Abstract
Traditionally, autistic spectrum disorder (ASD) and specific language impairment (SLI) are regarded as distinct conditions with separate etiologies. Yet these disorders co-occur at above chance levels, suggesting shared etiology. Simulations, however, show that additive pleiotropic genes cannot account for observed rates of language impairment in relatives, which are higher for probands with SLI than for those with ASD + language impairment. An alternative account is in terms of ‘phenomimicry’, i.e., language impairment in comorbid cases may be a consequence of ASD risk factors, and different from that seen in SLI. However, this cannot explain why molecular genetic studies have found a common risk genotype for ASD and SLI. This paper explores whether nonadditive genetic influences could account for both family and molecular findings. A modified simulation involving G × G interactions obtained levels of comorbidity and rates of impairment in relatives more consistent with observed values. The simulations further suggest that the shape of distributions of phenotypic trait scores for different genotypes may provide evidence of whether a gene is involved in epistasis.
Keywords: Autism, Specific language impairment, Comorbidity, Epistasis
Introduction
Specific language impairment (SLI) refers to a condition where a child fails to develop spoken language on the normal schedule, for no obvious reason (Bishop and Norbury 2008). Potential causes such as hearing loss, low general ability, or physical impairment of articulators are excluded. Development in areas such as skills of daily living and nonverbal ability is age-appropriate. Autism is also excluded, and the textbook picture of SLI is of a child with normal social interaction and nonverbal communication, but with specific difficulties in mastering structural aspects of language, especially syntax (grammatical devices such as word order and inflectional endings) and phonological skills (identification and production of speech sounds). Autistic disorder also involves impairments of communication, but these are much broader, affecting pragmatics, i.e., the appropriate use of language in context, as well as nonverbal communication. In addition, there are impairments in social interaction and understanding, and the repertoire of behaviour and interests is unusual and often restricted (Dover and Le Couteur 2007). The diagnostic manuals ICD-10 and DSM-IV (American Psychiatric Association 1994; World Health Organization 1993) use different terminology, but both make a clear diagnostic distinction between a specific developmental disorder affecting language and the more pervasive difficulties seen in autism.
The past few decades have seen two major changes in our conceptualisation of the etiology of both autism and SLI. The first breakthrough was the abandonment of purely environmental explanations for these conditions as it became clear from twin studies that they were both highly heritable. Several studies showed that identical, monozygotic (MZ) twins were significantly more concordant than fraternal, dizygotic (DZ) twins for autism (Bailey et al. 1995; Folstein and Rutter 1977; Hallmayer et al. 2002; Steffenburg et al. 1989) and SLI (Bishop et al. 1995; Bishop and Hayiou-Thomas 2008; Lewis and Thompson 1992; Tomblin and Buckwalter 1998), despite growing up together and sharing many environmental influences. The next shift in understanding was from a focus on single genes of large effect to an etiological model of these conditions as complex and multifactorial, resulting from the influence of many genes of small effect, combined with environmental influences (see review by Bishop 2009). There were several reasons for this development. First, the high heritability estimates from twin studies did not translate into discoveries of common single gene mutations of large effect, as might have been expected. In the field of SLI, discovery of a mutation of the FOXP2 gene in one multigenerational family led to a fascinating series of studies exploring the gene’s evolution and mode of action (see Fisher, 2007, for review and Konopka et al. 2009 for more recent work), but it has become clear that it is a rare cause of speech and language impairments (Newbury et al. 2002). Second, pedigree studies showed that in both autism and SLI, rates of impairment in first degree relatives are higher than in the general population, but it is unusual to observe a classic Mendelian pattern of inheritance (Lewis et al. 1993; Rutter 2005a). In short, these conditions aggregate but do not segregate (Sing and Reilly 1993). Third, family studies indicated that first degree relatives of affected individuals often manifest subthreshold symptoms, such as subtle phonological difficulties in relatives of children with SLI (Barry et al. 2007), or mild social and communicative difficulties in relatives of those with autism (see review by Bailey et al. 1998). This suggested that these conditions correspond to points on a continuum of impairment, rather than all-or-none diseases. Finally, studies in clinical medicine led to growing awareness that complex multifactorial etiology is the rule rather than the exception for disorders that are common in the general population, consistent with arguments by evolutionary scientists that genetic variants that had large effects on reproductive fitness in ancestral humans would be unlikely to persist in modern humans (Keller and Miller 2006).
