Abstract
Protein glycosylation is the most versatile and common protein modification and plays important roles in various biological processes and disease progression. In this review, the development of microarray technology for protein glycosylation analysis is described. Three types are discussed: carbohydrate, lectin and natural glycoprotein microarrays. The advantages of microarray technology to study protein glycosylation are high-volume throughput coupled with a highly miniaturized platform. These techniques show great promise for detecting interactions that involve carbohydrates and as a screening tool to detect glycan patterns important for the early diagnosis of disease.
Keywords: Carbohydrate microarray, glycoprotein microarray, glycosylation, lectin, lectin microarray
Introduction
Glycosylation is perhaps the most extensive and complex form of protein post-translational modification (PTM), characteristic of various cell surface and secreted eukaryotic proteins [1•,2]. Both N-linked (Asn-linked) and O-linked (Ser- or Thr-linked) glycan variants, in the form of glycopeptides, glycolipids, glycosaminoglycans, or other glycoconjugates on the cell surface and in plasma, play important roles in various biological functions, including immune responses and cell-to-cell interactions. Alterations in protein glycosylation, which occur through varying the heterogeneity of glycosylation sites or changing the glycan structures of proteins on the cell surface and in body fluids, have been shown to correlate with the development or progression, or both, of cancer and other disease states.
Over the past decade, microarrays have emerged as an important tool for the characterization of cancer cells and are generated by spotting the analytes on a chemically derivatized surface. The slides are usually probed with a detection reagent conjugated to a fluorescent tag, which are quantified by laser scanning. Microarrays enable simultaneous screening of multiple samples, to produce a probe-based profile of the sample. The advantages of microarray technology include its high-throughput capacity coupled with a highly miniaturized platform and its potential as both an analytical and diagnostic tool.
Lectins are a group of proteins or glycoproteins that have unique affinities to carbohydrates, for example, they can reversibly and specifically interact with certain glycan structural motifs [3]. The ability to profile diverse glycan structures present on proteins has been aided by the availability of naturally occurring lectins and the availabilty of commercial antibodies. In this review, the development of microarray technology for protein glycosylation analysis is described. Three types of microarrays are discussed: carbohydrate, lectin and natural glycoprotein microarrays. Because carbohydrates isolated from biological systems are typically present in low amounts, miniaturized array methodology is particularly well suited for the analysis of protein glycosylation. The advantages and potential applications of these methodologies are discussed.
Protein glycosylation in cancer and disease
Glycans can regulate different aspects of tumor progression, including proliferation, invasion and metastasis [4–10]. Changes in glycosylation patterns have been observed in prostate cancer [11], colorectal cancer [12,13], and breast cancer [14]. Glycoproteins have also provided an ideal source for discovering biomarkers for disease detection. For example, various clinical cancer biomarkers and therapeutic targets are glycoproteins [9–14,15••], including cancer antigen 19–9 in gastrointestinal cancer [16], cluster of differentiation 340 (Her2/neu) in breast cancer [17], and prostate-specific antigen (PSA) in prostate cancer [11]. Alterations of glycan structures observed in cancer cells include increased glycan branching [15••,18,19•], and increased Lewis antigen expression [20]. In addition, glycosylation of PSA secreted by the prostate tumor cell line LNCaP differs significantly from PSA present in seminal plasma (normal control) [11]. These carbohydrate differences allow a distinction to be made between PSA from a normal origin and that from a tumor origin and can provide a valuable biochemical tool for diagnosis. Characterization of N-linked glycans from human pancreatic ribonuclease 1 isolated from healthy pancreas and pancreatic adenocarcinoma tumor cells (Capan-1 and MDAPanc-3), revealed different glycosylation patterns [6]. Furthermore, in a systematic analyses of serum concentrations of haptoglobin (Hp) and its glycoforms in patients with hepatocellular carcinoma (HCC) and non-cancer patients with chronic liver disease, a unique pattern of Hp glycoforms with altered sialylation and fucosylation specific to HCC was associated with tumor progression [21]. Another common glycan alteration observed in cancer is the truncation of O-linked glycans, particularly on mucins [22,23].
Differentially expressed glycosylation in serum taken from patients with pancreatic cancer has also been observed using liquid phase separation coupled with mass spectrometry (MS) analysis [7]. These glycosylation changes in tumor-secreted proteins reflect fundamental changes in the enzyme levels (or enzyme activities) of glycosyltransferases, such as fucosyltransferases and mannosidases, involved in the glycosylation pathway. Increased activity of specific glycosyltransferases (which may lead to increased expression of certain terminal glycans such as sialic acid and fucosyl residues) or decreased mannosidase activity (leading to decreased trimming of high mannose structures, with a corresponding increased branching of the high mannose core), can lead to the overexpression of highly-branched glycosylation modifications observed in cancer [24].
Glycans are heterogeneous and diverse because of the possible combination of available monosaccharides, linkages, branching and variable lengths of glycan chains. This diversity combined with the difficulty in stabilizing the branched structures means that characterizing the glycoproteome is challenging. Sugars are typically present as complex mixtures of similar, but distinct, sequences. Since sugars cannot be replicated into large quantities for analysis, the amount of carbohydrates present in a clinical sample are limited. Therefore, characterization of glycosylation is challenging and time consuming and, consequently, the development of a fast and sensitive technique is essential for glycosylation analysis in cancer research.
The importance of lectins in the study of protein glycosylation
The strength of the binding event between lectins and carbohydrates increases with the number of molecular interactions. In general, lectin-carbohydrate interactions are weaker than antigen-antibody complexes and their affinity constants are in the range of Kd 10−6 to 10−7 mol/l for glycoproteins [25]. There are several major commercially available lectins, some of which are listed in Table 1, which can be used to enrich glycoproteins with specific glycan structures. Lectin capture strategies have been combined with other techniques, such as MS, for the discovery of serum glycoprotein biomarkers. Hancock and coworkers developed a multi-lectin affinity column, which combines concanavalin A (ConA), wheat germ agglutinin (WGA), and jacalin, to capture the majority of glycoproteins present in the sera of breast cancer patients and controls [26]. Biomarker candidates in these groups were quantified by peak area measurements. In similar studies, Qui and Regnier utilized serial lectin affinity chromatography for fractionation and comparison of glycosylation heterogeneity on glycoproteins derived from human serum [27••] and Madera et al combined silica-based lectin microcolumns with high-resolution separation techniques for enrichment of glycoproteins and glycopeptides [28•]. In addition, a strategy was developed to identify sialylated glycoprotein markers in human cancer serum [5]. This method consisted of three steps: lectin affinity enrichment, liquid phase separation and characterization of the glycoprotein markers using MS. Utilization of different lectin columns allowed the distribution of α(2,3) and α(2,6) linkage type sialylation in cancer serum versus that in normal samples to be determined. Using this strategy, sialylated plasma protease C1 inhibitor was observed to be downregulated in pancreatic cancer serum [5].
