Abstract
Definitive identification of promoters, their cis-regulatory motifs, and their trans-acting proteins requires experimental analysis. To define the HNRNPK promoter and its cognate DNA–protein interactions, we performed a comprehensive study combining experimental approaches, including luciferase reporter gene assays, chromatin immunoprecipitations (ChIP), electrophoretic mobility shift assays (EMSA), and mass spectrometry (MS). We discovered that out of the four potential HNRNPK promoters tested, the one containing the palindromic motif TCTCGCGAGA exhibited the highest activity in a reporter system assay. Although further EMSA and MS analyses, performed to uncover the identity of the palindrome-binding transcription factor, did identify a complex of DNA-binding proteins, neither method unambiguously identified the pertinent direct trans-acting protein(s). ChIP revealed similar chromatin states at the promoters with the palindromic motif and at housekeeping gene promoters. A ChIP survey showed significantly higher recruitment of PARP1, a protein identified by MS as ubiquitously attached to DNA probes, within heterochromatin sites. Computational analyses indicated that this palindrome displays features that mark nucleosome boundaries, causing the surrounding DNA landscape to be constitutively open. Our strategy of diverse approaches facilitated the direct characterization of various molecular properties of HNRNPK promoter bearing the palindromic motif TCTCGCGAGA, despite the obstacles that accompany in vitro methods.
Keywords: HNRNPK promoter, palindromic motif, EMSA, mass spectrometry, chromatin immunoprecipitation
1. Introduction
HNRNPK is an abundant protein factor found in the nucleus, cytoplasm, mitochondria, and plasma membrane that belongs to the family of heterogeneous nuclear ribonucleoproteins (hnRNPs). These hnRNPs are involved in a variety of biological processes, including telomere biogenesis, cellular signalling, DNA repair, and the regulation of expression on both the transcriptional and translational levels.1 HNRNPK has been found to activate and repress gene expression, and its activity is mainly regulated by covalent modifications.2–4 Through its binding to single-stranded (ss) and double-stranded (ds) DNA, HNRNPK regulates gene expression in both CT element-dependent and -independent fashions.
Several studies have demonstrated the aberrant increase in HNRNPK expression in cancers.5–9 Other studies have shown that HNRNPK is a functional constituent of a cellular structure, the spreading initiation centre,10 and that it is indispensable for cellular migration.11 Overall, HNRNPK has been implicated as a potential key player in carcinogenesis, making it an attractive target for anticancer therapies. Although HNRNPK has been the subject of numerous studies, the mechanisms regulating its expression are still largely unknown.
The palindromic motif TCTCGCGAGA has been found in TATA-less promoters, and is a potential cis-regulatory element in ∼5% of human genes including cell cycle, transcription regulators, chromatin structure modulators, translation initiation factors, and ribosomal protein genes.12 Here, we present experimental and computational evidence that the TCTCGCGAGA motif represents a critical element in the regulation of HNRNPK expression.
2. Materials and methods
2.1. Reporter plasmid construction
Sequences obtained from annotated databases (GenBank and dbEST, available at the National Center for Biotechnology Information; http://www.ncbi.nlm.nih.gov) were analysed by bioinformatic tools, leading to the identification of four potential HNRNPK promoters (Supplementary Table S1). Promoter 1 contains the palindromic motif TCTCGCGAGA.
All analysed HNRNPK promoter sequences were amplified from human genomic DNA using Pfu Turbo DNA polymerase (Agilent Technologies, Inc., Santa Clara, CA, USA) and primers, as listed in Table 1; graphical location of analysed fragments is shown in Fig. 1A. For promoter fragments 1, 2, and 3, nested polymerase chain reactions (PCRs) were used, with pre-amplification of long fragments with the Promoter 1 forward and Promoter 3 reverse primers pair. For Fragment 4, a standard PCR reaction was used. The PCR-amplified DNA fragments were T/A subcloned into the pCR 2.1-TOPO vector using the TOPO TA Cloning kit (Invitrogen, Carlsbad, CA, USA), and the resulting vectors were transformed into chemically competent TOP10 cells. Plasmid DNA was isolated from randomly selected bacterial clones and sequenced using the ABI Prism 377 automated DNA sequencing system (Applied Biosystems, Foster City, CA, USA). Plasmids were digested with KpnI and EcoRV (Promoters: 1, 2, 3) or with SacI and EcoRV (Promoter 4), separated by agarose gel electrophoresis, and DNA fragments isolated from the gel were inserted into the corresponding restriction sites of the pGL4.10[luc2] luciferase reporter vector (Promega Corporation, Madison, WI, USA). Site-directed mutagenesis was performed using the QuikChange Multi Site-Directed Mutagenesis kit (Agilent Technologies, Inc.) in the context of pGL4.10[luc2]-promoter-1, according to the manufacturer's protocol. All final DNA constructs were confirmed by sequencing.
Table 1.
Oligonucleotide sequences used in this study
| Primer pairs used for amplification of promoter fragments | ||
| Promoter | Forward primer 5′–>3′ | Reverse primer 5′–>3′ |
| Promoter 1 | CTCCAACCCCAAAGGACTC* | ATAATGGCGTCTGCAGTGCT |
| Promoter 2 | GTTGGGGCTCCCTGAAAG | CGGTCGGAGCTAGACAGAAA |
| Promoter 3 | CCAGTTTTTCCGCTTGAGG | CCACTGCCCGAGACAAAG* |
| Promoter 4 | TTGGTGGTGTGGGAATTTTT | TGGCATTGTACTGCATTGGT |
| Primer pairs used in ChIP | ||
| Gene name | Forward primer 5′–>3′ | Reverse primer 5′–>3′ |
| Intergenic | GCCACATTCAGTTCTCGCTTT | TTCTCCTTTCCCTGGCAACA |
| RHO_intron2 | TGGGTGGTGTCATCTGGTAA | GGATGGAATGGATCAGATGG |
| RHO_exon5 | GGACATCCACCAAGACCTACTG | ATATGTTCTCCCTTCCCATTCA |
| HBB | TTATGCTGGTCCTGTCCTCC | CTGAAAGAGATGCGGTGG |
| HNRNPK | TCGTTCGCGCACTCACTA | CGCGTGAACTAGGCAGTTGT |
| TUBB6 | AAGGGGCAGCGAAACGGTGG | GGAGCCGGGCGGAAAAGGAC |
| GAPDH | TGAGCAGTCCGGTGTCACTA | AGGACTTTGGGAACGACTGA |
| HNRNPH | ACAGCTGCGCAACCCACCAC | CGCGAGACGGGAAAGTCGGT |
| RPS7 | CCGCCTCTTGCCTTCGGACG | CCGAGCGCCGGCTTAGGAAG |
| HSP70 | GGCGAAACCCCTGGAATATTCCCGA | AGCCTTGGGACAACGGGAG |
| EGR-1 | GGTCCTGCCATATTAGGGCTT | TGGGATCTCTCGCGACTCC |
| Primers used in mutagenesis | ||
| Primer name | Primer sequence | |
| P1M1 | 5′-CAGTTGTTAGATCTCGTGAGAGGTTCGCCCCCTAG-3′ | |
| P1M2 | 5′-CAGTTGTTAGATCTCGTAAGAGGTTCGCCCCCTAG-3′ | |
| P1M3 | 5′-CAGTTGTTAGATCTCGTAGGAGGTTCGCCCCCTAG-3′ | |
| Probes used in EMSA and MS | ||
| P1-30-F | 5′-AGTTGTTCTATCTCGCGAGAGGTTCGCCCC-3′ (+/− biotin) | |
| P1-30-R | 5′-GGGGCGAACCTCTCGCGAGATAGAACAACT-3′ | |
| P1M-30-F | 5′-AGTTGTTCTATCTCGTAGGAGGTTCGCCCC-3′ | |
| P1M-30-R | 5′-GGGGCGAACCTCCTACGAGATAGAACAACT-3′ | |
| P1-16-F | 5′-CTATCTCGCGAGAGGT-3′ (+/− biotin) | |
| P1-16-R | 5′-ACCTCTCGCGAGATAG-3′ | |
| P1M-16-F | 5′-CTATCTCGTAGGAGG-3′ | |
| P1M-16-R | 5′-ACCTCCTACGAGATAG-3′ | |
*Primers used for pre-amplification in nested PCR.
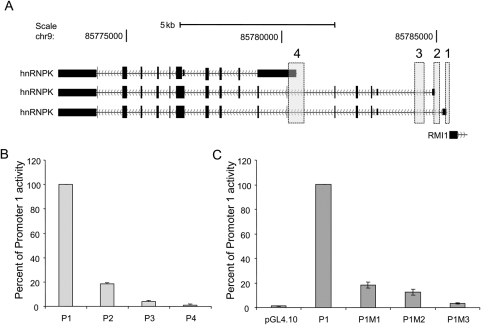
Figure 1.
