Summary
Enterotoxigenic Escherichia coli (ETEC) use colonization factors to attach to the human intestinal mucosa, followed by enterotoxin expression that induces net secretion and diarrhoeal illness. ETEC strain H10407 expresses CFA/I fimbriae, which are composed of multiple CfaB structural subunits and a CfaE tip subunit. Currently, the contribution of these individual fimbrial subunits in intestinal binding remains incompletely defined. To identify the role of CfaE in attachment in the native ETEC background, an R181A single-amino-acid substitution was introduced by recombination into the H10407 genome. The substitution of R181A eliminated haemagglutination and binding of intestinal mucosa biopsies in in vitro organ culture assays, without loss of CFA/I fimbriae expression. Wild-type in trans plasmid-expressed cfaE restored the binding phenotype. In contrast, in trans expression of cfaE containing amino acid 181 substitutions with similar amino acids, lysine, methionine and glutamine did not restore the binding phenotype, indicating that the loss of the binding phenotype was due to localized areas of epitope disruption. R181 appears to have an irreplaceable role in the formation of a receptor-binding feature on CFA/I fimbriae. The results specifically indicate that the CfaE tip protein is a required binding factor in CFA/I-mediated ETEC colonization, making it a potentially important vaccine antigen.
Introduction
Enterotoxigenic Escherichia coli (ETEC) contribute significantly to the global endemic diarrhoeal disease burden, particularly in less-developed countries, and are leading causes of traveller's diarrhoea. The development of a safe and efficacious vaccine against ETEC is a public health priority. Development would be expedited if the specific epitopes against which protective immune responses must be directed were defined. ETEC use fimbrial CFAs to colonize the small intestine as a preliminary to heat-stable (ST) and/or heat-labile (LT) enterotoxin production that induces net intestinal secretion culminating clinically in watery diarrhoeal illness. Animal models using porcine ETEC pathogens expressing K88 or K99 fimbriae demonstrated an association between the presence of fimbriae and the ability to cause diarrhoea. In contrast, fimbriae-negative strains lost both the ability to colonize and to cause diarrhoea in piglets (Jones and Rutter, 1972; Rutter and Jones, 1973; Moon et al., 1977). The importance of fimbriae from human isolates as critical virulence factors for the elicitation of diarrhoeal illness was demonstrated in volunteer studies, as strains that lacked fimbriae lost the ability to cause diarrhoeal disease (Satterwhite et al., 1978). In this study, six out of seven volunteers ingesting wild-type strain H10407 experienced significant diarrhoeal illness. No volunteer who ingested 1 × 108 cfu of ETEC strain H10407P, which lacks the plasmid encoding the CFA/I fimbrial operon, experienced a diarrhoeal episode. In addition, all H10407P recipients showed a significantly reduced duration of bacterial shedding (Evans et al., 1978; Satterwhite et al., 1978). Correspondingly, the protective efficacy of immune responses targeted to fimbriae has been demonstrated in epidemiologic as well as clinical studies. Volunteers immunized with an oral whole-cell CFA/I-expressing ETEC vaccine and challenged with CFA/I-positive ETEC of a heterologous O : H serotype were protected against diarrhoea (Evans et al., 1988). Therefore, vaccine development strategies have focused on using the most clinically relevant CFAs to elicit immune responses that block attachment of ETEC to the mucosa of the small intestine, thereby inhibiting colonization at a critical site in the host.
At least 22 different types of antigenically distinct fimbrial CFAs have been identified among ETEC strains. One of the most commonly identified antigenic types identified in humans with diarrhoea in numerous epidemiological field surveys is CFA/I (Levine et al., 1993; Sommerfelt et al., 1996; Viboud et al., 1999; Qadri et al., 2005). CFA/I are composed of two types of protein: thousands of CfaB major structural subunits form a stalk that supports one or a few CfaE minor tip subunits. This ratio is based upon data showing that the closely related CFA/I-like fimbria, CS1, has a molecular ratio of structural protein subunits to minor tip protein of about 1800:1 (Sakellaris et al., 1996). Modelling of CFA/I has placed the CfaB subunits in a recurring interaction pattern that forms a helical structure, with the tip protein at the end acting as an adhesin (Buhler et al., 1991; Mu et al., 2008). While data supporting a role for CfaB and/or CfaE in mediating specific target cell binding are somewhat conflicting, results of recent studies increasingly indicate that CfaE is the critical binding subunit. A role for CfaB was suggested by studies showing that CfaB-specific monoclonal antibodies were able to inhibit CFA/I-mediated haemagglutination of human red blood cells (Buhler et al., 1991). On the other hand, in vitro studies of CFA/I fimbriae expressed in DH5α suggested that CfaE, and not CfaB, mediated haemag-glutination. When CFA/I was expressed in DH5α with a single-amino-acid mutation R181A in CfaE, fimbriae were expressed, but haemagglutination activity was abolished (Sakellaris et al., 1999). In another study, examination of purified individual protein subunits revealed that CfaE expressed and stabilized by donor strand complementation was able to haemagglutinate human red blood cells similarly to purified whole wild-type fimbriae while CfaB was not (Poole et al., 2007). As this relates to colonization blocking immunity, the Fab fraction of antibodies generated against the N-terminal fraction of CfaE(23-211) containing R181 were able to inhibit haemagglutination and Caco2 cell adherence to a much greater degree than antibodies to CfaB monomers or the CfaE(212–360) C-terminal fraction (Anantha et al., 2004).
The recent publication of the structure of CfaE allowed definition of which amino acids may be contributing to the conformation of the CfaE N-terminal domain, and thus might be playing a role in receptor binding (Li et al., 2007). However, the identification of receptor binding epitope(s) responsible for colonization of the human intestinal mucosa by wild-type ETEC have not been specifically characterized, nor have the immunogenic epitopes essential for generation of protective immune responses. In order to evaluate the contribution of R181 and CfaE to binding within the context of the native wild-type ETEC background, we generated the R181A mutation in cfaE within the genomic background of H10407, a prototype clinical ETEC isolate that expresses CFA/I. Our results demonstrate that CfaE is required for CFA/I fimbrial assembly and for recognition of receptors to mediate attachment of wild-type ETEC to human erythrocytes and to human small intestinal mucosa (obtained by biopsy). Our biochemical results that the R181 amino acid possesses characteristics that are a critical and an irreplaceable part of the conformation of a functional CfaE advance the understanding of how conformational, functional CfaE is formed.
Results
Transcription and translation of CFA/I operon genes in H10407 mutant derivatives
To further evaluate the role of the CfaE tip protein in the wild-type ETEC background, and the adhesion-mediating arginine amino acid 181, we generated H10407 ETEC derivatives containing mutations in the cfaE gene that allowed assessment of binding in the context of other potential ETEC-specific factors. The CFA/I operon contains four genes sufficient for fimbrial biogenesis including cfaA encoding a chaperone, cfaB encoding the structural subunit, cfaC encoding an usher and cfaE encoding the tip adhesion (Fig. 1). The H10407Kan derivative of H10407 was generated by insertion of a kanamycin-resistance gene into cfaE at base pair 5228 within the CFA/I operon (according to NCBI Accession #M55661) (Fig. 1). CFA/I-like fimbriae require expression of the tip protein to assemble fimbriae (Froehlich et al., 1994), and disruption of the cfaE gene abolished CFA/I surface assembly. H10407Kan served as the recipient strain for homologous recombination with the suicide plasmid, pCACcfaE, containing the AGA to GCA base pair mutations converting arginine 181 to alanine. Following transformation, colonies were screened for successful allelic exchange by loss of kanamycin resistance and confirmed by PCR amplification of the genomic region. Primers CFAE1 and CFAE2 flank the aph insertion site and were used to amplify cfaE (Fig. 1). These primers amplified an 841 bp fragment in wild-type H10407 and a 1901 bp fragment in H10407Kan (Fig. 2). Successful allelic exchange mediated by pCACcfaE within the H10407Kan background resulted in a PCR fragment of 841 base pairs lacking the aph insertion (Fig. 2, lane 4), identical to wild-type H10407. Sequencing confirmed that anti-CFA/I agglutination-positive clone KB101 possessed the AGA to GCA conversion without other inadvertently introduced mutations within cfaE or the upstream regions in the 3′ end of cfaC.
Fig. 1.