Table 1.
Biotinylated lectins used for glycan detection and their specificities.
| Biotinylated lectin | Glycan structure detected |
|---|---|
| ConA | α-Linked mannose |
| MAL | Sialic acid in an (α-2,3) linkage |
| AAL | Fucose linked (α-1,6) to N-acetylglucosamine or to Fucose linked (α-1,3) to N-acetyllactosamine |
| SNA | Sialic acid attached to terminal galactose in (α-2,6), and to a lesser degree, (α-2,3), linkage |
| PNA | Galactosyl (β-1,3) N-acetylgalactosamine |
AAL aleuria aurantia, ConA, concanavalin A, MAL maackia amurensis II, PNA peanut agglutinin, SNA sambucus nigra (elderberry) bark.
(Reprinted with permission from Patwa TH, Zhao J, Anderson MA, Simeone DM, Lubman DM: Screening of glycosylation patterns in serum using natural glycoprotein microarrays and multi-lectin fluorescence detection. Anal Chem (2006) 78(18):6411-6421–2007 American Chemical Society) [19•].
Carbohydrate microarrays
Carbohydrate microarrays are developing into a standard tool to screen large numbers of sugars and elucidate the role of carbohydrates in biological systems [29•,30,31]. Various glycan-type structures are arrayed on a range of surface chemistries such as nitrocellulose, glass, and dextran, followed by screening for parallel binding (Figure 1a). Covalent attachment of chemically modified carbohydrates onto a derivatized surface, for example, hydrazide-linked carbohydrates onto an epoxide-derivatized slide, is the most general method for carbohydrate immobilization. Carbohydrates can also be attached by site-specific non-covalent binding to underivatized surfaces [31]. Non-specific and non-covalent attachment of carbohydrates on surfaces can become size-dependent [31]; therefore, targeted methods involving derivatized slides are desirable. The interactions of different carbohydrate structures with a wide variety of biological targets, including proteins, RNA, viruses, and whole cells have been investigated using this technique; for example, the profiling of carbohydrate-binding proteins [32], characterization of carbohydrate-cell recognition [29•], as well as the detection of specific antibodies for disease diagnosis [33]. The virus receptor specificity of influenza viruses, based on the species-specific nature of the interaction between the virus and host glycans, has also been characterized by glycan microarray technology [34]. Seeberger and coworkers prepared carbohydrate slides to identify HIV vaccine candidate antigens [35], detect pathogenic bacteria [36], and determine the binding profile of heparin-binding proteins [37]. Instead of intact glycans ‘neoglycolipids’, oligosaccharides covalently attached to lipid molecules have been used in glycan microarrays [38]. Their advantage is that they are easier to immobilize by hydrophobic forces on the chemical surface. These glycans can react with lectins, anti-glycan mAbs, carbohydrate–binding cytokines or chemokines [38]. Wang et al demonstrated that microbial polysaccharides could be immobilized on a surface-modified glass slide without chemical conjugation, thereby allowing simultaneous detection of a broad spectrum of antibody specificities with as little as a few microliters of serum [39].
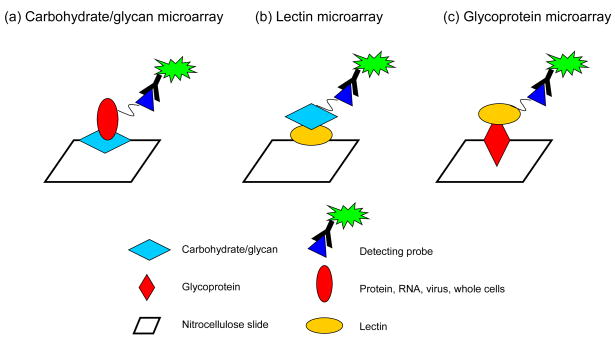
Figure 1. Different array formats for glycosylation studies.
(a) Carbohydrate/glycan microarrays. Multiple different glycan structures are arrayed on slides and probed with a variety of biological molecules to determine glycans’ interaction characteristics. (b) Lectin microarrays. A variety of lectins are spotted on array surfaces to assess their binding affinities as well as to distinguish the binding characteristics of the lectins with different probing media. (c) Glycoprotein microarrays. Antibodies or naturally occurring glycoproteins are arrayed on slides which are then probed with lectins to assess the glycan structures on these proteins.
The ability to test arrays of glycans in a single experiment would clearly increase the rate at which lectin can be characterized. There are approximately 100 lectins with known structures, but only half of their binding specificities are known. Glycan arrays have been used to confirm the binding specificity of lectins and a comprehensive database of glycan binding proteins is currently available from the Consortium of Functional Glycomics, a large research initiative aimed at understanding the role of carbohydrate-protein interactions in cell-cell communication [40]. The sugar-binding activity and specificity of the interactions in novel proteins can be determined using glycan arrays [31]. Despite their involvement in various cellular processes and ubiquitous distribution in biological systems, relatively little is known about the function of specific carbohydrates. This lack of understanding is mainly caused by the challenges of their chemical synthesis as well as the purification and characterization of complex sugars.