Graphical localization of HNRNPK promoters and their activity in HeLa cells. (A) Information from the annotated sequence databases GenBank and dbEST was analysed by computational tools, permitting the identification of four potential HNRNPK promoters. Localization of potential promoters sequences (vertical bars) are superimposed on three HNRNPK mRNA transcripts derived from the UCSC browser (hg18 genome assembly) http://genome.ucsc.edu (chr9:85772500-85786000). (B) HeLa cells were transiently transfected with constructs of four human HNRNPK promoters (P1, Promoter 1, containing the TCTCGCGAGA motif; P2, Promoter 2; P3, Promoter 3; P4, Promoter 4) fused to a luciferase reporter gene (pGL4.10) and cotransfected with the phrl-CMV plasmid encoding the Renilla luciferase gene as an internal control. (C) HeLa cells were transfected with mutated forms of the HNRNPK promoter 1 (P1M1, single mutation; P1M2, double mutation; P1M3, triple mutation; pGL4.10, null plasmid). Luciferase activities were measured 24 h after transfection. Mean firefly luciferase activities were normalized to Renilla luciferase activities measured for different promoter reporter plasmids. Data are expressed as a percentage of promoter fragment 1 activity, and represent the mean ± standard deviation of three independent experiments.
2.2. Transient transfection of HeLa cells
HeLa cells were grown at 37°C in 6% CO2 humidified atmosphere in DME medium supplemented with 10% foetal bovine serum (FBS), 2 mM glutamine, 100 units/ml penicillin, and 0.01% streptomycin in plastic cell culture flasks. Cells were routinely subcultured using a trypsin solution. Prior to transfection, cells were plated in ViewPlate-96 White plates (PerkinElmer Wellesley, MA, USA) at 90% confluency and cultured overnight. Each well was transfected for 4 h using Lipofectamine 2000 Reagent (Invitrogen), with 170 ng of the pGL4.10-promoter construct or the empty pGL4.10 vector encoding the firefly luciferase gene, and co-transfected with 30 ng of the phrl-CMV plasmid (Promega) encoding the Renilla luciferase gene. To counterbalance minute differences in construct length and to retain constant particle numbers, the quantity of plasmid DNA was adjusted relative to construct size. The quantity of phrl-CMV plasmid DNA remained unchanged. Lipofectamine–DNA complexes were prepared according to the manufacturer's instructions, except that MEM was used instead of Opti-MEM medium. To maximize transfection efficiency during the 4 h incubation, the serum complement was reduced to 0.5%. Afterward the medium was changed to MEM containing 4% serum and all standard complements. Twenty-four hours following transfection, cells were harvested and assayed for luciferase activity in a Victor 2 luminometer (PerkinElmer) using the Dual-Glow Luciferase Assay System reagents (Promega) in accordance with the manufacturer's instructions. To normalize non-specific variations in transfection efficiency and cell number, all promoter activities were expressed as the ratio of firefly luciferase to Renilla luciferase luminescence in each well. Three independent transfection experiments were conducted with six replicates of each plasmid construct in each experiment. All values are presented as mean ± standard deviation.
2.3. Analyses of protein–DNA interactions
Non-histone nuclear protein extracts (NEs) were isolated as previously reported.13 Protein Desalting Spin Columns (Pierce, Thermo Fisher Scientific, Waltham, MA, USA) were used for buffer exchange of protein samples. Two methods were applied for analysing interactions between the palindromic motif and DNA-binding proteins: electrophoretic mobility shift assay (EMSA) and mass spectrometry (MS). The sequences of oligonucleotides used in the study are shown in Table 1.
2.3.1. Ku80 immunodepletion
NEs were immunodepleted of Ku80 protein using monoclonal anti-Ku80 antibody (Abcam, Cambridge, UK; ab3107) and Immunoprecipitation Kit—Dynabeads® Protein G (Invitrogen) according to the manufacturer's protocol.
2.3.2. Electrophoretic mobility shift assay
EMSA were performed as described previously.12 DNA probes were phosphorylated with [γ-32P]ATP using T4 polynucleotide kinase (Fermentas International Inc., Burlington, Canada) according to the manufacturer's protocol. The reaction was carried out in a total volume of 15 µl (30 min at 25°C) containing 2.5 µg of NE proteins and 1.4 pmol of oligonucleotide in 100 mM NaCl-binding buffer (BB) (100 mM NaCl, 1 mM EDTA, 0.5 mM DTT, 10 mM Tris/HCl, pH 7.5). EMSA was performed using 4 µl of the product on an 8% non-denaturing polyacrylamide gel (37.5:1, Promega) for 1 h at 7.5 V/cm. Autoradiograms were obtained using Imaging screen K and Molecular Imager FX (BioRad, Hercules, CA, USA).
2.3.3. Ms
Fifty micrograms of NE diluted in 50 mM NaCl-BB [50 mM NaCl, 1 mM EDTA, 0.05% NP-40, 5 mM Tris–HCl, pH 7.5, containing protease inhibitors (Roche, Basel, Switzerland)] were incubated at 4°C for 60 min with rotation, in the presence of 200 pM biotinylated wild-type DNA probe with or without a 10-fold excess of competitor (non-biotinylated DNA probe containing the mutated motif). To precipitate the DNA–protein complexes, 1 mg of Dynabeads® M-280 Streptavidin (Invitrogen) was added and the mixture was incubated with rotation at 4°C for 15 min. After magnetic separation, the beads were extensively washed with 150 mM NaCl-BB, and the bound proteins were eluted from the beads with 300 mM NaCl, 500 mM NaHCO3, 1 mM EDTA, 5 mM Tris–HCl, pH 7.5. Eluted proteins were subjected to reduction, alkylation, tryptic digestion, and MS identification.
2.3.4. LC-MS settings
LC-MS analyses were carried out using the nano-Acquity (Waters Corporation, Milford, MA, USA) LC system coupled to an LTQ FTICR (Thermo Fisher Scientific, Waltham, MA, USA) mass spectrometer. Spectrometer parameters were as follows: capillary voltage—2.5 kV, cone—40 V, N2 gas flow—0, and m/z range—300–2000. The spectrometer was calibrated on a weekly basis with Calmix (Thermo Fisher Scientific). Samples were loaded from the autosampler tray (cooled to 10°C) to the precolumn [Symmetry C18, 180 µm × 20 mm, 5 µm (Waters)] using a mobile phase of 100% MilliQ water acidified with 0.1% formic acid. Peptides were transferred to the nano-UPLC column [BEH130 C18, 75 µm × 250 mm, 1.7 µm (Waters)] by a gradient of 5–30% ACN in 0.1% formic acid in 45 min (250 nl/min), then directly eluted to the ion source of the mass spectrometer. Each LC run was preceded by a blank run to ensure lack of carry-over of the material from the previous run. For qualitative analyses (peptide and protein identification), the spectrometer was run in data-dependent MS to MS/MS switch mode, and up to ten MS/MS processes were allowed for each MS scan. Quantitative analyses were carried out in separate profile type survey scan LC-MS runs using the same ACN gradient.
2.3.5. Qualitative analysis (LC-MS/MS)
LTQ-FT MS raw data files were processed to peak lists with the Mascot Distiller software (version 2.2.1, Matrix Science, London, UK). The pre-processed parent and daughter ion lists were used to search the Swissprot (http://www.expasy.org/sprot/) protein database, with taxonomy restriction to human (20 332 sequences, 11 229 110 residues). The Mascot search engine (version 2.2.03, Matrix Science, London, UK) was used to search the database with the following parameters: enzyme specificity—semi-trypsin, permitted number of missed cleavages—1, fixed modification—carbamidomethylation (C), variable modifications—oxidation (M), phospho (ST), phospho (Y), and carbamidomethyl (K), protein mass—unrestricted. The peptide and fragment mass tolerance settings were established separately for individual experiments after measured mass recalibration (Supplementary Method 1).
Statistical assessment of peptide assignments was based on the joined target/decoy database search strategy. This procedure (Supplementary Method 2) provided q-value estimates for each peptide spectrum match (PSM) in the dataset. All PSMs with q-values >0.001 were removed from further analysis. A protein was regarded as confidently identified if at least two peptides of this protein were found. Proteins identified by a subset of peptides from another protein were excluded from analysis. Proteins matching exactly the same set of peptides were joined into a single group.
