Abstract
Aims
A significant increase in congestive heart failure (CHF) was reported when the anti-ErbB2 antibody trastuzumab was used in combination with the chemotherapy drug doxorubicin (Dox). The aim of the present study was to investigate the role(s) of miRNAs in acute Dox-induced cardiotoxicity.
Methods and results
Neuregulin-1-ErbB signalling is essential for maintaining adult cardiac function. We found a significant reduction in ErbB4 expression in the hearts of mice after Dox treatment. Because the proteasome pathway was only partially involved in the reduction of ErbB4 expression, we examined the involvement of microRNAs (miRs) in the reduction of ErbB4 expression. miR-146a was shown to be up-regulated by Dox in neonatal rat cardiac myocytes. Using a luciferase reporter assay and overexpression of miR-146a, we confirmed that miR-146a targets the ErbB4 3′UTR. After Dox treatment, overexpression of miR-146a, as well as that of siRNA against ErbB4, induced cell death in cardiomyocytes. Re-expression of ErbB4 in miR-146a-overexpressing cardiomyocytes ameliorated Dox-induced cell death. To examine the loss of miR-146a function, we constructed ‘decoy’ genes that had tandem complementary sequences for miR-146a in the 3′UTR of a luciferase gene. When miR-146a ‘decoy’ genes were introduced into cardiomyocytes, ErbB4 expression was up-regulated and Dox-induced cell death was reduced.
Conclusion
These findings suggested that the up-regulation of miR-146a after Dox treatment is involved in acute Dox-induced cardiotoxicity by targeting ErbB4. Inhibition of both ErbB2 and ErbB4 signalling may be one of the reasons why those patients who receive concurrent therapy with Dox and trastuzumab suffer from CHF.
Keywords: MicroRNA, Neuregulin, Cardiomyocyte
1. Introduction
Neuregulin-1 (NRG-1) is an agonist for receptor tyrosine kinases of the epidermal growth factor receptor (EGFR) family, consisting of EGFR/HER1/ErbB1, ErbB2/HER2, ErbB3/HER3, and ErbB4/HER4.1 With the exception of ErbB2, which has no ligand, the ErbB proteins have a ligand-binding ectodomain, a single transmembrane segment, an intracellular tyrosine kinase domain, and a tyrosine-rich carboxy terminus.2 Among them, ErbB2 and ErbB4 are expressed in differentiated cardiomyocytes.3 Binding of NRG-1 to ErbB4 increases its kinase activity and leads to heterodimerization with ErbB2 or homodimerization with ErbB4 and stimulation of intracellular signal transduction pathways.4 Heterodimers with ErbB2 appear to be a more potent signalling complex than homodimers.5
Clinical observations clearly suggested increased cardiotoxicity of anthracyclines in patients receiving trastuzumab, a monoclonal antibody against ErbB2.6 Therefore, we first examined changes in ErbB2 and ErbB4 expression levels after stimulation with doxorubicin (Dox). Interestingly, Dox significantly reduced the protein expression of ErbB4 in the heart, although it did not affect ErbB2 expression. Several reports indicated that WW domain containing E3 ubiquitin protein ligase 1 targets the full-length ErbB4 for ubiquitin-mediated degradation in breast cancer;7 therefore, we pre-treated neonatal rat cardiac myocytes (NRCMs) with MG132, a proteasome inhibitor. However, MG132 could not fully abolish the reduction in ErbB4 expression after stimulation with Dox. This suggested that there may be other mechanisms for the reduction in ErbB4 protein expression.
MicroRNAs (miRNAs or miRs) are small, non-coding RNAs that modulate mRNA stability and post-transcriptional translation.8 miRNAs are of crucial importance for the heart to develop and function properly. It has already been shown that loss of cardiac miRNA-mediated regulation by Dgcr8 or Dicer gene deletion lead to dilated cardiomyopathy and heart failure.9,10 Furthermore, specific deletion of cardiac miR-1 or miR-133 lead to embryonic lethality and heart failure.11,12 Recently, DNA-damage-induced up-regulation of pre-miRNAs and mature miRNAs was demonstrated in vitro.13 Therefore, we speculated that anthracycline-induced injury is partly mediated by the induction of miRNAs that targets ErbB4.
It is well known that reactive oxygen species (ROS) are involved in Dox-induced cardiotoxicity.14,15 Previous reports suggested that miR-146a is up-regulated by nuclear factor (NF)-κB, which is a downstream mediator of ROS.16–18 Because miR-146a is abundantly expressed in the heart and ErbB4 has three potential miR-146a-binding sites in the 3′UTR, we examined the function of miR-146a in the heart after Dox treatment. In the present study, we found that (i) Dox reduced ErbB4 expression in the heart; (ii) miR-146a was up-regulated by Dox and overexpression of miR-146a reduced ErbB4 expression; (iii) miR-146a enhanced Dox-induced cell death as well as siRNA against ErbB4; (iv) re-expression of ErbB4 in miR-146a-overexpressing cardiomyocytes suppressed the increase in Dox-induced cell death; and (v) the loss of miR-146a function with ‘decoy’ genes up-regulated ErbB4 expression and reduced Dox-induced cell death.
These findings suggest that the up-regulated expression of miR-146a after Dox treatment is involved in acute Dox-induced cardiotoxicity by targeting ErbB4.
2. Methods
2.1. Reagents
Antibodies and reagents used in this study are summarized in the Supplementary methods.
2.2. Isolation of NRCMs
The investigation conformed to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication No. 85-23, revised 1996) and was approved by Kyoto University Ethics Review Board (Med Kyo 09280). NRCMs were prepared as described previously.19
2.3. Mice
Male C57BL/6 mice were purchased from Japan SLC Inc. (Shizuoka, Japan) and maintained in a specific pathogen-free facility.