Lectin microarrays
Alterations in carbohydrate structure of glycoproteins are known to be related to cellular regulation and tumor growth. Although carbohydrate arrays yield valuable data on carbohydrate-interacting proteins, they do not allow direct assessment of changes in glycosylation. Other technologies available for glycan analysis, such as chromatography and MS, tend to be time consuming and less suitable for high-throughput and systematic evaluation of protein glycosylation. Although detailed information on glycan structures may be obtained using MS, a higher level of expertise is required for the time-consuming data analysis. For these reasons, the use of lectins in an array format has gained increased interest (Figure 1b). Studies utilizing lectin arrays have focused on assessing the specificity of lectin binding to carbohydrates and have been useful in determining the most appropriate lectins for glycoprotein enrichment as well as for the removal of undesirable glycoproteins [41]. Lectin microarrays can also provide a rapid and simple analysis of protein glycosylation, although all glycans in a complex sample can not be detected and complete structure assignment is not provided. This technology is based on binding of intact glycoproteins or glycopeptides to the arrayed lectins, resulting in a characteristic fingerprint that is highly sensitive to changes in a protein’s glycan composition. Hsu et al described a lectin array protocol for high-throughput evaluation of cell-surface microbial sugars [41]. The binding patterns of fluorescent bacteria to these arrays provided a simple method to fingerprint bacteria based on their surface carbohydrates. A beneficial aspect of lectin arrays is that the large number of lectins, each with their specific recognition pattern, is highly sensitive to changes in the glycosylation pattern [42]. Yamada and Hirabayashi and colleagues have developed a lectin microarray procedure based on an evanescent-field fluorescence detection principle, allowing sensitive, real-time observation of multiple lectin-carbohydrate interactions [43,44]. Quantitative measurements of lectin-carbohydrate interactions and specific signal patterns for various cyanine 3-labeled glycoproteins, glycopeptides and tetramethylrhodamine-labeled oligosaccharides were determined using this platform [43,44]. Rosenfeld et al have also described a lectin array-based method using Qproteome GlycoArray kits for rapid analysis of glycosylation profiles of glycoproteins [45].
Although lectin arrays and carbohydrate arrays provide valuable data on carbohydrate-interacting proteins and lectin-glycoprotein interactions, they do not allow whole glycoproteomes to be screened in a manner that enables both changes in an individual protein’s glycan expression within that glycoproteome and changes in overall glycoprotein patterns to be investigated.
Glycoprotein microarrays
Protein microarrays can be utilized to screen whole cell lysates [46], fractionated proteomes and antigen-antibody reactions [47]. Unlike oligonucleotides, proteins are broadly heterogeneous in size, shape and chemistry. To optimize the quality of data obtained, choosing the surface with best signal-to-noise ratio is critical. Glass-coated surfaces with specific chemical functionalities are popular as background fluorescence is eliminated [48]. Deposition of proteins on the surface, such as an epoxide-coated surface, results in covalent linkage of the proteins to the glass. Nitrocellulose slides are among the most popular substrates because of their low cost; the ultra-thin nitrocellulose layer provides a lower fluorescence background, while maintaining the binding capacity [48]. Angeloni et al developed thin film-coated photoactivatable surfaces (dextran-coated glass slides) suitable for covalent immobilization of glycans, glycoconjugates, and lectins in microarray formats [49]. Standard glycoproteins were covalently immobilized and exposed glycans were successfully profiled with lectins for fucose, sialic acid and galactose. These platforms were also suitable for glycans and lectin immobilization, which demonstrates the feasibility of such microarray platforms for biomolecule binding and illustrates the versatility of microarray-based tools for different applications [49].
Antibodies have also been utilized to target and study cancer-associated carbohydrates, such as Lewis blood-group structures [50,51]. Chen et al developed a method to allow the efficient, multiplexed study of glycans on individual proteins from complex mixtures using antibody microarray capture of multiple proteins followed by detection with lectins or glycan-binding antibodies [52•]. Following capture of specific proteins from biological samples by immobilized antibodies, a biotinylated lectin was used to bind glycans on the captured proteins. Chemical derivatization of the glycans on the spotted antibodies prevented lectin binding to those glycans. By profiling both protein and glycan variation in multiple samples using parallel sandwich and glycan-detection assays, cancer-associated glycan alterations were identified on the proteins mucin 1 and carcinoembryonic antigen in the serum of patients with pancreatic cancer. The multiplexed detection of glycans on specific proteins allowed the use of small sample amounts and high-throughput sample processing [52•].
One promising strategy is the use of multidimensional fractionation techniques to simplify cell lysates into less complex fractions, which can be used to produce natural glycoprotein microarrays (Figure 1c) [19•]. Briefly, cellular proteins from a cancer cell line are first resolved by iso-electric point-based fractionation using chromatofocusing or isoelectric focusing. Each fraction is then separated using reversed-phase (RP)-HPLC; the fractionated proteins are lyophilized, resuspended in a suitable buffer, and printed onto a nitrocellulose-based microarray. The protein microarray is then screened using sera or a modification-specific detection reagent. This technique was used to study the humoral response and identify potential serum biomarkers for prostate cancer; specific fractions were immunoreactive against prostate cancer serum, but not against serum from healthy individuals [46].