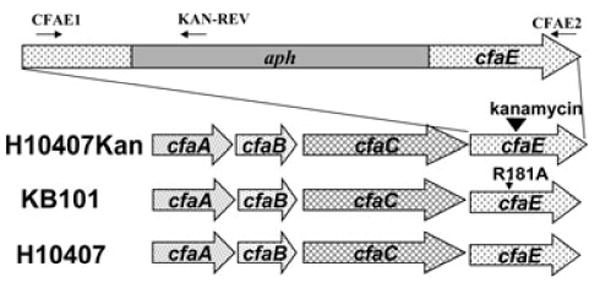
Genetic outline of the CFA/I operon. H10407 contains the wild-type operon. H10407Kan possesses an aph insertion at base pair 5228 within the cfaE gene of the CFA/I operon (NCBI M55661). KB101 contains base pair substitutions of AGA to GCA (base pairs 5323–5325). PCR and RT-PCR analyses were performed with primers CFAE1 and CFAE2 flanking the R181 region and the KAN-REV primer that anneals in aph.
Fig. 2.
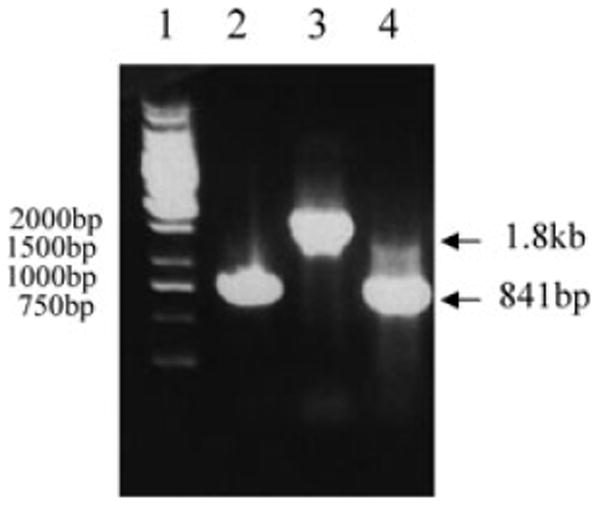
Genotypic confirmation of H10407 and derivatives by PCR. PCR was conducted with primers CFAE1 and CFAE2 using the following bacterial strains as templates: Lane 1, Fermentas 1 kb Generuler; lane 2, H10407; lane 3, H10407Kan and lane 4, KB101.
As CfaE is present on CFA/I fimbriae at very low copy numbers, it is not detected in Western blots using antifimbriae antibody (Sakellaris et al., 1996). Instead, the CfaB subunit is the predominant protein observed in Western blots and is used as a marker for translation of CFA/I operon proteins in general, in this case as a proxy for expression of CfaE. To examine whether the conversion of alanine to arginine in CfaE disrupted translation of CFA/I proteins, Western blot analysis using polyclonal anti-CFA/I fimbrial antibody was used to probe whole-cell lysates for detection of CfaB. Negative control strain H10407P, which lacks the 60 kDa plasmid bearing the CFA/I operon, lacked CfaB expression (Fig. 3, lane 3). CfaB was detected at equivalent levels in wild-type H10407 and KB101, while H10407Kan had little to no expression of the CfaB protein (Fig. 3, lanes 2, 5 and 4). Thus, the insertion mutation, but not the R181A mutation, in CfaE disrupted expression or stability of the CfaB structural protein and presumably, CfaE.
Fig. 3.
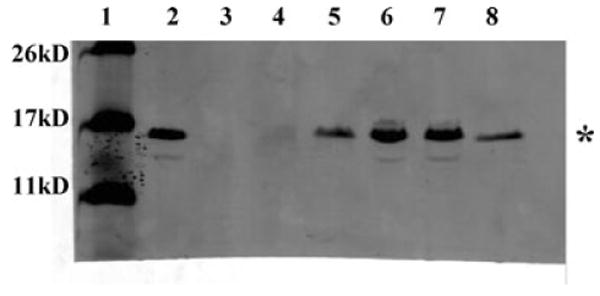
Western blot analysis of CfaB expression. Whole-cell lysates of ETEC strains were probed with anti-CFA/I polyclonal antibody. Lanes are as follows: 1, molecular weight markers; 2, H10407; 3, H10407P; 4, H10407Kan; 5, KB101; 6, H10407Kan(pcfaEgent); KB101(pcfaE). The asterisk (*) indicates CfaB major structural protein band at 15 kDa.
Reverse transcriptase PCR (RT-PCR) allowed evaluation of cfaE transcription from wild type and mutant ETEC derivatives. RT-PCR was conducted using the primers CFAE1 and CFAE2 that bind the region of the cfaE gene flanking the aph or R181A mutations (Fig. 1). The amplified fragment from KB101 was the same size as that from H10407 wild type, indicating that transcription was not disrupted by the amino acid substitution (Fig. 4A, lanes 7 and 4 respectively). Although transcription of cfaE from H10407Kan was not arrested, cfaE mRNA was disrupted with the insertion of aph resulting in a 1.9 kb fragment (Fig. 4A, lane 6). The KAN-REV primer was designed to anneal within the aph gene and amplified a 303-base-pair fragment in conjunction with primer CFAE1 confirming transcriptional fusion (Fig. 4B, lane 6). This band was not detected within KB101 confirming that allelic exchange resulted in loss of the kanamycin-resistance gene (Fig. 4B, lane 7). Controls lacking either RNA or reverse transcriptase enzyme, and the H10407P ETEC strain lacked cfaE transcript altogether, confirming that the RNA was uncontaminated and the primers specific (Fig. 4A, lanes 2, 3 and 5).
Fig. 4.
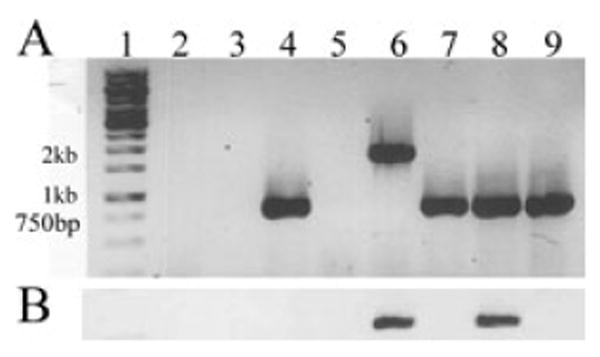
RT-PCR analysis of cfaE transcription in H10407 and derivative strains. Amplification of cDNA products was performed with primer pairs CFAE1 and CFAE2 (A), or CFAE1 and Kan-rev (B). Lanes are as follows: 1, Fermentas 1 kb Generuler marker; 2, control lacking reverse transcriptase enzyme; 3, control lacking RNA; 4, H10407; 5, H10407P; 6, H10407Kan; 7, KB101; 8, H10407Kan(pcfaEgent) and 9, KB101(pcfaE).
KB101 expresses CFA/I fimbriae
Point mutations in cfaE did not disrupt transcription of cfaE, or expression of CfaB protein, so strains were evaluated for the ability to assemble fimbriae on the bacterial surface. KB101 positively agglutinated with polyclonal anti-CFA/I fimbriae antibody (Table 1) and transmission electron microscopy revealed that KB101 expressed numerous hair-like fimbriae on its surface that were morphologically identical to CFA/I fimbriae found on wild-type H10407 (Table 1, Fig. 5A and C). Unequivocal recognition of fimbriae with anti-CFA/I antibody in an agglutination assay and in an immunogold EM (Fig. 6) confirmed the specificity of the fimbriae on KB101 as CFA/I. In contrast, H10407Kan was unable to assemble CFA/I fimbriae on the surface of the bacteria, consistent with the lack of CfaE and CfaB protein available (Fig. 5B, Table 1). Taken together, the data confirm that the single-amino-acid substitution of arginine 181 to an alanine did not affect transcription of cfaE, translation of CfaB protein or surface assembly of CFA/I fimbriae.
Table 1.
Anti-CFA/I agglutination and MRHA phenotype for ETEC strains.
| ETEC strain | α-CFA/I agglutination | Mrha of human erythrocytes |
|---|---|---|
| H10407 | + | + |
| H10407p | − | − |
| H10407kan | − | − |
| Kb101 | + | − |
| H10407KAN(pcfaEgent) | + | + |
| KB101(pcfaE) | + | + |
| H10407KAN(pcfaEgent-R181K) | + | − |
| KB101(pcfaE-R181K) | + | − |
| H10407KAN(pcfaEgent-R181M) | + | − |
| KB101(pcfaE-R181M) | + | − |
| H10407KAN(pcfaEgent-R181Q) | + | − |
| KB101(pcfaE-R181Q) | + | − |
| H10407KAN(pcfaEgent-R181A) | + | − |
| KB101(pcfaE-R181A) | + | − |
Fig. 5.