2.3.6. Quantitative feature extraction
The lists of peptides matching the acceptance criteria from all the LC-MS/MS runs were merged into one common list, which was next overlaid onto 2-D heat maps generated from the LC-MS profile data. This list was used to tag the corresponding peptide-related ion spectra on the basis of mass difference, deviation from the predicted elution time, and the match between the theoretical and observed isotopic envelopes. A more detailed description of the quantitative feature extraction procedure implemented by our in-house software is available.14 The relative abundance of each peptide was determined as the volume of a 2-D fit to the two most prominent peaks of the tagged isotopic envelope.
Multiple-charge states of the same peptide were combined by summing their relative abundances. Missing abundance values (signals below the detection level of the instrument) were replaced by a reference value equal to the smallest abundance observed in the entire LC-MS analysis. Normalization was carried out on the log-transformed peptide abundances by fitting a robust locally weighted regression smoother (LOESS) between the individual samples and a median pseudo-sample.
2.3.7. Statistical analysis of quantitative MS results
Principal components analysis (PCA) was used for graphical summarization and evaluation of the relationships among the studied samples. The statistical significance of protein abundance ratios was assessed by a one-sample t-test of the log-transformed peptide ratios for each protein. The resulting P-values were adjusted for multiple hypothesis testing using the Benjamini–Hochberg procedure that controls the false discovery rate (FDR).15 All statistical analyses were performed using proprietary software implemented in the MATLAB (MathWorks) environment.
2.4. Chromatin immunoprecipitation
Chromatin cross-linking and cell harvesting were performed as previously described.16 Chromatin was sheared for seven rounds of 20 s (1 s pulse, 0.5 s gap) on-off pulses with a 500-Watt Ultrasonic Processor (Cole-Parmer Instrument Co., Vernon Hills, IL, USA) equipped with a microtip and set to 30% of maximum power. Chromatin immunoprecipitation (ChIP) assays were performed using the Matrix-ChIP platform as previously described.17 Briefly, in-house prepared polypropylene 96-well PCR plates (washed once with 200 µl PBS/well) were incubated overnight at room temperature with 0.2 mg of Protein A in 100 µl PBS/well. After washing with 200 µl PBS/well, the wells were blocked with 200 µl blocking buffer for 30 min at room temperature. The wells were cleared, then incubated with 0.5 µg antibody [anti-RNAPII-CTD (Santa Cruz Biotechnology, Santa Cruz, CA, USA), sc-47701; anti-Histone H3, Abcam, ab1791; anti-Histone H3K4me3, Abcam, ab8580; anti-Histone H3K27me3 (Millipore, Billerica, MA, USA), 07-449; anti-Histone H3Ac(Lys9/18), Millipore, 07-593; anti-PARP, Abcam, ab6079] diluted in 100 µl blocking buffer/well for 60 min at room temperature. Chromatin samples (4 µl chromatin/100 µl blocking buffer) were added to the wells (100 µl/well), and the plates were floated in a 4°C ultrasonic water bath for 1 h in order to accelerate protein-antibody binding. Wells were washed three times with 150 µl IP buffer and once with 150 µl TE buffer. Wells were incubated with 100 µl elution buffers in a thermocycler for 15 min at 56°C, followed by 15 min at 95°C. DNA samples were stored at −20°C in the original Matrix-ChIP plates for repeated use.
The PCR mixture contained 2.5 µl 2× SYBR Green PCR master mix (SensiMix, Bioline, London, UK), 2.4 µl DNA template, and 0.1 µl primers (200 nM each) in 5 µl final volume. The reactions were run in 384-Well Optical Reaction Plates (Applied Biosystems). Amplification (two step, 40 cycles), data acquisition, and analysis were performed using the 7900HT Real-Time PCR system (Applied Biosystems). All PCR reactions were performed in triplicate. ChIP data are expressed as percent of input DNA as described previously,17 or as an input ratio of modified histone to total histone H3. The RNAPII/H3Ac ratio was calculated for each site tested by dividing the averaged RNAPII percent of input and the H3Ac/H3 level for all time points. The ChIP data are deposited in Supplementary Table S5. The primer sequences are listed in Table 1.
2.5. Electrophoresis and immunoblotting
Equal amounts of sample proteins (5 µg) were separated by 10% SDS–PAGE, electrotransfered to the Polyvinylidene Difluoride (PVDF) membrane and immunostained by standard methods as previously described.18
2.6. Extraction of an open chromatin ENCODE data for the promoters with the TCTCGCGAGA motif
The DNase-seq tracks at HNRNPK promoter (chr9:85785291-85785515) for HepG2, HeLa, k562, and GM12878 cell lines were obtained from the ENCODE Open Chromatin, Duke DNase-seq Base Overlap Signal datasets19 using the UCSC browser (hg18 genome assembly) http://genome.ucsc.edu. For acquiring an averaged open chromatin profile of 50 promoters bearing the palindromic motif the CisRED (http://www.cisred.org) Human 9 database was queried with TCTCGCGAGA sequence and the resulting coordinates of 50 promoters were used to extract the DNase-seq tracks for HeLa cell line (The ENCODE Open Chromatin, Duke DNase-seq Base Overlap Signal dataset). The open chromatin profiles were superimposed over the TCTCGCGAGA sequence present in each of the 50 promoters and extended over a 150 bp window.
3. Results and discussion
3.1. TCTCGCGAGA is the primary regulatory element of the HNRNPK promoter
Although computational approaches permit identification of gene regulatory and coding sequences, definitive characterization of promoters and their cis-regulatory motifs requires experimental investigation.20 To identify which bioinformatically identified potential HNRNPK promoter(s) preferentially activates transcription, a series of promoter region fragments fused to a reporter gene were generated, and these constructs were transiently transfected into HeLa cells. Reporter plasmids contained four fragments of an HNRNPK promoter region (Fig. 1A). Sequences were identical to the human reference sequence (GenBank, NC_000009.10), with the exception of two single nucleotide polymorphisms: rs7859578 in promoter fragment 1, and rs796 004 in promoter fragment 3. The highest luciferase activity resulted from the reporter with HNRNPK promoter fragment 1. Fragment 2 produced <20% of the luciferase activity obtained with reporter fragment 1, while the luciferase activities generated by constructs containing Fragments 3 and 4 were marginal (Fig. 1B).
Thus, promoter region 1 can be assumed to be the major HNRNPK promoter. It contains the palindromic motif TCTCGCGAGA, which has been recently grouped with the most-conserved motifs in a genome-wide human–mouse assessment of six to eight nucleotide segments.12 Palindromic motifs constitute an important group of regulatory elements. To further study the significance of this particular motif, we designed a series of Promoter 1 reporter constructs with mutated sequences at one, two, and three conserved nucleotide positions. Transcription assays revealed marked luciferase activity decreases in cells transfected with constructs containing the mutant motifs. A single point mutation in the palindromic motif resulted in a >80% loss of promoter activity, whereas triple mutations reduced the activity to near basal levels, demonstrating that this single regulatory site is essential for the majority of HNRNPK promoter activity (Fig. 1C). Similar results were demonstrated by Guo et al.,21 who investigated the role of this motif in FBN1 gene activity; the activity of a reporter gene decreased significantly when two middle nucleotides in the palindrome were replaced with two non-canonical nucleotides.
3.2. In vitro searches for the palindromic motif's interacting protein(s)
Regulatory elements that are concentrated in promoters (cis-elements) are recognized by trans-acting proteins that control transcriptional efficiency. To analyse the interactions between nuclear proteins and the palindromic motif in vitro, we performed EMSA and MS-based analyses.
In order to characterize pertinent binding interactions, we used NEs, ds 16 and 30 bp DNA probes, and ss 16 and 30 oligonucleotide (nt) probes containing either the wild-type palindromic motif or a similar motif mutated at three nucleotide positions (Table 1). As shown in Fig. 2, a protein complex bound to 16 bp (Fig. 2A) and 30 bp (Fig. 2B) ds probes, containing either the wild-type or mutated motif, caused a similar DNA mobility shift in the form of a single band (Fig. 2, left panels). The wild-type ss oligonucleotide probes, on the other hand, formed two main protein–DNA complexes represented by the typically shifted band and a slowly migrating band (Fig. 2, right panels). When the wild-type 16 nt probe was replaced with a mutated oligonucleotide, the binding activity of the protein complex was substantially abolished (Fig. 2C). In contrast, replacing the wild-type 30 nt sequence with a mutated probe did not change the intensity of the upper band, and significantly increased the intensity of the lower band (Fig. 2D).