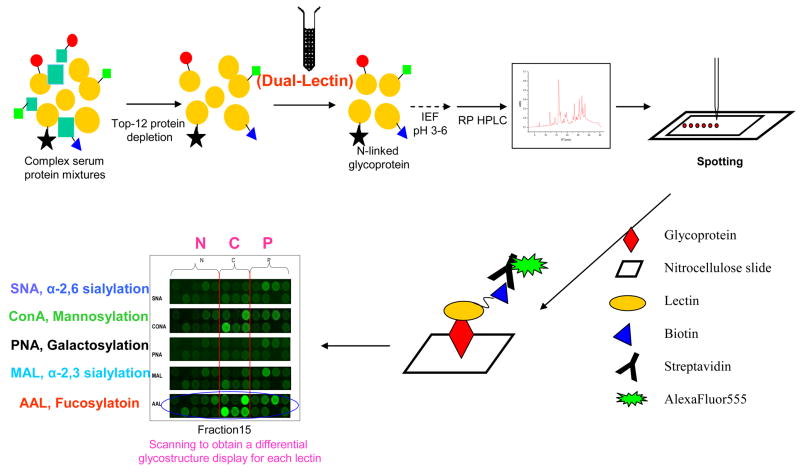
Based on the natural protein microarray approach, a method for global analysis of glycosylation patterns and detection of glycosylation alterations in cancer serum has been developed [19•]. This strategy used an all-liquid phase enrichment and pre-fractionation methodology coupled to glycoprotein microarray technology and a multiple lectin-based, biotin-streptavidin detection scheme. Selective detection of glycan structures was made possible by utilizing multiple lectins to screen serum samples for glycoproteins from normal individuals or patients with chronic pancreatitis or pancreatic cancer. The general strategy is shown in Figure 2, where a complex serum sample was first depleted of the top twelve most abundant proteins in human serum, using an antibody column, to facilitate detection of proteins present in lower concentrations. Glycoproteins were then enriched from depleted fractions using a dual-lectin column that contained lectin ConA and WGA. ConA recognizes N-linked mannose, including high-mannose-type and mannose core structures, and WGA recognizes terminal N-acetylglucosamine (N-GlcNAc). By using these two broad specificity lectins, the entire pool of glycoproteins could potentially be isolated. The glycoprotein pool was further fractionated by one-dimensional or two-dimensional liquid phase separation, such as isoelectric focusing, coupled with RP-HPLC. The purified glycoproteins were then spotted onto nitrocellulose slides and probed with multiple biotinylated lectins followed by streptavidin conjugated to a fluorescent tag. The five lectins used and their specificities are listed in Table 1. Both maackia amurensis lectin (MAL) and sambucus nigra lectin (SNA) recognize sialic acid on the terminal branches. While SNA binds preferentially to sialic acid attached to terminal galactose in an (α-2,6) and, to a lesser degree, an (α-2,3) linkage, MAL detects glycans containing N-acetylneuraminic acid-Gal-N-GlcNAc with sialic acid at the 3-position of galactose. In contrast, peanut agglutinin binds desialylated exposed galactosyl (α-1,3) N-acetylgalactosamine. Aleuria aurantia lectin (AAL) recognizes fucose linked (α-1,6) to N-GlcNAc or (α-1,3) to N-acetyllactosamine. The combination of these five lectins covers the majority of N-linked glycan types identified and differentiates them according to their specific structures. The printed glycoproteins were incubated with biotinylated lectins for binding and the bound biotinylated lectins were subsequently detected with streptavidin conjugated to AlexaFluor555. The biotin-streptavidin interaction is highly specific and the use of a sandwich-type detection scheme improved the signal to-noise ratio significantly.
Figure 2. Experimental strategy for the study of serum glycoproteins.
Proteins in serum samples are first depleted (top-12 protein depletion) to remove abundant proteins. The sample is then enriched for N-linked glycoproteins using a dual-lectin affinity column. Glycoproteins are resolved on a non-porous silica reversed-phase high-pressure liquid chromatography (RP-HPLC) column. Resulting fractions are arrayed or spotted onto nitrocellulose slides via a non-contact piezoelectric arrayer, after which the slides are probed for specific glycan structures utilizing biotinylated lectins on a glycoprotein microarray. Scanned images of the lectin-glycoprotein interaction are visualized by secondary detection with streptavidin conjugated to a fluorescent probe. AAL aleuria aurantia, ConA concanavalin A, IEF isoelectric focusing, MAL maackia amurensis II, PNA peanut agglutinin, SNA sambucus nigra (elderberry) bark.
(Reprinted with permission from Zhao J, Patwa TH, Qiu W, Shedden K, Hinderer R, Misek DE, Anderson MA, Simeone DM, Lubman DM: Glycoprotein microarrays with multi-lectin detection: Unique lectin binding patterns as a tool for classifying normal, chronic pancreatitis and pancreatic cancer sera. J Proteome Res (2007) 6(5):1864–1874. © 2007 American Chemical Society) [53].
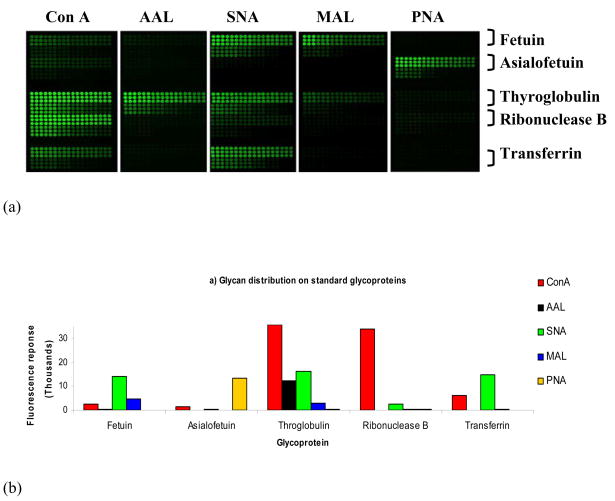
The specificity, reproducibility and sensitivity of this strategy were assessed using five standard glycoproteins: fetuin, asialofetuin, thyroglobulin, ribonuclease B, and transferrin [19•]. The images obtained when slides were probed with each of the lectins are shown in Figure 3a [19•]. These data correlated well with reported glycan structures corresponding to the standard glycoproteins used in this study (Figure 3b) [19•]. Detection sensitivity was approximately 5 fentomoles and the variance was within 10% as determined by nine replicate spotting events for each standard protein. This method has been applied to the glycosylation profiling of sera from patients with pancreatic cancer, colon cancer and esophageal cancer [53,54]. Data shown in Figure 4 demonstrate its utility for biomarker detection in pancreatic cancer [53].
Figure 3. Glycoprotein microarray proof-of-concept experiment utilizing glycoprotein standards.
(A) Scanned images of printed standard glycoproteins (AAL aleuria aurantia, ConA concanavalin A, MAL maackia amurensis II, PNA peanut agglutinin, SNA sambucus nigra [elderberry] bark) probed with various lectins [19•]. Each section contains a standard glycoprotein stated to the right of the section in a dilution series from 2 mg/ml to 0.025 mg/ml. Each dilution was printed as 9 replicates in a 3 × 3 block. (B) Glycan distribution (x-axis) on standard glycoproteins printed at a concentration of 1 mg/ml. Levels are represented by fluorescence response on the y-axis.
