Abstract
Purpose
We had previously demonstrated that there is familial aggregation of JIA. Using a large JIA cohort, we sought to find additional JIA case-clusters and to calculate robust estimates of relative risks (RR) for siblings and cousins of JIA probands, and to estimate the population attributable risk (PAR) of familial factors in JIA.
Methods
A probabilistic record-linking analysis was performed by matching records of 862 JIA cases with records of ~7 million individuals in the Utah Population Database (UPDB), a computerized genealogical database. For each case, 5 controls matched on birth year and gender were selected from the UPDB. Specialized software (Kinship Analysis Tools) was used to test for familial aggregation of disease and to estimate the magnitude of familial risks, and identify families at high risk for disease.
Results
We identified 22 founders with significantly more descendants with JIA than expected (5–13 descendants; p<0.0001 to <0.008). The PAR for familial factors for JIA was ~13%. The RR of JIA in siblings of cases was significantly elevated; (11.6; 4.9–27.5; p<3×10−8). The RR of JIA in first-cousins was also elevated (5.8; 2.5–13.8; p<6×10−5).
Conclusions
We have identified the largest sets of JIA pedigrees described to date. Approximately 13% of cases of JIA can be attributed to familial factors. Siblings and first-cousins of probands with JIA have an increased risk of JIA. The observed decline in the magnitude of risk between siblings and cousins suggests JIA is influenced by shared genetic factors.
Keywords: genetics, juvenile idiopathic arthritis, familial, relative risk
Juvenile idiopathic arthritis (JIA) refers to a collection of chronic arthropathies in children. The etiology of JIA is complex, but genetic factors are thought to play a role in susceptibility to JIA.(1) While several genetic variants that predispose to JIA susceptibility have been identified, it is likely there are more variants that contribute to JIA susceptibility. Most investigations of genetic factors in JIA have utilized candidate gene studies of unrelated cases and controls.(2) This has been due, in part, to the lack of large multiplex JIA families. Identification of extended multiplex families might facilitate linkage studies to identify other genetic factors that play a role in susceptibility to JIA.
While affected twin and sibling pairs with JIA have been described, there are very few reports of extended multiplex families with JIA. One of the measures of the magnitude of genetic contribution to disease susceptibility is the relative risk (RR) to siblings and other relatives of probands with JIA, compared with that for the general population. Only three estimates of RR for siblings of JIA probands have been published, with values ranging from 15 to 30.(1, 3, 4) Besides RR for siblings and twins, there have been no published reports of RR for other classes of relatives such as cousins. Earlier we demonstrated that there was familial aggregation of JIA.(3) Using a larger cohort of JIA we sought to 1) identify additional extended multiplex JIA pedigrees, 2) provide robust estimates of RR of JIA for siblings and cousins of probands with JIA and 3) calculate the population attributable risk (PAR) of familial factors in JIA.
Methods
This study was approved by the University of Utah IRB and the administrative body for the Utah Population Database (UPDB), the Utah Resource for Genetic and Epidemiological Research. This is a population based study using data from the UPDB, which includes data from several sources, including the Family History Library maintained by the Church of Jesus Christ of Latter-day Saints, vital records from Utah State Department of Health, and other statewide data sets. The resources in UPDB include family history data for approximately ~7 million individuals, representing pedigrees spanning as many as 11 generations.(5) The majority of families living in Utah are represented in this database, with a special emphasis on genealogical records of the founders of Utah and their descendants.
Cases were 862 children with JIA in the Intermountain States Database of Childhood Rheumatic Diseases (ISDCRD), a database that contains clinical and demographic information for subjects with JIA seen in the pediatric Rheumatology Clinics at the University of Utah. All patients had been examined by one of three pediatric rheumatologists, and patients were diagnosed with JIA according to the ILAR criteria (6). There were 405 children with oligoarticular JIA, 161 with rheumatoid factor (RF)-negative polyarticular JIA, 108 with enthesitis-related arthritis, 77 with systemic JIA, 53 with RF-positive polyarticular JIA, 8 with psoriatic JIA and 50 with undifferentiated JIA. A majority (67%) of the subjects were female. The mean age of onset of JIA was 6.8 years. For each case, 5 controls matched by birth year and gender were randomly selected from the UPDB.