2.4. Plasmids
Expression vectors for the negative control (miR-control) and the miRNAs were generated using BLOCK-iT™ PolIImiR RNAi Expression Vector Kits (Invitrogen). The sequences of all constructs were analysed using an ABI 3100 genetic analyzer. All of the constructs were inserted correctly into a pLenti6/V5-D-TOPO vector (Invitrogen) driven by a CMV promoter to stably express genes in NRCMs.20
2.5. siRNA-mediated knockdown of rat ErbB4
The oligonucleotides used for siRNA of ErbB4 are indicated in the Supplementary methods.
Randomly shuffled forms of the ErbB4 siRNA1 were used as a control (control-siRNA). Every siRNA construct was made using a pSINsi-mU6 vector (Takara Bio Inc.) and the siRNA constructs were introduced to a lentivirus vector plasmid and used for transfection of NRCMs.20
2.6. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay
Cells were labelled with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) at a final concentration of 0.5 mg/mL for at least 4 h at 37°C.21 Viability was then evaluated by measuring the absorbance at 595 nm using an El × 800 Microplate Reader (BIO-TEK Instruments, Inc.). The absorbance of the control cells without treatment was considered as 100% viability, and the results were expressed as the per cent viability in comparison with the control cells.
2.7. Western blotting
Cell lysates were prepared as described previously and subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis followed by standard western blotting procedures.19
2.8. Quantitative real-time-polymerase chain reaction for mRNA and miRNA
Total RNA was isolated using TRIzol® reagent (Invitrogen). cDNA was synthesized using SuperScript II reverse transcriptase (Invitrogen) and polymerase chain reaction (PCR) was performed with a SYBR Green PCR master mix (Applied Biosystems), normalized with GAPDH. Gene-specific primers are indicated in the Supplementary methods. miRNAs were quantified in accordance with TaqMan MicroRNA Assays (Applied Biosystems), normalized with U6 snRNA. An ABI Prism 7900HT sequence detection system was used as a thermal cycler.
2.9. Dual-luciferase assays (3′UTR assay)
PCR fragments were amplified from human or rat cDNAs and subcloned downstream of a CMV-driven Firefly luciferase cassette in a pMIR-REPORT vector (Ambion). To make wild-type or mutant ErbB4 3′UTR luciferase reporters, about 100 bp oligonucleotides around binding site 3 were annealed and inserted into a pMIR-REPORT vector. An internal control reporter, Renilla reniformis luciferase, driven by the thymidine kinase (TK) promoter (pRL-TK: Promega) was also co-transfected to normalize the transfection efficiency.
2.10. Measurement of mitochondrial membrane potential by flow cytometry
TMRE dye (100 nM) was added and staining was performed at 37°C for 30 min. Then, the cells were washed once with phosphate-buffered saline (PBS), re-suspended in PBS at 4°C, and kept on ice. Flow cytometry was performed immediately using a FACS Aria (Beckman Dickinson). Appropriate compensation was set. For each sample, data from >30 000 cells were collected. The ratio of TMRE intensity of cardiomyocytes with Dox compared with cardiomyocytes without Dox for each group was calculated as a percentage and plotted on the graph.
2.11. Measurement of apoptosis by flow cytometry
AnnexinV and propidium iodide (PI) staining was performed using a Vybrant® Apoptosis Assay kit #2 (Molecular Probes) in accordance with the manufacturer's protocol. The proportions of apoptotic cells (AnnexinV-positive and PI-negative: Q2), and the total number of dead cells (AnnexinV-positive: Q2 + Q4) and live cells (AnnexinV-negative and PI-negative: Q3) were analysed by flow cytometry using a FACS Aria. Appropriate compensation was set. For each sample, data from >30 000 cells were collected.
2.12. Statistics
Data are presented as means ± SE. Statistical comparisons were performed using unpaired two-tailed Student's t-tests or one-way analysis of variance with Bonferonni's post hoc test where appropriate, with a probability value of <0.05 taken to indicate significance.
3. Results
3.1. ErbB4 expression was decreased by Dox treatment in vivo and in vitro
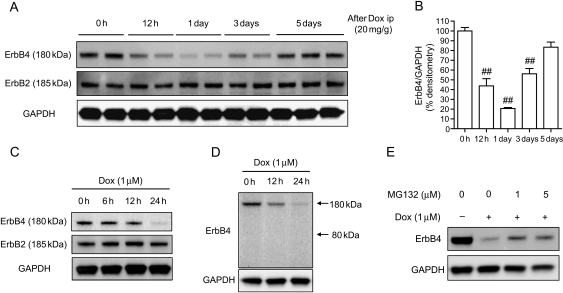
We examined the expression of ErbB2 and ErbB4 in the hearts of mice that had received a single intraperitoneal injection of Dox (20 mg/g).22 We found that ErbB4 expression was significantly reduced 1 day after Dox injection and recovered to the control level 5 days after injection. On the other hand, ErbB2 expression was not affected (Figure 1A and B). The reduction of ErbB4 expression was also observed in NRCMs in a time-dependent manner, whereas ErbB2 expression was not altered (Figure 1C). Figure 1D shows that Dox did not increase the level of cleaved-ErbB4 (80 kDa) in NRCMs. This was the same in the hearts of mice after Dox injection (see Supplementary material online, Figure S1). These data indicated that the acute reduction in ErbB4 level after Dox was not the result of protein shedding. We also examined the possible involvement of the ubiquitin–proteasome pathway in the reduction of ErbB4 level after Dox treatment. As shown in Figure 1E, MG132, a proteasome inhibitor, ameliorated the Dox-induced reduction in ErbB4 levels. However, the effect was limited and ErbB4 expression did not recover to the basal level. These findings suggested that there might be other pathways that regulate ErbB4 expression.
Figure 1.
