Abstract
Graft-versus-host disease (GVHD) is a frequent complication after hematopoietic cell transplant (HCT). Tissue damage as a result of chemo-radiation injury is the initiating event in the pathogenesis of acute GVHD. Variations in DNA repair can influence the amount of tissue damage in response to alkylating agents and ionizing radiation used as conditioning during HCT. Since DNA damage caused by these agents is repaired by the Base Excision Repair (BER) pathway, we hypothesized that single nucleotide polymorphisms (SNPs) in BER pathway will be associated with GVHD after HCT. Hence, we analyzed 179 SNPs in BER pathway in 470 recipients of allogeneic HCT for association with acute and chronic GVHD. In multivariate analysis, one SNP (rs6844176) in RFC1 gene was independently associated with a higher risk of grade II-IV acute GVHD (RR:1.39, 95% CI:1.14-1.70, p=0.001), and showed a trend towards higher risk of grade III-IV acute GVHD (RR:1.33, 95% CI:0.95-1.85, p=0.09). One SNP in PARP1 gene (rs1805410) was associated with a higher risk of chronic GVHD (RR:1.81, 95% CI:1.29-2.54, p=0.001). These results show that SNPs in the BER pathway can be used as genetic biomarkers to predict those at high risk for GVHD towards whom novel prophylactic strategies could be targeted.
Keywords: graft-versus-host disease, polymorphism, SNP, BER, DNA repair, GVHD
Introduction
Graft versus host disease (GVHD) remains a major cause of morbidity and mortality post hematopoietic cell transplant (HCT). The pathophysiology of acute GVHD involves priming of immune response by pre-transplant conditioning leading to tissue damage and induction of cytokine release, T cell activation and co-stimulation, alloreactive T cell expansion, differentiation and trafficking and destruction of target tissues by effector T cells (1). The pathophysiology of chronic GVHD is more complex. Four theories have been postulated based on experimental studies including thymic damage and defective negative selection of T cells generated from marrow progenitors after HCT, aberrant production of transforming growth factor-ß, auto-antibody production, and deficiency of T-regulatory cells (2).
The main mechanism of action of alkylator based conditioning regimens and ionizing radiation is induction of DNA damage. This damage is repaired by the DNA repair pathways (3). We have recently demonstrated that single nucleotide polymorphisms (SNPs) in the Base Excision Repair (BER) pathway are significantly associated with transplant related mortality post allogeneic transplant (4). We further hypothesized that SNPs in the BER pathway can lead to altered repair (after conditioning induced tissue damage) and altered T cell function (secondary to thymic damage) and may be one of the pathways implicated in the pathophysiology of GVHD. The BER pathway was selected as it plays an important role in repairing damage caused by ionizing radiation and alkylating agents commonly used in conditioning regimens for HCT. We analyzed 179 SNPs in 27 genes in the BER pathway in 470 recipients of allogeneic HCT using alkylating agents and ionizing radiation based conditioning regimes for hematologic malignancies at University of Minnesota, for association with acute and chronic GVHD.
Patients and Methods
All consecutive patients who underwent an allogeneic HCT from an HLA-identical sibling donor, matched or mismatched unrelated donor (URD) or single umbilical cord blood (UCB) transplant for hematological malignancies from January 1, 1998 to December 31, 2007 were eligible (n=587). All patients received a conditioning regimen including an alkylating agent and/or ionizing radiation. DNA samples were available for 480 (82%) recipients. This study was approved by the Institutional Review Board at the University of Minnesota and waiver of informed consent from individual patients was obtained for this study. Demographic and transplant related information regarding all patients undergoing a HCT is prospectively entered in the University of Minnesota Bone Marrow Transplant Database. This was supplemented with chart reviews where necessary. This included patient and donor age at transplant, patient and donor gender, type of donor (related, URD or UCB), graft source (bone marrow versus peripheral blood stem cell versus cord blood), race, patient’s underlying disease, disease status at transplant, HLA matching between donor and recipient, conditioning intensity (myeloablative or reduced intensity), conditioning regimen used for transplant, GVHD prophylaxis, cytomegalovirus (CMV) serological status of recipient and donor prior to transplant, date of diagnosis and grade of acute GVHD, date of diagnosis and development of chronic GVHD, malignancy relapse and survival at time of last follow up. HLA-matching status for URD transplants was categorized as well matched, partially matched or mismatched based on classification proposed by Weisdorf et al (5). HLA-matching status for cord blood transplants was based on antigen level HLA-A, B and allele level HLA-DRB1 typing.
DNA samples from patients were obtained from peripheral blood samples collected prior to undergoing HCT. All patients with acute leukemia had no evidence of leukemic blasts at the time of obtaining blood for DNA extraction. DNA was extracted from buffy coat using either the Puregene DNA extraction method (for samples collected prior to 2001) (Gentra Systems, Minneapolis, MN) or the Qiagen Mini Blood kit (all samples collected during or after 2001) (Qiagen Inc., San Jose, CA) as per manufacturer recommendations. All DNA samples were collected and stored at 4°C in the Molecular Diagnostics Laboratory at University of Minnesota Medical Center, Fairview prior to use in the study.