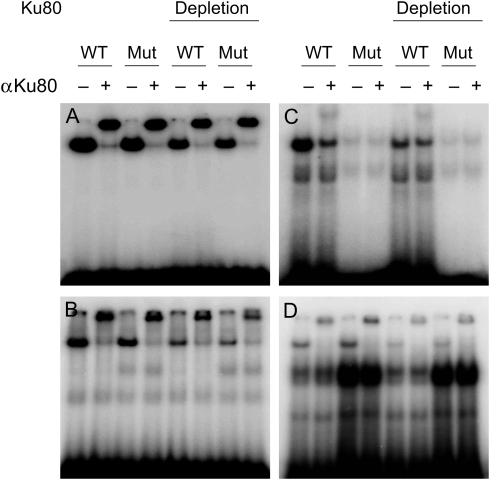
Figure 2.
Electrophoretic mobility shift assay of in vitro binding of TCTCGCGAGA motif probes. Intact or Ku80-depleted NEs, prepared from proliferating HeLa cells, were incubated with a 32P-labelled (A) ds 16 bp probe, (B) ds 30 bp probe, (C) ss 16 nt probe, or (D) ss 30 nt probe and resolved on the gel. WT, wild-type probe, containing proper palindrome motif; MT, mutated probe.
With the exception of the ss 16 nt probes, the other probes predominantly captured proteins in a sequence-non-specific manner; most of the signal originating from the shifted band was supershifted by antibodies directed against ATP-dependent DNA helicase 2 subunit 2 (Ku80), one of the proteins forming DNA double-strand break repair complexes.22 Immunodepletion of Ku80 from the NEs had no effect on the supershift pattern, although the overall binding signal was weaker (Fig. 2). On the other hand, the EMSA with a 16 nt ss probe produced quite a different picture of affinity to nuclear factors when compared with the activity of the other DNA probes. The supershift signal introduced by the anti-Ku80 antibody was much weaker in this assay, and competitor binding was diminished. This result suggests that the palindromic sequence in a single-stranded conformation may represent a functional cis-element. Since a palindromic motif may form a hairpin structure only when single-stranded, it is plausible that this state preferentially favours certain nuclear factors, or facilitates changes in local chromatin structure that drive transcription from promoters bearing that sequence.
To gain a broader view of the protein repertoire that might exist in complexes with the DNA probes, MS-based analyses were applied. We used NEs isolated from resting and proliferating cells in binding reactions with four 5′-biotinylated ss and ds probes, of two lengths, containing the palindromic motif sequence TCTCGCGAGA. Paired binding reactions were performed with and without a 10-fold excess of a competitor with similar characteristics (ss or ds, shorter or longer, non-biotinylated probe with a mutated motif). In addition, binding reactions were repeated using NEs that were depleted by immunoprecipitation with anti-Ku80 antibodies. Altogether, quantitative MS analyses of proteins bound to the DNA probes were carried out on 32 individual samples.
Database searches were performed on 135 592 MS/MS spectra, resulting in the identification of 15 150 PSMs with q-values ≤ 0.001 (corresponding to 2268 unique peptides). In total, 522 proteins were identified. Some proteins were represented by multiple peptides in all analytical MS runs, whereas other proteins appeared in individual runs as singular peptides; 297 proteins were identified by at least two peptides. Two proteins were detected by the MS/MS analysis only in the depleted samples, while 33 proteins were found only in the samples using crude NEs. Two hundred and sixty-two proteins were common to both experimental conditions. For statistical analysis we selected a subset of 132 proteins identified by at least three peptides that were annotated with the GO term nucleus (GO: 0005634), after exclusion of ribosomal proteins and histones (Supplementary Table S2).
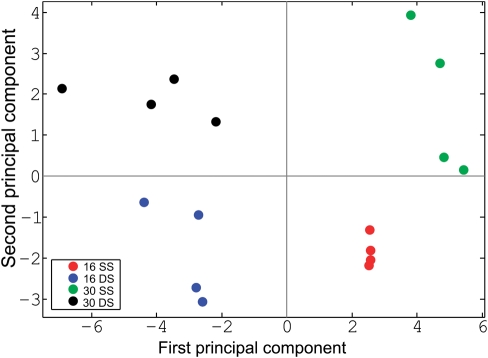
Normalized relative protein abundances were first transformed to corresponding principal-component scores. In the resulting plot (Fig. 3), the first component divided the analysed samples into two distinct groups by discriminating between the binding of ss and ds sequences, although the second component discriminated based on probe length.
Figure 3.
Principal component analysis of the relative abundances of 132 proteins selected for MS-based quantitative analysis. Experiments are colour-coded as follows: red, 16 nt single-stranded (ss) DNA probe, starving cells; blue, 16 bp ds DNA probe; green, 30 nt ss DNA probe; black, 30 bp ds DNA probe.
Proteins may interact with a DNA sequence directly and/or indirectly, in both sequence-specific and -non-specific manners. Assuming that a competitor competes out non-specific DNA–protein interactions, we constructed a protein abundance ratios matrix. The elements of the matrix were equal to the ratios of the protein abundances measured in the paired binding reactions conducted with and without DNA competitor molecules. We assumed that lack of statistically significant changes in the protein abundance, or its enrichment within protein complexes existing in the presence of a competitor, may indicate that proteins are bound to DNA probes in a sequence-specific manner.
As already demonstrated by PCA, the relative abundances of DNA probe-associated proteins, and the protein compositions of DNA–bound complexes, were mostly dependent on probe sequence. Although the relative levels of 75 proteins, extracted from the nuclei of both resting and proliferating cells and associated with ss 16 nt and 30 nt probes, were not changed or were enriched in the binding reactions conducted in the presence of a competitor, the same was true only for 21 proteins associated with ds DNA probes (Supplementary Table S3). Further depletion of NEs by anti-Ku80 antibodies treatment additionally reduced the numbers of proteins which interaction with wild-type motif was unaffected by competitors to 40 and 8, respectively (Supplementary Table S4). Only three proteins (P46 013, Antigen KI-67; Q09666, Neuroblast differentiation-associated protein AHNAK; Q5SSJ5, Heterochromatin protein 1-binding protein 3) were common to all experimental settings.
Therefore, analysis by EMSA and MS of the in vitro formation of DNA–protein complexes revealed compositional complexity of proteins interacting with the DNA probes, but did not select trans-acting protein(s) that unambiguously directly recognize and bind the palindromic motif TCTCGCGAGA.
EMSA is a well-established method of monitoring the ability of a protein to bind to a DNA sequence in vitro. Despite the fact that it has been successfully employed to characterize transcription factors and the transcriptional regulation of various genes,23 it also responds to non-specific protein binding, as in the case of the abundant DNA repair proteins. In particular, a heterodimer of Ku80 and Ku70 binds with high affinity to DNA ends, attracting the other components of the DNA double-strand break repair machinery.22 We have encountered this inherent technical flaw during our EMSA of oligonucleotides containing the palindromic motif. Recently, Hu et al.24 used a protein microarray-based strategy to systematically characterize the repertoire of proteins bound to 752 predicted DNA motifs from previously published studies; the palindrome TCTCGCGAGA was characterized among them. Using their data deposited in the Human Protein-DNA Interactome database,25 we obtained a list of 21 proteins with reported in vitro affinity to this motif: LIG1, MTCP1, HOXB9, TRIP6 PHOX2A, TFAP2E, RBPMS2, NEIL2, MYOD1, ETV7, CREB5, RBM19, TCF7, C1orf25 LIG3, ACTL6B, MAFB, ZNF498, SORCS3, POU6F1, IRF3, and PARP3. We also generated a list of the 10 best matching proteins (RPS4X, PLG, H1FX, HES5, TFEB, USF2, GRHPR, TP73, ZMAT2, and LARP4) when the database was queried with the TCTCGCGAGA motif. Additionally, the hDREF protein, which was shown to regulate the expression of ribosomal proteins genes, is another protein with reported in vitro affinity to this motif.26 Surprisingly, none of these 32 proteins were found within the group of 132 proteins taken through quantitative statistical analysis of our MS data. This discrepancy again underlines the difficulties inherent in in vitro methods, where slightly different conditions (salt, detergent, protein, and DNA probe concentrations) may significantly affect the experimental readout and the ultimate data interpretation.
3.3. Cellular proliferation state increases non-specific DNA-binding
We discovered that the composition of protein complexes interacting with the DNA probes relates to the proliferation status of the cells used for NE preparation. Using a FDR threshold equal to 0.05, our analysis identified 13 proteins that bind to ds 16 and 30 bp DNA probes with significantly higher affinities during proliferation, compared with quiescent cells (Table 2). All these proteins are components of DNA repair process according to the GO annotation analysis (GO: 0006281). The fold change values of their relative abundances were significantly (P < 0.02; Mann–Whitney test) decreased by a competitor and by anti-Ku80 antibody depletion of NEs used in the binding reactions.