Probabilistic record-linking was performed by matching records in the ISDCRD with records of individuals in the UPDB, as previously described.(3, 7) The goal of the record-linking was to determine if the individuals in the ISDCRD were also in the UPDB. This was accomplished by using patient’s identifiers to match data present in the UPDB (for example, to match to their birth certificate). Once records were linked, specialized software, Kinship Analysis Tools(KAT; University of Utah Salt Lake City, UT), was used to estimate the following measures of familial risk.(8)
Familial Standardized Incidence Ratio (FSIR)
The FSIR permits the quantification of a person’s familial risk of disease, and it takes into account the number of biological relatives, degree of relatedness to the proband, and person-time at risk among family members.(8) FSIR is calculated by tabulating the observed and expected numbers of cases of disease among all relatives of an individual, weighting the contribution of each relative by the probability that the relative shares an allele with the subject by common descent. FSIR computes a similar familial risk as Familial Risk (FR: used in our 2004 paper) but is further refined by stratifying the incidence calculations on age and sex.
Pedigree p-values
The KAT was used to find families with excess JIA. The probability of a family having some observed number of cases (x) under the null hypothesis of no familial disease aggregation is the Poisson probability of X>x, given an expected number, μ.
Population attributable risk (PAR)
PAR was calculated using a conditional logistic regression method as described previously.(9) First, a conditional logistic regression model is used to predict relative risk as a function of FSIR. The FSIR was modified using an empirical Bayes’ adjustment for measurement error.(10) From this model, individual probabilities of causation (PAC) for each case are computed, where PAC = (RR−1)/RR, where RR is the relative risk estimated from the model for the observed level of FSIR. The PAR is calculated as the mean PAC across all the cases.
Relative risks
RRs for siblings, first cousins, and second cousins of the cases and controls were also calculated using unconditional logistic regression, following the method described by Bai et al.(11)
Results
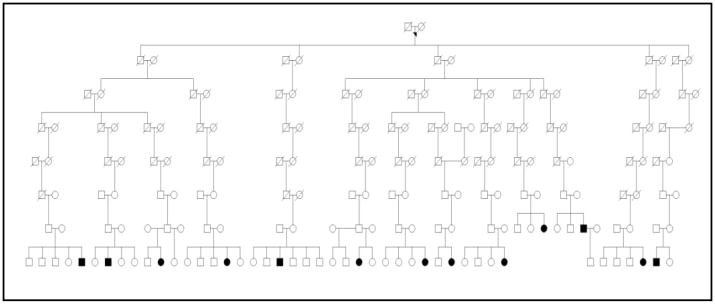
All of the 862 probands could be linked to the UPDB since they had at least one record in the UPDB. However, 94 probands (11%) did not have familial records and therefore were not used for further analysis. Using the KAT, we identified 22 founders with families that had 5 to 13 affected descendants, and pedigree sizes ranging from 2058 to 21,517 descendants (Table 1). Founders are the earliest generation in the database, i.e., they do not have ancestral genealogical relationships in the UPDB. These 22 ancestors had significantly more descendants with JIA than expected (5–13 descendants; p <0.0001 to <0.008), (Table 1). There were 84 affected descendants in the 22 founder families. in A proband may appear as a descendant of more than one founder; descendants with JIA number 141. The mean FSIR in these families was 4.64 (2.48–9.51; p <0.01) suggesting that members in these families had over a fourfold higher clustering of JIA than would expected by a uniform distribution. An example of a large family cluster is shown in figure 1.
Table 1.