SNP selection and genotyping
We used data from Environmental Genome Project (6,7) to identify and select Linkage Disequilibrium (LD) tagSNPs that had a frequency of ≥ 5% in the Caucasian population in 27 BER genes. If genes did not have tagSNPs information in the Environmental Genome Project, we used HapMap data for Caucasian populations to identify tagSNPs that had ≥5% frequency in Caucasian populations using the Genome Variation Server software (http://gvs.gs.washington.edu/GVS/). We then supplemented this list with non-synonymous SNPs that were identified to be functionally important using three software programs; PolyPhen (8), SIFT (9) and SNP3D (10). This approach allowed us to select 263 SNPs (210 tagSNPs and 53 SNPs that may be functionally important) in 27 genes with known function in the BER pathway. All DNA samples were genotyped using the Sequenom iPLEX system at the Biomedical Genomics Center at University of Minnesota. All primers and probes were designed using Sequenom primer design software. We were able to design primers and probes for 239 SNPs. Of these, 179 SNPs were successfully genotyped after assuring quality control by selecting only SNPs with call rates >80%. High quality genotyping was also assured by only selecting samples with a concordance rate of 95% or greater (using 61 pairs of blinded duplicates) for further analysis. Hence 179 SNPs and 470 samples were analyzed. Among SNPs that were successfully genotyped (n=179) 80% were tagSNPs (n=143) and 20% were functionally important SNPs (n=36). Detailed description of number of SNPs genotyped within individual genes is given in table 1.
Table 1. Completion rates for genes in the base excision repair pathway.
| Gene | SNPs with genotyping assays designed (N = 239) |
SNPs without genotyping assays designed (N=24) |
Total SNPs (N=263) |
|
|---|---|---|---|---|
| SNPS with genotype (N=179) (%) |
SNPs without genotype (N=60) (%) |
|||
| ADPRT | 14 (100) | 0 (0) | 2 | 16 |
| APEX1 | 1 (25) | 3 (75) | 0 | 4 |
| FEN1 | 0 (0) | 3 (100) | 3 | 6 |
| LIG1 | 21 (88) | 3 (12) | 1 | 25 |
| LIG3 | 4 (80) | 1 (20) | 0 | 5 |
| MBD4 | 6 (75) | 2 (25) | 0 | 8 |
| MPG | 8 (80) | 2 (20) | 0 | 10 |
| MUTYH | 8 (100) | 0 (0) | 0 | 8 |
| NTHL1 | 1 (100) | 0 (0) | 1 | 2 |
| OGG1 | 5 (83) | 1 (17) | 2 | 8 |
| PCNA | 2 (50) | 2 (50) | 0 | 4 |
| PNKp | 4 (100) | 0 (0) | 1 | 5 |
| POLb | 4 (80) | 1 (20) | 0 | 5 |
| POLd1 | 8 (100) | 0 (0) | 0 | 8 |
| POLd2 | 4 (100) | 0 (0) | 0 | 4 |
| POLe | 15 (75) | 5 (25) | 1 | 21 |
| POLi | 6 (46) | 7 (54) | 0 | 13 |
| POLl | 2 (75) | 1 (25) | 2 | 5 |
| RCF5 | 9 (64) | 5 (36) | 0 | 14 |
| RFC1 | 10 (63) | 6 (37) | 4 | 20 |
| RFC2 | 2 (40) | 3 (60) | 0 | 5 |
| RFC3 | 6 (67) | 3 (33) | 1 | 10 |
| RFC4 | 4 (50) | 4 (50) | 0 | 8 |
| SMUG1 | 6 (86) | 1 (14) | 0 | 7 |
| TDG | 15 (88) | 2 (12) | 2 | 19 |
| UNG | 3 (75) | 1 (25) | 0 | 4 |
| XRCC1 | 11 (73) | 4 (27) | 4 | 19 |
Statistical Analysis
All statistical analyses were performed using SAS software version 9.1 (SAS Institute, Inc., Cary, NC) and R Statistical software version 2.4.1 (11). Minor allele frequencies (MAF) and Hardy Weinberg proportions of SNPs genotyped were estimated. Linkage disequilibrium among SNPs in the RFC1 gene was estimated using Haploview software (12). Assuming an additive model for the SNPs (where each SNP was treated as a continuous variable with the homozygous major, heterozygote and homozygous minor genotypes indicating 0, 1 or 2 copies of the minor allele being present), the association between each of these SNPs and acute and chronic GVHD was evaluated using a proportional hazards model with death as the competing risk as described by Fine and Gray (13) after adjustment for clinical covariates. Covariates included recipient age, race, donor type, diagnosis, disease status at transplant, gender mismatch, CMV serostatus of recipient and donor, stem cell source, conditioning regimen (myeloablative or reduced intensity) and GVHD prophylaxis. A p value ≤ 0.01 was considered to be significant to select a SNP for multivariate analysis and individual SNPs that were significant were evaluated in a backward stepwise multiple regression model after adjustment for clinical covariates.