ErbB4 expression was decreased after Dox treatment in vivo and in vitro. (A) Immunoblotting for ErbB4 and ErbB2 in the hearts of mice after intraperitoneal injection of Dox (20 mg/g). (B) Densitometry for ErbB4 after intraperitoneal injection of Dox. Values are the means ± SE of three independent experiments (##P < 0.01 vs. 0 h). (C) Immunoblotting for ErbB4 and ErbB2 in NRCMs after Dox treatment. (D) Immunoblotting for ErbB4 in NRCMs after Dox treatment. Because the antibody against ErbB4 recognizes the C-terminus of ErbB4, full length of ErbB4 is present at 180 kDa and cleaved-ErbB4 is present at 80 kDa. Note that cleaved-ErbB4 was not seen at 80 kDa. (E) NRCMs were pre-treated with or without proteasome inhibitor MG132 for 1 h and then treated with Dox for 24 h.
3.2. miR-146a was up-regulated in Dox-treated NRCMs
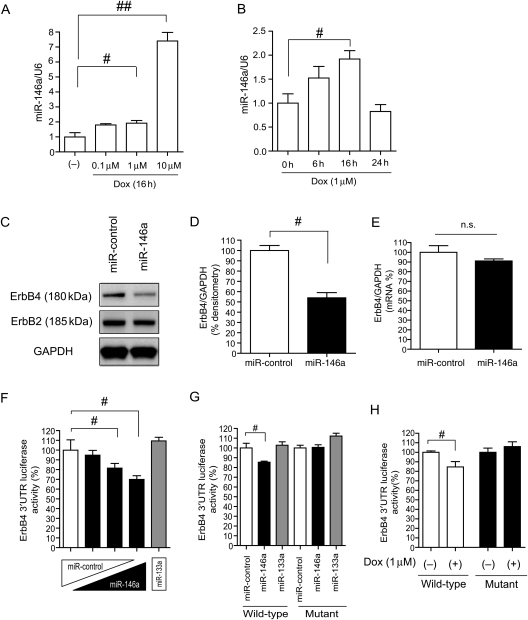
It is well known that ROS are involved in Dox-induced cardiotoxicity.14,15 Previous reports suggested that miR-146a is up-regulated by NF-κB, which is a downstream mediator of ROS.16–18 Because miR-146a is abundantly expressed in the heart23 and ErbB4 has three potential miR-146a-binding sites in the 3′UTR (TargetScan4.0: http://www.targetscan.org/; see Supplementary material online, Figure S2A), we examined the function of miR-146a in the heart after Dox treatment. We compared the expression of miR-146a between NRCMs and cardiac fibroblasts and confirmed that miR-146a was predominantly expressed in NRCMs as well as miR-133a (see Supplementary material online, Figure S2B and C). As shown in Figure 2A and B, miR-146a was significantly up-regulated in a dose-dependent manner, and its level peaked at 16 h after Dox treatment. Next, we overexpressed miR-146a using a lentivirus vector in NRCMs.20 The transduction efficiency was always over 90%, as shown in the Supplementary material online, Figure S2D. The protein level of ErbB4 was significantly reduced in miR-146a-overexpressing NRCMs, whereas ErbB2 levels were not altered (Figure 2C and D). mRNA levels of ErbB4 are shown in Figure 2E. In 293T cells, a human embryonic kidney cell line, miR-146a overexpression also reduced ErbB4 expression (see Supplementary material online, Figure S2E). To verify which binding site was most important, we performed a luciferase reporter assay in 293T cells. We amplified a ∼500 bp fragment of the ErbB4 3′UTR from either human or rat cDNAs and cloned these into downstream of a CMV-driven luciferase reporter gene. We found that the most conserved binding site (site 3) was important in both human and rat (see Supplementary material online, Figure S2F and G). Figure 2F shows that miR-146a overexpression reduced the ErbB4 3′UTR luciferase activity (site 3), whereas miR-133a, one of the most abundant miRNAs in the heart, did not affect luciferase activity. Introduction of mutations in the miR-146a-binding site abolished the miR-146a-mediated inhibition of ErbB4 3′UTR luciferase activity (Figure 2G; see Supplementary material online, Figure S2H). Dox reduced wild-type ErbB4 3′UTR luciferase activity in NRCMs. Conversely, this reduction was not observed in an ErbB4 3′UTR reporter that had mutations in the miR-146a-binding site (Figure 2H). These results suggested that up-regulation of endogenous miR-146a after Dox treatment can affect myocardial ErbB4 expression post-transcriptionally.
Figure 2.
ErbB4 is a target for miR-146a. (A) qRT-PCR for miR-146a after Dox for 16 h at the indicated concentrations in NRCMs. Values are the means ± SE of three to four independent experiments (#P < 0.05, ##P < 0.01). (B) qRT-PCR for miR-146a after Dox (1 µM) for the indicated times in NRCMs. Values are the means ± SE of three to four independent experiments (#P < 0.05). (C) Immunoblotting for ErbB4 and ErbB2 in miR-control or miR-146a-overexpressing NRCMs. (D) Densitometry for ErbB4 in miR-control or miR-146a-overexpressing NRCMs (#P < 0.05). (E) qRT-PCR analysis for ErbB4 in miR-control or miR-146a-overexpressing NRCMs. Values are the means ± SE of four independent experiments (n.s.; not significant). (F) ErbB4 3′UTR Firefly luciferase (F-luc) activity at binding site 3 in 293T cells. Values are the means ± SE of four independent experiments (#P < 0.05). (G) ErbB4 3′UTR F-luc activity at binding site 3 in 293T cells. Values are the means ± SE of four independent experiments (#P < 0.05). (H) ErbB4 3′UTR F-luc activity at binding site 3 in NRCMs. Values are the means ± SE of six independent experiments (#P < 0.05).