Founders with excessive risk of descendants with JIA
| Founder | FSIR | P value | Descendants | JIA subtypes | Onset age Mean (range) | Female N (%) | ANA + N | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | with JIA | expected | S | R | P | O | E | Ps | U | ||||||
| 1 | 4.03 | 0.004 | 6266 | 6 | 1.5 | 3 | 3 | 4.4 (0.9–8.6) | 6 (100) | 3 | |||||
| 2 | 4.03 | 0.004 | 5272 | 5 | 1.5 | 2 | 2 | 1 | 4.3 (0.9–8.6) | 4 (80) | 3 | ||||
| 3 | 2.48 | 0.002 | 21517 | 13 | 5.6 | 1 | 2 | 2 | 5 | 3 | 8.1 (1.7–15.7) | 8 (62) | 4 | ||
| 4 | 3.50 | 0.005 | 5878 | 6 | 2.0 | 1 | 2 | 2 | 1 | 9.5 (1.1–15.7) | 5 (83) | 1 | |||
| 5 | 4.32 | 0.007 | 3915 | 5 | 1.2 | 2 | 2 | 1 | 9.5 (1.7–15.7) | 4 (80) | 1 | ||||
| 6 | 2.49 | 0.008 | 17389 | 10 | 4.0 | 3 | 4 | 2 | 1 | 9.2 (1.1–15.3) | 5 (50) | 1 | |||
| 7 | 2.88 | 0.008 | 10875 | 8 | 2.8 | 1 | 1 | 1 | 5 | 6.8 (2.2–12.5) | 3 (50) | 3 | |||
| 8 | 5.42 | 0.001 | 4646 | 6 | 1.1 | 1 | 2 | 2 | 1 | 9.9 (2.7–15.6) | 5 (83) | 3 | |||
| 9 | 7.04 | 0.0003 | 3455 | 6 | 0.9 | 1 | 2 | 2 | 1 | 9.9 (2.7–15.6) | 5 (83) | 3 | |||
| 10 | 6.85 | 0.0001 | 5185 | 7 | 1.0 | 1 | 2 | 3 | 1 | 8.1 (1.3–15.8) | 6 (86) | 3 | |||
| 11 | 4.23 | 0.002 | 7536 | 7 | 1.7 | 1 | 2 | 3 | 1 | 8.1 (1.3–15.8) | 6 (86) | 3 | |||
| 12 | 4.43 | 0.006 | 5726 | 5 | 1.1 | 1 | 1 | 3 | 4.8 (1.3–13.1) | 4 (80) | 2 | ||||
| 13 | 9.51 | 0.0002 | 2058 | 5 | 0.5 | 1 | 3 | 1 | 6.3 (1.4–13.6) | 5 (100) | 3 | ||||
| 14 | 5.40 | 0.003 | 2868 | 5 | 0.9 | 2 | 2 | 1 | 7.2 (0.3–13.3) | 3 (60) | 4 | ||||
| 15 | 4.30 | 0.007 | 4944 | 5 | 1.2 | 1 | 1 | 1 | 2 | 6.1 (1.3–13.3) | 4 (80) | 2 | |||
| 16 | 3.36 | 0.002 | 12763 | 9 | 2.7 | 1 | 1 | 2 | 4 | 1 | 7.6 (0.8–15.4) | 7 (78) | 5 | ||
| 17 | 3.63 | 0.007 | 7251 | 5 | 1.7 | 4 | 1 | 6.1 (2.6–9.0) | 3 (60) | 1 | |||||
| 18 | 4.29 | 0.003 | 6120 | 5 | 1.4 | 4 | 1 | 6.1 (2.6–9.0) | 3 (60) | 1 | |||||
| 19 | 3.73 | 0.003 | 7151 | 7 | 1.9 | 1 | 3 | 3 | 7.8 (2.2–14.0) | 3 (43) | 2 | ||||
| 20 | 7.00 | 0.0009 | 2266 | 5 | 0.7 | 1 | 1 | 2 | 1 | 7.7 (3.7–10.0) | 4 (80) | 1 | |||
| 21 | 3.97 | 0.005 | 5491 | 6 | 1.5 | 1 | 2 | 1 | 1 | 1 | 8.1 (3.6–13.2) | 5 (83) | 2 | ||
| 22 | 5.31 | 0.003 | 3735 | 5 | 0.9 | 1 | 1 | 1 | 1 | 1 | 6.7 (1.3–13.6) | 2 (40) | 2 | ||
Founders are defined as individuals for whom there are no ancestral genealogical relationships in the Utah Population Database. Thus, they are the earliest generation in the database. FSIR: familial standardized incidence ratio. JIA subtypes: S = systemic JIA, R = RF + polyarticular JIA; P = RF negative polyarticular JIA, O = oligoarticular JIA, E = enthesitis related arthritis, Ps = psoriatic JIA, U = undifferentiated JIA. ANA: Anti nuclear antibody.
