Abstract
Microtubules are nucleated in vivo by γ-tubulin complexes. The 300 kDa γ-tubulin small complex (γTuSC), consisting of two molecules of γ-tubulin and one copy each of the accessory proteins Spc97p and Spc98p, is the conserved, essential core of the microtubule nucleating machinery1,2. In metazoa multiple γTuSCs assemble with other proteins into γ-tubulin ring complexes (γTuRCs). The structure of γTuRC suggested that it functions as a microtubule template2–5. Because each γTuSC contains two molecules of γ-tubulin, it was assumed that the γTuRC-specific proteins are required to organize γTuSCs to match thirteen-fold microtubule symmetry. Here, we show that γTuSC forms rings even in the absence of other γTuRC components. The yeast adaptor protein Spc110p stabilizes the rings into extended filaments and is required for oligomer formation under physiological buffer conditions. The 8Å cryo-EM reconstruction of the filament reveals thirteen γ-tubulins per turn, matching microtubule symmetry, with plus ends exposed for interaction with microtubules, implying that one turn of the filament constitutes a microtubule template. The domain structures of Spc97p and Spc98p suggest functions for conserved sequence motifs, with implications for the γTuRC-specific proteins. The γTuSC filaments nucleate microtubules at a low level, and the structure provides a strong hypothesis for how nucleation is regulated, converting this less active form to a potent nucleator.
Microtubules assembled in vitro have a broad distribution of protofilament numbers centred around fourteen6. However, microtubules nucleated in cells have mostly thirteen protofilaments7, suggesting that γ-tubulin complexes constrain microtubule geometry. Thirteen-fold symmetry is likely preferred as it allows the protofilaments to run straight along the microtubule (as opposed to being twisted in other protofilament symmetries), allowing motor proteins tracking processively to remain on one face of the microtubule. It has generally been assumed that γTuRC-specific proteins form a cap-like structure that establishes thirteen-fold symmetry by providing a scaffold for γTuSC assembly. How γTuSC is organized in organisms like Saccharomyces cerevisiae, which lack all of the γTuRC-specific proteins, has remained an open question.
The sequence and structural similarity between γ- and α/β-tubulin suggested that nucleation results from microtubule-like contacts between γ- and α/β-tubulin8. The microtubule lattice consists of lateral and longitudinal contacts9,10; the stronger longitudinal contacts define microtubule polarity, with “plus” and “minus” ends (Supplementary Fig. 1). The template model for microtubule nucleation predicts that γ-tubulins interact laterally to form a ring which makes longitudinal contacts with α/β-tubulin2–5 minus ends; alternative models that predict lateral interactions between γ- and α/β-tubulin11 have not been definitively ruled out.
Our previous 25 Å structure of Saccharomyces γTuSC was determined in buffer conditions that yielded predominantly monomeric complexes12,13. Here, we show that buffer conditions that promote microtubule growth (BRB80: low salt, pH 6.9) also promote spontaneous assembly of γTuSCs into rings similar to Drosophila γTuRCs2,4 (Fig. 1a,b). Ring formation was sensitive to both salt and pH (Supplementary Fig. 2a–c). The γTuSC rings bound microtubules, and many of these microtubule ends are capped (Fig. 1c), similar to microtubules nucleated in vivo14,15 or from γTuRCs in vitro3,4,16. Spontaneous assembly suggests that ring formation is an intrinsic property of γTuSC, and not dependent on γTuRC-specific proteins.
Figure 1. γTuSC oligomers form spontaneously and are stabilized by Spc110p.