3.3. Both miR-146a overexpression and ErbB4 knockdown reduced NRCM survival after Dox treatment
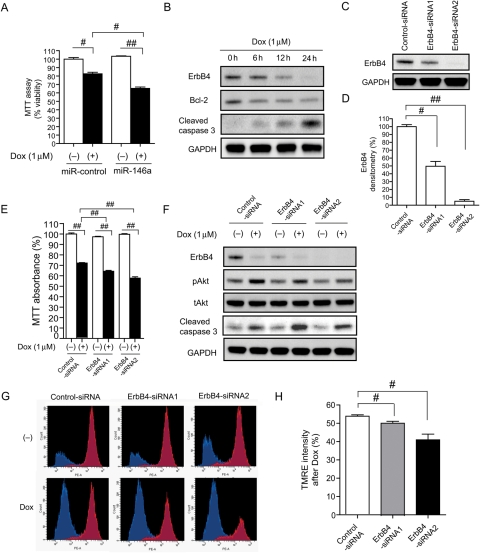
To evaluate the effect of miR-146a induction after Dox treatment on cardiac myocytes, we stimulated miR-146a-overexpressing NRCMs using Dox. Dox induced more cell death in miR-146a-overexpressing NRCMs than miR-control (negative control) NRCMs, which was shown in microscopy images (see Supplementary material online, Figure S3A) and in cell viability assays using MTT (Figure 3A). These data suggested that the up-regulation of miR-146a may be harmful in cardiac myocytes. Dox reduced ErbB4 expression and increased cleaved caspase 3 expression, which is a sensitive apoptosis marker (Figure 3B), and miR-146a levels increased at the same time (Figure 2A and B). Therefore, we speculated that Dox-induced apoptosis resulted from the down-regulation of ErbB4 by miR-146a. To verify the function of ErbB4 in cardiomyocytes, two kinds of siRNA against ErbB4 were introduced into NRCMs. The knockdown efficiency of these siRNAs was 50 and 10%, respectively, at the protein level (Figure 3C and D). Knockdown of ErbB4 exaggerated Dox-induced cell death in the MTT assay, according to the knockdown efficiency (Figure 3E). Knockdown of ErbB4 reduced Dox-induced Akt activation and increased the amount of cleaved caspase 3 (Figure 3F). We also evaluated mitochondrial function using TMRE dye. TMRE is a cell-permeant, cationic, red-orange fluorescent dye that is readily sequestered by active mitochondria.24 Fluorescence microscopy images of NRCMs with or without Dox are shown in the Supplementary material online, Figure S3B. We measured the intensity of TMRE staining, which indicates mitochondrial membrane potential, using flow cytometry. In the basal state, about 80% of cells resided in the TMRE high population (red) and 20% cells in the TMRE low population (blue). After Dox treatment, the TMRE high population and TMRE intensity were reduced in ErbB4 knocked-down NRCMs, according to the knock-down efficiency (Figure 3G and H).
Figure 3.
Both miR-146a overexpression and ErbB4 knockdown reduced NRCMs survival after Dox treatment. (A) MTT assay of NRCMs transduced with miR-control or miR-146a with or without Dox treatment for 24 h. Values are the means ± SE of six independent experiments (#P < 0.05, ##P < 0.01). (B) Immunoblotting for ErbB4, bcl-2, and cleaved caspase 3 after Dox in NRCMs. (C) Immunoblotting for ErbB4 in NRCMs transduced with control-siRNA (scrambled) or ErbB4-siRNAs. (D) Densitometry for ErbB4 in NRCMs transduced with ErbB4-siRNA. Values are the means ± SE of three independent experiments (#P < 0.05, ##P < 0.01). (E) MTT assay of NRCMs transduced with control-siRNA or ErbB4-siRNA with or without Dox treatment for 24 h. Values are the means ± SE of six independent experiments (##P < 0.01). (F) Immunoblotting for ErbB4, pAkt, tAkt, and cleaved caspase 3 in NRCMs, infected with control-siRNA or ErbB4-siRNAs with or without Dox treatment for 24 h. (G) Flow cytometric analysis of TMRE in NRCMs transduced with control-siRNA or ErbB4-siRNA with or without Dox treatment for 24 h. The X-axis represents TMRE intensity (PE intensity) and the Y-axis represents cell number. Red populations indicate the TMRE high group and blue populations indicate the TMRE low group. (H) The ratio of TMRE intensity with Dox compared with without Dox for each group. Values are the means ± SE of three to four independent experiments (#P < 0.05).
3.4. miR-146a enhanced Dox-induced apoptosis in NRCMs
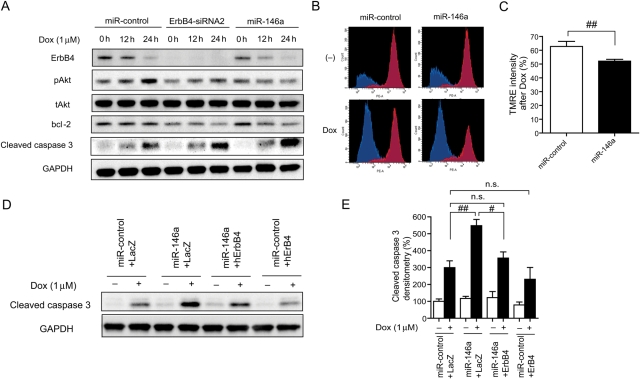
Both overexpression of miR-146a and ErbB4 siRNA2 reduced levels of ErbB4 expression, Akt phosphorylation, and bcl-2 expression and increased cleaved caspase 3 level after Dox treatment in NRCMs (Figure 4A). The TMRE high population and TMRE intensity after Dox were more reduced in miR-146a-overexpressing NRCMs than in control cells (Figure 4B and C). These results indicated that miR-146a enhanced Dox-induced apoptosis in NRCMs. Following the transduction of ErbB4 without a 3′UTR in miR-146a-overexpressing NRCMs, the increase in Dox-induced cleaved caspase 3 was reduced to the control level (Figure 4D and E). The intensity of TMRE staining also recovered to the control level (see Supplementary material online, Figure S4A and B).