One hundred and thirty five of 355 patients who were alive by day 100 developed chronic GVHD and were analyzed in a similar manner. Grade II-IV acute GVHD was evaluated as a time dependant covariate in the model for chronic GVHD.
Results
Table 2 shows the demographic and transplant characteristics of the study population. The majority underwent myeloablative HCT from an HLA-identical sibling donor (n=231), 72% of the cohort were ≥ 21 years at the time of transplant and 86% of the cohort were Caucasian. Acute leukemia (acute myeloid leukemia and acute lymphoid leukemia) (51%) was the most frequent diagnosis. Among 75 URD recipients, 27 (36%) of recipients received a transplant from a well matched donor, 36 (48%) from a partially matched donor and 10 (13%) from a mismatched donor. Insufficient data was available to categorize two patients. Among 99 UCB recipients, HLA-matching status for UCB transplants was based on antigen level HLA-A, B and allele level HLA-DRB1 typing. Of these, 47% were mismatched at a single locus (5/6), 43% were mismatched at 2 loci (4/6) and 9% received a 6/6 matched transplant.
Table 2. Demographic and transplant characteristics.
| Variable | N (%) |
|---|---|
| N | 470 (100) |
| Recipient Age at Transplant (Years) | |
| <20 | 133 (28) |
| 21 – 40 | 109 (23) |
| >40 | 228 (49) |
| Median age (range), years | 35.7 (0.7 - 69.8) |
| Race | |
| African American | 14(3) |
| Asian | 18(4) |
| Caucasian | 404(86) |
| Hispanic | 13(3) |
| Native American | 4(1) |
| Unknown | 10(2) |
| Mixed | 7(1) |
| Diagnosis | |
| Acute lymphoid leukemia | 92 (20) |
| Acute myeloid leukemia | 151 (31) |
| Chronic myeloid leukemia | 73 (16) |
| Lymphoma | 78 (16) |
| Myelodysplastic syndrome | 40 (9) |
| Others | 36 (8) |
| Disease Status at Transplant | |
| Complete response | 288 (62) |
| Partial response | 24(5) |
| Relapse / primary induction failure | 118 (25) |
| Myelodysplastic syndrome | 38 (8) |
| Missing | 2 (<1) |
| CMV Serological status | |
| Recipient positive / donor positive | 101 (22) |
| Recipient positive / donor negative | 146 (31) |
| Recipient negative / donor positive | 62 (13) |
| Recipient negative / donor negative | 161 (34) |
| Conditioning | |
| Myeloablative | 378 (80) |
| Reduced Intensity | 92 (20) |
| Donor Type | |
| Sibling | 296 (63) |
| Unrelated donor | 75 (16) |
| Cord blood | 99 (21) |
| Stem Cell Source | |
| Marrow | 145 (31) |
| PBSC | 226 (48) |
| Cord blood | 99 (21) |
| GVHD Prophylaxis | |
| CSA/MMF | 125 (27) |
| CSA/MTX | 244 (52) |
| T-cell depletion | 43 (9) |
| Other | 58 (12) |
|
Follow up in years (Survivors), median
(range) |
3.3 (0.9-8.6) |
Abbreviations CMV: cytomegalovirus; PBSC: peripheral blood stem cell; CSA: cyclosporine; MMF: mycophenolate mofetil; MTX: methotrexate
The cumulative incidence of grade II-IV acute GVHD was 40.6% (95% CI 35.8-45.4%) and the cumulative incidence of chronic GVHD was 34% (95% CI 28.8-39.3%). The overall survival was 48.5% (95% CI 43.8-53.2%) at three years and 44.4% (95% CI 39.4-49.5%) at five years.
Association between grade II-IV and grade III-IV acute GVHD and BER SNPs
Univariate analysis was performed by entering each SNP in a model adjusted for all clinical covariates. In univariate analysis, three SNPs located in RFC1 gene (rs1057807, rs4975003 and rs6844176) were significantly associated with grade II-IV acute GVHD (Table 3a). Other significant variables included donor type, diagnosis and disease status at transplant. However, all three SNPs were in strong linkage disequilibrium with each other (r2 ≥0.88) as shown in figure 1.