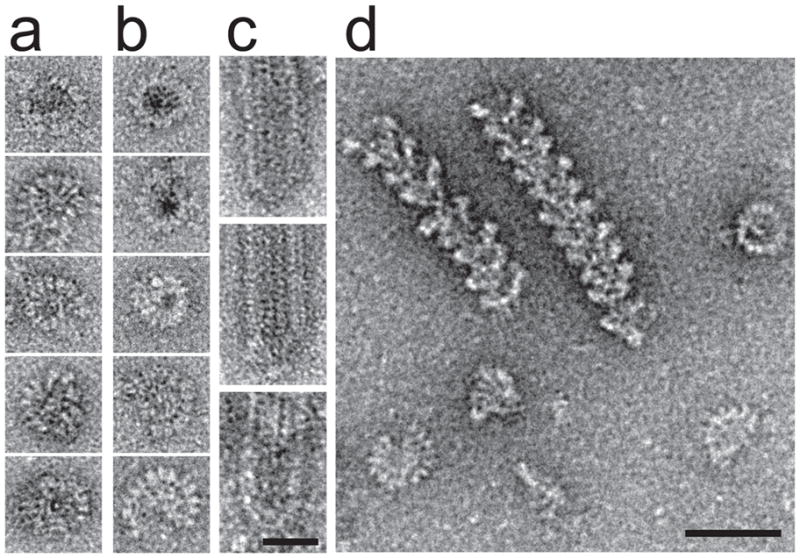
a) Ring-like structures were observed in negative stain electron micrographs of Saccharomyces γTuSC at pH 6.9. b) γTuRC purified from Drosophila embryos is similar in shape and size to the γTuSC rings. c) Capped ends were observed on microtubules grown in the presence of pre-formed γTuSC rings. Scalebar for a, b, and c, 25 nm. d) Negative stain images of γTuSC filaments formed upon copurification with Spc110p1-220. Scalebar, 50 nm.
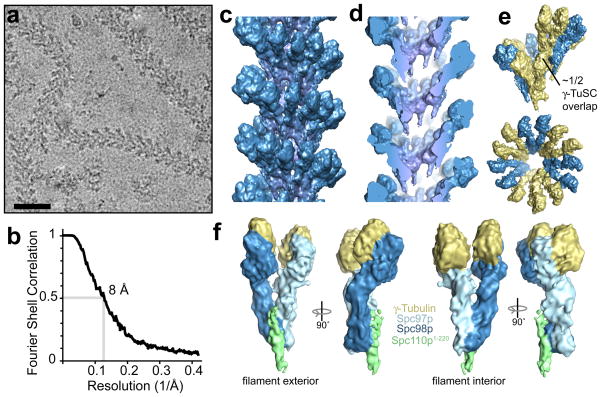
The N-terminal 220 residues of Spc110p (Spc110p1-220), which attaches γTuSC to the nuclear face of the yeast spindle pole body, dramatically increased the stability and length of γTuSC assemblies (Fig. 1d). Copurification with Spc110p1-220 yielded a continuum of γTuSC oligomers ranging from dimers to long, well-ordered helical filaments, even under conditions where γTuSC alone fails to assemble (Supplementary Fig. 2d–f). We determined the three-dimensional structure of γTuSC filaments from cryo-electron micrographs (Fig. 2a), using iterative helical real space reconstruction, a single particle approach to helical structure determination17. The resolution of the reconstruction, which included about 25,000 γTuSC subunits, was estimated at 8 Å by the Fourier shell correlation 0.5 cutoff (Fig. 2b).
Figure 2. γTuSC filament structure.
a) Cryo-electron micrograph of γTuSC filaments. Scalebar, 50 nm. b) The resolution of the structure is estimated at 8 Å by the Fourier shell correlation. c) A segment of the reconstructed filament filtered to 8 Å. d) A cutaway view of the filament, illustrating the lack of connection between helical layers. e) One turn of the helix, coloured by γTuSC. The filament has six and a half γTuSCs per turn, with a half γTuSC overlap. f) A single γTuSC/Spc110p1-220 subunit from the filament, with the approximate boundaries between the individual proteins indicated by colour.
The filament is a single spiral of laterally associated γTuSCs, without contact between layers (Fig. 2c,d; Supplementary Movie 1). The helical symmetry (54.3° rotation and 22.2 Å rise per subunit) gives rise to just over six and a half γTuSCs – or thirteen γ-tubulins – per turn, with a half γTuSC overlap. Each turn of helix forms a lock-washer shape similar to γTuRC4 (Fig. 2e). The thirteen-fold γ-tubulin symmetry of the filament is dictated largely by the extensive lateral interactions between Spc97p and Spc98p of adjacent γTuSCs (Figure 2e, Supplementary Fig. 6a), locking in the lateral tubulin contacts which on their own are flexible enough to accommodate a range of different symmetries. We propose that a γTuSC assembly very similar to a single ring from the filament provides the constraint that limits microtubules to 13 protofilaments in all eukaryotes in vivo7.