Figure 4.
miR-146a enhanced Dox-induced apoptosis in NRCMs. (A) Immunoblotting for ErbB4, pAkt, tAkt, bcl-2, and cleaved caspase 3 in NRCMs transduced with miR-control, miR-146a, or ErbB4-siRNA2 in the presence or absence of Dox for the indicated time periods. (B) Flow cytometric analysis of TMRE in NRCMs transduced with miR-control of miR-146a with or without Dox for 24 h. (C) The ratio of TMRE intensity with Dox compared with without Dox for each group. Values are the means ± SE of three to four independent experiments (##P < 0.01). (D) Immunoblotting for cleaved caspase 3 in miR-control or miR-146a-overexpressing NRCMs infected with either lacZ or ErbB4 with or without Dox for 24 h. (E) Densitometry for cleaved caspase 3 in miR-control or miR-146a-overexpressing NRCMs infected with either lacZ or ErbB4 with or without Dox for 24 h. Values are the means ± SE of four independent experiments (#P < 0.05, ##P < 0.01).
3.5. Reduction of endogenous miR-146a ameliorated Dox-induced apoptosis in NRCMs
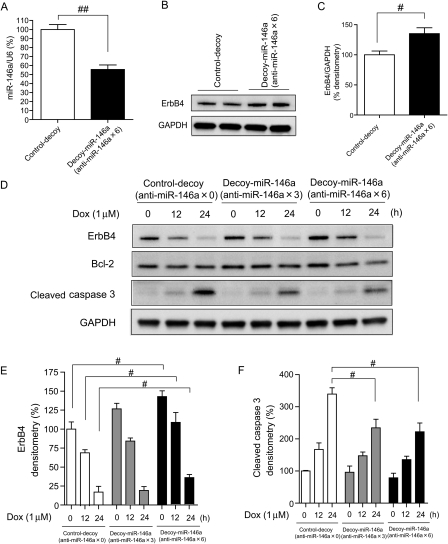
To assess the functional consequences of silencing endogenous miR-146a in vitro, NRCMs were infected with a lentivirus vector, in which three or six tandem sequences partially complementary to miR-146a were linked to a luciferase or GFP reporter gene in their 3′UTRs (see Supplementary material online, Figure S5A and F). The complementary sequences acted as a decoy, sequestering endogenous miR-146a and other miRNAs with the same seed sequences (see Supplementary material online, Figure S5H).20,25–27 Transfection with a luciferase-decoy gene along with miR-146a reduced the luciferase activity and the extent was more prominent with anti-miR-146a × 6 than that with anti-miR-146a × 3 in 293T cells, whereas transfection along with miR-control or miR-133a did not reduce luciferase activity (see Supplementary material online, Figure S5B–D). When the same amount of plasmids of control and decoy gene were transduced into NRCMs, luciferase activity was significantly reduced (see Supplementary material online, Figure S5E). NRCMs infected with GFP-decoy (see Supplementary material online, Figure S4F) reduced the intensity of GFP in flow cytometry (see Supplementary material online, Figure S5G). These results indicated that endogenous miR-146a specifically bound to the decoy gene and inhibited the translation of luciferase or GFP in NRCMs. The expression of miR-146a was significantly reduced in NRCMs infected with decoy-miR-146a × 6 (Figure 5A). ErbB4 expression was increased significantly in NRCMs infected with decoy-miR-146a × 6 (Figure 5B and C). Decoy-miR-146a ameliorated the Dox-induced reduction of ErbB4 and Dox-induced increase in cleaved caspase 3 (Figure 5D–F). TMRE intensity was significantly increased in NRCMs infected with decoy-miR-146a × 6 compared with control cells (see Supplementary material online, Figure S5I and J).
Figure 5.
Reduction in endogenous miR-146a ameliorated Dox-induced apoptosis in NRCMs. (A) qRT-PCR analysis for miR-146a in NRCMs, infected with control-decoy (no binding site) or decoy-miR-146a (anti-miR-146a × 6) using lentivirus vector. Values are the means ± SE of three independent experiments (##P < 0.05). (B) Immunoblotting for ErbB4 in NRCMs infected with control-decoy or decoy-miR-146a. (C) Densitometry for ErbB4 in NRCMs infected with control-decoy or decoy-miR-146a. Values are the means ± SE of four independent experiments (#P < 0.05). (D) Immunoblotting for ErbB4, bcl-2, and cleaved caspase 3 in NRCMs infected with control or decoy gene in the presence or absence of Dox for the indicated time periods. (E) Densitometry for ErbB4 in NRCMs infected with control or decoy gene in the presence or absence of Dox. Values are the means ± SE of four independent experiments (#P < 0.05). (F) Densitometry for cleaved caspase 3 in NRCMs infected with control or decoy gene in the presence or absence of Dox. Values are the means ± SE of four independent experiments (#P < 0.05).
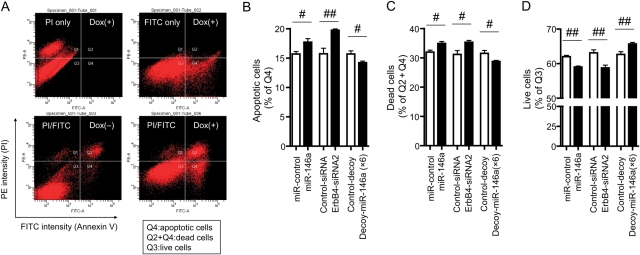
3.6. AnnexinV and PI staining of NRCMs
Finally, we stained NRCMs with AnnexinV/PI and then measured the numbers of apoptotic cells, dead cells, and live cells using flow cytometry. Figure 6A shows that the numbers of apoptotic and dead cells were increased after Dox treatment. Transfection of miR-146a or ErbB4 siRNA induced apoptosis and cell death, whereas transfection of decoy-miR-146a (anti-miR-146a × 6) reduced these in NRCMs after treatment with Dox (Figure 6B–D).