Table 2.
Relative protein abundances in proliferating vs. resting cells, bound to ds DNA probes
| 16 bp ds DNA probe |
30 bp ds DNA probe |
|||||||
|---|---|---|---|---|---|---|---|---|
| Anti-Ku80 depletion |
Anti-Ku80 depletion |
|||||||
| Competitor | Competitor | Competitor | Competitor | |||||
| FC | FC | FC | FC | FC | FC | FC | FC | |
| P09874 Poly [ADP-ribose] polymerase 1 | 5.51 | 1.66 | 1.10 | 1.05 | 8.68 | 2.30 | 0.93 | 1.37 |
| P12956 ATP-dependent DNA helicase 2 subunit 1 | 3.66 | 1.35 | 1.23 | 1.65 | 2.60 | 1.67 | 1.05 | ND |
| P13010 ATP-dependent DNA helicase 2 subunit 2 | 3.47 | UP | 1.06 | ND | 6.91 | 0.57 | 1.58 | ND |
| P18887 DNA repair protein XRCC1 | 12.24 | 4.46 | 1.58 | 0.66 | 7.70 | 5.80 | 1.41 | 0.98 |
| P29372 DNA-3-methyladenine glycosylase | 3.26 | 1.05 | 1.57 | 0.96 | 3.19 | ND | 1.24 | ND |
| P35249 Replication factor C subunit 4 | 4.78 | 1.95 | ND | ND | 6.18 | 3.06 | ND | ND |
| P35250 Replication factor C subunit 2 | ND | 1.68 | ND | ND | 11.88 | 1.98 | ND | ND |
| P35251 Replication factor C subunit 1 | 4.22 | 2.58 | ND | ND | 7.37 | 2.88 | ND | ND |
| P49916 DNA ligase 3 | 3.89 | 2.45 | 1.19 | 0.84 | 6.58 | 3.04 | 0.58 | 1.2 |
| P78527 DNA-dependent protein kinase catalytic subunit | 3.31 | 1.21 | 1.39 | 1.24 | 3.66 | 1.00 | 1.93 | 1.00 |
| P78549 Endonuclease III-like protein 1 | 5.01 | ND | ND | ND | 8.35 | ND | ND | ND |
| Q7Z2E3 Aprataxin | 7.48 | 1.81 | ND | ND | 7.64 | 2.02 | ND | ND |
| Q96T60 Bifunctional polynucleotide phosphatase/kinase | 7.85 | 3.74 | 1.35 | 1.06 | 9.89 | 6.65 | 0.83 | ND |
The binding reactions with intact or Ku80-immunodepleted NEs from both proliferating and resting HeLa cells were performed with and without competitor. The eluted proteins were subjected to reduction, alkylation, tryptic digestion, and MS identification, followed by quantitative analysis of MS spectra. FC, fold change in protein abundance; ND, not detected; UP, detected only in proliferating HeLa cells, bold digits refer to statistically significant differences (adjusted P ≤ 0.05).
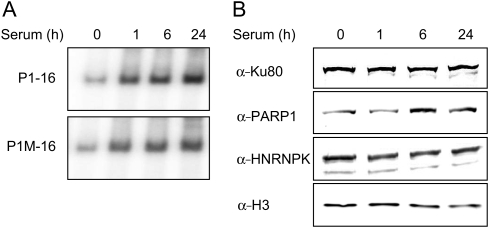
To further characterize the changes in the DNA-binding affinities of non-specific binders in response to mitogenic signalling, quiescent cells were treated with 15% FBS for 0, 1, 6, and 24 h, after which NEs were prepared and used in EMSA. Again, the amount of the shifted ds probes increased with extension of time of serum induction of cells used for NE preparation. Results of a representative experiment with ds 16 bp DNA probes are presented in Fig. 4A. These results indicate that either expression of the non-specifically interacting proteins in proliferating cells is increased, or that there is an increased interaction with non-specific DNA-binding proteins, or both. To differentiate between these possibilities, we analysed equal amounts of proteins from NEs by western blotting, using anti-Ku80, anti-PARP1, and anti-hnRNP K antibodies. This analysis revealed equal amounts of Ku80 and hnRNP K in extracts from both resting and proliferating cells and moderately increased signal of PARP1 after 6 h of serum stimulation (Fig. 4B).
Figure 4.
Cellular proliferation state increases the affinity of non-specific DNA binders. (A) Electrophoretic mobility shift assay of in vitro binding of 16 bp ds DNA (P1-16), and a corresponding mutated probe (P1M-16), to HeLa NEs after 0, 1, 6, and 24 h of serum stimulation. (B) NEs (5 µg) as in (A) were resolved by SDS–PAGE and electrotransferred to a PVDF membrane. Blotted proteins were immunostained with antibodies against Ku80, PARP1, hnRNP K, and Histone H3.
3.4. Histone modification at promoters with the palindromic motif TCTCGCGAGA
Chromatin is composed of nucleosomes, in which ∼147 bp of DNA are wrapped 1.7 times around a core of two copies each of histone proteins H2A, H2B, H3, and H4. In vivo positioning and covalent modifications of nucleosomes play an important role in transcriptional regulation. Post-translational covalent modifications of histone proteins include methylation, acetylation, phosphorylation, and ubiquitination. Such processes recruit and bind chromatin remodelling complexes which alter the configuration of the chromatin, and in turn, control gene transcription.27 There is a high degree of complexity in the number of enzymes, histone modifications, and variant histones involved in chromatin regulation.28 The spatial and temporal intricacies of the interactions of these factors with the genome suggest that chromatin regulation is at least as important to the transcriptional state of a gene as the DNA cis-elements and trans-factors present in its promoter. Thus, any complete study of the factors regulating gene expression must include the chromatin state at that gene's locus.
The promoters of expressed genes are characterized by decreased nucleosome occupancy (particularly a nucleosome-free region around the transcription start site), acetylation of H3 and H4 histones (H3Ac and H4Ac), trimethylation of histone H3 lysine 4 (H3K4me3), and the presence of histone destabilizing chromatin remodelling complexes.29 The overall effect of these modifications is a general decreased presence and stability of nucleosomes, and as a result, increased accessibility to transcription factors and the RNA polymerase II (RNAPII) complex. On the other hand, silenced genes are characterized by a different set of histone modifications and, in general, by denser chromatin architecture. Histone H3 lysine 9 and lysine 27 trimethylation levels (H3K9me3 and H3K27me3) are highest at silent genes.29
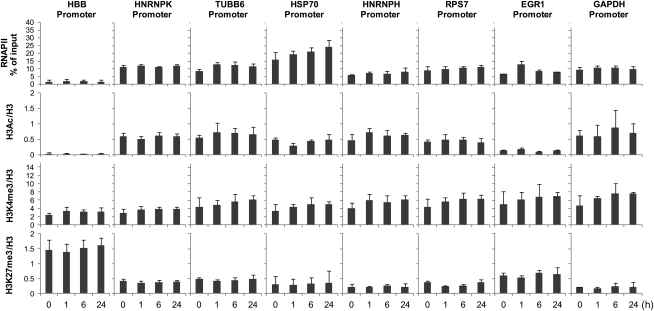
To characterize the HNRNPK promoter in greater detail, we compared it to other promoters bearing the palindromic motif TCTCGCGAGA (HNRNPH—heterogeneous ribonucleoprotein H and RPS7—ribosomal protein S7), well-established constitutively active housekeeping genes (HGs) (TUBB6—Tubulin beta-6, GAPDH—glyceraldehyde-3-phosphate dehydrogenase), an immediate early gene (EGR1—early growth response 1), a heat shock-induced gene (HSP70—heat shock 70 kDa protein), and to a silent gene (HBB—beta globin). We measured histone modification and RNAPII complex levels during serum stimulation in a ChIP assay (Fig. 5).
Figure 5.
Promoter levels of RNA polymerase II and histone modifications. HeLa resting (0 h) and serum-treated cells (1, 6, and 24 h) were used in ChIP assays with antibodies against RNAPII, H3, H3Ac(K9/K18), H3K4me3, and H3K27me3. Isolated DNA was used in real-time PCR with primers to the promoters of HBB, HNRNPK, TUBB6, GAPDH, HNRNPH, RPS7, HSP70, and EGR1. Data are expressed as percent of input chromatin for RNAPII, or as ratios of modified histone marks to total H3 ChIP signals. Data represent the mean ± sd of four independent experiments.