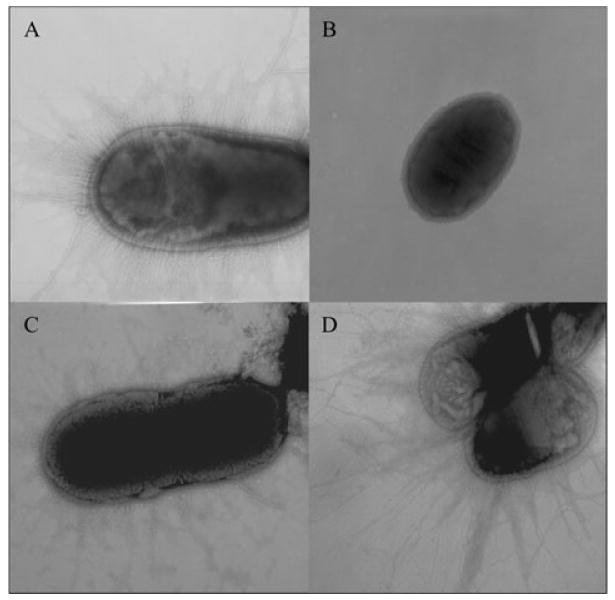
Expression of CFA/I fimbriae assessed by transmission electron microscopy. ETEC strains were stained with 2% ammonium molybdate, and photographed at 30× magnification.
A. H10407.
B. H10407Kan.
C. KB101.
D. H10407Kan(pcfaEgent).
Fig. 6.
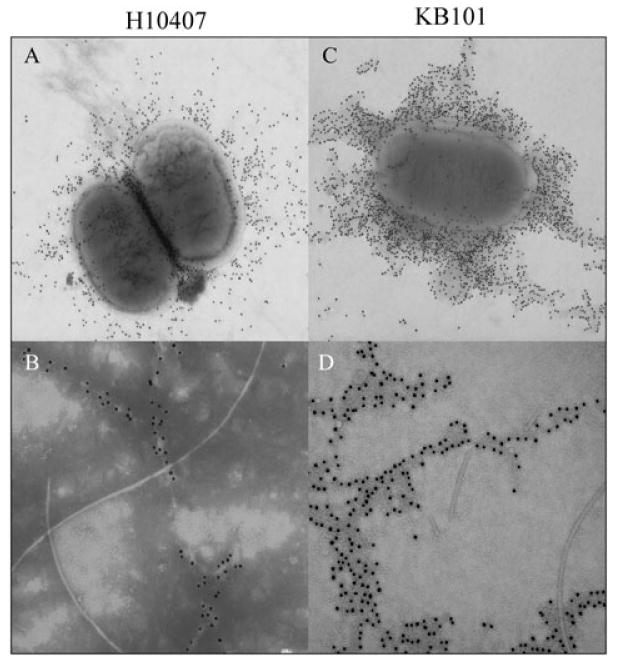
Confirmation of CFA/I fimbrial expression by immunogold electron microscopy. WT and mutant ETEC derivatives were stained with anti-CFA/I antibody.
A. H10407 at 30× magnification.
B. H10407 at 50× magnification.
C. KB101 at 25× magnification.
D. KB101 80× magnification.
Expression of cfaE in trans restores fimbrial assembly in H10407Kan
To confirm that no inadvertent mutations could be responsible for the observed phenotypes, H10407Kan and KB101 were transformed with a plasmid, pcfaE, containing wild-type cfaE, and tested for complementation. As H10407Kan already possessed kanamycin resistance, the gene for gentamicin resistance was cloned into pcfaE, generating pcfaEgent. Transformation of the plasmids into KB101 and H10407Kan generated KB101(pcfaE) and H10407Kan(pcfaEgent). It was expected that expression of a wild-type cfaE gene in trans would be able to complement mutations made in cfaE that disrupted fimbrial assembly in H10407Kan.
Western blot analysis confirmed that CfaB protein expression was restored in H10407Kan(pcfaEgent) and maintained in KB101(pcfaE) (Fig. 3, lanes 7 and 8). The restoration of CfaB protein expression in KB101 and the complemented strains suggests that CfaE is required for interaction with, and stabilization of, CfaB protein, resulting in initiation of fimbriae assembly. This is dissimilar to closely related CS1 fimbriae, wherein expression of the minor tip protein was not required to stabilize the major structural subunit but was required for initiation of fimbriae assembly (Froehlich et al., 1994). In spite of the homology between the two fimbriae, protein folding facilitates differing levels of stability for the protein subunits. This suggests that different mechanisms of regulation at a post-transcriptional level exist for homologous fimbriae.
Primers CFAE1, CFAE2 and KAN-REV were used in RT-PCR assays to evaluate whether cfaE was being transcribed in the complemented strains. As expected, the KB101(pcfaE) cfaE RNA transcript was present and the amplified fragment was identical in size to H10407 and KB101, suggesting that two types of CfaE, differing by one amino acid, might be simultaneously expressed (Fig. 4A, lane 9). Introduction of the wild-type cfaE gene on a plasmid in H10407Kan(pcfaEgent) resulted in amplification of the 841 bp wild-type size fragment. A second fragment of 1.9 kb was expected from the genomic cfaE copy containing the aph insertion. However, the polymerase appeared to preferentially amplify only the smaller cDNA copies in the PCR with CFAE1 and CFAE2 (Fig. 4A, lane 8). In a separate reaction, primers KAN-REV and CFAE1 amplified the expected 303 bp fragment following RT-PCR with the template H10407Kan(pcfaEgent) confirming cotranscription from the genomic cfaE∷aph locus. This fragment was only amplified from H10407Kan and H10407Kan(pcfaEgent), but not in wild-type H10407, KB101 or KB101(pcfaE). This suggests that the lack of a 1.9 kb fragment in the H10407Kan(pcfaEgent) strain was a false negative, and that the pcfaEgent plasmid did not usurp RNA transcription from the CFA/I operon. The preferential bias towards amplification of smaller fragments was found as well when the CFAE1, CFAE2 and KAN-REV primers were combined in a single reaction. Only the 303 bp fragment was amplified following RT-PCR using the H10407Kan and H10407Kan(pcfaEgent) templates, and not the expected 841 bp or 1.9 kb fragments (data not shown).
To test whether wild-type CfaE can complement fimbrial assembly, KB101(pcfaE) and H10407Kan (pcfaEgent) were examined by agglutination with anti-CFA/I, and by EM. As expected, KB101(pcfaE) demonstrated strong agglutination with anti-CFA/I (Table 1) and typical CFA/I fimbriae on the surface of the bacteria (data not shown). H10407Kan(pcfaEgent) also exhibited strong agglutination with anti-CFA/I, indicating complementation of fimbrial assembly (Table 1). Numerous fimbriae were visualized on the bacterial surface (Fig. 5D) that were identified as CFA/I by immunogold (Fig. 6D). Thus, restoration of wild-type CfaE protein by plasmid expression provided a functional tip protein that could interact with CfaB to initiate fimbrial assembly, like wild-type H10407.
R181 is required for binding in CFA/I-mediated mannose-resistant haemagglutination
CFA/I fimbrial expression on wild-type H10407 ETEC mediate specific adherence to human type A erythrocytes in a mannose-resistant manner. Unlike the rapid and strong H10407 mannose-resistant haemagglutination (MRHA) response to human erythrocytes, KB101 lacked haemagglutination activity, in spite of the CFA/I fimbriae assembled on the bacterial surface (Fig. 7D, and Table 1). This result was consistent with the previously published reports that CFA/I fimbriae bearing CfaE-R181A tip protein expressed from the pEU2124 plasmid in DH5α had a negative MRHA response (Sakellaris et al., 1999). Specific binding was restored when wild-type cfaE was provided in trans in KB101(pcfaE) (Fig. 7F, and Table 1). This supports the hypothesis that arginine 181 is critical for formation of a receptor binding epitope on CfaE. The loss of fimbrial surface expression on H10407Kan resulted in a loss of receptor binding to human type A red blood cells (Fig. 7C, and Table 1). This binding phenotype was also completely restored following expression of cfaE in trans, and thus fimbriae assembly, in H10407Kan (pcfaEgent) (Fig. 7E).
Fig. 7.
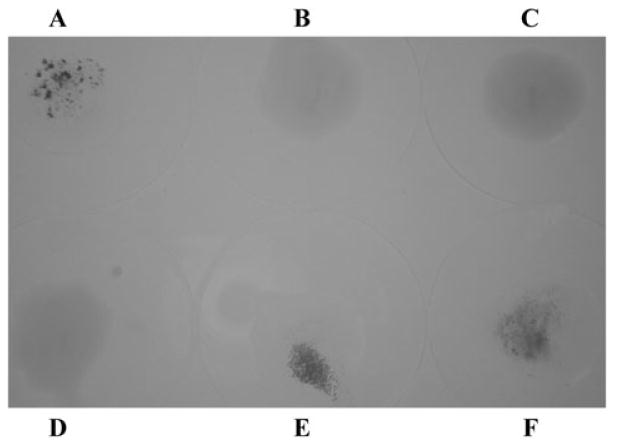
MRHA testing ETEC strain binding of human erythrocytes.
