variety of eukaryotes.49–51 The consensus sequence is YYANWYY in mammalian49,52 and TCAKTY in Drosophila,50,53 and the tgINRs were similar to both of them. It is known that TAFII250 and TAFII150 interact with INR.54 Their homologous genes are identified in both Plasmodium falciparum12 and T. gondii (TGME49_076180 and TGME49_014240). These observations suggest that the transcription machinery involving INR was conserved in a wide variety of organisms, including Apicomplexan parasites. On the other hand, DTC was a unique motif localized from approximately +3 to +14. Unlike other core promoter motifs, this element did not show strict positional specificity, and its complementary pattern, the adenine cluster, was also observed in the same region even with much lower frequency (Fig. 3A, Table 5). Furthermore, it is reported that a similar motif, the Y patch, are distributed around the TSSs in plants.47,48 The Y patch consists of T or C, and there is no clear pattern in their order, as is the case of DTC in T. gondii. Therefore, it was implied that DTC and Y patch were involved in transcriptional regulation even though their function at the molecular level remains unknown.
Interestingly, other major core promoter elements reported in mammalians were not observed in the T. gondii core promoter. It has been reported that there is no TATA box in SAG1,23 GRA1, 2, 5, and 6,17 NTP1, 2, and 3,24 MIC4,25 DHFR-TS,18 and ENO1 and 2,26 and the functional TATA box has not yet been described in T. gondii. These observations were determined by our exhaustive analysis (Fig. 3B), demonstrating that there was no significant localization of the TATA box consensus sequence in the proper (around −30 from TSS) position for 6,898 core promoter sequences and also there were not any distinguishable peaks from background when we extended scanned areas by −200 nucleotides from TSSs (data not shown). In yeast, it is reported that TATA boxes are distributed over a wide area (from −120 to −40)55 and it seems to be difficult to find specific peaks from such a wide area with the scanning method. Therefore it was suggested that T. gondii did not utilize functional TATA boxes around −30 positions even though those distributed over a wide area could not be excluded. According to other motifs, BREu, BREd, DPE, and MTE, we could not find a significant localization in T. gondii core promoters (data not shown). Although core promoters were originally thought to be invariant, they have been found to possess considerable structural and functional diversity. Furthermore, it appears that core promoter diversity makes an important contribution to the regulation of gene expression.22 However, our finding implied relative low complicity in the T. gondii core promoter; even though, it was not clear whether this was because of the nature of the parasite or motifs that had been undiscovered. Therefore, subsequent elaborative studies are desirable.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This study was partly supported by ‘Asia-Africa Science and Technology Strategic Cooperation Promotion Program’, Special Coordination Funds for Promotion of Science and Technology, MEXT and KAKENHI (Grant-in-Aid for Scientific Research) on Priority Areas ‘Comparative Genomics’ from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Supplementary Material
Acknowledgements
We are grateful to Ms. Kazumi Abe for expertise of tss-seq. Full-Toxoplasma database has been constructed and maintained by a Grant-in-Aid for Publication of Scientific Research Results from Japan Society for Promotion of Science.