Figure 1.
A pedigree showing a family with increased risk of JIA in which there are thirteen descendents with JIA. Circle: female; square: male; black: JIA. The founder of this pedigree is founder 3 in table 1.
The case-control analysis was used to calculate the PAR. Using the Bayes adjusted PAR, the risk for JIA attributable to familial factors was ~13% (95 % CI 9–17%). The RRs for each kinship class were computed using conditional logistic regression. The RR for siblings of JIA probands was significantly elevated compared to controls (11.6; 4.9–27.5; p <3×10−8) (Table 2). The RR for first-cousins was also elevated compared to controls. (5.82; 2.5–13.8; p 6 ×10−5). The RR for second-cousins was not significantly elevated compared to controls (1.05; ns). There were 63 individuals with JIA among >50,000 siblings, first and second cousins of healthy controls, yielding a prevalence of 1.2/1000 for JIA (Table 2).
Table 2.
Recurrence risk of JIA among siblings and cousins of cases with JIA and matched controls.
| Relative class | Cases | Controls | RR (95% CI) | P value | ||||
|---|---|---|---|---|---|---|---|---|
| Relatives | JIA | Rate | Relatives | JIA | Rate | |||
| Siblings | 544 | 14 | 0.0257 | 3578 | 8 | 0.0022 | 11.6 (4.9–27.5) | < 2.59 × 10−8 |
| First cousins | 1896 | 10 | 0.0053 | 10969 | 11 | 0.0010 | 5.82 (2.5–13.8) | < 6.07 × 10−5 |
| Second cousins | 6348 | 8 | 0.0012 | 36247 | 44 | 0.0012 | 1.05 (0.5–2.23) | 0.89 |
| Total | 8788 | 32 | 0.0038 | 50794 | 63 | 0.0012 | ||
The rate of JIA among relatives of cases was compared with the rate of JIA among relatives of controls to yield the recurrence risk of JIA among relatives of probands with JIA. Significant results are shown in bold. RR = recurrence risk.
We repeated the analyses with common JIA subtypes. For oligoarticular JIA the PAR was 10 % (7–13%), and that for polyarticular JIA was 6 % (2–9%). When we repeated the conditional logistic regression analysis after stratifying by JIA subtype, the RR for siblings of probands with oligoarticular JIA was statistically significantly elevated compared to controls (RR 22.6; 5.3–95.8; p 2.37 × 10−5). The RR for first cousins was non-significantly elevated compared to controls (RR 3.03; 0.64–14.3; p ns). The small sample sizes of siblings and cousins with polyarticular JIA precluded estimation of RR for poly JIA.
Discussion
Although the etiology of JIA is complex, genetic factors have long been implicated in its causation.(1) Besides descriptions of twins(12, 13) and sibling pairs(14) with JIA, there are very few descriptions of extended multiplex families of JIA. The few case-series of twins concordant for JIA indicate a monozygotic twin concordance rate of 25–40%(4, 12), which implies a RR of 250–400 for a monozygotic twin of a proband with JIA.(4) A limited number of studies have estimated RR for siblings of JIA probands, ranging from ~15 (1) from the United States, to ~20 in Finland (4). The denominators in these studies used extrapolated figures, and these studies did not provide confidence intervals or statistical significance levels for the RR estimates.