Table 3. Univariate analysis showing association between individual SNPs in BER genes and grade II-IV acute GVHD (table 3a), grade III-IV acute GVHD (table 3b) and chronic GVHD (table 3c) after adjustment for clinical covariates.
| *a: Grade II-IV Acute GVHD | ||||||||
|---|---|---|---|---|---|---|---|---|
| SNP | Gene | Alleles | MAF | Location | N evaluable | RR | 95% CI | P- value |
| rs1057807 | RFC1 | T/C | 0.41 | EXON 25:3′ untranslated region |
454 | 0.77 | 0.62 -0.95 | 0.014 |
| rs4975003 | RFC1 | G/C | 0.44 | INTRON11 | 461 | 0.76 | 0.62 - 0.93 | 0.008 |
| rs6844176 | RFC1 | T/C | 0.48 | INTRON1 | 465 | 1.39 | 1.14 - 1.70 | 0.001 |
| ** b: Grade III-IV Acute GVHD | ||||||||
| rs1057807 | RFC1 | T/C | 0.41 | EXON 25:3′ untranslated region |
454 | 0.69 | 0.50 - 0.95 | 0.022 |
| rs4975003 | RFC1 | G/C | 0.44 | INTRON11 | 461 | 0.70 | 0.50 - 0.97 | 0.032 |
| rs6844176 | RFC1 | T/C | 0.48 | INTRON1 | 465 | 1.38 | 0.99 - 1.91 | 0.056 |
| *** c: Chronic GVHD | ||||||||
| rs1805410 | PARP1 | A/G | 0.51 | INTRON 9 | 352 | 1.81 | 1.29-2.54 | 0.001 |
Shown are SNPs significant in univariate analysis.
Other significant variables include donor type, diagnosis and disease stage at transplant. (Diagnosis was not significant for rs4975003).
Other significant variables include gender mismatch.
Other significant variables include year of transplant and prior grade II-IV acute GVHD.
Clinical covariates listed above were significant at <0.05 level.
Abbreviations: MAF minor allele frequency; RR Relative Risk
Figure 1.
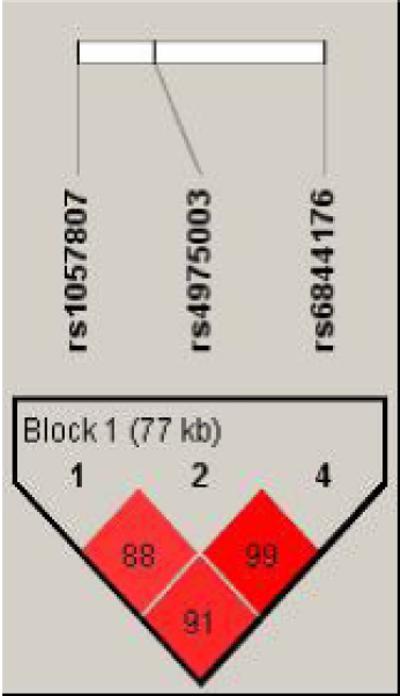
The 3 single nucleotide polymorphisms (SNPs); rs1057807 (exon 25), rs4975003 (intron 11) and rs6844176 (intron 1) located in a 77 kb region in the RFC1 gene showed strong linkage disequilibrium with each other. The 3 SNP pairs; rs1057807/ rs4975003, rs4975003/rs6844176 and rs1057807/rs6844176 showed linkage disequilibrium (r2) of 0.88, 0.99 and 0.91 respectively.
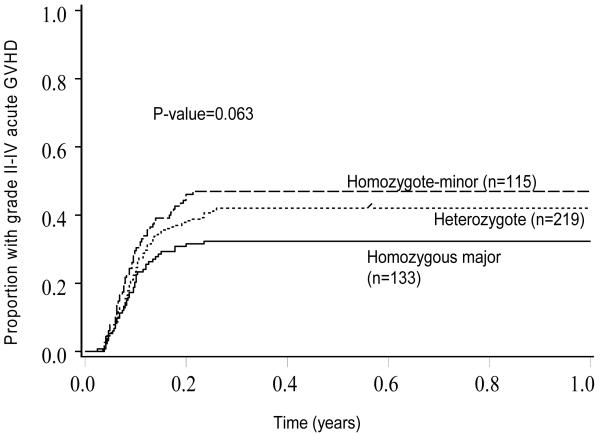
In multivariate analysis (table 4a), one SNP (rs6844176) remained independently associated with a higher risk of grade II-IV acute GVHD (RR 1.39, 95% CI 1.14-1.70, p=0.001). Other significant variables included donor type, diagnosis and disease status at transplant. The model was also evaluated for interactions between SNP and significant clinical co-variates. A significant interaction was noted between disease status at transplant and the SNP rs6844176. The SNP, rs6844176, was significantly associated with higher risk of grade II-IV acute GVHD in patients with relapse or primary induction failure at transplant (p=0.03), but not in those without relapse or primary induction failure (p=0.15). No other SNP-clinical covariate interactions were observed (all p>0.05). The cumulative incidence of grade II-IV acute GVHD was 47% (95% CI 37-56.9%) in 115 patients with homozygous minor genotype versus 42% (95% CI 34.9-49.1%) in 219 patients with heterozygous genotype versus 32.3% (95% CI 24-40.7%) in 133 patients with homozygous major genotype (p =0.06) (Figure 2).