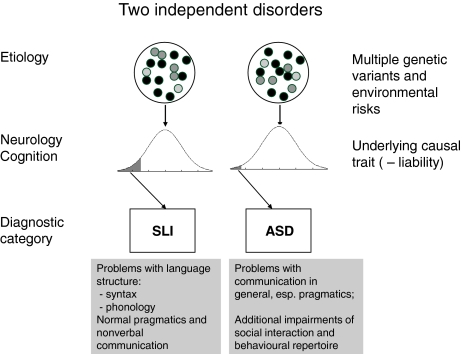
A model of the etiology of SLI and autism is shown in Fig. 1. This is based on the original approach to complex multifactorial disorders proposed by Falconer (1965), and subsequently developed to contrast different models of comorbidity by Neale and Kendler (1995). The model assumes that underlying a categorical diagnosis is a normally distributed liability continuum. Figure 1 differs from the traditional models by having disorder result when the value on the continuum falls below a threshold, whereas Falconer treated liability as a positive risk variable with disorder resulting from a high value. The reason for this change is to make the model more compatible with conceptualisations of developmental disorders, where disorder is typically recognised when a score on a normally distributed ability is unusually low. In effect, we work with negative liability, but this affects only the graphical depiction of the model, and has no computational consequences. In the classic Falconer model, liability is a theoretical construct, not directly observed, but inferred from the frequencies of disorders in probands and their relatives. Nevertheless, it should be possible to identify measures that are correlated with the causal trait, which will be termed ‘liability markers’ for disorder. These could be indices of neurological or cognitive function. The construct of liability marker overlaps with the notion of endophenotype (Gottesman and Gould 2003), but is more general because it is not restricted to genetically determined traits. An additional point to note is that although the model specifies a single causal trait, risk factors are not unitary: on the contrary, they can be partitioned into genetic and environmental risks, with the latter subdivided into those shared between relatives and those specific to the individual. For each type of risk, there are many influences, each of small impact, whose combined effect gives rise to the continuous distribution on the relevant causal trait.
Fig. 1.
Traditional model of specific language impairment (SLI) and autism spectrum disorder (ASD) as separate disorders with distinct etiologies. Both disorders are conceptualised as spectrum disorders, with disorder identified when a continuous underlying causal trait falls below a threshold
The cutoff on the causal trait will determine the frequency of disorder. The prevalence of SLI depends on the operational criteria that are used; an epidemiological study in the US estimated prevalence as 7% in kindergarten (Tomblin et al. 1997). This prevalence corresponds to a z-score threshold of around –1.5. Estimates of the prevalence of autism have mounted steadily (Rutter 2005b), from 4 per 10 000 in the 1960s up to 38.9 per 10 000 in a recent epidemiological survey in the UK (Baird et al. 2006). This is usually thought to reflect changing diagnostic criteria (King and Bearman 2009), although a genuine increase cannot be ruled out. In addition, autism is now regarded as a spectrum disorder rather than an all-or-none disease, with milder and partial forms of disorder being labelled as cases of ‘autism spectrum disorder’ (ASD) or pervasive developmental disorder not otherwise specified. When these cases are included, the prevalence rises to 116.1 per 10 000 (Baird et al. 2006), with corresponding z-score = −2.3. Nevertheless, even this estimate is lower than the prevalence of SLI and the cutoffs are shown as different in Fig. 1 to reflect this. In simulations discussed below we use the latter prevalence rate, and hence refer to ASD rather than autism.
The neat divide between independent disorders, shown in Fig. 1, has been questioned in recent years. It has been argued that the conditions of ASD and SLI may be less distinct than the textbooks imply. There are three lines of evidence that any etiological model has to account for: (a) apparently above chance levels of comorbidity between SLI and ASD; (b) rates of language impairment in relatives of probands with SLI and ASD; (c) molecular genetic findings of shared genetic risk factors for ASD and SLI.
Comorbidity between SLI and ASD
According to conventional diagnostic frameworks, SLI and ASD are mutually exclusive diagnoses—ASD is explicitly excluded when making a diagnosis of SLI, which is, by definition, a ‘specific’ developmental disorder. From this perspective it does not make sense to talk of comorbidity. Nevertheless, diagnostic frameworks do not necessarily reflect clinical reality, and there has been interest over many years in the idea that there might be overlapping language deficits in the two conditions.
In discussing overlaps between SLI and ASD, it is important to distinguish between different aspects of communication. On the one hand, children need to master the structural aspects of their language—phonology and syntax. These are the domains that are most often noted to be impaired in SLI. On the other hand, children need to use those skills to communicate with others—pragmatics. Although there are exceptions, most formal language tests focus either on vocabulary or structural aspects of language, but do not assess how effectively language is used to communicate in everyday situations. The conventional view of SLI maintains that pragmatic skill is intact and the child may communicate reasonably despite having limited structural language skills (Bishop 2000).
As demonstrated in a landmark study by Bartak et al. (1975), many children with ASD are poor at both structural and functional aspects of communication. These authors compared children with severe receptive SLI (termed ‘developmental dysphasia’), and children with autism. They documented similarities for the two groups on language milestones and measures of language structure, but striking differences in the functional use of language (Cantwell et al. 1978). Children with autism had much broader communicative difficulties than those with SLI, extending to encompass nonverbal as well as verbal communication.