A. H10407.
B. H10407P.
C. H10407Kan.
D. KB101.
E. H10407Kan(pcfaEgent).
F. KB101(pcfaE). All strains were normalized to an OD600 of 1.0. Drops were made by combining 50 μl each of bacteria, 1% D-Mannose and 3% human Type A red blood cells and mixing with gentle stirring at room temperature.
As a negative control, KB101 and H10407Kan were transformed with pcfaE or pcfaEgent containing cfaE-R181A, generating KB101(pcfaE-R181A) and H10407 Kan(pcfaEgent-R181A). Like H10407Kan(pcfaEgent), H10407Kan(pcfaEgent-R181A) was complemented for restoration of fimbriae assembly, but not for MRHA activity with human type A erythrocytes (Table 1). Likewise, the MRHA functional binding phenotype was complemented in KB101(pcfaE), but not in KB101(pcfaE-R181A). These data cumulatively indicate that the R181A mutation in CfaE allows for expression of a protein that can initiate fimbrial biogenesis. However, the mutation caused loss of the features that enable MRHA, or receptor binding on human erythrocytes.
KB101 is deficient in adherence to human intestinal biopsy tissue
To evaluate binding in an assay that more accurately represents the native environment of the human small intestine, we utilized an In vitro organ culture (IVOC) assay, which has been used previously to evaluate binding of several pathogenic bacteria strains to the human intestinal epithelium. Small intestinal mucosal biopsy tissue was incubated in triplicate with strains H10407, H10407P or KB101 for 8 h using IVOC conditions and examined by scanning electron microscopy. In 3/3 sets of intestinal mucosa tissue incubated with wild-type ETEC H10407, large aggregates of bacteria were seen adhering to the mucosal surface (Fig. 8A). Numerous fimbriae were clearly visible extending from the bacterial surface and adhering to the mucosa at higher magnification (Fig. 8A inset). By contrast, strain H10407P, which lacks CFA/I fimbriae, did not attach to intestinal tissue in biopsies from the three paediatric subjects tested (Fig. 8B). Furthermore, it was determined that the attachment was also distinctly CfaE-mediated, as no KB101 bacteria were bound to the tissue in 2/3 samples and only a single bacillus and a small colony of adhering bacteria (binding to an area of basement epithelium where mucosa surface had disintegrated) were found in the third sample (Fig. 8C and inset). On the other hand, when KB101(pcfaE) was incubated with tissue samples, binding activity was restored and was identical to that seen with wild-type H10407; that is, large numbers of bacteria were attached to the mucosa and to each other in clusters (Fig. 8D and inset). This indicates that CfaE is specifically required for binding to the human intestinal epithelium by ETEC bearing CFA/I.
Fig. 8.
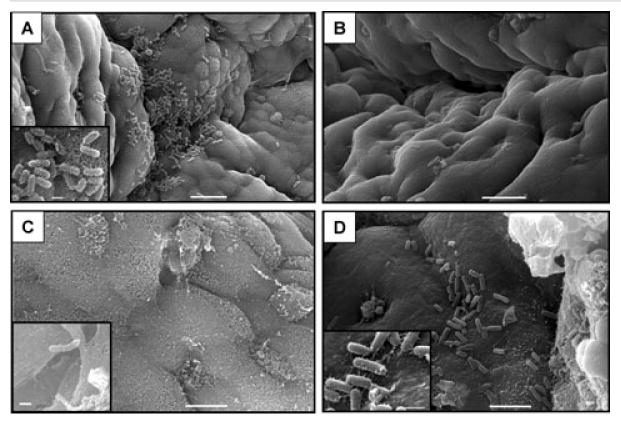
IVOC of ETEC strains with human small intestine.
A. H10407 adhering to mucosal surface (bar = 10 μm); inset shows bacterial detail (bar = 1 μm).
B. Surface detail of mucosa incubated with H10407P showing no evidence of bacterial adhesion (bar = 10 μm).
C. Mucosal surface without adhering bacteria in KB101 IVOC (bar = 5 μm); inset shows single adhering bacillus (bar = 1 μm).
D. KB101(pcfaE) demonstrating mucosal bacterial adhesion (bar = 5 μm); inset shows detail of adhering bacteria (bar = 1 μm).
R181 is required for formation of the binding epitope
The crystal structure of CfaE revealed that R181 lies buried within the pocket, perhaps acting to co-ordinate surrounding tertiary protein folding through its side-chains (Li et al., 2006). This suggests that the loss of MRHA for KB101 is caused either by an alteration in the binding epitope region through the single-amino-acid mutation, or by disruption of the integrity of a larger portion of the folded CfaE protein. Arginine and alanine are highly dissimilar amino acids, and it is possible that other amino acids with similar side-chain properties could substitute for arginine in the pocket at the tip of the CfaE protein on assembled CFA/I fimbriae. To evaluate the potential contribution of the arginine side-chain, single-amino-acid mutations of arginine 181 to lysine, glutamine or methionine (with alanine as a control for a plasmid-based expression effect) were generated in CfaE. The variant cfaE genes were cloned into the pSEC10 or pSEC10gent plasmids and transformed into KB101 or H10407Kan respectively, generating KB101(pcfaE-R181L), KB101(pcfaE-R181M), KB101(pcfaE-R181Q), KB101(pcfaE-R181A), H10407Kan(pcfaEgent-R181L), H10407Kan(pcfaEgent-R181M) and H10407Kan(pcfaEgent-R181Q), and H10407Kan(pcfaEgent-R181A).
Agglutination with anti-CFA/I antibody confirmed that expression of CfaE containing the R181L, R181M, R181Q and R181A mutations was still sufficient for restoration of CFA/I fimbrial assembly in the fimbriae-negative H10407Kan strain (Table 1). To compare the ability of these fimbriae to those of KB101 in formation of an epitope that binds receptors on human cells, the strains were evaluated for haemagglutination in a MRHA assay. Unlike in trans expression of the wild-type CfaE, neither lysine, methionine nor glutamine were capable of functionally replacing arginine for the complementation of functional binding in MRHA (Table 1). All KB101 and H10407Kan strains transformed with a non-wild-type cfaE lacked the ability to bind erythrocytes.
Discussion
Previous studies have presented compelling evidence for the role of the CfaE tip adhesin of CFA/I in mediating target cell binding (Sakellaris et al., 1999; Li et al., 2007; Poole et al., 2007). The data presented herein using derivatives of wild-type ETEC and a human IVOC binding assay provide direct evidence to support this hypothesis. Mutations were made in cfaE within the native ETEC genomic background, allowing assessment of the impact of an insertion mutation and an R181A amino acid mutation on attachment using a relevant new method. Using fresh human intestinal biopsy tissue to assess direct attachment of a clinically isolated ETEC strain adds relevance to the overall understanding of fimbriae-mediated colonization in the context of the complete milieu of other ETEC-specific potential binding factors. We demonstrated that a kanamycin gene inserted into cfaE resulted in the loss of protein expression from the CFA/I operon and a lack of fimbriae expression. In contrast, converting arginine 181 to an alanine in strain KB101 maintained expression of CFA/I proteins and allowed formation of a functional protein sufficiently capable of associating with the chaperone for initiation of fimbriae assembly and for proper interaction with the major structural subunit, as indicated by immunogold EM with anti-CFA/I antibody (Sakellaris and Scott, 1998). Thus, the mutation of arginine 181 to alanine in the ETEC background does result in expression of fimbriae like those characterized in the E. coli DH5α background (Sakellaris et al., 1999).
It has been previously demonstrated by X-ray crystallography that arginine 181 lies in the N-terminal domain of CfaE in a pocket found at the opposite end of the fimbrial subunit from where CfaE interacts with CfaB (Li et al., 2006). The CfaB structural protein stabilizes CfaE by cross-strand interaction of secondary structure with the CfaE C-terminal protein domain. R181 interacts with multiple side-chains of other amino acids in secondary loops buried within the terminal pocket, suggesting that the properties of its side-chains facilitate tertiary folding behaviour forming the human cell receptor-binding motif for CfaE. In terms of side-chain properties, the arginine to alanine substitution in KB101 eliminated the specific features of arginine that may have defined its role in protein folding. It is possible that the mutation caused complete disruption of the N-terminal domain, but sufficient proper folding of CfaE occurred to enable interaction with the CfaB structural subunit that resulted in assembly of full-length fimbriae on the bacterial surface. However, in spite of the formation of the fimbriae, the ability of strain KB101 to bind to receptors on human type A erythrocytes in MRHA assays, and more tellingly to human ileum, was completely lost.