Table 4. Multivariate analysis showing association of SNPs in BER genes with grade II-IV acute GVHD (table 4a), grade III-IV acute GVHD (table 4b) and chronic GVHD (table 4c).
| *a: Grade II-IV acute GVHD | |||||
|---|---|---|---|---|---|
| SNP | Gene | N eval | RR | 95% CI | P value |
| rs6844176 | RFC1 | 465 | 1.39 | 1.14-1.70 | P=0.001 |
| ** b: Grade III-IV acute GVHD | |||||
| rs6844176 | RFC1 | 465 | 1.33 | 0.95-1.85 | P=0.094 |
| *** c: Chronic GVHD | |||||
| rs1805410 | PARP1 | 352 | 1.81 | 1.29-2.54 | 0.001 |
Shown are SNPs significant in multivariate analysis.
Other significant co-variates included donor type, diagnosis and disease status at transplant. Significant interaction was observed between rs6844176 and disease status at transplant.
No other clinical co-variates were significant.
Other significant co-variates included year of transplant and prior grade II-IV acute GVHD as a time dependent variable. No interactions were observed between rs1805410 and clinical covariates.
Figure 2. Cumulative Incidence of grade II-IV acute GVHD in those with homozygous major, heterozygous or homozygous minor genotype for SNP rs6844176.
Footnote: Shown are the proportion of patients with grade II-IV acute GVHD across the three categories of SNP rs6844176.
The same three SNPs (rs1057807, rs4975003 and rs6844176) in RFC1 gene also showed similar but non significant association with grade III-IV acute GVHD in univariate analysis (all p ≤0.06) (Table 3b). As shown in table 4b, in multivariate analysis, the same SNP (rs6844176) showed a trend towards a higher risk of grade III-IV acute GVHD (p=0.09).
Association between chronic GVHD and BER SNPs
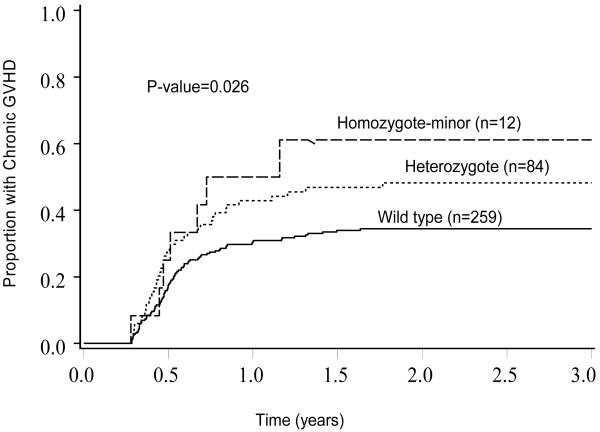
In univariate analysis, one SNP (rs1805410), located in the PARP1 gene was significantly associated with a higher risk of chronic GVHD with a relative risk of 1.81 (95% CI 1.29-2.54, p=0.001) (Table 3c). Other significant variables included year of transplant and prior grade II-IV acute GVHD. The cumulative incidence of chronic GVHD was 50% (95% CI 20.8-79.2%) in 12 patients with homozygous minor genotype versus 42.9% (95% CI 31.5-54.3%) in 84 patients with heterozygous genotype versus 30.9 (95% CI 24.9- 36.9%) in 259 patients with homozygous major genotype (p=0.026) (Figure 3). There was no significant gene-clinical covariate interaction (all p>0.05).
Figure 3. Cumulative Incidence of chronic GVHD in those with homozygous major, heterozygous or homozygous minor genotype for SNP rs1805410.
Footnote: Shown are the proportion of patients with chronic GVHD across the three categories of SNP rs1805410.
Discussion
GVHD remains a major barrier to successful transplantation. The pathophysiology of the disease (especially chronic GVHD) remains to be fully defined. We found three SNPs in RFC1 genes to be significantly associated with grade II-IV acute GVHD and one SNP in the PARP1 gene to be significantly associated with chronic GVHD.
In our prior report (Association between genetic variants in the BER pathway and outcomes after HCT, BBMT), we have identified three SNPs in TDG, LIG3 and MUTYH genes (rs167715, rs3135974, rs3219463) to be associated with transplant related mortality (TRM) in multiple regression analysis. These differ from the genetic variants identified in the current analysis, suggesting that the polymorphisms identified in the current analysis represent an association with occurrence of GVHD and not with mortality. Further analysis of TRM showed that the association between the 3 SNPs (rs167715, rs3135974, rs3219463) and TRM remained unchanged even after controlling for grade II-IV acute GVHD as a time dependent co-variate (data not shown), indicating that these SNPs affect TRM through mechanisms independent of GVHD.