The RNAPII complex levels measured on HNRNPK, HNRNPH, and RPS7 were similar to those at the HGs (∼10% of input), and remained relatively unchanged during serum stimulation. As expected, RNAPII levels at the HBB promoter were smallest, and levels at the EGR1 promoter after 1 h of serum treatment were 2-fold higher than quiescent cells, followed by a return to basal levels at 6 and 24 h. We also observed progressive accumulation of RNAPII at the HSP70 promoter during the serum time course, from 15.8% of input up to 24.1% in quiescent cells and after 24 h in serum, respectively (fold change = 1.52; Fig. 5, Supplementary Table S5).
We subsequently analysed levels of acetylation of histone H3 lysine 9/18 (H3K9/18Ac), trimethylation of H3 lysine 4 (H3K4me3), and trimethylation of H3 lysine 27 (H3K27me3) at the various promoters. Acetylation of lysines eliminates their positive charge which, when it occurs on certain histone tails, has been shown to have a negative effect on the higher order structure of chromatin, essentially making it more open. H3K9/18Ac is associated with actively transcribed genes.27 As expected, H3Ac levels were mirrored by the presence of RNAPII complexes, and remained similar during the serum stimulation on both palindromic and HG promoters. On the other hand, H3Ac levels measured at the EGR1 and HSP70 promoters were relatively smaller when compared with RNAPII amounts. The RNAPII/H3Ac ratios for EGR1 and HSP70 were 67:1 and 47:1, respectively, whereas the same measurement for the HBB promoter was 69:1. On the other hand, the ratios for both palindromic and HG promoters ranged from 11:1 for HNRNPH to 23:1 for RPS7 (Fig. 5, Supplementary Table S5). H3K4me3 is associated with the 5′ ends of actively transcribed genes; H3K4me3 has been suggested to mark an open chromatin state by recruiting ATP-dependent chromatin remodelling complexes.27 H3K27me3, however, marks silenced genes,29 and appears to be dominant over other activating marks; genes that are marked with both K3K27me3 and H3K4me3 are typically silent. As expected, H3K4me3 was detected at the promoters of all analysed genes, including HBB. Also as expected, the H3K27me3 levels for all promoters were lower in comparison to levels at the transcriptionally silent HBB. In sum, these results indicate that RNAPII levels and histone modification levels at the promoters containing the palindromic motif TCTCGCGAGA are similar to those of constitutively active HG promoters.
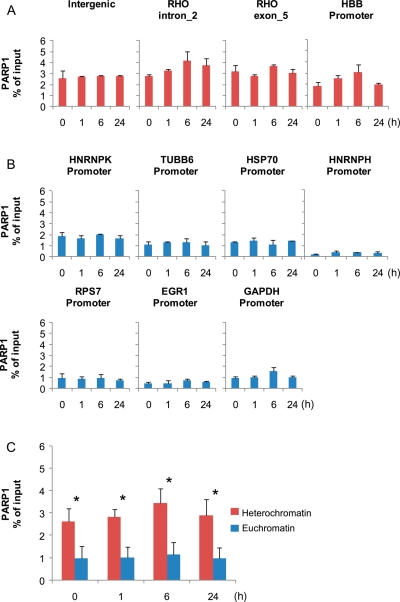
We also wished to determine whether any of the proteins belonging to the DNA double-strand break repair complexes, as determined by EMSA and MS (Table 2), were present at these promoters. Using ChIP, we measured the abundances of bound Ku80 and PARP1 at these promoters. PARP1 is responsible for the synthesis and attachment of polymers of ADP-ribose from nicotinamide adenine dinucleotide (NAD+) to the glutamic or aspartic acid residues of target proteins.30 PARP1 binds to the HSP70 promoter in vivo,31 and therefore we used this gene as a ChIP positive control for PARP1 binding. Although Ku80 did not bind any of the studied promoters (data not shown), unexpectedly, our ChIP experiment demonstrated PARP1 binding not only to the HSP70 promoter, but to all promoters tested, with the highest level of bound PARP1 protein at the HBB promoter (Fig. 6A and B). Additionally, we tested an intergenic region and two sites within the rhodopsin (RHO) gene, where we found similar levels of PARP1 as measured at the HBB promoter (Fig. 6B), along with the expected depletion of RNAPII abundance (data not shown). Overall, the ChIP assays revealed ubiquitous binding of PARP1 to various promoters, with significantly higher abundance at heterochromatin sites (Fig. 6C).
Figure 6.
Distribution of PARP1 at heterochromatic and euchromatic sites. HeLa quiescent (0 h) and serum-treated cells (1, 6, and 24 h) were used in ChIP assays with antibodies against PARP1. Isolated DNA was used in real-time PCR with primers to the intergenic site, two sites within the RHO gene, and the promoter of HBB (A), as well as the promoters of HNRNPK, TUBB6, GAPDH, HNRNPH, RPS7, HSP70, and EGR1 (B). Data are expressed as percent of input chromatin and represent the mean ± sd of four independent experiments. (C) The comparison of PARP1 abundance at heterochromatic and euchromatic sites. The data represent an averaged % of input for all heterochromatin and euchromatin genes tested separately for each time point. The differences between heterochromatin and euchromatin in PARP1 occupancy were statistically significant (*P < 0.005; unpaired t-test) for all time points tested.
A large body of evidence has accumulated which unambiguously supports the involvement of double-strand break repair complex proteins in transcriptional regulation.32–34 Ku80, Ku70, DNA-PK (DNA-dependent protein kinase), and PARP1 were among the most abundant proteins bound to DNA probes, as quantified by our MS assay (Table 2).
PARP1, a ubiquitous and abundant nuclear protein, functions as part of the DNA repair pathway, but also orchestrates transcription through interactions with transcription factors, nucleosomes, histone-modifying proteins, and specific binding to promoters, enhancers, and insulators.30 PARP1 was found to display an affinity not only to ds DNA ends, but also to DNA secondary structures like hairpins.35,36 In particular, the PARP1 promoter contains several imperfect repeats that are capable of forming a hairpin structure recognized by PARP1 itself, resulting in self-mediated transcriptional inhibition.36 Since the TCTCGCGAGA motif present in the HNRNPK promoter may form a hairpin structure in vivo, we speculate that such a spatial scaffold may attract PARP1. This possibility could explain the loss of signal in EMSA employing the mutated 16 nt ss probe, as the correct hairpin structure would be recognized by PARP1 both in vivo and in vitro. PARP1 was described as a structural component of heterochromatin that modulates transcription by self-modification in an NAD-dependent manner.37 Furthermore, it was shown in vitro that PARP1 is able to promote the compaction of nucleosomes into higher order structures when NAD+ is not present; on the contrary, saturating amounts of NAD+ caused decondensation.38 Overall, PARP1 acts as either an activator or a repressor of specific genes, depending on the physiological conditions,30 and we cannot rule out the possibility of its direct involvement in the regulation of promoters with the TCTCGCGAGA motif.
3.5. Chromatin accessibility at promoters with the palindromic motif TCTCGCGAGA
During gene activation, nucleosomes are displaced and/or repositioned in a process involving histone modification, ATP-dependent nucleosome remodelling complexes, histone chaperones, and the shuffling of histone variants.28 Thus, characterizing global nucleosome positions and their chemical and compositional modifications is key to unravelling the mechanism of transcriptional regulation.
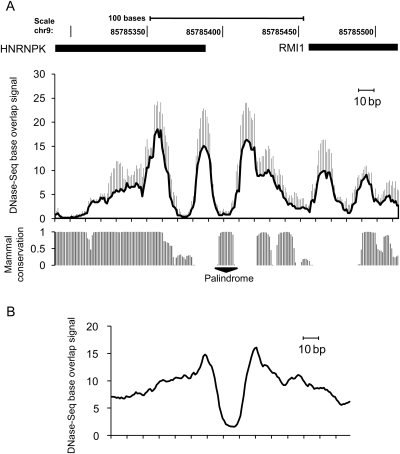
The advent of massively parallel DNA sequencing technology has allowed the localization of every nucleosome across the genomes with a high degree of accuracy.39 We made use of publicly available databases to gain additional insight into chromatin structures surrounding promoters bearing the TCTCGCGAGA palindromic motif. We explored datasets provided by the ENCODE consortium within the Open Chromatin project, which, among other datasets, offers DNase I hypersensitive site (HS) tracks for multiple cell lines.19 DNase I HS sites contain a mixture of cis-regulatory elements, and it has been shown that there is a striking co-occurrence of HS with regions of well-positioned nucleosomes.40
First, we examined the DNase I HS profile within the HNRNPK promoter (Fig. 7A), averaging the HS profile for four cell lines. This analysis revealed decreased values in the HS profile occurring at the palindromic motif, as well as an oscillating pattern of hypersensitive DNA that may represent a combination of trans-elements and positioned nucleosome occupancies within this region. Next, we extracted a HeLa dataset for 50 genes containing the palindromic motif TCTCGCGAGA, centred the dataset on that sequence, and superimposed all HS profiles averaged over a 150 bp window (Fig. 7B). This analysis revealed that TCTCGCGAGA may define the boundaries of open chromatin, as it is flanked by two distinct HS peaks corresponding to positioned nucleosomes. On the basis of this observation, we believe that the palindrome itself may demarcate nucleosomal positions within promoters bearing it, exhibiting features that prevent nucleosomal occupancy of this motif.