γTuSC in the filament is remarkably similar to free γTuSC13,18 (Fig. 2f, Supplementary Fig. 3), indicating that oligomerization does not induce large scale conformational changes. The 8 Å structure provides new insight into the domain architecture of Spc97p and Spc98p. Spc97p and Spc98p dimerize at their N-terminal ends nearest the helical axis, and have extended central domains connecting to C-terminal γ-tubulin binding domains. The central domain of Spc98p is kinked, at a position previously shown to be the site of limited hinge-like flexibility13. The masses of the domains determined from the cryo-EM map provide a rough estimate of their boundaries in each sequence, indicating the positions of the grip1 and grip2 motifs, conserved in all γ-tubulin complex proteins19 (Supplementary Fig. 4a,b). The grip2 motif covers nearly half of the C-terminal domains, strongly suggesting that it is important for γ-tubulin binding. The grip1 motif is in the central domain, near inter-γTuSC contacts and the kink in Spc98p. We tentatively assign Spc110p1-220 to a ridge of density running along the exterior face of γTuSC in the filament, making contacts primarily with Spc98p (Supplementary Fig. 3). The resolution of the reconstruction appears to be nonuniform, as tubes of alpha helical density are clear in the N-terminal domains of Spc97p and Spc98p at the core of the structure, while secondary structure features are not well defined in the peripheral density where γ-tubulin is located (Supplementary Fig. 5). The lower effective resolution in the γ-tubulin regions may be due to limited flexibility in the weak connections between the central and C-terminal domains of Spc97p and Spc98p.
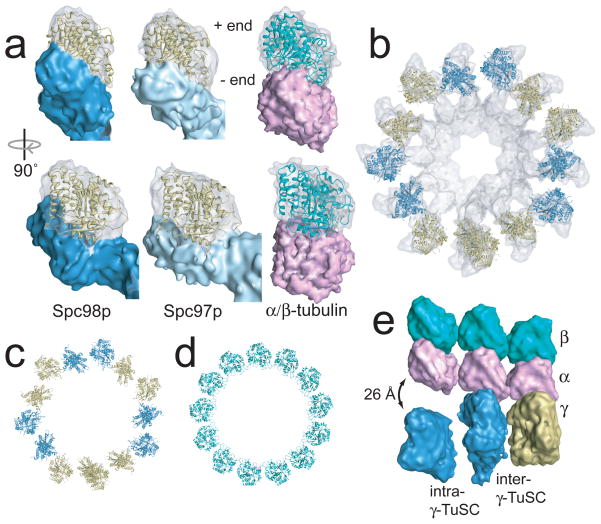
The human γ-tubulin crystal structure20 was fit into the density in the regions previously assigned to γ-tubulin13 (Fig. 3a,b). The minus end longitudinal surface of γ-tubulin is completely buried in the interface with Spc97p/Spc98p. The lateral contacts between γ-tubulins of neighbouring γTuSCs are nearly identical to microtubule lateral contacts. The two γ-tubulins within each γTuSC are skewed slightly apart, in a configuration incompatible with the microtubule lattice (Fig 3b,c), as observed in the free γTuSC structure13. This symmetry gives rise to an alternating pattern of γ-tubulin pairs with microtubule-like lateral spacing separated by gaps, generating a staggered mismatch with the microtubule lattice (Fig. 3c–e). The only microtubule lattice surface of γ-tubulin fully exposed in the filament is the plus end face, favouring a model in which γ-tubulin makes longitudinal contacts with α/β-tubulin. This, combined with the thirteen-fold γ-tubulin symmetry, provides the strongest evidence to date to support a γ-tubulin template mechanism for microtubule nucleation.