Nevertheless, while poor functional communication is a hallmark of ASD, not all cases are impaired on formal language tests. Kjelgaard and Tager-Flusberg (2001) noted substantial variation in language abilities in a group of 89 children with autism. On a wide-ranging battery of language tests commonly used to diagnose SLI, 76% of them performed in the impaired range. The remaining 24% had no evidence of structural language deficits. In an epidemiological sample, Loucas et al. (2008) found that 41 of 72 (57%) children with autism and normal nonverbal IQ had impaired performance on a language battery. Subsequently, Tager-Flusberg and Joseph (2003) noted that many children with ASD were particularly poor at repeating nonsense words, a measure that has been proposed as a marker of heritable SLI (Bishop et al. 1996). Furthermore, these children tended to make similar morphosyntactic errors to those seen in SLI, i.e., omission of verb inflectional endings.
These studies indicate that structural and pragmatic language deficits are logically separable, but often co-occur. Tager-Flusberg and Joseph (2003) argued that the existence of cases of ASD whose language features resembled those of SLI suggested overlaps between these disorders at a deeper level. In this paper, a distinction is drawn between pure ASD, SLI and the apparently ‘comorbid’ cases, referred to here as ASD+LI, who have classic autism together with language impairments of the kind seen in SLI.
Rates of language impairment in relatives of probands with SLI or ASD
There have been several studies of parents and siblings of people with autism on language measures, with rather mixed results (see Bailey et al. 1998). In general, relatives of people with autism are more likely than control relatives to report a personal history of language or literacy problems, but these have proved harder to demonstrate on formal testing, especially when the measures come from instruments that are sensitive to SLI (Bishop et al. 2004; Whitehouse et al. 2007). A study by Lindgren et al. (2009), which is noteworthy for its methodological rigour and relatively large sample size, explicitly compared parents and siblings from three groups of probands: pure SLI (N = 36), pure ASD (N =20), and comorbid ASD+LI (N = 32). Particular care was taken to exclude from the SLI group any individuals with autistic features. The probands with SLI and comorbid ASD+LI were similar in their language profiles, but their siblings and parents differed. The relatives of those with SLI had language deficits, in line with previous studies of SLI relatives (Barry et al. 2007), but relatives of those with ASD+LI had language scores in the normal range. Relatives of the pure ASD group tended to obtain higher scores than the ASD+LI parents on language measures, but both the language scores and nonverbal IQ of the pure ASD group were above average. A supplementary analysis in which relatives were categorised according to whether they met criteria for language impairment showed the following rates of language impairment in relatives of those with pure ASD, ASD+LI and SLI respectively: siblings, 11%, 16% and 42%; fathers, 21%, 35% and 54%; mothers, 5%, 29% and 60%. The rates of LI in relatives were significantly lower in ASD+LI than in SLI probands for all relatives except fathers.
Molecular genetic risks for SLI and ASD
Vernes et al. (2008) studied a sample of individuals with SLI and demonstrated association between nonword repetition skills (a marker of SLI) and polymorphisms of CNTNAP2, a gene on chromosome 7q35 that is a downstream target of FOXP2. CNTNAP2 encodes a neurexin and is expressed in the developing human brain. These authors noted that association with ASD had been demonstrated for the same locus (Arking et al. 2008), with the strength of association greatest when cases were restricted to probands with severely delayed language milestones (Alarcón et al. 2008).
Taken together, the evidence from comorbidity, impairments in relatives and molecular genetic risks presents a puzzling picture. The comorbidity and molecular genetic findings would appear to point to overlapping etiology, yet the data from relatives are inconsistent with that picture. In this paper, I shall first present a formal simulation of overlapping etiology through additive pleiotropic effects to demonstrate the problems this model has in accounting for observed data, before going onto consider two radically different accounts of the etiological relationship between the two disorders.
A simulation of overlapping etiology
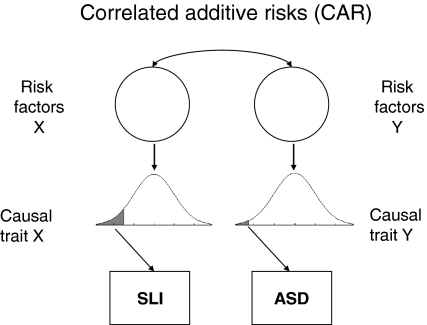
A ‘correlated additive risks’ model
The ‘correlated additive risks’ (CAR) model shown in Fig. 2 corresponds to Neale and Kendler’s (1995) ‘correlated liability model’ of comorbidity. It depicts the same causal routes as Fig. 1 except for the two-headed pathway between risk factors. According to the model, each condition has its own separate risk factors and causal traits, but the risk factors for the two conditions co-occur at above chance levels. Such a model predicts that relatives of individuals affected with one disorder will be at increased risk for the other; the simulation allows us to quantify this prediction depending on the extent of correlation between risk factors, and the relative frequency of the two disorders.