The IVOC has been used as a relevant ex vivo binding assay in the study of several other pathogenic bacteria, including Salmonella (Haque et al., 2004), Shiga-toxin-producing E. coli (Schuller et al., 2004), enteroaggregative E. coli (Henderson et al., 1999), enteropathogenic E. coli and Citrobacter rodentium (Schuller et al., 2007), which in many cases previously relied upon more indirect methods for evaluating pathogenesis. IVOC technology has been established before as an accurate relevant method for studying CFA/I-mediated adherence of ETEC to intact intestinal mucosal biopsies (Knutton et al., 1985; 1987; 1989). However, herein for the first time we describe its use to screen the role of individual CFA/I proteins in binding. Our results validate the fimbriae-mediated binding hypothesis and confirm the validity of using other more-rapid and less-complicated in vitro binding assays, like haemagglutination, as proxies. The significance of the use of the IVOC system (with human tissue) is underscored by the lack of good animal models to study ETEC pathogenesis. It is anticipated that this model could be used to study the adherence mediated by other ETEC CFAs and potentially binding inhibition strategies.
When full-length wild-type cfaE was expressed in trans in KB101, it restored the ability of the bacteria to bind both erythrocytes and ileal mucosa from human paediatric intestinal biopsies. This phenotype confirmed both that there were no inadvertent mutations introduced during recombination that altered other CFA/I fimbrial proteins, and that the arginine substitution in particular was the specific cause for loss of binding. This effect was seen even more clearly when cfaE was expressed in H10407Kan, due to restoration of both fimbriae expression and binding. Taken together, the evidence we generated confirms that CFA/I fimbriae are responsible for the direct binding of H10407 ETEC to the human intestinal mucosa and that CfaE, specifically, and not CfaB, is the subunit that actually mediates attachment to receptors on human cells. It also confirms that R181 is required for formation of the receptor recognizing epitope on the N-terminal domain of CfaE, either through direct receptor interaction, by co-ordinating minor secondary chain interactions and/or spatial occupation, or through stabilization of the globular structure of the N-terminal domain altogether.
To examine what properties of arginine might be important for its ability to form a binding epitope, we tested the ability of amino acids bearing similar biochemical, structural or functional properties for their ability to complement the arginine mutation and allow binding to human host cells. Arginine and lysine are most similar in that both are positively charged, polar amino acids that characteristically play an important role in protein structure and are frequently found in protein active or binding sites (Betts and Russell, 2003). Generally, the positively charged amino end of their side-chains results in surface localization, but the presence of the long hydrophobic carbon-containing backbone also permits part of the amino acid to be buried. Lysine is the most commonly substituted amino acid tolerated in place of arginine, with glutamine being second. Glutamine also is usually involved in protein active and binding sites, but is not amphipathic, more often localizing to protein surfaces and an aqueous environment. Its polar side-chain permits interaction with other polar or charged surfaces. In contrast, methionine is non-polar, uncharged and hydrophobic, so it is typically buried. It usually does not play a role in formation of functional protein features, but can occasionally be found substituting for arginine. However, the methionine side-chain does fill more space and thus is still distinctly different from the simple alanine substitution in KB101.
In H10407Kan, expression of CfaE containing substitutions of alanine, lysine, glutamine or methionine were all sufficient for restoring fimbriae expression, but unlike expression of wild-type CfaE in H10407Kan(pcfaEgent), they were not sufficient for formation of an epitope that binds receptors on human cells. Introduction of the R181 variants in KB101 also failed to restore functional binding, confirming that a particular feature of the arginine is specifically required for epitope formation or binding interactions. This suggests that individual characteristics of arginine, such as polarity, possession of an amphipathic side-chain, side-chain spatial occupancy or positively charged side-chains, are not individually controlling bonding interactions resulting in formation of the epitope, but rather arginine's properties are required for multiple interactions, resulting in a cumulative effect on folding of functional CfaE. Another possible important feature of arginine is the complex guanidinium group on the end of the side-chain that allows the formation of multiple hydrogen bonds with negatively charged phosphates. Lysine has only one amino group limiting the number of potential hydrogen bond formations with phosphates. The extra guanidinium amino group on arginine spatially occupies more area that could gap-fill in a manner that arranges surrounding interacting side-chains in an epitope-governing pattern.
Given the data provided herein confirming a critical role for CfaE in attachment to the mucosa, adapting vaccination strategies for development of a colonization blocking vaccine seems likely to be successful. In fact, the ability of antibodies developed specifically against the N-terminal domain of the CfaE tip protein to confer similar levels of adherence blocking activity in in vitro assays as antibodies to CFA/I whole fimbriae, CfaE or closely related hetero-logous tip adhesin antigens prompted Anantha and colleagues to suggest that attachment blocking vaccines might offer a broader yet more effectively targeted level of protection (Anantha et al., 2004). It is possible, however, that immune recognition of the CfaE tip protein of CFA/I fimbriae is not absolutely required for effective protective responses. Given the high ratio of CfaB to CfaE in the fimbriae, the immune system is much more likely to detect the CfaB structural protein. As anti-CFA/I antibodies composed of anti-CfaB and anti-CfaE fractions, as well as anti-CfaB monoclonal antibodies, are capable of inhibiting haemagglutination of CFA/I-expressing bacteria, natural immune responses against CfaB are very likely involved in blocking adherence of bacteria (Buhler et al., 1991). This could possibly be due to steric interaction of the Fc portion of the antibody as anti-CfaB Fab antibody was unable to block MRHA (Anantha et al., 2004). This cumulatively suggests that an anti-CFA/I vaccine response could trigger immune responses for clearing ETEC infections. This cumulative response would be most effective by taking advantage of anti-CfaE adherence blocking in combination with anti-CfaB epitope recognition. Understanding the importance of individual fimbrial proteins in attachment and in generation of protective immunity will ultimately lead to more effective vaccine development strategies.
Experimental procedures
Antiserum
Polyclonal antiserum against CFA/I fimbriae was prepared by immunizing rabbits with CFA/I fimbriae purified from H10407. Sera were stored at 4°C.
Bacterial strains and culture conditions
Plasmid constructs were maintained in one shot E. coli strains (Invitrogen, Carlsbad, CA). Strains were grown in Luria–Bertani (LB) medium alone or with agar containing appropriate antibiotics at concentrations as follows. Antibiotics added to liquid or solid media were kanamycin at a concentration of 50 μg ml−1, nalidixic acid at 20 μg ml−1, carbenicillin at 50 μg ml−1, gentamicin at 15 μg ml−1 or chloramphenicol at 25 μg ml−1. Strain H10407, isolated from a Bangladeshi patient with watery diarrhoea, is the prototype ETEC strain that expresses CFA/I fimbriae (Evans et al., 1972; Evans and Evans, 1973). H10407P is a derivative of H10407 that has lost the 60 Mdal plasmid containing the CFA/I operon. H10407P was generated by serial passage of H10407 over 4 years of time on artificial media (Evans et al., 1975). ETEC strains were grown on CFA solid media containing 1% Casamino acids, 0.15% Bacto Yeast Extract, 0.005% MgSO4, 0.0005% MnCl2, 1.5% Difco agar (Becton Dickinson, Franklin Lakes, NJ). Wild-type ETEC or E. coli carrying a plasmid encoding CFA/I were streaked from frozen master stocks stored at −78°C onto CFA agar with appropriate antibiotic(s), and were incubated at 37°C overnight to obtain isolated colonies. A spontaneous nalid-ixic acid resistant clone of H10407 was selected by streaking a heavy inoculum onto media with nalidixic acid. Resultant colonies were validated for CFA/I expression by agglutination and haemagglutination of human type A red blood cells. H10407Kan contains an aph gene insertion at a HindIII site at approximately the middle of cfaE. It was created by allelic exchange using the suicide plasmid pKTN-cfa∷kan. Plasmid pKTN-cfa∷kan was created by cloning the cfaE gene and flanking sequences from H10407 into the vector pKTN701 (Nishibuchi et al., 1991). The aph gene amplified from pGEN9-aph (Koprowski et al., 2000) as a 1.060 kb fragment with HindIII sites on the ends was inserted into the unique HindIII site at base pair 5228 of the CFA/I operon (NCBI M55661) in the cfaE gene. Plasmid pKTN701 contains the RP4 origin of transfer to allow conjugation, the oriR6K origin of replication that does not function in E. coli and a chloramphenicol-resistance marker. Plasmid pKTN-cfa∷kan was mobilized for conjugation into H10407-Nal. Nalidixic acid-, kanamycin- and chloramphenicol-resistant colonies were selected. Next, cultures were passed on media without chloram-phenicol and colonies that were kanamycin- and nalidixic acid-resistant but chloramphenicol-sensitive were identified. The insertion created a fimbriae-negative phenotype, as confirmed by the lack of agglutination with anti-CFA/I. H10407Kan is useful as a target strain for the introduction of unmarked mutations into cfaE.