(Reprinted with permission from Patwa TH, Zhao J, Anderson MA, Simeone DM, Lubman DM: Screening of glycosylation patterns in serum using natural glycoprotein microarrays and multi-lectin fluorescence detection. Anal Chem (2006) 78(18):6411-6421. © 2007 American Chemical Society).[19•]
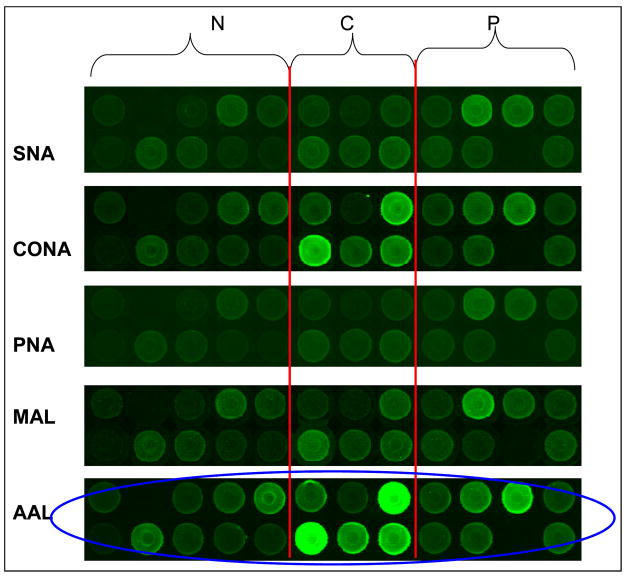
Figure 4. Sample microarray image demonstrating quality of lectin response.
Sections of a glycoprotein microarray in which a peak obtained from non-porous silica reversed-phase high-pressure liquid chromatography column separation is compared across all 24 samples [53]. The lectin probes are shown on the left side of the panel and each panel represents a section from similarly printed arrays. The predominant protein modifications identified in this fraction are mannosylation and fucosylation. AAL aleuria aurantia, ConA concanavalin A, MAL maackia amurensis II, PNA peanut agglutinin, SNA sambucus nigra (elderberry) bark.
(Reprinted with permission from Zhao J, Patwa TH, Qiu W, Shedden K, Hinderer R, Misek DE, Anderson MA, Simeone DM, Lubman DM: Glycoprotein microarrays with multi-lectin detection: Unique lectin binding patterns as a tool for classifying normal, chronic pancreatitis and pancreatic cancer sera. J Proteome Res (2007) 6(5):1864-1874 © 2007 American Chemical Society) [53].
Figure 4 shows a scanned image of the levels of glycosylation detected by the 5 lectins in an HPLC fraction derived from 10 normal, 8 pancreatitis and 6 pancreatic cancer serum samples [53]. The normalized glycoprotein microarray responses to lectins were visualized by principal component analysis (PCA). Figure 5 shows the response to AAL lectin, which binds to fucosylation sites and generally distinguishes the three clinical groups. Pancreatic cancer samples clustered further away from normal samples compared with chronic pancreatitis, especially in response to fucosylation and sialylation. Altered glycosylation patterns were observed in individual proteins such as hemopexin, antithrombin III and kininogen 1 in pancreatic cancer sera (p < 0.05). The advantage of this strategy is that the differential response can be tracked back to the original fraction from which the spot originated. Combined with chromatographic separation, MS, and other characterization techniques, the target glycome could potentially be fully defined. The altered glycosylation was determined using a combination of Western blotting and MS. The results from peptide mapping and Western blotting experiments confirmed the observations made in the glycoprotein microarray experiments. For example, elevated sialylation of a specific glycosylation site was detected on Hp-related protein by peptide mass mapping and increased fucosylation was detected by the lectin-blotting experiment [53]. These data are consistent with the microarray study in which both elevated levels of fucosylation and sialylation were observed in pancreatic cancer serum.
Figure 5. Principal component analysis data illustrating similarity between samples used.
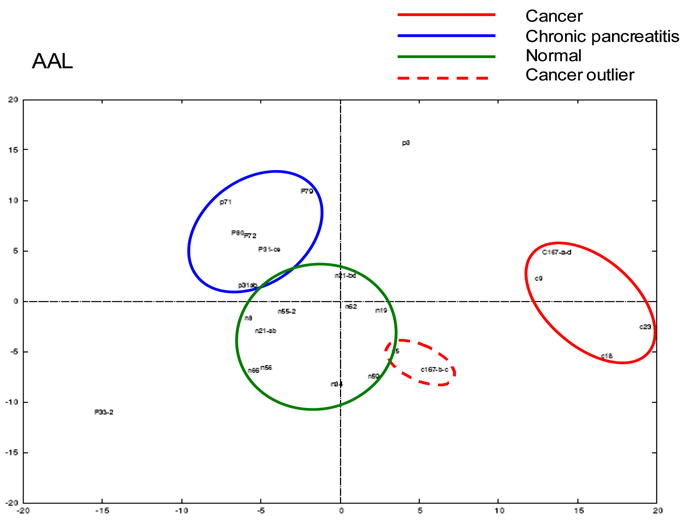
Principal component analysis was used to visualize the normalized glycoprotein microarray responses to the lectin AAL [53]. The following serum samples were evaluated: 6 pancreatic cancers, 8 chronic pancreatitis and 10 normal. AAL aleuria aurantia.
(Reprinted with permission from Zhao J, Patwa TH, Qiu W, Shedden K, Hinderer R, Misek DE, Anderson MA, Simeone DM, Lubman DM: Glycoprotein microarrays with multi-lectin detection: Unique lectin binding patterns as a tool for classifying normal, chronic pancreatitis and pancreatic cancer sera. J Proteome Res (2007) 6(5):1864-1874. © 2007 American Chemical Society) [53].
Therefore, protein glycosylation screening analysis may allow the detection of alterations in protein glycosylation in samples from patients with different clinical conditions. This includes not only alterations in absolute protein levels, but, importantly, changes in PTMs, such as glycosylation, which could suggest the presence or absence of disease. The ability to rapidly and sensitively screen protein glycosylation patterns and detect glycosylation alterations may provide an efficient method to screen patients for cancer, such as pancreatic cancer and possibly other cancers.
Conclusions
The microarray system enables the investigation of various types of binding events. The advantage of microarray technology is attributed to the high-throughput, highly miniaturized platform and the small amount of sample required for the investigation of a broad range of possible binding partners. Therefore, this technique is highly suited for probe-based profiling of multiple samples. Alterations in the carbohydrate structure of glycoproteins can be detected by probing the arrayed carbohydrate or glycoprotein with lectins or antibodies. Lectin/antibody microarrays provide rapid glycosylation pattern analysis with moderate structural information, but are limited by the number of lectins or antibodies available. As a new discovery tool for biomarkers and a potential diagnostic tool, glycoprotein microarrays provide the ability to screen a multitude of naturally occurring glycosylations in a single experiment. Furthermore, this method is not limited by the complexities of carbohydrate synthesis and the availability or high cost of antibodies. A variety of applications and platform designs of microarrays are currently under development and additional improvements in selectivity, reproducibility and sensitivity are expected to enable improved characterization of glycosylation and related biological effects and lead to the discovery of novel biomarkers.