Footnotes
Edited by Masahira Hattori
References
- 1.Dawson D. Foodborne protozoan parasites. Int. J. Food Microbiol. 2005;103:207–27. doi: 10.1016/j.ijfoodmicro.2004.12.032. doi:10.1016/j.ijfoodmicro.2004.12.032. [DOI] [PubMed] [Google Scholar]
- 2.Gajria B., Bahl A., Brestelli J., et al. ToxoDB: an integrated Toxoplasma gondii database resource. Nucleic Acids Res. 2008;36:D553–6. doi: 10.1093/nar/gkm981. doi:10.1093/nar/gkm981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ajioka J.W., Boothroyd J.C., Brunk B.P., et al. Gene discovery by EST sequencing in Toxoplasma gondii reveals sequences restricted to the Apicomplexa. Genome Res. 1998;8:18–28. doi: 10.1101/gr.8.1.18. [DOI] [PubMed] [Google Scholar]
- 4.Manger I.D., Hehl A., Parmley S., et al. Expressed sequence tag analysis of the bradyzoite stage of Toxoplasma gondii: identification of developmentally regulated genes. Infect. Immun. 1998;66:1632–7. doi: 10.1128/iai.66.4.1632-1637.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawase O., Nishikawa Y., Bannai H., et al. Proteomic analysis of calcium-dependent secretion in Toxoplasma gondii. Proteomics. 2007;7:3718–25. doi: 10.1002/pmic.200700362. doi:10.1002/pmic.200700362. [DOI] [PubMed] [Google Scholar]
- 6.Xia D., Sanderson S.J., Jones A.R., et al. The proteome of Toxoplasma gondii: integration with the genome provides novel insights into gene expression and annotation. Genome Biol. 2008;9:R116. doi: 10.1186/gb-2008-9-7-r116. doi:10.1186/gb-2008-9-7-r116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cleary M.D., Singh U., Blader I.J., Brewer J.L., Boothroyd J.C. Toxoplasma gondii asexual development: identification of developmentally regulated genes and distinct patterns of gene expression. Eukaryot. Cell. 2002;1:329–40. doi: 10.1128/EC.1.3.329-340.2002. doi:10.1128/EC.1.3.329-340.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh U., Brewer J.L., Boothroyd J.C. Genetic analysis of tachyzoite to bradyzoite differentiation mutants in Toxoplasma gondii reveals a hierarchy of gene induction. Mol. Microbiol. 2002;44:721–33. doi: 10.1046/j.1365-2958.2002.02903.x. doi:10.1046/j.1365-2958.2002.02903.x. [DOI] [PubMed] [Google Scholar]
- 9.Matrajt M., Donald R.G., Singh U., Roos D.S. Identification and characterization of differentiation mutants in the protozoan parasite Toxoplasma gondii. Mol. Microbiol. 2002;44:735–47. doi: 10.1046/j.1365-2958.2002.02904.x. doi:10.1046/j.1365-2958.2002.02904.x. [DOI] [PubMed] [Google Scholar]
- 10.Radke J.R., Behnke M.S., Mackey A.J., Radke J.B., Roos D.S., White M.W. The transcriptome of Toxoplasma gondii. BMC Biol. 2005;3:26. doi: 10.1186/1741-7007-3-26. doi:10.1186/1741-7007-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seeber F. Consensus sequence of translational initiation sites from Toxoplasma gondii genes. Parasitol. Res. 1997;83:309–11. doi: 10.1007/s004360050254. doi:10.1007/s004360050254. [DOI] [PubMed] [Google Scholar]
- 12.Callebaut I., Prat K., Meurice E., Mornon J.P., Tomavo S. Prediction of the general transcription factors associated with RNA polymerase II in Plasmodium falciparum: conserved features and differences relative to other eukaryotes. BMC Genomics. 2005;6:100. doi: 10.1186/1471-2164-6-100. doi:10.1186/1471-2164-6-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balaji S., Babu M.M., Iyer L.M., Aravind L. Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Res. 2005;33:3994–4006. doi: 10.1093/nar/gki709. doi:10.1093/nar/gki709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Silva E.K., Gehrke A.R., Olszewski K., et al. Specific DNA-binding by apicomplexan AP2 transcription factors. Proc. Natl Acad. Sci. USA. 2008;105:8393–8. doi: 10.1073/pnas.0801993105. doi:10.1073/pnas.0801993105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iyer L.M., Anantharaman V., Wolf M.Y., Aravind L. Comparative genomics of transcription factors and chromatin proteins in parasitic protists and other eukaryotes. Int. J. Parasitol. 2008;38:1–31. doi: 10.1016/j.ijpara.2007.07.018. doi:10.1016/j.ijpara.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 16.Soldati D., Boothroyd J.C. A selector of transcription initiation in the protozoan parasite Toxoplasma gondii. Mol. Cell. Biol. 1995;15:87–93. doi: 10.1128/mcb.15.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mercier C., Lefebvre-Van Hende S., Garber G.E., Lecordier L., Capron A., Cesbron-Delauw M.F. Common cis-acting elements critical for the expression of several genes of Toxoplasma gondii. Mol. Microbiol. 1996;21:421–8. doi: 10.1046/j.1365-2958.1996.6501361.x. doi:10.1046/j.1365-2958.1996.6501361.x. [DOI] [PubMed] [Google Scholar]
- 18.Matrajt M., Platt C.D., Sagar A.D., Lindsay A., Moulton C., Roos D.S. Transcript initiation, polyadenylation, and functional promoter mapping for the dihydrofolate reductase-thymidylate synthase gene of Toxoplasma gondii. Mol. Biochem. Parasitol. 2004;137:229–38. doi: 10.1016/j.molbiopara.2003.12.015. doi:10.1016/j.molbiopara.2003.12.015. [DOI] [PubMed] [Google Scholar]
- 19.Van Poppel N.F., Welagen J., Vermeulen A.N., Schaap D. The complete set of Toxoplasma gondii ribosomal protein genes contains two conserved promoter elements. Parasitology. 2006;133:19–31. doi: 10.1017/S0031182006009954. doi:10.1017/S0031182006009954. [DOI] [PubMed] [Google Scholar]
- 20.Behnke M.S., Radke J.B., Smith A.T., Sullivan W.J., Jr, White M.W. The transcription of bradyzoite genes in Toxoplasma gondii is controlled by autonomous promoter elements. Mol. Microbiol. 2008;68:1502–18. doi: 10.1111/j.1365-2958.2008.06249.x. doi:10.1111/j.1365-2958.2008.06249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mullapudi N., Joseph S.J., Kissinger J.C. Identification and functional characterization of cis-regulatory elements in the apicomplexan parasite Toxoplasma gondii. Genome Biol. 2009;10:R34. doi: 10.1186/gb-2009-10-4-r34. doi:10.1186/gb-2009-10-4-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smale S.T., Kadonaga J.T. The RNA polymerase II core promoter. Annu. Rev. Biochem. 2003;72:449–79. doi: 10.1146/annurev.biochem.72.121801.161520. doi:10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- 23.Burg J.L., Perelman D., Kasper L.H., Ware P.L., Boothroyd J.C. Molecular analysis of the gene encoding the major surface antigen of Toxoplasma gondii. J. Immunol. 1988;141:3584–91. [PubMed] [Google Scholar]
- 24.Nakaar V., Bermudes D., Peck K.R., Joiner K.A. Upstream elements required for expression of nucleoside triphosphate hydrolase genes of Toxoplasma gondii. Mol. Biochem. Parasitol. 1998;92:229–39. doi: 10.1016/s0166-6851(97)00220-x. doi:10.1016/S0166-6851(97)00220-X. [DOI] [PubMed] [Google Scholar]
- 25.Brecht S., Carruthers V.B., Ferguson D.J., et al. The toxoplasma micronemal protein MIC4 is an adhesin composed of six conserved apple domains. J. Biol. Chem. 2001;276:4119–27. doi: 10.1074/jbc.M008294200. doi:10.1074/jbc.M008294200. [DOI] [PubMed] [Google Scholar]
- 26.Kibe M.K., Coppin A., Dendouga N., et al. Transcriptional regulation of two stage-specifically expressed genes in the protozoan parasite Toxoplasma gondii. Nucleic Acids Res. 2005;33:1722–36. doi: 10.1093/nar/gki314. doi:10.1093/nar/gki314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Javahery R., Khachi A., Lo K., Zenzie-Gregory B., Smale S.T. DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol. Cell. Biol. 1994;14:116–27. doi: 10.1128/mcb.14.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kadonaga J.T. The DPE, a core promoter element for transcription by RNA polymerase II. Exp. Mol. Med. 2002;34:259–64. doi: 10.1038/emm.2002.36. [DOI] [PubMed] [Google Scholar]