Base excision repair is the major DNA repair pathway for repair of non-bulky damaged bases, abasic sites and single stranded breaks caused by ionizing radiation and alkylating agents (3). The three SNPs associated with grade II-IV acute GVHD were all located in RFC1 gene. Though in multivariate analysis, only rs6844176 remained significantly associated with acute GVHD, all three SNPs (rs6844176, rs4975003 and rs1057807) were in strong LD, hence, it is likely that the SNP with the strongest association may in fact be a surrogate for the other two SNPs strongly correlated and located in the region. Independent validation and correlative functional studies are needed to prove these results. The protein encoded by this gene is the large subunit of replication factor C, a DNA-dependent ATPase that is required for eukaryotic DNA replication and repair. The protein acts as an activator of DNA polymerases and promotes coordinated synthesis of both strands (14). These SNPs showed a similar (though not significant) association with grade III-IV acute GVHD, possibly due to limited sample size when evaluating grade III-IV acute GVHD.
PARP1 (poly [ADP-ribose] polymerase 1) gene encodes an enzyme that works by modifying nuclear proteins by poly ADP-ribosylation (15). PARP1 also binds with single strand breaks through its N-terminal zinc fingers and recruits XRCC1, DNA polβ, and DNA ligase III in the short patch BER pathway; and recruits XRCC1, flap endonuclease 1, and DNA ligase I in long-patch BER pathway (16). PARP-1 overactivation in response to extensive DNA damage results in necrosis (17-20). In addition to inducing necrosis, PARP-1 also has a profound modulatory effect on the inflammatory response. PARP-1 can form stable complexes with transcription factors such as p53 and AP-2 (21,22). PARP-1 can also act as a coactivator of NF-kb (21-23) and, therefore, may be involved in the induction of TNF-α. Polymorphisms resulting in altered activity in this gene could modulate cellular necrosis and altered inflammatory response and hence be implicated in the pathogenesis of GVHD.
These results also need to be confirmed in patients receiving non-alkylator based reduced intensity regimens where the observed higher risk associated with these SNPs should not be present. This analysis could not be performed in the current study as all conditioning regimens at our institution (myeloablative as well as reduced intensity) are alkylator based.
Several studies have evaluated the impact of polymorphisms involving minor histocompatibility antigens, cytokine genes and innate immunity genes towards GVHD. A recent study reported a higher risk of acute GVHD when donor and recipient were mismatched for homozygous deletion of UGT2B17, a gene expressed in GVHD-affected tissues and giving rise to multiple histocompatibility antigens (24). Other minor histocomatibility antigens including HY, HA-1, HA-2 and HA-3 have been evaluated in the etiology of both GVHD and disease relapse (25-35). Polymorphisms in donor and recipient genes for cytokines have been demonstrated to be risk factors for GVHD. Tumor Necrosis Factor (TNF)-α, Interleukin 10 (IL-10), Interferon-γ (IFNγ) variants have correlated with GVHD in some, but not all, studies (36-39). Genetic polymorphisms of proteins involved in innate immunity, such as nucleotide oligomerization domain 2 and Keratin 18 receptors, have also been associated with GVHD (40). These studies provide support to the hypothesis that tissue injury and resulting cytokine release are implicated in the pathogenesis of graft versus host disease. Polymorphisms in DNA repair pathways in conjunction with other pathways could also be implicated by impeding or accelerating repair of injured tissue and hence affecting GVHD.
This study represents the first analysis of SNPs in BER DNA repair pathway towards acute and chronic GVHD. Our results are novel and hypothesis generating. If confirmed, they represent an important step towards understanding the pathophysiology of GVHD as well as identifying subgroups at high risk for GVHD towards whom novel prophylactic strategies could be targeted.
Acknowledgments
This work was supported by NIAID (R21 AI079354-01) and the Leukemia Research Fund, University of Minnesota.
This project was supported by grants from the following: Leukemia Research Fund, University of Minnesota, NIAID: R21 AI079354-01
Footnotes
Conflicts of interest: Dr. Mukta Arora has received funding from Leukemia Research Fund, University of Minnesota; and NIAID for this project and Dr. Bharat Thyagarajan has received funding from NIAID; the other co-authors declare no conflicts of interest.