Figure 3. γ-tubulin in the γTuSC filament has a geometry similar to 13-protofilament microtubules.
a) The EM density of γ-tubulin (transparent) and the Spc97p/Spc98p binding domains (opaque) are shown with the γ-tubulin crystal structure fit in the density. The density is tilted relative to the helical axis so that in each case the plus end is vertical. An α/β-tubulin heterodimer is shown with simulated EM density for comparison. b) Thirteen γ-tubulins fit into one turn of the filament. Neighbouring γ-tubulins in the same γTuSC are the same colour. c) The γ-tubulin symmetry is similar to the symmetry of a 13-protofilament microtubule (d), but separated intra-γTuSC γ-tubulins alternate with contacting inter-γTuSC γ-tubulins. e) To illustrate the mismatch between geometries laterally-interacting α/β-tubulin heterodimers were aligned to the filament so that the central α/β-tubulin makes longitudinal contacts to γ-tubulin.
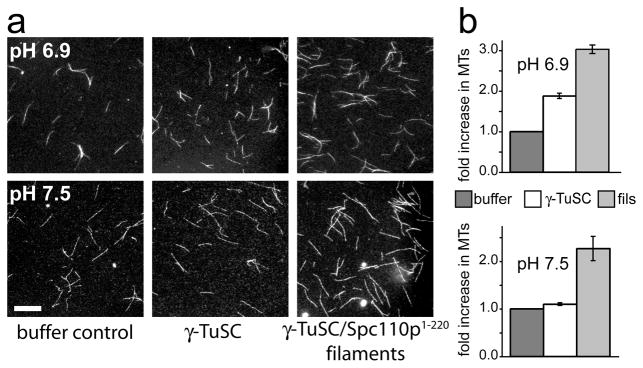
We tested the capacity of γTuSC oligomers to nucleate microtubules (Fig. 4). At pH 6.9 both γTuSC alone and γTuSC filaments provided modest levels of nucleation, slightly higher for γTuSC filaments than γTuSC alone. At pH 7.5 γTuSC alone does not nucleate microtubules, whereas the filaments retain a low level of nucleation. As γTuSC rings do not form at pH 7.5, but γTuSC filaments remain intact (Supplementary Fig. 2b,c), these results suggest that γTuSC nucleation activity is assembly-dependent. The levels of nucleation observed are consistent with previous measurements for γTuSC2,21, but less robust than seen with γTuRC2,5, suggesting that assembly alone is insufficient to fully activate γTuSC nucleating activity. The arrangement of γ-tubulin in γTuSC oligomers provides a structural explanation for their relatively modest nucleating activity. Nucleation likely arises from the inter-γTuSC γ-tubulin pairs, which have the correct microtubule lattice spacing. Simulations indicate that a γTuSC assembly in which all of the γ-tubulins make lateral microtubule-like contacts would provide greatly enhanced nucleation (L. Rice, personal communication).
Figure 4. γTuSC oligomers nucleate microtubules at low levels.
a) Fluorescence micrographs of rhodamine labelled microtubules assembled at pH 6.9 or 7.5 in the presence of buffer, γTuSC, or γTuSC/Spc110p1-220 filaments (final γTuSC concentration 150 nM). Scalebar, 5 μM. b) The mean number of microtubules per field is shown for nucleation at pH 6.9 (n=5) and pH 7.5 (n=3). Error bars represent the standard error of the mean.
The structure provides a clear hypothesis for how nucleation could be fully activated. We previously predicted that bending at the flexible kink in Spc98p is required to bring the intra-γTuSC γ-tubulins to the microtubule spacing13,18. We speculated that γTuSC assembly might drive this change, but that clearly is not the case – a similar conformational change is still required in γTuSC rings. In the lower resolution γTuSC structure we predicted that the movement would be a closure of the gap between γ-tubulins; here we see that the movement must be more perpendicular to the edge of the ring, bringing γ-tubulin in toward the helical axis. A rotation of 23° about the kink in Spc98p would reposition γ-tubulin by 26 Å, bringing it to the microtubule lattice spacing (Supplementary Fig. 7, Movies 2 and 3). The staggered γ-tubulin arrangement likely serves a regulatory function, maintaining γTuSC oligomers in a low activity state until a signal (protein binding, post-translational modification, etc.) directs the rearrangements in Spc98p necessary to form a template with exact microtubule lattice geometry. Although less likely, rearrangement of the γ-tubulins could be induced by binding of α/β-tubulin. In such a model γTuSC would function primarily as a cap for stabilizing and localizing microtubule minus ends, rather than as a strong nucleator.