- 29.Lim C.Y., Santoso B., Boulay T., Dong E., Ohler U., Kadonaga J.T. The MTE, a new core promoter element for transcription by RNA polymerase II. Genes Dev. 2004;18:1606–17. doi: 10.1101/gad.1193404. doi:10.1101/gad.1193404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki Y., Sugano S. Construction of a full-length enriched and a 5′-end enriched cDNA library using the oligo-capping method. Methods Mol. Biol. 2003;221:73–91. doi: 10.1385/1-59259-359-3:73. [DOI] [PubMed] [Google Scholar]
- 31.Tsuchihara K., Suzuki Y., Wakaguri H., et al. Massive transcriptional start site analysis of human genes in hypoxia cells. Nucleic Acids Res. 2009;37:2249–63. doi: 10.1093/nar/gkp066. doi:10.1093/nar/gkp066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin V.X., Singer G.A., Agosto-Perez F.J., Liyanarachchi S., Davuluri R.V. Genome-wide analysis of core promoter elements from conserved human and mouse orthologous pairs. BMC Bioinformatics. 2006;7:114. doi: 10.1186/1471-2105-7-114. doi:10.1186/1471-2105-7-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hertz G.Z., Stormo G.D. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15:563–77. doi: 10.1093/bioinformatics/15.7.563. doi:10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- 34.Ueda H.R., Hayashi S., Matsuyama S., et al. Universality and flexibility in gene expression from bacteria to human. Proc. Natl Acad. Sci. USA. 2004;101:3765–9. doi: 10.1073/pnas.0306244101. doi:10.1073/pnas.0306244101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barabasi A.L., Bonabeau E. Scale-free networks. Sci. Am. 2003;288:60–9. doi: 10.1038/scientificamerican0503-60. doi:10.1038/scientificamerican0503-60. [DOI] [PubMed] [Google Scholar]
- 36.Jeong H., Tombor B., Albert R., Oltvai Z.N., Barabasi A.L. The large-scale organization of metabolic networks. Nature. 2000;407:651–4. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 37.Yook S.H., Oltvai Z.N., Barabasi A.L. Functional and topological characterization of protein interaction networks. Proteomics. 2004;4:928–42. doi: 10.1002/pmic.200300636. doi:10.1002/pmic.200300636. [DOI] [PubMed] [Google Scholar]
- 38.Davuluri R.V., Suzuki Y., Sugano S., Plass C., Huang T.H. The functional consequences of alternative promoter use in mammalian genomes. Trends Genet. 2008;24:167–77. doi: 10.1016/j.tig.2008.01.008. doi:10.1016/j.tig.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 39.Manger I.D., Hehl A.B., Boothroyd J.C. The surface of Toxoplasma tachyzoites is dominated by a family of glycosylphosphatidylinositol-anchored antigens related to SAG1. Infect. Immun. 1998;66:2237–44. doi: 10.1128/iai.66.5.2237-2244.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang S., Parmley S.F. Toxoplasma gondii expresses two distinct lactate dehydrogenase homologous genes during its life cycle in intermediate hosts. Gene. 1997;184:1–12. doi: 10.1016/s0378-1119(96)00566-5. doi:10.1016/S0378-1119(96)00566-5. [DOI] [PubMed] [Google Scholar]
- 41.Dzierszinski F., Mortuaire M., Dendouga N., Popescu O., Tomavo S. Differential expression of two plant-like enolases with distinct enzymatic and antigenic properties during stage conversion of the protozoan parasite Toxoplasma gondii. J. Mol. Biol. 2001;309:1017–27. doi: 10.1006/jmbi.2001.4730. doi:10.1006/jmbi.2001.4730. [DOI] [PubMed] [Google Scholar]