Figure 6.
AnnexinV and PI staining of NRCMs. (A) Dot-plot analysis of AnnexinV/PI staining showed that apoptotic cells (Q4) and dead cells (Q2 + Q4) were increased after Dox treatment for 24 h. (B) The percentages of apoptotic cells after Dox treatment. (C) The percentages of dead cells after Dox treatment. (D) The percentages of live cells after Dox treatment. Values are the means ± SE of six independent experiments (#P < 0.05, ##P < 0.01).
4. Discussion
Intensive investigations into Dox-induced cardiotoxicity have been ongoing for decades. In the present study, we have clarified for the first time that miR-146a-mediated suppression of ErbB4 is a substantial causal mechanism of Dox-induced cardiac toxicity.
NRG-1/ErbB signalling is best known for its indispensable role during cardiac and neuronal development. The importance of the physiological role of ErbB signalling in the post-natal heart was demonstrated by analysing the hearts of conditional mutant mice. ErbB2-deficient conditional mutant mice were viable; however, physiological analysis revealed the onset of multiple independent parameters of dilated cardiomyopathy.28,29 Conditional inactivation of ErbB4 in ventricular muscle cells also led to a severe dilated cardiomyopathy.30 Heterozygous knockout of NRG-1 in mice worsens survival and left ventricular function in the presence of Dox-induced cardiac injury.31 These reports showed the importance of NRG-1/ErbB signalling not only during development but also after birth.
Observations in patients treated with trastuzumab (Herceptin), an inhibitory antibody against ErbB2, which is administered with or following anthracyclines in breast cancer, suggested that ErbB-mediated myocardial protection against cardiotoxic drugs is also prominent in the human heart.6 Despite extensive research, a significant increase in cardiotoxic effects of concurrent treatment with an anthracycline and trastuzumab has remained difficult to explain. Trastuzumab cardiotoxicity appears to be dose independent and largely reversible, suggesting a different mechanism from that of anthracyclines.32 From the present results, it is possible that the transient reduction in ErbB4 expression via miR-146a up-regulation and/or ubiquitin–proteasome activation by Dox worsens the cardiotoxic effects of trastuzumab, an inhibitor of ErbB2, by complete shutdown of the NRG-1/ErbB pathway. Because the reduction of ErbB4 by Dox was reversible in the present experiments, changing the timing of Dox and trastuzumab administration may reduce their cardiotoxic effects. Other strategies for the prevention of side effects may include limits on the amount of Dox and administration of Dox in combination with NRG-1.
It is well known that ROS are involved in Dox-induced cardiotoxicity, and the generation of ROS can be interpreted by the chemical structure of Dox, which possesses a tendency to generate ROS during drug metabolism.14,15 Recent findings indicate that the endothelial nitric oxide synthase reductase domain converts Dox to an unstable semiquinone intermediate that favours ROS generation.15 Because ROS serve as an upstream mediator for the activation of NF-κB,33,34 and a number of previous reports have suggested that miR-146a transcription is regulated by NF-κB,16–18 miR-146a induction by Dox in our experiment may be due to ROS-mediated NF-κB activation. Several reports suggested that there are inhibitory κB kinase-dependent or independent pathways for the activation of NF-κB by Dox in vitro.35,36 Further examinations are required in order to determine the precise mechanisms for the induction of miR-146a by Dox in vivo.
NRG-1 is reported to be predominantly expressed in the cardiac microvascular endothelium and to have beneficial effects on cardiomyocytes through the Akt and Erk pathway.37 NRG-1β has been reported to be 10–100 times more active than NRG-1α.38,39 We performed immunoblotting and quantitative real-time (qRT)-PCR for NRG-1β and detected NRG-1β in NRCMs. Therefore, NRG-1 is one of the factors that protect cardiomyocytes in an autocrine or paracrine manner, although it is possible that NRCMs may express much more NRG-1β than adult cardiac myocytes. Dox reduced the expression of NRG-1β (see Supplementary material online, Figure S6A). miR-146a and knockdown of ErbB4 increased NRG-1β expression at the mRNA and protein levels (see Supplementary material online, Figure S6B–E). We do not know the reason why NRG-1β was up-regulated following miR-146a overexpression or knockdown of ErbB4, but there may be a negative feedback loop between ErbB4-NRG-1β to compensate for the reduction in ErbB4 expression.
In conclusion, the present results suggest that the up-regulated expression of miR-146a after Dox treatment is involved in acute Dox-induced cardiotoxicity by targeting ErbB4. There is emerging evidence for the involvement of NRG-1/ErbB signals in heart failure. Therefore, the development of new therapeutic strategies based on NRG-1, such as the delivery of nucleotides that inhibit miR-146a, is promising for treating heart failure.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Funding
This work was supported in part by the Global COE Program ‘Center for Frontier Medicine’ K.O., K.H., T.K., and T.K. Funding to pay the open access publication charge for this article was provided by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan to K.O.
Supplementary Material
Acknowledgements
Authors would line to thank Naoya Sowa for excellent technical assistance.
Conflict of interest: none declared.