We have also previously calculated RR for siblings of JIA probands, with an estimated value of 30.(3). However, the confidence intervals of this estimate were wide (7.0 to infinity) reflecting the small sample size of only 8 individuals with JIA among relatives of probands. Furthermore, in our earlier study we stratified relatives by degree of relationship which combines individuals of different generations in a kinclass. This is likely to have biased our estimates since only children seen in the pediatric rheumatology clinics were included in the ISDCRD, and if earlier generations such as parents, aunts or grandparents had JIA, they would not have been included in the database. By using classes of relatives who are all in the same generation such as siblings and cousins, we have minimized this potential bias. Thus we have now provided reliable estimates with robust confidence intervals for the RR of JIA among siblings. We have also shown that first-cousins of probands with JIA also have a significant excessive risk of JIA, (RR = 6). We believe ours is the first description of RR for relatives beyond siblings and twins. Such estimates would be difficult if not impossible to obtain without resources such as the UPDB, making our study unique. The observed decline in the magnitude of risk between siblings and cousins supports the notion that JIA is influenced by at least several shared genetic factors. Our study also provides a population-based estimate of the prevalence of JIA at 1.2/1000 individuals.
The majority of studies investigating genetic factors in JIA have been case-control association studies. This has been due in part to a lack of large extended multiplex families with multiple JIA cases. Besides reports of affected sibling pairs with JIA, there have only been limited descriptions of JIA multiplex families. Earlier, we had described four clusters of 5 cases of JIA each.(3) The UPDB is a dynamic database and receives annual updates of new records. Adding more individuals to a hierarchical database has a multiplicative effect on connections. The present study used twice the number of JIA cases. These factors increased the number of familial links and increased the probability of clustering beyond random associations which allowed to us pick more clusters of interest. Thus our present study expands our earlier findings with the identification of several additional and larger case-clusters of JIA which could aid in the search for genes underlying susceptibility to this condition. The affected relatives in these clusters share common ancestors and in many instances are distantly related.
We also calculated, to our knowledge, the first estimates of PAR for familial factors in JIA. Familial factors account for ~13% of JIA. This is modest compared to 39% in carcinoma of the breast, or 35% in colon carcinoma, and is similar to the PAR of 10% estimated in Mullerian anomalies.(7, 15) We are not aware of PAR estimates for other rheumatic diseases.
While it is possible that there are additional cases of JIA that were not included in our database, we believe our study includes most cases of JIA given that they were seen in the only tertiary pediatric center in the State of Utah. The diagnosis of JIA was made by pediatric rheumatologists who examined these children, thereby assuring the validity of the diagnosis. Our well characterized JIA cohort resembles other published JIA cohorts, suggesting that our results can be generalized to other JIA cohorts.
We acknowledge that our cases come from one hospital system and hence our RR estimates might be inflated by referral bias. However, our PAR calculation is population-based and computes the percent of risk for JIA associated with being a member of an extended family. Also, since we included only JIA cases seen at the Pediatric Rheumatology clinics at the Univeristy of Utah, our observed PAR might be lower than the actual value as some cases with JIA in the population may have sought care at other facilities.
The subtypes of JIA have distinct clinical and genetic associations. However, since clinically distinct autoimmune phenotypes share common susceptibility factors, we treated JIA as a combined phenotype. We did analyze the oligoarticular and polyarticular JIA subtypes separately, and demonstrate that the sibling RR of oligoarticular JIA is also statistically significantly elevated.
Our study provides insight into the familial distribution of JIA based on the analysis of a large number of affected individuals using a unique resource, the Utah Population Database. Comprehensive identification and characterization of JIA within kinships would allow genetic linkage analysis to help identify potentially important genes underlying this common pediatric rheumatic disease.
Acknowledgments
Supported by, The National Institute of Arthritis and Musculoskeletal and Skin Diseases (K23-AR50177), The Arthritis Foundation, and The Val A Browning Charitable Foundation, and The Primary Children’s Medical Center Foundation, Salt Lake City, UT, and the Rooms To Go Foundation, Atlanta, GA. Partial support for the Utah Population Database is provided by Huntsman Cancer Institute at the University of Utah.