- 42.Bohne W., Gross U., Ferguson D.J., Heesemann J. Cloning and characterization of a bradyzoite-specifically expressed gene (hsp30/bag1) of Toxoplasma gondii, related to genes encoding small heat-shock proteins of plants. Mol. Microbiol. 1995;16:1221–30. doi: 10.1111/j.1365-2958.1995.tb02344.x. doi:10.1111/j.1365-2958.1995.tb02344.x. [DOI] [PubMed] [Google Scholar]
- 43.Parmley S.F., Weiss L.M., Yang S. Cloning of a bradyzoite-specific gene of Toxoplasma gondii encoding a cytoplasmic antigen. Mol. Biochem. Parasitol. 1995;73:253–7. doi: 10.1016/0166-6851(95)00100-f. doi:10.1016/0166-6851(95)00100-F. [DOI] [PubMed] [Google Scholar]
- 44.Dzierszinski F., Popescu O., Toursel C., Slomianny C., Yahiaoui B., Tomavo S. The protozoan parasite Toxoplasma gondii expresses two functional plant-like glycolytic enzymes. Implications for evolutionary origin of apicomplexans. J. Biol. Chem. 1999;274:24888–95. doi: 10.1074/jbc.274.35.24888. doi:10.1074/jbc.274.35.24888. [DOI] [PubMed] [Google Scholar]
- 45.Butler J.E., Kadonaga J.T. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 2002;16:2583–92. doi: 10.1101/gad.1026202. doi:10.1101/gad.1026202. [DOI] [PubMed] [Google Scholar]
- 46.Deng W., Roberts S.G. Core promoter elements recognized by transcription factor IIB. Biochem. Soc. Trans. 2006;34:1051–3. doi: 10.1042/BST0341051. [DOI] [PubMed] [Google Scholar]
- 47.Yamamoto Y.Y., Ichida H., Matsui M., et al. Identification of plant promoter constituents by analysis of local distribution of short sequences. BMC Genomics. 2007;8:67. doi: 10.1186/1471-2164-8-67. doi:10.1186/1471-2164-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamamoto Y.Y., Yoshitsugu T., Sakurai T., Seki M., Shinozaki K., Obokata J. Heterogeneity of Arabidopsis core promoters revealed by high-density TSS analysis. Plant. J. 2009;60:350–62. doi: 10.1111/j.1365-313X.2009.03958.x. doi:10.1111/j.1365-313X.2009.03958.x. [DOI] [PubMed] [Google Scholar]
- 49.Corden J., Wasylyk B., Buchwalder A., Sassone-Corsi P., Kedinger C., Chambon P. Promoter sequences of eukaryotic protein-coding genes. Science. 1980;209:1406–14. doi: 10.1126/science.6251548. doi:10.1126/science.6251548. [DOI] [PubMed] [Google Scholar]
- 50.Hultmark D., Klemenz R., Gehring W.J. Translational and transcriptional control elements in the untranslated leader of the heat-shock gene hsp22. Cell. 1986;44:429–38. doi: 10.1016/0092-8674(86)90464-2. doi:10.1016/0092-8674(86)90464-2. [DOI] [PubMed] [Google Scholar]
- 51.Struhl K. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell. 1987;49:295–7. doi: 10.1016/0092-8674(87)90277-7. doi:10.1016/0092-8674(87)90277-7. [DOI] [PubMed] [Google Scholar]
- 52.Smale S.T., Baltimore D. The ‘initiator’ as a transcription control element. Cell. 1989;57:103–13. doi: 10.1016/0092-8674(89)90176-1. doi:10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 53.Arkhipova I.R. Promoter elements in Drosophila melanogaster revealed by sequence analysis. Genetics. 1995;139:1359–69. doi: 10.1093/genetics/139.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chalkley G.E., Verrijzer C.P. DNA binding site selection by RNA polymerase II TAFs: a TAF(II)250-TAF(II)150 complex recognizes the initiator. EMBO J. 1999;18:4835–45. doi: 10.1093/emboj/18.17.4835. doi:10.1093/emboj/18.17.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuehner J.N., Brow D.A. Quantitative analysis of in vivo initiator selection by yeast RNA polymerase II supports a scanning model. J. Biol. Chem. 2006;281:14119–28. doi: 10.1074/jbc.M601937200. doi:10.1074/jbc.M601937200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.