References
- 1.Li JY, Wang H, May S, Song X, Fueyo J, Fuller GN. Constitutive activation of c-Jun N-terminal kinase correlates with histologic grade and EGFR expression in diffuse gliomas. J Neurooncol. 2008;88:11–17. doi: 10.1007/s11060-008-9529-1. doi:10.1007/s11060-008-9529-1. [DOI] [PubMed] [Google Scholar]
- 2.Spector NL, Blackwell KL. Understanding the mechanisms behind trastuzumab therapy for human epidermal growth factor receptor 2-positive breast cancer. J Clin Oncol. 2009;27:5838–5847. doi: 10.1200/JCO.2009.22.1507. doi:10.1200/JCO.2009.22.1507. [DOI] [PubMed] [Google Scholar]
- 3.Zhao YY, Sawyer DR, Baliga RR, Opel DJ, Han X, Marchionni MA, et al. Neuregulins promote survival and growth of cardiac myocytes. Persistence of ErbB2 and ErbB4 expression in neonatal and adult ventricular myocytes. J Biol Chem. 1998;273:10261–10269. doi: 10.1074/jbc.273.17.10261. doi:10.1074/jbc.273.17.10261. [DOI] [PubMed] [Google Scholar]
- 4.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. doi:10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 5.Pinkas-Kramarski R, Soussan L, Waterman H, Levkowitz G, Alroy I, Klapper L, et al. Diversification of Neu differentiation factor and epidermal growth factor signaling by combinatorial receptor interactions. EMBO J. 1996;15:2452–2467. [PMC free article] [PubMed] [Google Scholar]
- 6.Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. doi:10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 7.Feng SM, Muraoka-Cook RS, Hunter D, Sandahl MA, Caskey LS, Miyazawa K, et al. The E3 ubiquitin ligase WWP1 selectively targets HER4 and its proteolytically derived signaling isoforms for degradation. Mol Cell Biol. 2009;29:892–906. doi: 10.1128/MCB.00595-08. doi:10.1128/MCB.00595-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schroen B, Heymans S. MicroRNAs and beyond: the heart reveals its treasures. Hypertension. 2009;54:1189–1194. doi: 10.1161/HYPERTENSIONAHA.109.133942. doi:10.1161/HYPERTENSIONAHA.109.133942. [DOI] [PubMed] [Google Scholar]
- 9.Chen JF, Murchison EP, Tang R, Callis TE, Tatsuguchi M, Deng Z, et al. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci USA. 2008;105:2111–2116. doi: 10.1073/pnas.0710228105. doi:10.1073/pnas.0710228105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rao PK, Toyama Y, Chiang HR, Gupta S, Bauer M, Medvid R, et al. Loss of cardiac microRNA-mediated regulation leads to dilated cardiomyopathy and heart failure. Circ Res. 2009;105:585–594. doi: 10.1161/CIRCRESAHA.109.200451. doi:10.1161/CIRCRESAHA.109.200451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. doi:10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 12.Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, et al. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. doi:10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. doi:10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 14.Iarussi D, Indolfi P, Casale F, Coppolino P, Tedesco MA, Di Tullio MT. Recent advances in the prevention of anthracycline cardiotoxicity in childhood. Curr Med Chem. 2001;8:1649–1660. doi: 10.2174/0929867013371888. [DOI] [PubMed] [Google Scholar]
- 15.Neilan TG, Blake SL, Ichinose F, Raher MJ, Buys ES, Jassal DS, et al. Disruption of nitric oxide synthase 3 protects against the cardiac injury, dysfunction, and mortality induced by doxorubicin. Circulation. 2007;116:506–514. doi: 10.1161/CIRCULATIONAHA.106.652339. doi:10.1161/CIRCULATIONAHA.106.652339. [DOI] [PubMed] [Google Scholar]
- 16.Hurst DR, Edmonds MD, Scott GK, Benz CC, Vaidya KS, Welch DR. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res. 2009;69:1279–1283. doi: 10.1158/0008-5472.CAN-08-3559. doi:10.1158/0008-5472.CAN-08-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perry MM, Williams AE, Tsitsiou E, Larner-Svensson HM, Lindsay MA. Divergent intracellular pathways regulate interleukin-1beta-induced miR-146a and miR-146b expression and chemokine release in human alveolar epithelial cells. FEBS Lett. 2009;583:3349–3355. doi: 10.1016/j.febslet.2009.09.038. doi:10.1016/j.febslet.2009.09.038. [DOI] [PubMed] [Google Scholar]
- 18.Lukiw WJ, Zhao Y, Cui JG. An NF-kappaB-sensitive micro RNA-146a-mediated inflammatory circuit in Alzheimer disease and in stressed human brain cells. J Biol Chem. 2008;283:31315–31322. doi: 10.1074/jbc.M805371200. doi:10.1074/jbc.M805371200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horie T, Ono K, Nagao K, Nishi H, Kinoshita M, Kawamura T, et al. Oxidative stress induces GLUT4 translocation by activation of PI3-K/Akt and dual AMPK kinase in cardiac myocytes. J Cell Physiol. 2008;215:733–742. doi: 10.1002/jcp.21353. doi:10.1002/jcp.21353. [DOI] [PubMed] [Google Scholar]
- 20.Nishi H, Ono K, Iwanaga Y, Horie T, Nagao K, Takemura G, et al. MICRORNA-15B modulates cellular ATP levels and degenerates mitochondria via ARL2 in neonatal rat cardiac myocytes. J Biol Chem. 2010;285:4920–4930. doi: 10.1074/jbc.M109.082610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagao K, Ono K, Iwanaga Y, Tamaki Y, Kojima Y, Horie T, et al. Neural cell adhesion molecule is a cardioprotective factor up-regulated by metabolic stress. J Mol Cell Cardiol. 2010;48:1157–1168. doi: 10.1016/j.yjmcc.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 22.Li K, Sung RY, Huang WZ, Yang M, Pong NH, Lee SM, et al. Thrombopoietin protects against in vitro and in vivo cardiotoxicity induced by doxorubicin. Circulation. 2006;113:2211–2220. doi: 10.1161/CIRCULATIONAHA.105.560250. doi:10.1161/CIRCULATIONAHA.105.560250. [DOI] [PubMed] [Google Scholar]