References
- 1.Socie G, Blazar BR. Acute graft-versus-host disease; from the bench to the bedside. Blood. 2009;114:4327–36. doi: 10.1182/blood-2009-06-204669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin PJ. Biology of chronic graft-versus-host disease: implications for a future therapeutic approach. Keio J Med. 2008;57:177–83. doi: 10.2302/kjm.57.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lieberman HB. DNA damage repair and response proteins as targets for cancer therapy. Curr Med Chem. 2008;15:360–367. doi: 10.2174/092986708783497328. [DOI] [PubMed] [Google Scholar]
- 4.Bharat Thyagarajan, Bruce Lindgren, Saonli Basu, Sriharsha Nagaraj, Myron Gross, Daniel Weisdorf, et al. Association between genetic variants in the base excision repair pathway and outcomes after hematopoietic cell transplant. Biol Blood Marrow Transplant. 2010 Mar 10; doi: 10.1016/j.bbmt.2010.03.001. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weisdorf D, Spellman S, Haagenson M, Horowitz M, Lee S, Anasetti C, et al. Classification of HLA-matching for retrospective analysis of unrelated donor transplantation: revised definitions to predict survival. Biol Blood Marrow Transplant. 2008;14:748–58. doi: 10.1016/j.bbmt.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. http://egp.gs.washington.edu/ber.html.
- 7.NIEHSSNPs . NIEHS Environmental Genome Project. University of Washington; Seattle, WA: Jan, 2008. URL: http://egp.gs.washington.edu. [Google Scholar]
- 8.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng PC, Henikoff S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 2002;12:436–446. doi: 10.1101/gr.212802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yue P, Melamud E, Moult J. SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinformatics. 2006;7:166. doi: 10.1186/1471-2105-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.R D, Core, Team . R: A Language and Environment for Statistical Computing. Foundation for Statistical Computing; Vienna, Austria: 2008. [Google Scholar]
- 12.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 13.Fine J, Gray R. A proportional hazards model for the sub distribution of a competing risk. Journal of The American Statistical Association. 1999;94:496–509. [Google Scholar]
- 14.Entrez Gene: RFC1 replication factor C (activator 1) 1, 145kDa. [Google Scholar]
- 15.Entrez Gene: PARP1 poly (ADP-ribose) polymerase family, member 1. [Google Scholar]
- 16.Jagtap P, Szabo C. Poly (ADP-ribose) polymerase and the therapeutic effects of its inhibitors. Nat Rev Drug Dis. 2005;4:421–40. doi: 10.1038/nrd1718. [DOI] [PubMed] [Google Scholar]
- 17.Zong WX, Ditsworth D, Bauer DE, Wang ZQ, Thompson CB. Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes Dev. 2004;18:1272–82. doi: 10.1101/gad.1199904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edinger AL, Thompson CB. Death by design: apoptosis, necrosis and autophagy. Curr. Opin. Cell Biol. 2004;16:663–669. doi: 10.1016/j.ceb.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 19.Berger NA, Berger SJ. Metabolic consequences of DNA damage: the role of poly (ADP-ribose) polymerase as mediator of the suicide response. Basic Life Sci. 1986;38:357–363. doi: 10.1007/978-1-4615-9462-8_39. [DOI] [PubMed] [Google Scholar]
- 20.Ha HC, Snyder SH. Poly(ADP-ribose) polymerase is a mediator of necrotic cell death by ATP depletion. Proc. Natl. Acad. Sci. USA. 1999;96:13978–13982. doi: 10.1073/pnas.96.24.13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Virag L, Szabo C. The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol. Rev. 2002;54:375–429. doi: 10.1124/pr.54.3.375. [DOI] [PubMed] [Google Scholar]
- 22.Chiarugi A. Poly(ADP-ribose) polymerase: killer or conspirator? The ‘suicide hypothesis’ revisited. Trends Pharmacol. Sci. 2002;23:122–129. doi: 10.1016/S0165-6147(00)01902-7. [DOI] [PubMed] [Google Scholar]
- 23.Hassa PO, Hottiger MO. A role of poly (ADP-ribose) polymerase in NF-_B transcriptional activation. Biol. Chem. 1999;380:953–959. doi: 10.1515/BC.1999.118. [DOI] [PubMed] [Google Scholar]
- 24.McCarroll SA, Bradner JE, Turpeinen H, Volin L, Martin PJ, Chilewski SD, et al. Donor-recipient mismatch for common gene deletion polymorphisms in graft-versus-host disease. Nat Genet. 2009;41:1341–4. doi: 10.1038/ng.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bleakley MR, Riddell SR. Molecules and mechanisms of the graft-versus-leukaemia effect. Nature Rev Cancer. 2004;4:371–380. doi: 10.1038/nrc1365. [DOI] [PubMed] [Google Scholar]