Molecular genetic constructs
Standard techniques were used to construct the plasmids. Taq DNA polymerase or Platinum Pfx or Pfu DNA polymerase was used in PCR amplification according to manufacture's recommendations (Invitrogen). One shot E. coli were electro-porated according to manufacture's recommendations. ETEC were electroporated with recombinant plasmids at 2.5 kV, 200 ohms. All transformed cultures were grown overnight on selective media with appropriate antibiotic(s) at 37°C, unless otherwise indicated. Isolated transformants were frozen as master stocks using LB containing 50% glycerol and stored at −78°C.
Plasmid construction and recombination of R181A ETEC strain
Primers used for amplification of the cloned alleles are described in Table 2. The plasmid pEU2124 (kindly provided by June Scott) contains the entire CFA/I operon including an AGA to GCA base pair mutation that converts arginine 181 within cfaE to an alanine (Sakellaris et al., 1999). An 841 bp (base pairs 5041–5865 according to NCBI accession #M55661) fragment of the cfaE-R181A gene was amplified from pEU2124 using primers CFAEPstI and CFAEBamHI, which included 5′ and 3′ terminal restriction enzyme sites for PstI and BamHI respectively. The fragment was cloned into TOPO pCR Blunt according to manufacturer's recommendations (Invitrogen). The resultant plasmid, named pTO-POcfaE, was digested in parallel with the temperature-sensitive suicide vector, pCactus (Eisenstein, 1991), using PstI and XbaI restriction enzymes. The 841 bp fragment of cfaE-R181A and the 7962 bp fragment of pCactus were ligated overnight, and then transformed into DH5α for overnight growth at 30°C. One chloramphenicol-resistant clone, pCACcfaE, was sequenced to confirm that the cfaE insert contained the GCA base pairs encoding alanine instead of arginine.
Table 2.
Primer sequences.
| Primers | Sequence |
|---|---|
| CFAEPstI | ggtaccctgcaggtgatgcccctggcactgctac |
| CFAEBamHI | ggtaccggatccctagagtgtttgactacttggtg |
| CFAB1 | gcaagctgatggcaatgctctg |
| CFAB2 | tcctgaatagtttcctgcagttg |
| CFAE1 | gtgatgcccctggcactgctac |
| CFAE2 | ctagagtgtttgactacttggtg |
| CFAE3f | gtacggtgattattcgctagggagcaacg |
| CFAE4r | atgagcaaggctgcgttattgctgc |
| CFAE5r | gagatgagttaagtgctagtttagatgggc |
| CFAEBamHIf | ggatccaaaggataaacgatgaata |
| CFAENheIr | gctagcgttatctagagtgtttgact |
| XHOGENf | cccctcgagcaccgtggaaacggatgaaggcacg |
| XHOGENr | cccctcgagttaggtggcggtacttgggtcgata |
| pSECf | gagaatggacttgccgactga |
| pSECr | cggtgcctgactgcgttagcaat |
| CFAER181Lf | gaagctaaatgtaaaaaaacgatatgatacaacc |
| CFAER181Mf | gaagctaaatgtaaaaatgcgatatgatacaacc |
| CFAER181Qf | gaagctaaatgtaaaacaacgatatgatacaacc |
| CFAER181Lr | ggttgtatcatatcgtttttttacatttagcttc |
| CFAER181Mr | ggttgtatcatatcgcatttttacatttagcttc |
| CFAER181Qr | ggttgtatcatatcgttgttttacatttagcttc |
| KAN-REV | gctgaagagcttggcggcgaatg |
The nalidixic acid-resistant strain of H10407Kan was used as the recipient strain for conjugation with the suicide vector bearing mutagenic cfaE to allow for screening of allelic exchange via kanamycin sensitivity. The sequenced pCACcfaE plasmid was transformed into SM10lpir. SM10λpir(pCACcfaE) and H10407Kan were grown separately to an OD600 of 0.6 and mixed in equal volumes. After incubation for 1 h at 37°C, the bacterial culture was transferred to LB agar for overnight conjugation. Colonies from the overnight conjugation were patched onto nalidixic acid and chloramphenicol-containing solid media for overnight culture at 42°C to isolate nalidixic acid-resistant H10407Kan with pCACcfaE integrated into the ETEC genome. Three resultant ETEC colonies were transferred to LB agar containing 10% sucrose plus nalidixic acid and grown at 30°C over 2 days to select for loss of the plasmid from the genomic back-bone. Resultant colonies were then screened in triplicate on sucrose plus nalidixic acid, kanamycin or chloramphenicol plates to screen colonies for successful recombination by loss of kanamycin resistance, and for curing of the plasmid by restoration of chloramphenicol sensitivity.
DNA sequence analysis of one resultant clone, named KB101, using primers CFAE1 and CFAE2 confirmed that KB101 possessed an intact cfaE gene with no alterations other than the AGA to GCA recombination mutation (Fig. 1). Validation of the conservation of the surrounding genomic sequence in the region up- and downstream of the cfaE gene was verified by further sequencing of amplified PCR products of KB101 between 4278 and 6185 bp (M55661) using forward primer CFAE3f and reverse primer CFAE4r, as well as reverse primer CFAE5r. Further sequence analysis using primers CFAB1 and CFAB2 confirmed that no alterations occurred during recombination to the cfaB upstream region.
Plasmid construction for in trans expression of cfaE
The pSEC10 low-copy-number stable expression plasmid was a gift from J. Galen (Galen et al., 2004). The plasmid used to study complementation of the wild-type cfaE gene in KB101, pcfaE, was constructed by amplifying the cfaE allele from base pairs 4771 to 5870 in the CFA/I operon from wild-type H10407 ETEC using primers CFAEBamHIf and CFAENheIr. The 1112 bp PCR product was cloned into TOPO pCR Blunt, followed by restriction enzyme digestion of the fragment and ligation into BamHI and NheI sites on the pSEC10 backbone. Colonies were screened by restriction enzyme digestion and by PCR using the CFAEBamHIf and CFAENheIr primers.
As H10407Kan possesses the aph gene conferring resistance to kanamycin, it was necessary to include an alternate antibiotic selection marker prior to transformation of the plasmid into H10407Kan. Cloning a gene for gentamicin resistance into the XhoI site of pSEC10 and pcfaE generated the pSEC10gent and pcfaEgent plasmids respectively. Gentamicin primers were designed with 5′ XhoI sites: XHOGENf and XHOGENr. PCR was used to amplify the gene encoding gentamicin resistance from pSS1129 (Stibitz and Yang, 1991), which was then subcloned into TOPO pCR Blunt. Digestion of the plasmid with XhoI released the gentamicin-resistance gene for ligation into a XhoI cleavage site in pSEC10 and pcfaE. Colonies were cultured in LB media containing gentamicin for the purification of plasmid with the Qiagen miniprep kit (Qiagen, Valencia, CA). Plasmids were further screened by PCR using primers flanking the insertion site: pSECf and pSECr, followed by sequencing of both pcfaE and pcfaEgent plasmids to confirm integrity of the amplification and cloning process. The pcfaE plasmid was then transformed into KB101 and the pcfaEgent plasmid was transformed into H10407Kan. Isolated plasmid-containing ETEC colonies were isolated following overnight growth on CFA agar with either kanamycin or gentamicin respectively, and named KB101(pcfaE) and H10407Kan(pcfaEgent).
Plasmid construction for in trans expression of cfaE amino acid mutants
The PCR site-directed mutagenesis was used to generate single-amino-acid mutations from arginine to lysine, methionine or glutamine in cfaE for cloning and expression within the pSEC10 plasmid backbone. Forward and reverse primers containing site-directed base pair mutations of the R181 base pairs (AGA) to L181 (AAA), M181 (ATG) or Q181 (CAA) were engineered to create overlapping oligonucleotides during two-step PCR of cfaE when used with CFAEBamHIf and CFAENheIr. Overlapping primers used during two-step PCR were: for R181L conversion, CFAER181L and CFAER181Lr; for R181M conversion, CFAER181Mf and CFAER181Mr; and for R181Q conversion, CFAER181Qf and CFAER181Qr. CFAEBamHIf was used with each of the reverse primers, and CFAENheIr was used with the matching overlapping forward primer to amplify cfaE* in two parts from the original pCFAE plasmid. Those overlapping PCR products were then combined during the second step by amplification with CFAEBamHIf and CFAENheIr generating full-length cfaE-R181L, cfaE-R181M or cfaE-R181Q. These PCR fragments were subcloned through Zero Blunt TOPO (Invitrogen), excised with BamHI and NheI and ligated into compatible sites within pSEC10 or pSEC10gent, creating: pcfaE-R181L, pcfaE -R181M, pcfaE-R181Q or pcfaEgent-R181L, pcfaEgent-R181M and pcfaEgent-R181Q respectively. Clones were screened on media containing kanamycin or gentamicin and sequenced, prior to transformation into ETEC. KB101 were transformed individually with the pSEC10 plasmids creating: KB101(pcfaE-R181L), KB101(pcfaE-R181M), KB101(pcfaE-R181Q). H10407Kan were transformed individually with the pSEC10gent plasmids creating H10407Kan(pcfaEgent-R181L), H10407Kan(pcfaEgent-R181M) and H10407Kan(pcfaEgent-R181Q).