- 23.Cheng Y, Ji R, Yue J, Yang J, Liu X, Chen H, et al. MicroRNAs are aberrantly expressed in hypertrophic heart: do they play a role in cardiac hypertrophy? Am J Pathol. 2007;170:1831–1840. doi: 10.2353/ajpath.2007.061170. doi:10.2353/ajpath.2007.061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon LI, Burke MA, Singh AT, Prachand S, Lieberman ED, Sun L, et al. Blockade of the erbB2 receptor induces cardiomyocyte death through mitochondrial and reactive oxygen species-dependent pathways. J Biol Chem. 2009;284:2080–2087. doi: 10.1074/jbc.M804570200. doi:10.1074/jbc.M804570200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horie T, Ono K, Nishi H, Iwanaga Y, Nagao K, Kinoshita M, et al. MicroRNA-133 regulates the expression of GLUT4 by targeting KLF15 and is involved in metabolic control in cardiac myocytes. Biochem Biophys Res Commun. 2009;389:315–320. doi: 10.1016/j.bbrc.2009.08.136. doi:10.1016/j.bbrc.2009.08.136. [DOI] [PubMed] [Google Scholar]
- 26.Brown BD, Naldini L. Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nat Rev Genet. 2009;10:578–585. doi: 10.1038/nrg2628. doi:10.1038/nrg2628. [DOI] [PubMed] [Google Scholar]
- 27.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. doi:10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ozcelik C, Erdmann B, Pilz B, Wettschureck N, Britsch S, Hubner N, et al. Conditional mutation of the ErbB2 (HER2) receptor in cardiomyocytes leads to dilated cardiomyopathy. Proc Natl Acad Sci USA. 2002;99:8880–8885. doi: 10.1073/pnas.122249299. doi:10.1073/pnas.122249299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crone SA, Zhao YY, Fan L, Gu Y, Minamisawa S, Liu Y, et al. ErbB2 is essential in the prevention of dilated cardiomyopathy. Nat Med. 2002;8:459–465. doi: 10.1038/nm0502-459. doi:10.1038/nm0502-459. [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Rivello H, Taranda J, Said M, Cabeza-Meckert P, Vila-Petroff M, Scaglione J, et al. Dilated cardiomyopathy in Erb-b4-deficient ventricular muscle. Am J Physiol Heart Circ Physiol. 2005;289:H1153–H1160. doi: 10.1152/ajpheart.00048.2005. doi:10.1152/ajpheart.00048.2005. [DOI] [PubMed] [Google Scholar]
- 31.Liu FF, Stone JR, Schuldt AJ, Okoshi K, Okoshi MP, Nakayama M, et al. Heterozygous knockout of neuregulin-1 gene in mice exacerbates doxorubicin-induced heart failure. Am J Physiol Heart Circ Physiol. 2005;289:H660–H666. doi: 10.1152/ajpheart.00268.2005. doi:10.1152/ajpheart.00268.2005. [DOI] [PubMed] [Google Scholar]
- 32.Ewer MS, Vooletich MT, Durand JB, Woods ML, Davis JR, Valero V, et al. Reversibility of trastuzumab-related cardiotoxicity: new insights based on clinical course and response to medical treatment. J Clin Oncol. 2005;23:7820–7826. doi: 10.1200/JCO.2005.13.300. doi:10.1200/JCO.2005.13.300. [DOI] [PubMed] [Google Scholar]
- 33.Wang S, Kotamraju S, Konorev E, Kalivendi S, Joseph J, Kalyanaraman B. Activation of nuclear factor-kappaB during doxorubicin-induced apoptosis in endothelial cells and myocytes is pro-apoptotic: the role of hydrogen peroxide. Biochem J. 2002;367:729–740. doi: 10.1042/BJ20020752. doi:10.1042/BJ20020752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schulze-Osthoff K, Los M, Baeuerle PA. Redox signalling by transcription factors NF-kappa B and AP-1 in lymphocytes. Biochem Pharmacol. 1995;50:735–741. doi: 10.1016/0006-2952(95)02011-z. doi:10.1016/0006-2952(95)02011-Z. [DOI] [PubMed] [Google Scholar]
- 35.Tergaonkar V, Bottero V, Ikawa M, Li Q, Verma IM. IkappaB kinase-independent IkappaBalpha degradation pathway: functional NF-kappaB activity and implications for cancer therapy. Mol Cell Biol. 2003;23:8070–8083. doi: 10.1128/MCB.23.22.8070-8083.2003. doi:10.1128/MCB.23.22.8070-8083.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tergaonkar V, Pando M, Vafa O, Wahl G, Verma I. p53 stabilization is decreased upon NFkappaB activation: a role for NFkappaB in acquisition of resistance to chemotherapy. Cancer Cell. 2002;1:493–503. doi: 10.1016/s1535-6108(02)00068-5. doi:10.1016/S1535-6108(02)00068-5. [DOI] [PubMed] [Google Scholar]
- 37.Lemmens K, Doggen K, De Keulenaer GW. Role of neuregulin-1/ErbB signaling in cardiovascular physiology and disease: implications for therapy of heart failure. Circulation. 2007;116:954–960. doi: 10.1161/CIRCULATIONAHA.107.690487. doi:10.1161/CIRCULATIONAHA.107.690487. [DOI] [PubMed] [Google Scholar]
- 38.Jones JT, Akita RW, Sliwkowski MX. Binding specificities and affinities of egf domains for ErbB receptors. FEBS Lett. 1999;447:227–231. doi: 10.1016/s0014-5793(99)00283-5. doi:10.1016/S0014-5793(99)00283-5. [DOI] [PubMed] [Google Scholar]
- 39.Pinkas-Kramarski R, Shelly M, Glathe S, Ratzkin BJ, Yarden Y. Neu differentiation factor/neuregulin isoforms activate distinct receptor combinations. J Biol Chem. 1996;271:19029–19032. doi: 10.1074/jbc.271.32.19029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.