- 26.Goulmy E, Schipper R, Pool J, Blockland E, Falkenburg JH, Vossen J, et al. Mismatches of minor histocompatibility antigens between HLA-identical donors and recipients and the development of graft-versus-host disease after bone marrow transplantation. N Engl J Med. 1996;334:281–285. doi: 10.1056/NEJM199602013340501. [DOI] [PubMed] [Google Scholar]
- 27.Tseng LH, Lin MT, Hansen JA, Gooley T, Pei J, Smith AG, et al. Correlation between disparity for the minor histocompatibility antigen HA-1 and the development of acute graft-versus-host disease after allogeneic marrow transplantation. Blood. 1999;94:2911–2914. [PubMed] [Google Scholar]
- 28.Gallardo D, Arostegui JI, Balas A, Torres A, Caballero D, Carreras E, et al. Disparity for the minor histocompatibility antigen HA-1 is associated with an increased risk of acute graft-versus-host disease (GvHD) but it does not affect chronic GvHD incidence, disease-free survival or overall survival after allogeneic human leucocyte antigenidentical sibling donor transplantation. Br J Haematol. 2001;114:931–936. doi: 10.1046/j.1365-2141.2001.03013.x. [DOI] [PubMed] [Google Scholar]
- 29.Socie G, Loiseau P, Tamouza R, Janin A, Busson N, Gluckman E, et al. Both genetic and clinical factors predict the development of graftversus-host disease after allogeneic hematopoietic stem cell transplantation. Transplantation. 2001;72:699–706. doi: 10.1097/00007890-200108270-00024. [DOI] [PubMed] [Google Scholar]
- 30.Nesci S, Buffi O, Iliescu A, Andreani M, Lucarelli G. Recipient mHag-HA1 disparity and aGVHD in thalassemic-transplanted patients. Bone Marrow Transplant. 2003;31:575–578. doi: 10.1038/sj.bmt.1703880. [DOI] [PubMed] [Google Scholar]
- 31.Tait BD, Maddison R, McCluskey J, Deayton S, Heatley S, Lester S, et al. Clinical relevance of the minor histocompatibility antigen HA-1 in allogeneic bone marrow transplantation between HLA identical siblings. Transplant Proc. 2001;33:1760–1761. doi: 10.1016/s0041-1345(00)02816-5. [DOI] [PubMed] [Google Scholar]
- 32.Miklos DB, Kim HT, Zorn E, Hochberg EP, Guo L, Mattes-Ritz A, et al. Antibody response to DBY minor histocompatibility antigen is induced after allogeneic stem cell transplantation and in healthy female donors. Blood. 2004;103:353–359. doi: 10.1182/blood-2003-03-0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zorn E, Miklos DB, Floyd BH, Mattes-Ritz A, Guo L, Soiffer RJ, et al. Minor histocompatibility antigen DBY elicits a coordinated B and T cell response after allogeneic stem cell transplantation. J Exp Med. 2004;199:1133–1142. doi: 10.1084/jem.20031560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishida T, Akatsuka Y, Morishima Y, Hamajima N, Tsujimura K, Kuzushima K, et al. Clinical relevance of a newly identified HLA-A24-restricted minor histocompatibility antigen epitope derived from BCL2A1, ACC-1, in patients receiving HLA genotypically matched unrelated bone marrow transplant. Br J Haematol. 2004;124:629–635. doi: 10.1111/j.1365-2141.2004.04823.x. [DOI] [PubMed] [Google Scholar]
- 35.Marijt WA, Heemskerk MH, Kloosterboer FM, Goulmy E, Kester MG, van der Hoorn MA, et al. Hematopoiesis-restricted minor histocompatibility antigens HA-1- or HA-2-specific T cells can induce complete remissions of relapsed leukemia. Proc Natl Acad Sci U S A. 2003;100:2742–2747. doi: 10.1073/pnas.0530192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Antin JH, Ferrara JL. Cytokine dysregulation and acute graft-versus-host disease. Blood. 1992;80:2964–2968. [PubMed] [Google Scholar]
- 37.Cavet J, Middleton PG, Segall M, Noreen H, Davies SM, Dickinson AM. Recipient tumor necrosis factor-alpha and interleukin-10 gene polymorphisms associate with early mortality and acute graft-versus-host disease severity in HLA-matched sibling bone marrow transplants. Blood. 1999;94:3941–32946. [PubMed] [Google Scholar]
- 38.Lin MT, Storer B, Martin PJ, Tseng LH, Gooley T, Chen PJ, et al. Relation of an interleukin-10 promoter polymorphism to graft-versus-host disease and survival after hematopoietic-cell transplantation. N Engl J Med. 2003;349:2201–2210. doi: 10.1056/NEJMoa022060. [DOI] [PubMed] [Google Scholar]
- 39.Dickinson AM, Charron D. Non-HLA immunogenetics in hematopoietic stem cell transplantation. Curr Opin Immunol. 2005;17:517–525. doi: 10.1016/j.coi.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 40.Holler E, Rogler G, Brenmoehl J, Hahn J, Herfarth H, Greinix H, et al. Prognostic significance of NOD2/CARD15 variants in HLA-identical sibling hematopoietic stem cell transplantation: effect on long-term outcome is confirmed in 2 independent cohorts and may be modulated by the type of gastrointestinal decontamination. Blood. 2006;107:4189–4193. doi: 10.1182/blood-2005-09-3741. [DOI] [PubMed] [Google Scholar]