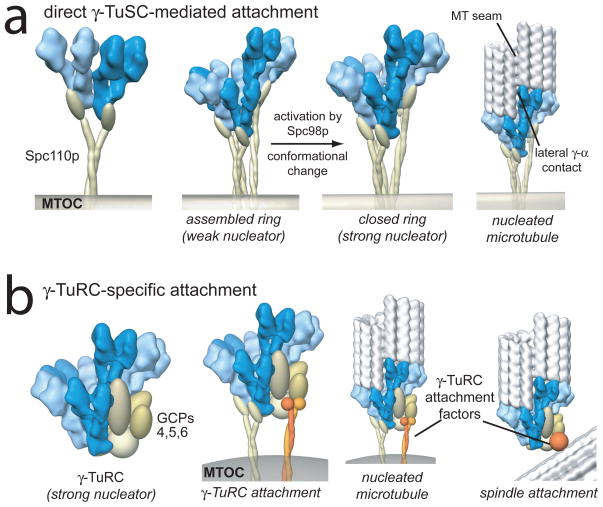
The dramatic enhancement of γTuSC oligomer stability by Spc110p, combined with its role in γTuSC localization, likely serves to ensure that microtubule template assembly in yeast occurs only at the spindle pole body. We propose a general model for microtubule nucleation in which Spc110p or its functional equivalent directly attaches γTuSC to microtubule organizing centres, promoting template assembly. A subsequent activation step then fully activates nucleation by rearranging the γ-tubulin network (Fig. 5a). In a template with seven γTuSCs, the location of the half γTuSC overlap defines the position of the 13 protofilament microtubule seam; a single lateral contact between γ-tubulin and α-tubulin would be made at the overlap, as well. It is unclear how many γTuSCs are required to nucleate a microtubule – an incomplete ring may be sufficient to initiate growth.
Figure 5. Models of nucleation complex attachment and activation.
a) In the absence of γTuRC-specific components, as in Saccharomyces, Spc110p, or its equivalent, directly attaches γTuSC to microtubule organizing centres, promoting ring assembly. We hypothesize a conformational change in Spc98p promotes nucleation by rearranging γ-tubulin into an exact microtubule template. b) In organisms with complete γTuRCs, active complexes attach to organizing centres directly via γTuSCs, or potentially through unique sites in the γTuRC-specific components. Localization of γTuRCs at non-MTOC locations, for example within the mitotic spindle, is mediated through the γTuRC-specific proteins. In both scenarios, γTuSC interactions define the geometry of the nucleating template.
The γTuSC filament structure provides unique insight into the roles of γTuRC-specific proteins. Our results clearly show that γTuSC assembly alone establishes thirteen-fold γ-tubulin symmetry, indicating that the γTuRC-specific proteins are not required as a scaffold. This is consistent with the observation that all of the γTuRC-specific proteins can be depleted without affecting centrosomal microtubule nucleation of thirteen protofilament microtubules 22,23. While not required as a scaffold, the γTuRC-specific proteins may serve to stabilize the ring structure or fully activate nucleation activity, and they are essential for γTuRC localization at non-centrosomal sites, as in augmin-dependent binding within the mitotic spindle24 (Fig. 5b).
We suggested above that the grip1 and grip2 motifs, conserved in all the γTuRC-specific proteins, are involved in ring assembly contacts and γ-tubulin binding, respectively. This raises the intriguing possibility that the γTuRC-specific proteins may each bind γ-tubulin and substitute for Spc97p or Spc98p in the ring itself. To do this, they might form hybrid γTuSCs with one of the γTuRC-specific proteins plus Spc97p or Spc98p, alternative γTuSCs with two different γTuRC-specific proteins, or unique half γTuSCs (Supplementary Fig. 8). Such alternative γTuSCs might serve to initiate or terminate γTuSC oligomerization, or to stabilize the ring at the overlapping ends, while providing unique attachment sites in the structure of the ring itself.