Acknowledgments
This work was supported in part by National Institutes of Health grants R01GM49500, R21CA124441 and R01CA106402 (DML), and a Michigan Economic Development Grant MEDC03-622 (DMS).
References
•• of outstanding interest
• of special interest
- 1•.Bertozzi CR, Kiessling LL. Chemical glycobiology. Science. 2001;291(5512):2357–2364. doi: 10.1126/science.1059820. This review provides a detailed explanation of the importance of glycobiology in the modulation of key cellular processes. It also describes chemically how such processes can be probed using synthetic strategies and how they can be applied for potential therapeutic-type applications. [DOI] [PubMed] [Google Scholar]
- 2.Rudd PM, Elliott T, Cresswell P, Wilson IA, Dwek RA. Glycosylation and the immune system. Science. 2001;291(5512):2370–2376. doi: 10.1126/science.291.5512.2370. [DOI] [PubMed] [Google Scholar]
- 3.Osawa T, Tsuji T. Fractionation and structural assessment of oligosaccharides and glycopeptides by use of immobilized lectins. Annu Rev Biochem. 1987;56:21–42. doi: 10.1146/annurev.bi.56.070187.000321. [DOI] [PubMed] [Google Scholar]
- 4.Durand G, Seta N. Protein glycosylation and diseases: Blood and urinary oligosaccharides as markers for diagnosis and therapeutic monitoring. Clin Chem. 2000;46(6):795–805. [PubMed] [Google Scholar]
- 5.Zhao J, Simeone DM, Heidt D, Anderson MA, Lubman DM. Comparative serum glycoproteomics using lectin selected sialic acid glycoproteins with mass spectrometric analysis: Application to pancreatic cancer serum. J Proteome Res. 2006;5(7):1792–1802. doi: 10.1021/pr060034r. [DOI] [PubMed] [Google Scholar]
- 6.Peracaula R, Royle L, Tabares G, Mallorqui-Fernández G, Barrabés S, Harvey DJ, Dwek RA, Rudd PM, de Llorens R. Glycosylation of human pancreatic ribonuclease: Differences between normal and tumor states. Glycobiology. 2003;13(4):227–244. doi: 10.1093/glycob/cwg019. [DOI] [PubMed] [Google Scholar]
- 7.Zhao J, Qiu W, Simeone DM, Lubman DM. N-linked glycosylation profiling of pancreatic cancer serum using capillary liquid phase separation coupled with mass spectrometric analysis. J Proteome Res. 2007;6(3):1126–1138. doi: 10.1021/pr0604458. [DOI] [PubMed] [Google Scholar]
- 8.Kobata A, Amano J. Altered glycosylation of proteins produced by malignant cells, and application for the diagnosis and immunotherapy of tumours. Immunol Cell Biol. 2005;83 (4):429–439. doi: 10.1111/j.1440-1711.2005.01351.x. [DOI] [PubMed] [Google Scholar]
- 9.Dube DH, Bertozzi CR. Glycans in cancer and inflammation – potential for therapeutics and diagnostics. Nat Rev Drug Discov. 2005;4(6):477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 10.An HJ, Miyamoto S, Lancaster KS, Kirmiz C, Li B, Lam KS, Leiserowitz GS, Lebrilla CB. Profiling of glycans in serum for the discovery of potential biomarkers for ovarian cancer. J Proteome Res. 2006;5(7):1626–1635. doi: 10.1021/pr060010k. [DOI] [PubMed] [Google Scholar]
- 11.Peracaula R, Tabarés G, Royle L, Harvey DJ, Dwek RA, Rudd PM, de Llorens R. Altered glycosylation pattern allows the distinction between prostate-specific antigen (PSA) from normal and tumor origins. Glycobiology. 2003;13(6):457–470. doi: 10.1093/glycob/cwg041. [DOI] [PubMed] [Google Scholar]
- 12.Kasbaoui L, Harb J, Bernard S, Meflah K. Differences in glycosylation state of fibronectin from two rat colon carcinoma cell lines in relation to tumoral progressiveness. Cancer Res. 1989;49 (19):5317–5322. [PubMed] [Google Scholar]
- 13.Kellokumpu S, Sormunen R, Kellokumpu I. Abnormal glycosylation and altered Golgi structure in colorectal cancer: Dependence on intra-golgi pH. FEBS Lett. 2002;516(1–3):217–224. doi: 10.1016/s0014-5793(02)02535-8. [DOI] [PubMed] [Google Scholar]
- 14.Ng RC, Roberts AN, Wilson RG, Latner AL, Turner GA. Analyses of protein extracts of human breast cancers: Changes in glycoprotein content linked to the malignant phenotype. Br J Cancer. 1987;55(3):249–254. doi: 10.1038/bjc.1987.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15• •.Orntoft TF, Vestergaard EM. Clinical aspects of altered glycosylation of glycoproteins in cancer. Electrophoresis. 1999;20(2):362–371. doi: 10.1002/(SICI)1522-2683(19990201)20:2<362::AID-ELPS362>3.0.CO;2-V. A variety of changes to O- and N-linked glycoproteins observed in cancer cells are described. Mechanisms by which glycoproteins interact and can be potentially controlled are also discussed. [DOI] [PubMed] [Google Scholar]
- 16.Duffy MJ. CA 19-9 as a marker for gastrointestinal cancers: A review. Ann Clin Biochem. 1998;35 (Pt 3):364–370. doi: 10.1177/000456329803500304. [DOI] [PubMed] [Google Scholar]
- 17.Ferretti G, Felici A, Papaldo P, Fabi A, Cognetti F. HER2/neu role in breast cancer: From a prognostic foe to a predictive friend. Curr Opin Obstet Gynecol. 2007;19(1):56–62. doi: 10.1097/GCO.0b013e328012980a. [DOI] [PubMed] [Google Scholar]