Figure 7.
An open chromatin profile at the promoters with the TCTCGCGAGA motif. (A) Averaged profile of DNase-seq tracks surrounding the HNRNPK promoter (chr9:85785291-85785515) acquired for HepG2, HeLa, k562, and GM12878 cell lines, sd as whiskers. Data were obtained from the ENCODE Open Chromatin, Duke DNase-seq Base Overlap Signal datasets. The mammal conservation data were taken from Placental Mammal Basewise Conservation track deposited in the UCSC browser (hg18 genome assembly) http://genome.ucsc.edu. (B) Averaged HeLa cell profile of DNase-seq tracks surrounding 50 promoters bearing the TCTCGCGAGA motif. The CisRED (http://www.cisred.org) Human 9 database was queried with TCTCGCGAGA, and the resulting coordinates of 50 promoters were used to extract the DNase-seq tracks from the ENCODE Open Chromatin, Duke DNase-seq Base Overlap Signal dataset. The open chromatin profiles were superimposed over the TCTCGCGAGA sequence present in each of the 50 promoters and extended over a 150 bp window. The plot represents an averaged value of DNase-seq signal for 50 promoters.
The control of transcription is one of the most important and highly regulated steps in eukaryotic gene expression, and thus is instrumental in orchestrating the metabolic, growth, replicative, and differentiation states of cells. It is not surprising then, that transcription is carried out by a complex sequence of events, any combination of which may be regulated. Studies that investigate gene regulation are greatly facilitated by methods that characterize the interactions between proteins and DNA. Several methods have been developed to identify the direct binding of proteins to DNA sequences. In the classical DNA footprinting protocol, sequences that bind to a protein of interest are determined by identifying regions of DNA that are protected from digestion by exonuclease III or DNase I. The protected sequence corresponds to the ‘footprint’ the protein leaves on the DNA during digestion.41 Another classical method, EMSA, involves the retardation of a radiolabelled DNA probe by the binding of a protein of interest (or by a protein within a complex), indicating a molecular interaction.23 All of these methods involve the in vitro interaction between a recombinant or NE-extracted protein with a naked segment of DNA, eliminating the chromatin context and the possible modification state(s) of the protein of interest. Similarly, assays utilizing reporter genes fail to take into account the local chromatin context, whereas potential cis-elements (promoters and enhancers) are tested using transient transfections, in which the test plasmid remains unintegrated in the nucleus.20 Despite improvements in both computational prediction of functional DNA sites and experimental verification techniques, current methods allow only snapshots of the transcription process, and as such favour indirect conclusions.
Recent advancements in high-throughput microarray (ChIP-chip) and sequencing methods (ChIP-Seq) have increased our understanding of chromatin composition, revealing transcription factor binding sites (TFBS),42,43 chromatin accessibility,44 histone modifications,45 and nucleosome positions39 on a genome-wide scale. These varied experimental datasets augment computational TFBS prediction methods, and aid in the description of transcriptional regulation on the scales of single genes to the entire genome.
3.6. Conclusions
In the present study, we have characterized the molecular features of the palindromic motif TCTCGCGAGA, found not only in the HNRNPK promoter but also, as described in our previous work,12 a potential cis-regulatory element of ∼5% of human genes. We used a reporter system assay to show that out of four identified HNRNPK promoters, the one containing the palindromic motif exhibits the highest expression of a reporter gene. Although EMSA and MS analyses, performed to discover the palindrome-binding transcription factor, did identify a complex of DNA-binding proteins, neither method unambiguously identified the relevant direct trans-acting protein(s). Moreover, for a collection of proteins functionally implicated in DNA repair processes we observed that changes in their affinity to ds DNA probes are related to the proliferation status of the cells.
To analyse the chromatin configuration at the HNRNPK promoter, we performed comparative ChIP experiments of RNAPII occupancy and histone modification density among the promoters of housekeeping, inducible, and silent genes. Our analysis revealed that promoters with the TCTCGCGAGA motif display properties of the promoters of HGs (GAPDH and TUBB6), containing similar RNAPII levels and relatively more H3Ac modifications than inducible and silent genes.
The analyses of open chromatin data provided by the ENCODE project revealed that the TCTCGCGAGA motif has features that promote constitutive openness of the chromatin and favor DNA accessibility. This function could be due to the binding of an unknown transcription factor, a hairpin spatial structure preventing nucleosome occupancy, or both. In sum, experimental observations using in vitro methods and analyses of open chromatin data support the function of this sequence in local nucleosomal organization, resulting in the regulation of transcription of the HNRNPK gene. Our findings suggest that the TCTCGCGAGA motif may play a critical role in regulating and establishing nucleosome patterns at the promoters containing it. Further advancements in analytical methods should elucidate the comprehensive picture of the function of this motif in gene expression regulation.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by grants from the Polish Ministry of Science and Higher Education: N401193 32/4033 and R13010 03 (MS software development).
Supplementary Material
Acknowledgements
We thank Lucjan Wyrwicz for computational identification of potential HNRNPK promoters.
Footnotes
Edited by Minoru Ko
References
- 1.Bomsztyk K., Denisenko O., Ostrowski J. hnRNP K: one protein multiple processes. Bioessays. 2004;26:629–38. doi: 10.1002/bies.20048. doi:10.1002/bies.20048. [DOI] [PubMed] [Google Scholar]
- 2.Habelhah H., Shah K., Huang L., et al. ERK phosphorylation drives cytoplasmic accumulation of hnRNP-K and inhibition of mRNA translation. Nat. Cell biol. 2001;3:325–30. doi: 10.1038/35060131. doi:10.1038/35060131. [DOI] [PubMed] [Google Scholar]
- 3.Michelotti E.F., Michelotti G.A., Aronsohn A.I., Levens D. Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol. Cell. Biol. 1996;16:2350–60. doi: 10.1128/mcb.16.5.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moumen A., Masterson P., O'Connor M.J., Jackson S.P. hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell. 2005;123:1065–78. doi: 10.1016/j.cell.2005.09.032. doi:10.1016/j.cell.2005.09.032. [DOI] [PubMed] [Google Scholar]
- 5.Carpenter B., McKay M., Dundas S.R., Lawrie L.C., Telfer C., Murray G.I. Heterogeneous nuclear ribonucleoprotein K is over expressed, aberrantly localised and is associated with poor prognosis in colorectal cancer. Br. J. Cancer. 2006;95:921–7. doi: 10.1038/sj.bjc.6603349. doi:10.1038/sj.bjc.6603349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L.C., Hsueh C., Tsang N.M., et al. Heterogeneous ribonucleoprotein k and thymidine phosphorylase are independent prognostic and therapeutic markers for nasopharyngeal carcinoma. Clin. Cancer Res. 2008;14:3807–13. doi: 10.1158/1078-0432.CCR-08-0155. doi:10.1158/1078-0432.CCR-08-0155. [DOI] [PubMed] [Google Scholar]
- 7.Hatakeyama H., Kondo T., Fujii K., et al. Protein clusters associated with carcinogenesis, histological differentiation and nodal metastasis in esophageal cancer. Proteomics. 2006;6:6300–16. doi: 10.1002/pmic.200600488. doi:10.1002/pmic.200600488. [DOI] [PubMed] [Google Scholar]
- 8.Pino I., Pio R., Toledo G., et al. Altered patterns of expression of members of the heterogeneous nuclear ribonucleoprotein (hnRNP) family in lung cancer. Lung Cancer. 2003;41:131–43. doi: 10.1016/s0169-5002(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 9.Roychoudhury P., Chaudhuri K. Evidence for heterogeneous nuclear ribonucleoprotein K overexpression in oral squamous cell carcinoma. Br. J. Cancer. 2007;97:574–5. doi: 10.1038/sj.bjc.6603911. author reply 576. doi:10.1038/sj.bjc.6603911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Hoog C.L., Foster L.J., Mann M. RNA and RNA binding proteins participate in early stages of cell spreading through spreading initiation centers. Cell. 2004;117:649–62. doi: 10.1016/s0092-8674(04)00456-8. doi:10.1016/S0092-8674(04)00456-8. [DOI] [PubMed] [Google Scholar]