Methods Summary
γTuSC was co-expressed with GST-Spc110p1-220 as described13,21, except that complexes were eluted by cleavage of the GST tag with TEV protease as the final purification step. γTuSC rings were formed by 30 minute incubation on ice after dilution to 0.2 μM in BRB80 (80 mM PIPES pH 6.9, 1mM EGTA, 1mM MgCl2). Nucleation assays were performed essentially as described21. The cryo-EM reconstruction was performed essentially as described by Egelman17 and Sasche, et al.25.
Supplementary Material
Acknowledgments
We are grateful to M. Braunfeld and A. Avila-Sakar for microscopy assistance; Y. Cheng, E. Muller, M. Moritz and K. Huang for helpful discussions; and B. Carragher, C. Potter and J. Quispe for the use of their EM facilities and technical assistance with data collection. Some of the work presented here was conducted at the National Resource for Automated Molecular Microscopy which is supported by the National Institutes of Health though the National Center for Research Resources’ P41 program. This work was supported by the National Institutes of Health (D.A.A. and T.N.D.) and the Howard Hughes Medical Institute (D.A.A.). J.M.K. was a NIH Ruth L. Kirschstein National Research Service Award (NRSA) postdoctoral fellow.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions J.M.K. purified and prepared samples for EM, collected cryo-EM data, determined the structure, and performed microtubule nucleation experiments. J.K.P. explored γ-TuSC assembly conditions and prepared and imaged capped microtubules. A.Z. designed and cloned expression constructs, and generated and tested baculovirus strains. D.A.A and J.M.K. designed experiments and analyzed data. J.M.K., D.A.A. and T.N.D. wrote the paper. All the authors discussed the results and commented on the manuscript.
Author Information The cryo-EM reconstruction has been deposited with the Electron Microscopy Database with the accession code 1731. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Knop M, Schiebel E. Spc98p and Spc97p of the yeast gamma-tubulin complex mediate binding to the spindle pole body via their interaction with Spc110p. Embo J. 1997;16 (23):6985–6995. doi: 10.1093/emboj/16.23.6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oegema K, et al. Characterization of two related Drosophila gamma-tubulin complexes that differ in their ability to nucleate microtubules. J Cell Biol. 1999;144 (4):721–733. doi: 10.1083/jcb.144.4.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keating TJ, Borisy GG. Immunostructural evidence for the template mechanism of microtubule nucleation. Nat Cell Biol. 2000;2 (6):352–357. doi: 10.1038/35014045. [DOI] [PubMed] [Google Scholar]
- 4.Moritz M, Braunfeld MB, Guenebaut V, Heuser J, Agard DA. Structure of the gamma-tubulin ring complex: a template for microtubule nucleation. Nat Cell Biol. 2000;2 (6):365–370. doi: 10.1038/35014058. [DOI] [PubMed] [Google Scholar]
- 5.Zheng Y, Wong ML, Alberts B, Mitchison T. Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature. 1995;378 (6557):578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]
- 6.Chretien D, Wade RH. New data on the microtubule surface lattice. Biol Cell. 1991;71 (1–2):161–174. doi: 10.1016/0248-4900(91)90062-r. [DOI] [PubMed] [Google Scholar]
- 7.Evans L, Mitchison T, Kirschner M. Influence of the centrosome on the structure of nucleated microtubules. J Cell Biol. 1985;100 (4):1185–1191. doi: 10.1083/jcb.100.4.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oakley BR, Oakley CE, Yoon Y, Jung MK. Gamma-tubulin is a component of the spindle pole body that is essential for microtubule function in Aspergillus nidulans. Cell. 1990;61 (7):1289–1301. doi: 10.1016/0092-8674(90)90693-9. [DOI] [PubMed] [Google Scholar]
- 9.Nogales E, Wolf SG, Downing KH. Structure of the alpha beta tubulin dimer by electron crystallography. Nature. 1998;391 (6663):199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- 10.Nogales E, Whittaker M, Milligan RA, Downing KH. High-resolution model of the microtubule. Cell. 1999;96 (1):79–88. doi: 10.1016/s0092-8674(00)80961-7. [DOI] [PubMed] [Google Scholar]