- 18.Demetriou M, Nabi IR, Coppolino M, Dedhar S, Dennis JW. Reduced contact-inhibition and substratum adhesion in epithelial cells expressing GlcNAc-transferase V. J Cell Biol. 1995;130(2):383–392. doi: 10.1083/jcb.130.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19•.Patwa TH, Zhao J, Anderson MA, Simeone DM, Lubman DM. Screening of glycosylation patterns in serum using natural glycoprotein microarrays and multi-lectin fluorescence detection. Anal Chem. 2006;78(18):6411–6421. doi: 10.1021/ac060726z. Initial studies in which naturally occurring glycoprotein arrays were probed with lectins to detect changes in glycosylation that may be occurring as a function of disease were described. The experimental design is of particular interest as novel changes could potentially be detected using the approach; such changes may not be highlighted in an antibody array if an antibody of a potentially novel candidate is not present. [DOI] [PubMed] [Google Scholar]
- 20.Orntoft TF, Greenwell P, Clausen H, Watkins WM. Regulation of the oncodevelopmental expression of type 1 chain ABH and Lewis(b) blood group antigens in human colon by α-2-L-fucosylation. Gut. 1991;32(3):287–293. doi: 10.1136/gut.32.3.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ang IL, Poon TC, Lai PB, Chan AT, Ngai SM, Hui AY, Johnson PJ, Sung JJ. Study of serum haptoglobin and its glycoforms in the diagnosis of hepatocellular carcinoma: A glycoproteomic approach. J Proteome Res. 2006;5(10):2691–2700. doi: 10.1021/pr060109r. [DOI] [PubMed] [Google Scholar]
- 22.Burchell JM, Mungul A, Taylor-Papadimitriou J. O-linked glycosylation in the mammary gland: Changes that occur during malignancy. J Mammary Gland Biol Neoplasia. 2001;6(3):355–364. doi: 10.1023/a:1011331809881. [DOI] [PubMed] [Google Scholar]
- 23.Muller S, Hanisch FG. Recombinant MUC1 probe authentically reflects cell-specific O-glycosylation profiles of endogenous breast cancer mucin. High density and prevalent core 2-based glycosylation. J Biol Chem. 2002;277(29):26103–26112. doi: 10.1074/jbc.M202921200. [DOI] [PubMed] [Google Scholar]
- 24.Dall’Olio F, Chiricolo M. Sialyltransferases in cancer. Glycoconj J. 2001;18(11–12):841–850. doi: 10.1023/a:1022288022969. [DOI] [PubMed] [Google Scholar]
- 25.Drake RR, Schwegler EE, Malik G, Diaz J, Block T, Mehta A, Semmes OJ. Lectin capture strategies combined with mass spectrometry for the discovery of serum glycoprotein biomarkers. Mol Cell Proteomics. 2006;5(10):1957–1967. doi: 10.1074/mcp.M600176-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Yang Z, Harris LE, Palmer-Toy DE, Hancock WS. Multilectin affinity chromatography for characterization of multiple glycoprotein biomarker candidates in serum from breast cancer patients. Clin Chem. 2006;52(10):1897–1905. doi: 10.1373/clinchem.2005.065862. [DOI] [PubMed] [Google Scholar]
- 27••.Qiu R, Regnier FE. Comparative glycoproteomics of N-linked complex-type glycoforms containing sialic acid in human serum. Anal Chem. 2005;77(22):7225–7231. doi: 10.1021/ac050554q. This study described an emerging technique that used glycoprotein enrichment and separation in the liquid phase. This facilitated easy coupling to MS, the method of choice to characterize glycopeptides and glycoproteins. [DOI] [PubMed] [Google Scholar]
- 28•.Madera M, Mechref Y, Novotny MV. Combining lectin microcolumns with high-resolution separation techniques for enrichment of glycoproteins and glycopeptides. Anal Chem. 2005;77(13):4081–4090. doi: 10.1021/ac050222l. This study described the use of lectin columns to isolate proteins with specific glycan structures. The column characteristics and the use of HPLC in such enrichment and high-resolution separation are described in great depth. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29•.Nimrichter L, Gargir A, Gortler M, Altstock RT, Shtevi A, Weisshaus O, Fire E, Dotan N, Schnaar RL. Intact cell adhesion to glycan microarrays. Glycobiology. 2004;14(2):197–203. doi: 10.1093/glycob/cwh022. This study described one method in which glycoprotein function can be investigated using a microarray approach. In particular, glycan arrays were used to monitor cellular adhesion. Extension of this method to CD4+ human T-cells on a 45-glycan diversity array revealed interesting adhesion patterns to very specific Lewis structures. [DOI] [PubMed] [Google Scholar]
- 30.Wang D, Lu J. Glycan arrays lead to the discovery of autoimmunogenic activity of SARS-CoV. Physiol Genomics. 2004;18(2):245–248. doi: 10.1152/physiolgenomics.00102.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feizi T, Fazio F, Chai W, Wong CH. Carbohydrate microarrays - a new set of technologies at the frontiers of glycomics. Curr Opin Struct Biol. 2003;13(5):637–645. doi: 10.1016/j.sbi.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 32.Huang CY, Thayer DA, Chang AY, Best MD, Hoffmann J, Head S, Wong CH. Carbohydrate microarray for profiling the antibodies interacting with Globo H tumor antigen. Proc Natl Acad Sci USA. 2006;103(1):15–20. doi: 10.1073/pnas.0509693102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manimala JC, Li Z, Jain A, VedBrat S, Gildersleeve JC. Carbohydrate array analysis of anti-Tn antibodies and lectins reveals unexpected specificities: Implications for diagnostic and vaccine development. Chembiochem. 2005;6(12):2229–2241. doi: 10.1002/cbic.200500165. [DOI] [PubMed] [Google Scholar]