Reverse transcriptase PCR
Bacteria were grown on CFA agar plates, and resuspended in PBS to an OD600 of 0.5, followed by lysis and RNA isolation using the Qiagen RNeasy kit (Qiagen). Reverse transcription was conducted using the Qiagen One-Step RT-PCR kit according to the manufacturer's protocols. Primers CFAE1 and CFAE2 were used with the resultant cDNA template to detect cfaE. The expected PCR product for wild-type cfaE was an 841 bp product and for cfaE∷aph, a 1901 bp product. CFAE1 and KAN-REV primers were used to confirm the genomic H10407Kan background for the presence of the aph insertion. A PCR product of 303 bp indicated positive RNA transcript for the cfaE gene containing aph at the 5228 bp location in the ETEC genome.
Western blot analysis
Bacteria were grown on CFA agar plates and resuspended in PBS to an OD600 of 0.5, followed by centrifuging at 6000 g for 1 min and resuspension in 2× Laemmli protein loading buffer (Bio-Rad, Hercules, CA) plus 0.5% β-mercaptoethanol. Boiling for 10 min lysed the bacteria. Samples of 10 μl were loaded on a 10% SDS polyacrylamide gel and electrophoresed at 150 mV. Gels were either stained for 1 h with Gelcode Blue and destained in water, or were transferred to PVDF membrane at 200 mA for 1 h. Membranes were blocked in a 1:1 dilution of Odyssey Blocking Solution (Li-Cor Biosciences, Nebraska) in PBS for 1 h, and then incubated overnight in a 1:7000 dilution of anti-CFA/I polyclonal antibody in blocking solution with vigorous shaking at 4°C. Membranes were washed four times for 5 min each in PBS, plus 0.1% Tween-20. Alexa-Fluor 680 goat anti-rabbit IgG (H and L) secondary antibody from Invitrogen was diluted 1:5000 in blocking solution and incubated for 1 h at room temperature, then washed again. Blots were scanned using an Odyssey Infrared Imaging System (Li-Cor).
Transmission electron microscopy
Bacteria were grown overnight on CFA agar and resuspended in PBS to an OD600 of 1.0. Drops of 50 μl bacterial suspensions were incubated with 300 mesh Formvar coated copper grids (Electron Microscopy Services, Hatfield, PA) for 20 min. Grids were gently blotted and placed on 30 μl drops of 2% Ammonium molybdate (Sigma-Aldrich, St Louis, MO) for 2 min followed by blotting. After air-drying, grids were examined using a JEOL electron microscope JEM-1200EX (JEOL, Toyko, Japan).
Immunogold EM
Bacteria were grown overnight on CFA agar and resuspended in PBS. EMS 200 mesh Formvar coated nickel grids were incubated with 50 μl droplets of the bacterial sample. Grids were blotted and incubated for 30 min with blocking solution composed of 5% BSAc (EMS), 0.1% CWFS gelatin (EMS). Grids were blotted and incubated with 1:500 dilutions of polyclonal anti-CFA/I in 5% BSAc for 1 h. Grids were blotted and washed six times in 50 μl droplets of 5% BSAc for 5 min each. Grids were then incubated with 50 μl drops of a 1:20 dilution of 10 nm gold-labelled goat anti-rabbit IgG (H and L) in 5% BSAc for 1 h. Grids were blotted and washed six times in 50 μl droplets of 5% BSAc for 5 min each, followed by negative staining with 2% Ammonium molybdate for 2 min 30 s. Grids were examined in a JEOL electron microscope JEM-1200EX.
Agglutination
A slide agglutination test was used to confirm expression of CFA/I fimbriae prior to all assays. Fimbriated bacteria were agglutinated by gently mixing 20 μl of a 1:10 dilution of anti-CFA/I polyclonal rabbit sera with 40 μl of resuspended bacteria at an OD600 of 1.0 on a glass microscope slide.
Haemagglutination
Bacteria were harvested from CFA/Nal agar into 0.15 M NaCl and standardized to an OD600 of 1.0. Fresh human type A red blood cells were washed 3× and finalized in a 3% (v/v) solution in 0.15 M NaCl. Drops of 50 μl each of 3% red blood cells and 1% D-Mannose were mixed with 50 μl of bacteria on a Falcon six-well polystyrene tissue culture plate at room temperature.
Human intestinal IVOC
Paediatric tissue was obtained with fully informed parental consent and local ethical committee approval using grasp forceps during routine endoscopic investigation of intestinal disorders. Small intestinal mucosal biopsies that appeared macroscopically normal were taken for organ culture experiments as described previously (Hicks et al., 1998). Biopsies were infected with wild-type H10407, H10407P, KB101 and KB101(pcfaE) for 8 h. An uninfected biopsy was included in each experiment to exclude endogenous bacterial infection.
Acknowledgments
We are grateful to Drs Franco Torrente and Camilla Salvestrini for providing endoscopic biopsies for human intestinal IVOC experiments, and to the Crohn's and Colitis in Childhood Charity for funding J. Morison. This work was funded by Grants RO1-AI059223 to E.M.B. and RO1-AI29471 to M.M.L.
References
- Anantha RP, McVeigh AL, Lee LH, Agnew MK, Cassels FJ, Scott DA, et al. Evolutionary and functional relationships of colonization factor antigen i and other class 5 adhesive fimbriae of enterotoxigenic Escherichia coli. Infect Immun. 2004;72:7190–7201. doi: 10.1128/IAI.72.12.7190-7201.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts MJ, Russell RB. Bioinformatics for Geneticists. Hoboken, NJ: John Wiley & Sons; 2003. [Google Scholar]
- Buhler T, Hoschutzky H, Jann K. Analysis of colonization factor antigen I, an adhesin of enterotoxigenic Escherichia coli O78: H11: fimbrial morphology and location of the receptor-binding site. Infect Immun. 1991;59:3876–3882. doi: 10.1128/iai.59.11.3876-3882.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenstein E. Cloning, expression, purification, and characterization of biosynthetic threonine deaminase from Escherichia coli. J Biol Chem. 1991;266:5801–5807. [PubMed] [Google Scholar]
- Evans DJ, Jr, Evans DG. Three characteristics associated with enterotoxigenic Escherichia coli isolated from man. Infect Immun. 1973;8:322–328. doi: 10.1128/iai.8.3.322-328.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DJ, Jr, Chen LC, Curlin GT, Evans DG. Stimulation of adenyl cyclase by Escherichia coli enterotoxin. Nat New Biol. 1972;236:137–138. doi: 10.1038/newbio236137a0. [DOI] [PubMed] [Google Scholar]
- Evans DG, Silver RP, Evans DJ, Jr, Chase DG, Gorbach SL. Plasmid-controlled colonization factor associated with virulence in Escherichia coli enterotoxigenic for humans. Infect Immun. 1975;12:656–667. doi: 10.1128/iai.12.3.656-667.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DG, Satterwhite TK, Evans DJ, Jr, DuPont HL. Differences in serological responses and excretion patterns of volunteers challenged with enterotoxigenic Escherichia coli with and without the colonization factor antigen. Infect Immun. 1978;19:883–888. doi: 10.1128/iai.19.3.883-888.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DG, Evans DJ, Jr, Opekun AR, Graham DY. Non-replicating oral whole cell vaccine protective against enterotoxigenic Escherichia coli (ETEC) diarrhea: stimulation of anti-CFA (CFA/I) and anti-enterotoxin (anti-LT) intestinal IgA and protection against challenge with ETEC belonging to heterologous serotypes. FEMS Microbiol Immunol. 1988;1:117–125. doi: 10.1111/j.1574-6968.1988.tb02363.x. [DOI] [PubMed] [Google Scholar]
- Froehlich BJ, Karakashian A, Melsen LR, Wakefield JC, Scott JR. CooC and CooD are required for assembly of CS1 pili. Mol Microbiol. 1994;12:387–401. doi: 10.1111/j.1365-2958.1994.tb01028.x. [DOI] [PubMed] [Google Scholar]