References
- 1.Glass DN, Giannini EH. Juvenile rheumatoid arthritis as a complex genetic trait. Arthritis Rheum. 1999;42:2261–8. doi: 10.1002/1529-0131(199911)42:11<2261::AID-ANR1>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 2.Prahalad S, Glass DN. A comprehensive review of the genetics of juvenile idiopathic arthritis. Pediatr Rheumatol Online J. 2008;6:11. doi: 10.1186/1546-0096-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prahalad S, O’Brien E, Fraser AM, Kerber RA, Mineau GP, Pratt D, et al. Familial aggregation of juvenile idiopathic arthritis. Arthritis Rheum. 2004;50:4022–7. doi: 10.1002/art.20677. [DOI] [PubMed] [Google Scholar]
- 4.Savolainen A, Saila H, Kotaniemi K, Kaipianen-Seppanen O, Leirisalo-Repo M, Aho K. Magnitude of the genetic component in juvenile idiopathic arthritis. Ann Rheum Dis. 2000;59:1001. doi: 10.1136/ard.59.12.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wylie JE, Mineau GP. Biomedical databases: protecting privacy and promoting research. Trends Biotechnol. 2003;21:113–6. doi: 10.1016/S0167-7799(02)00039-2. [DOI] [PubMed] [Google Scholar]
- 6.Petty RE, Southwood TR, Manners P, Baum J, Glass DN, Goldenberg J, et al. International League of Associations for Rheumatology classification of juvenile idiopathic arthritis: second revision, Edmonton, 2001. J Rheumatol. 2004;31:390–2. [PubMed] [Google Scholar]
- 7.Hammoud AO, Gibson M, Peterson CM, Kerber RA, Mineau GP, Hatasaka H. Quantification of the familial contribution to mullerian anomalies. Obstet Gynecol. 2008;111:378–84. doi: 10.1097/01.AOG.0000267219.10869.9f. [DOI] [PubMed] [Google Scholar]
- 8.Kerber RA. Method for calculating risk associated with family history of a disease. Genet Epidemiol. 1995;12:291–301. doi: 10.1002/gepi.1370120306. [DOI] [PubMed] [Google Scholar]
- 9.Bruzzi P, Green SB, Byar DP, Brinton LA, Schairer C. Estimating the population attributable risk for multiple risk factors using case-control data. Am J Epidemiol. 1985;122:904–14. doi: 10.1093/oxfordjournals.aje.a114174. [DOI] [PubMed] [Google Scholar]
- 10.Boucher KM, Kerber RA. Measures of familial aggregation as predictors of breast-cancer risk. J Epidemiol Biostat. 2001;6:377–85. doi: 10.1080/135952201753336960. [DOI] [PubMed] [Google Scholar]
- 11.Bai Y, Sherman S, Khoury MJ, Flanders WD. Bias associated with study protocols in epidemiologic studies of disease familial aggregation. Am J Epidemiol. 2000;151:927–37. doi: 10.1093/oxfordjournals.aje.a010297. [DOI] [PubMed] [Google Scholar]
- 12.Ansell BM, Bywaters EG, Lawrence JS. Familial aggregation and twin studies in Still’s disease. Juvenile chronic polyarthritis. Rheumatology. 1969;2:37–61. [PubMed] [Google Scholar]
- 13.Prahalad S, Ryan MH, Shear ES, Thompson SD, Glass DN, Giannini EH. Twins concordant for juvenile rheumatoid arthritis. Arthritis Rheum. 2000;43:2611–2. doi: 10.1002/1529-0131(200011)43:11<2611::AID-ANR33>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 14.Moroldo MB, Chaudhari M, Shear E, Thompson SD, Glass DN, Giannini EH. Juvenile rheumatoid arthritis affected sibpairs: extent of clinical phenotype concordance. Arthritis Rheum. 2004;50:1928–34. doi: 10.1002/art.20292. [DOI] [PubMed] [Google Scholar]
- 15.Kerber RA, O’Brien E. A cohort study of cancer risk in relation to family histories of cancer in the Utah population database. Cancer. 2005;103:1906–15. doi: 10.1002/cncr.20989. [DOI] [PubMed] [Google Scholar]