- 34.Stevens J, Blixt O, Paulson JC, Wilson IA. Glycan microarray technologies: Tools to survey host specificity of influenza viruses. Nat Rev Microbiol. 2006;4(11):857–864. doi: 10.1038/nrmicro1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams EW, Ratner DM, Bokesch HR, McMahon JB, O’Keefe BR, Seeberger PH. Oligosaccharide and glycoprotein microarrays as tools in HIV glycobiology; glycan-dependent gp120/protein interactions. Chem Biol. 2004;11(6):875–881. doi: 10.1016/j.chembiol.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 36.Disney MD, Magnet S, Blanchard JS, Seeberger PH. Aminoglycoside microarrays to study antibiotic resistance. Angew Chem Int Ed Engl. 2004;43(12):1591–1594. doi: 10.1002/anie.200353236. [DOI] [PubMed] [Google Scholar]
- 37.de Paz JL, Noti C, Seeberger PH. Microarrays of synthetic heparin oligosaccharides. J Am Chem Soc. 2006;128(9):2766–2767. doi: 10.1021/ja057584v. [DOI] [PubMed] [Google Scholar]
- 38.Feizi T. Progress in deciphering the information content of the ‘glycome’ - a crescendo in the closing years of the millennium. Glycoconj J. 2000;17(7–9):553–565. doi: 10.1023/a:1011022509500. [DOI] [PubMed] [Google Scholar]
- 39.Wang D, Liu S, Trummer BJ, Deng C, Wang A. Carbohydrate microarrays for the recognition of cross-reactive molecular markers of microbes and host cells. Nat Biotechnol. 2002;20 (3):275–281. doi: 10.1038/nbt0302-275. [DOI] [PubMed] [Google Scholar]
- 40.Nature CFG. Functional glycomics gateway: Consortium for functional glycomics. The Scripps Research Institute; La Jolla, CA, USA: 2008. www.functionalglycomics.org/static/consortium/consortium.shtml. [Google Scholar]
- 41.Hsu KL, Mahal LK. A lectin microarray approach for the rapid analysis of bacterial glycans. Nat Protoc. 2006;1(2):543–549. doi: 10.1038/nprot.2006.76. [DOI] [PubMed] [Google Scholar]
- 42.Pilobello KT, Krishnamoorthy L, Slawek D, Mahal LK. Development of a lectin microarray for the rapid analysis of protein glycopatterns. Chembiochem. 2005;6(6):985–989. doi: 10.1002/cbic.200400403. [DOI] [PubMed] [Google Scholar]
- 43.Kuno A, Uchiyama N, Koseki-Kuno S, Ebe Y, Takashima S, Yamada M, Hirabayashi J. Evanescent-field fluorescence-assisted lectin microarray: A new strategy for glycan profiling. Nat Methods. 2005;2(11):851–856. doi: 10.1038/nmeth803. [DOI] [PubMed] [Google Scholar]
- 44.Uchiyama N, Kuno A, Koseki-Kuno S, Ebe Y, Horio K, Yamada M, Hirabayashi J. Development of a lectin microarray based on an evanescent-field fluorescence principle. Methods Enzymol. 2006;415:341–351. doi: 10.1016/S0076-6879(06)15021-1. [DOI] [PubMed] [Google Scholar]
- 45.Rosenfeld R, Bangio H, Gerwig GJ, Rosenberg R, Aloni R, Cohen Y, Amor Y, Plaschkes I, Kamerling JP, Maya RB. A lectin array-based methodology for the analysis of protein glycosylation. J Biochem Biophys Methods. 2007;70(3):415–426. doi: 10.1016/j.jbbm.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 46.Yan F, Sreekumar A, Laxman B, Chinnaiyan AM, Lubman DM, Barder TJ. Protein microarrays using liquid phase fractionation of cell lysates. Proteomics. 2003;3(7):1228–1235. doi: 10.1002/pmic.200300443. [DOI] [PubMed] [Google Scholar]
- 47.Gembitsky DS, Lawlor K, Jacovina A, Yaneva M, Tempst P. A prototype antibody microarray platform to monitor changes in protein tyrosine phosphorylation. Mol Cell Proteomics. 2004;3 (11):1102–1118. doi: 10.1074/mcp.M400075-MCP200. [DOI] [PubMed] [Google Scholar]
- 48.Tomizaki KY, Usui K, Mihara H. Protein-detecting microarrays: Current accomplishments and requirements. Chembiochem. 2005;6(5):782–799. doi: 10.1002/cbic.200400232. [DOI] [PubMed] [Google Scholar]
- 49.Angeloni S, Ridet JL, Kusy N, Gao H, Crevoisier F, Guinchard S, Kochhar S, Sigrist H, Sprenger N. Glycoprofiling with micro-arrays of glycoconjugates and lectins. Glycobiology. 2005;15 (1):31–41. doi: 10.1093/glycob/cwh143. [DOI] [PubMed] [Google Scholar]
- 50.Hanisch FG, Hanski C, Hasegawa A. Sialyl Lewis(x) antigen as defined by monoclonal antibody AM-3 is a marker of dysplasia in the colonic adenoma-carcinoma sequence. Cancer Res. 1992;52(11):3138–3144. [PubMed] [Google Scholar]
- 51.Lucka L, Fernando M, Grunow D, Kannicht C, Horst AK, Nollau P, Wagener C. Identification of Lewis x structures of the cell adhesion molecule CEACAM1 from human granulocytes. Glycobiology. 2005;15(1):87–100. doi: 10.1093/glycob/cwh139. [DOI] [PubMed] [Google Scholar]
- 52•.Chen S, Laroche T, Hamelinck D, Bergsma D, Brenner D, Simeone D, Brand RE, Haab BB. Multiplexed analysis of glycan variation on native proteins captured by antibody microarrays. Nat Methods. 2007;4(5):437–444. doi: 10.1038/nmeth1035. This study described the use of an antibody array to highlight critical glycosylation differences in cancer. Such an experimental scheme is particularly interesting because both protein abundance as well as specific glycan changes can be monitored on the same microarray by using difference detection schemes. [DOI] [PubMed] [Google Scholar]
- 53.Zhao J, Patwa TH, Qiu W, Shedden K, Hinderer R, Misek DE, Anderson MA, Simeone DM, Lubman DM. Glycoprotein microarrays with multi-lectin detection: Unique lectin binding patterns as a tool for classifying normal, chronic pancreatitis and pancreatic cancer sera. J Proteome Res. 2007;6(5):1864–1874. doi: 10.1021/pr070062p. [DOI] [PubMed] [Google Scholar]
- 54.Qiu Y, Patwa TH, Xu L, Shedden K, Misek DE, Tuck M, Jin G, Ruffin MT, Turgeon DK, Synal S, Bresalier R, et al. Plasma glycoprotein profiling for colorectal cancer biomarker identification by lectin glycoarray and lectin blot. J Proteome Res. 2008;7(4):1693–1703. doi: 10.1021/pr700706s. [DOI] [PMC free article] [PubMed] [Google Scholar]