- 11.Erickson HP. Gamma-tubulin nucleation: template or protofilament? Nat Cell Biol. 2000;2 (6):E93–96. doi: 10.1038/35014084. [DOI] [PubMed] [Google Scholar]
- 12.Aldaz H, Rice LM, Stearns T, Agard DA. Insights into microtubule nucleation from the crystal structure of human gamma-tubulin. Nature. 2005;435 (7041):523–527. doi: 10.1038/nature03586. [DOI] [PubMed] [Google Scholar]
- 13.Kollman JM, et al. The structure of the gamma-tubulin small complex: implications of its architecture and flexibility for microtubule nucleation. Mol Biol Cell. 2008;19 (1):207–215. doi: 10.1091/mbc.E07-09-0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Byers B, Shriver K, Goetsch L. The role of spindle pole bodies and modified microtubule ends in the initiation of microtubule assembly in Saccharomyces cerevisiae. J Cell Sci. 1978;30:331–352. doi: 10.1242/jcs.30.1.331. [DOI] [PubMed] [Google Scholar]
- 15.Moritz M, et al. Three-dimensional structural characterization of centrosomes from early Drosophila embryos. J Cell Biol. 1995;130 (5):1149–1159. doi: 10.1083/jcb.130.5.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiese C, Zheng Y. A new function for the gamma-tubulin ring complex as a microtubule minus-end cap. Nat Cell Biol. 2000;2 (6):358–364. doi: 10.1038/35014051. [DOI] [PubMed] [Google Scholar]
- 17.Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85 (4):225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 18.Choy RM, Kollman JM, Zelter A, Davis TN, Agard DA. Localization and orientation of the gamma-tubulin small complex components using protein tags as labels for single particle EM. J Struct Biol. 2009;168 (3):571–574. doi: 10.1016/j.jsb.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gunawardane RN, et al. Characterization and reconstitution of Drosophila gamma-tubulin ring complex subunits. J Cell Biol. 2000;151 (7):1513–1524. doi: 10.1083/jcb.151.7.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rice LM, Montabana EA, Agard DA. The lattice as allosteric effector: structural studies of alphabeta- and gamma-tubulin clarify the role of GTP in microtubule assembly. Proc Natl Acad Sci U S A. 2008;105 (14):5378–5383. doi: 10.1073/pnas.0801155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vinh DB, Kern JW, Hancock WO, Howard J, Davis TN. Reconstitution and characterization of budding yeast gamma-tubulin complex. Mol Biol Cell. 2002;13 (4):1144–1157. doi: 10.1091/mbc.02-01-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goshima G, et al. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316 (5823):417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verollet C, et al. Drosophila melanogaster gamma-TuRC is dispensable for targeting gamma-tubulin to the centrosome and microtubule nucleation. J Cell Biol. 2006;172 (4):517–528. doi: 10.1083/jcb.200511071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goshima G, Mayer M, Zhang N, Stuurman N, Vale RD. Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J Cell Biol. 2008;181 (3):421–429. doi: 10.1083/jcb.200711053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sachse C, et al. High-resolution electron microscopy of helical specimens: a fresh look at tobacco mosaic virus. J Mol Biol. 2007;371 (3):812–835. doi: 10.1016/j.jmb.2007.05.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohi M, Li Y, Cheng Y, Walz T. Negative Staining and Image Classification -Powerful Tools in Modern Electron Microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quispe J, et al. An improved holey carbon film for cryo-electron microscopy. Microsc Microanal. 2007;13 (5):365–371. doi: 10.1017/S1431927607070791. [DOI] [PubMed] [Google Scholar]
- 28.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142 (3):334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 29.Frank J. Three-Dimensional Electron Microscopy of Macromolecular Assemblies. Academic Press, Inc; San Diego: 1996. [Google Scholar]
- 30.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25 (13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.