- 11.Inoue A., Sawata S.Y., Taira K., Wadhwa R. Loss-of-function screening by randomized intracellular antibodies: identification of hnRNP-K as a potential target for metastasis. Proc. Natl. Acad. Sci. USA. 2007;104:8983–8. doi: 10.1073/pnas.0607595104. doi:10.1073/pnas.0607595104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wyrwicz L.S., Gaj P., Hoffmann M., Rychlewski L., Ostrowski J. A common cis-element in promoters of protein synthesis and cell cycle genes. Acta Biochim. Pol. 2007;54:89–98. [PubMed] [Google Scholar]
- 13.Ostrowski J., Sims J.E., Sibley C.H., et al. A serine/threonine kinase activity is closely associated with a 65-kDa phosphoprotein specifically recognized by the kappa B enhancer element. J. Biol. Chem. 1991;266:12722–33. [PubMed] [Google Scholar]
- 14.Bakun M., Karczmarski J., Poznanski J., et al. An integrated LC-ESI-MS platform for quantitation of serum peptide ladders. Application for colon carcinoma study. Proteomics Clin. Appl. 2009;3:932–46. doi: 10.1002/prca.200800111. doi:10.1002/prca.200800111. [DOI] [PubMed] [Google Scholar]
- 15.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. Ser. B. 1995;50:289–300. [Google Scholar]
- 16.Mikula M., Hanusek K., Paziewska A., et al. Halogenated imidazole derivatives block RNA polymerase II elongation along mitogen inducible genes. BMC Mol. Biol. 2010;11:4. doi: 10.1186/1471-2199-11-4. doi:10.1186/1471-2199-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flanagin S., Nelson J.D., Castner D.G., Denisenko O., Bomsztyk K. Microplate-based chromatin immunoprecipitation method, Matrix ChIP: a platform to study signaling of complex genomic events. Nucleic Acids Res. 2008;36:e17. doi: 10.1093/nar/gkn001. doi:10.1093/nar/gkn001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ostrowski J., Woszczynski M., Kowalczyk P., Trzeciak L., Hennig E., Bomsztyk K. Treatment of mice with EGF and orthovanadate activates cytoplasmic and nuclear MAPK, p70S6k, and p90rsk in the liver. J. Hepatol. 2000;32:965–74. doi: 10.1016/s0168-8278(00)80101-4. doi:10.1016/S0168-8278(00)80101-4. [DOI] [PubMed] [Google Scholar]
- 19.Rosenbloom K.R., Dreszer T.R., Pheasant M., et al. ENCODE whole-genome data in the UCSC Genome Browser. Nucleic Acids Res. 2010;38:D620–D625. doi: 10.1093/nar/gkp961. doi:10.1093/nar/gkp961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elnitski L., Jin V.X., Farnham P.J., Jones S.J. Locating mammalian transcription factor binding sites: a survey of computational and experimental techniques. Genome Res. 2006;16:1455–64. doi: 10.1101/gr.4140006. doi:10.1101/gr.4140006. [DOI] [PubMed] [Google Scholar]
- 21.Guo G., Bauer S., Hecht J., Schulz M.H., Busche A., Robinson P.N. A short ultraconserved sequence drives transcription from an alternate FBN1 promoter. Int. J. Biochem. Cell Biol. 2008;40:638–50. doi: 10.1016/j.biocel.2007.09.004. doi:10.1016/j.biocel.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 22.Kanaar R., Hoeijmakers J.H., van Gent D.C. Molecular mechanisms of DNA double strand break repair. Trends Cell Biol. 1998;8:483–9. doi: 10.1016/s0962-8924(98)01383-x. doi:10.1016/S0962-8924(98)01383-X. [DOI] [PubMed] [Google Scholar]
- 23.Jiang D., Jarrett H.W., Haskins W.E. Methods for proteomic analysis of transcription factors. J. Chromatogr. 2009;1216:6881–9. doi: 10.1016/j.chroma.2009.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu S., Xie Z., Onishi A., et al. Profiling the human protein–DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell. 2009;139:610–22. doi: 10.1016/j.cell.2009.08.037. doi:10.1016/j.cell.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie Z., Hu S., Blackshaw S., Zhu H., Qian J. hPDI: a database of experimental human protein–DNA interactions. Bioinformatics. 2010;26:287–9. doi: 10.1093/bioinformatics/btp631. doi:10.1093/bioinformatics/btp631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamashita D., Sano Y., Adachi Y., et al. hDREF regulates cell proliferation and expression of ribosomal protein genes. Mol. Cell. Biol. 2007;27:2003–13. doi: 10.1128/MCB.01462-06. doi:10.1128/MCB.01462-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. doi:10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 28.Li B., Carey M., Workman J.L. The role of chromatin during transcription. Cell. 2007;128:707–19. doi: 10.1016/j.cell.2007.01.015. doi:10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 29.Mellor J., Dudek P., Clynes D. A glimpse into the epigenetic landscape of gene regulation. Curr. Opin. Genet. Dev. 2008;18:116–22. doi: 10.1016/j.gde.2007.12.005. doi:10.1016/j.gde.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 30.Kraus W.L. Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr. Opin. Cell Biol. 2008;20:294–302. doi: 10.1016/j.ceb.2008.03.006. doi:10.1016/j.ceb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petesch S.J., Lis J.T. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 2008;134:74–84. doi: 10.1016/j.cell.2008.05.029. doi:10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hossain M.B., Ji P., Anish R., Jacobson R.H., Takada S. Poly(ADP-ribose) polymerase 1 interacts with nuclear respiratory factor 1 (NRF-1) and plays a role in NRF-1 transcriptional regulation. J. Biol. Chem. 2009;284:8621–32. doi: 10.1074/jbc.M807198200. doi:10.1074/jbc.M807198200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ju B.G., Lunyak V.V., Perissi V., et al. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 34.Shi L., Qiu D., Zhao G., et al. Dynamic binding of Ku80, Ku70 and NF90 to the IL-2 promoter in vivo in activated T-cells. Nucleic Acids Res. 2007;35:2302–10. doi: 10.1093/nar/gkm117. doi:10.1093/nar/gkm117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Potaman V.N., Shlyakhtenko L.S., Oussatcheva E.A., Lyubchenko Y.L., Soldatenkov V.A. Specific binding of poly(ADP-ribose) polymerase-1 to cruciform hairpins. J. Mol. Biol. 2005;348:609–15. doi: 10.1016/j.jmb.2005.03.010. doi:10.1016/j.jmb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 36.Soldatenkov V.A., Chasovskikh S., Potaman V.N., Trofimova I., Smulson M.E., Dritschilo A. Transcriptional repression by binding of poly(ADP-ribose) polymerase to promoter sequences. J. Biol. Chem. 2002;277:665–70. doi: 10.1074/jbc.M108551200. doi:10.1074/jbc.M108551200. [DOI] [PubMed] [Google Scholar]
- 37.Kim M.Y., Mauro S., Gevry N., Lis J.T., Kraus W.L. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 2004;119:803–14. doi: 10.1016/j.cell.2004.11.002. doi:10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 38.Wacker D.A., Ruhl D.D., Balagamwala E.H., Hope K.M., Zhang T., Kraus W.L. The DNA binding and catalytic domains of poly(ADP-ribose) polymerase 1 cooperate in the regulation of chromatin structure and transcription. Mol. Cell. Biol. 2007;27:7475–85. doi: 10.1128/MCB.01314-07. doi:10.1128/MCB.01314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schones D.E., Cui K., Cuddapah S., et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–98. doi: 10.1016/j.cell.2008.02.022. doi:10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y., Shin H., Song J.S., Lei Y., Liu X.S. Identifying positioned nucleosomes with epigenetic marks in human from ChIP-Seq. BMC Genomics. 2008;9:537. doi: 10.1186/1471-2164-9-537. doi:10.1186/1471-2164-9-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu C. An exonuclease protection assay reveals heat-shock element and TATA box DNA-binding proteins in crude nuclear extracts. Nature. 1985;317:84–7. doi: 10.1038/317084a0. doi:10.1038/317084a0. [DOI] [PubMed] [Google Scholar]
- 42.Johnson D.S., Mortazavi A., Myers R.M., Wold B. Genome-wide mapping of in vivo protein–DNA interactions. Science. 2007;316:1497–502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 43.Valouev A., Johnson D.S., Sundquist A., et al. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat. Methods. 2008;5:829–34. doi: 10.1038/nmeth.1246. doi:10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boyle A.P., Davis S., Shulha H.P., et al. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–22. doi: 10.1016/j.cell.2007.12.014. doi:10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barski A., Cuddapah S., Cui K., et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. doi:10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.