- Galen JE, Zhao L, Chinchilla M, Wang JY, Pasetti MF, Green J, Levine MM. Adaptation of the endogenous Salmonella enterica serovar Typhi clyA-encoded hemolysin for antigen export enhances the immunogenicity of anthrax protective antigen domain 4 expressed by the attenuated live-vector vaccine strain CVD 908-htrA. Infect Immun. 2004;72:7096–7106. doi: 10.1128/IAI.72.12.7096-7106.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque A, Bowe F, Fitzhenry RJ, Frankel G, Thomson M, Heuschkel R, et al. Early interactions of Salmonella enterica serovar typhimurium with human small intestinal epithelial explants. Gut. 2004;53:1424–1430. doi: 10.1136/gut.2003.037382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Hicks S, Navarro-Garcia F, Elias WP, Philips AD, Nataro JP. Involvement of the enteroaggregative Escherichia coli plasmid-encoded toxin in causing human intestinal damage. Infect Immun. 1999;67:5338–5344. doi: 10.1128/iai.67.10.5338-5344.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks S, Frankel G, Kaper JB, Dougan G, Phillips AD. Role of intimin and bundle-forming pili in enteropathogenic Escherichia coli adhesion to pediatric intestinal tissue in vitro. Infect Immun. 1998;66:1570–1578. doi: 10.1128/iai.66.4.1570-1578.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones GW, Rutter JM. Role of the K88 antigen in the pathogenesis of neonatal diarrhea caused by Escherichia coli in piglets. Infect Immun. 1972;6:918–927. doi: 10.1128/iai.6.6.918-927.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutton S, Lloyd DR, Candy DC, McNeish AS. Adhesion of enterotoxigenic Escherichia coli to human small intestinal enterocytes. Infect Immun. 1985;48:824–831. doi: 10.1128/iai.48.3.824-831.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutton S, Lloyd DR, McNeish AS. Identification of a new fimbrial structure in enterotoxigenic Escherichia coli (ETEC) serotype O148: H28 which adheres to human intestinal mucosa: a potentially new human ETEC colonization factor. Infect Immun. 1987;55:86–92. doi: 10.1128/iai.55.1.86-92.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutton S, McConnell MM, Rowe B, McNeish AS. Adhesion and ultrastructural properties of human enterotoxigenic Escherichia coli producing colonization factor antigens III and IV. Infect Immun. 1989;57:3364–3371. doi: 10.1128/iai.57.11.3364-3371.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprowski H, 2nd, Levine MM, Anderson RJ, Losonsky G, Pizza M, Barry EM. Attenuated Shigella flexneri 2a vaccine strain CVD 1204 expressing colonization factor antigen I and mutant heat-labile enterotoxin of enterotoxigenic Escherichia coli. Infect Immun. 2000;68:4884–4892. doi: 10.1128/iai.68.9.4884-4892.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine MM, Ferreccio C, Prado V, Cayazzo M, Abrego P, Martinez J, et al. Epidemiologic studies of Escherichia coli diarrheal infections in a low socioeconomic level peri-urban community in Santiago, Chile. Am J Epidemiol. 1993;138:849–869. doi: 10.1093/oxfordjournals.aje.a116788. [DOI] [PubMed] [Google Scholar]
- Li YF, Poole S, Rasulova F, Esser L, Savarino SJ, Xia D. Crystallization and preliminary X-ray diffraction analysis of CfaE, the adhesive subunit of the CFA/I fimbriae from human enterotoxigenic Escherichia coli. Acta Crystallograph Sect F Struct Biol Cryst Commun. 2006;62:121–124. doi: 10.1107/S1744309105043198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YF, Poole S, Rasulova F, McVeigh AL, Savarino SJ, Xia D. A receptor-binding site as revealed by the crystal structure of CfaE, the colonization factor antigen I fimbrial adhesin of enterotoxigenic Escherichia coli. J Biol Chem. 2007;282:23970–23980. doi: 10.1074/jbc.M700921200. [DOI] [PubMed] [Google Scholar]
- Moon HW, Nagy B, Isaacson RE, Orskov I. Occurrence of K99 antigen on Escherichia coli isolated from pigs and colonization of pig ileum by K99+ enterotoxigenic E. coli from calves and pigs. Infect Immun. 1977;15:614–620. doi: 10.1128/iai.15.2.614-620.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu XQ, Savarino SJ, Bullitt E. The three-dimensional structure of CFA/I adhesion pili: traveler's diarrhea bacteria hang on by a spring. J Mol Biol. 2008;376:614–620. doi: 10.1016/j.jmb.2007.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishibuchi M, Kumagai K, Kaper JB. Contribution of the tdh1 gene of Kanagawa phenomenon-positive Vibrio parahaemolyticus to production of extracellular thermostable direct hemolysin. Microb Pathog. 1991;11:453–460. doi: 10.1016/0882-4010(91)90042-9. [DOI] [PubMed] [Google Scholar]
- Poole ST, McVeigh AL, Anantha RP, Lee LH, Akay YM, Pontzer EA, et al. Donor strand complementation governs intersubunit interaction of fimbriae of the alternate chaperone pathway. Mol Microbiol. 2007;63:1372–1384. doi: 10.1111/j.1365-2958.2007.05612.x. [DOI] [PubMed] [Google Scholar]
- Qadri F, Svennerholm AM, Faruque AS, Sack RB. Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clin Microbiol Rev. 2005;18:465–483. doi: 10.1128/CMR.18.3.465-483.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter JM, Jones GW. The K88 antigen of Escherichia coli – a model for vaccination with a virulence factor? J Med Microbiol. 1973;6:P8–P9. [PubMed] [Google Scholar]
- Sakellaris H, Balding DP, Scott JR. Assembly proteins of CS1 pili of enterotoxigenic Escherichia coli. Mol Microbiol. 1996;21:529–541. doi: 10.1111/j.1365-2958.1996.tb02562.x. [DOI] [PubMed] [Google Scholar]
- Sakellaris H, Munson GP, Scott JR. A conserved residue in the tip proteins of CS1 and CFA/I pili of enterotoxigenic Escherichia coli that is essential for adherence. Proc Natl Acad Sci USA. 1999;96:12828–12832. doi: 10.1073/pnas.96.22.12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakellaris H, Scott JR. New tools in an old trade: CS1 pilus morphogenesis. Mol Microbiol. 1998;30:681–687. doi: 10.1046/j.1365-2958.1998.01088.x. [DOI] [PubMed] [Google Scholar]
- Satterwhite TK, Evans DG, DuPont HL, Evans DJ., Jr Role of Escherichia coli colonisation factor antigen in acute diarrhoea. Lancet. 1978;2:181–184. doi: 10.1016/s0140-6736(78)91921-9. [DOI] [PubMed] [Google Scholar]
- Schuller S, Frankel G, Phillips AD. Interaction of Shiga toxin from Escherichia coli with human intestinal epithelial cell lines and explants: Stx2 induces epithelial damage in organ culture. Cell Microbiol. 2004;6:289–301. doi: 10.1046/j.1462-5822.2004.00370.x. [DOI] [PubMed] [Google Scholar]
- Schuller S, Chong Y, Lewin J, Kenny B, Frankel G, Phillips AD. Tir phosphorylation and Nck/N-WASP recruitment by enteropathogenic and enterohaemorrhagic Escherichia coli during ex vivo colonization of human intestinal mucosa is different to cell culture models. Cell Microbiol. 2007;9:1352–1364. doi: 10.1111/j.1462-5822.2006.00879.x. [DOI] [PubMed] [Google Scholar]
- Sommerfelt H, Steinsland H, Grewal HM, Viboud GI, Bhandari N, Gaastra W, et al. Colonization factors of enterotoxigenic Escherichia coli isolated from children in north India. J Infect Dis. 1996;174:768–776. doi: 10.1093/infdis/174.4.768. [DOI] [PubMed] [Google Scholar]
- Stibitz S, Yang MS. Subcellular localization and immunological detection of proteins encoded by the vir locus of Bordetella pertussis. J Bacteriol. 1991;173:4288–4296. doi: 10.1128/jb.173.14.4288-4296.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viboud GI, Jouve MJ, Binsztein N, Vergara M, Rivas M, Quiroga M, Svennerholm AM. Prospective cohort study of enterotoxigenic Escherichia coli infections in Argentinean children. J Clin Microbiol. 1999;37:2829–2833. doi: 10.1128/jcm.37.9.2829-2833.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]