Fig. 2.
Correlated additive risks (CAR) model. SLI specific language impairment, ASD autism spectrum disorder
Before describing the simulation, it is worth noting the different routes by which risk factors may be correlated: In the simulation presented here, genetic correlation is induced by including pleiotropic genes that lead to increased risk for both disorders. However, there are other possibilities: risk genes for the two disorders may be transmitted together because they are close together on a chromosome (linkage): in general, if linkage is tight, then predictions are similar to those from a pleiotropic model. Furthermore, nonrandom (assortative) mating could lead to different risk genes being contributed by each parent. In that case, predictions about affectedness in relatives may differ for parents and siblings. Environmental risks could also be correlated, but are not discussed here because evidence from twin studies suggests that environmental factors play a relatively minor role in the etiology of both SLI and ASD (Bishop 2006; Newschaffer et al. 2002).
Methods
A Matlab program that simulates the CAR model is available from http://psyweb.psy.ox.ac.uk/oscci/Miscellaneous.htm. The simulation starts by assigning a set of probands values for a set of genotypes, aa, aA and AA, whose frequency is determined by the user, with default minor allele frequency of .5. The ‘a’ allele is designated the risk allele, and has an additive impact on the causal trait for one or both disorders, with genotype aA having an effect intermediate between aa (low) and AA (high). The user specifies the number of probands, the number of genes affecting a causal trait, and the proportion of genes that affect both traits (pleiotropy). If we have 10 genes, when pleiotropy is set to zero, there are five genes affecting each trait, and none affecting both, and r g is zero (corresponding to Fig. 1). If the proportion of pleiotropic genes is set to .2, then genes 1, 2, 3 and 4 affect SLI only, genes 7, 8, 9 and 10 affect ASD only, and genes 5 and 6 affect both traits, giving a computed value for r g of .33. The model was designed to simulate twin data, and can be set to give estimated trait scores for MZ as well as DZ twins. The focus of interest here, however, is in similarities between probands and their first degree relatives, and so only the predictions for DZ twins are considered. In addition, environmental effects that are shared between relatives are modelled by a random normal variable that is identical for first degree relatives (and for MZ twins), and nonshared environmental effects (including measurement error) are modelled with a random normal variable that has no correlation between relatives. The genetic effects, shared and nonshared environmental effects are then combined in a weighted sum where the weights reflect the values of h 2, c 2 and e 2 input by the user. The diagnostic categories of probands and their relatives are then assigned depending on whether or not the standardized score on the liability distributions for ASD and SLI fall below threshold.
Results
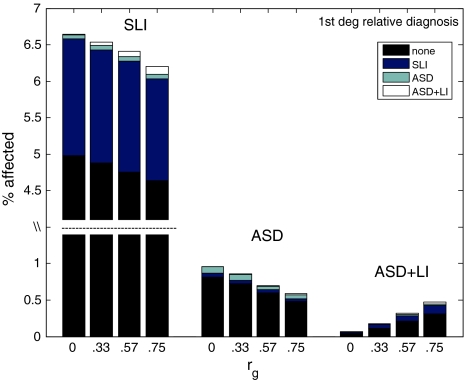
Figure 3 shows the predicted rates of pure and comorbid disorders in probands and their relatives, at different levels of pleiotropy, when settings for h 2, c 2 and e 2 are .7, .1 and .2, respectively for both traits, with 100,000 simulated probands, cutoffs of −1.5 z for SLI and −2.3 z for ASD, and 10 genes in total. With pleiotropy set to zero, five genes affect the SLI liability, and five affect ASD liability. When pleiotropy is greater than zero, a subset of genes affects both traits, with r g increasing with number of pleiotropic genes.
Fig. 3.
Simulated data from CAR model showing proportions of relatives of probands with SLI, ASD and ASD+LI (labelled above histograms), with diagnosis in relatives indicated by shading, for four levels of r g. Note that the scale for the relatives of SLI probands is broken so that the full range can be shown for all groups. SLI specific language impairment, ASD autism spectrum disorder, ASD+LI ‘comorbid’ cases
As shown in Fig. 3, the prevalence of comorbid cases increases as the genetic correlation increases. Regardless of the size of genetic correlation, pure disorders tend to ‘breed true’, but the less extreme threshold for SLI means that probands with ASD+LI will have more relatives with SLI than with pure or comorbid ASD. The proportion of relatives with language impairment is similar for the SLI and comorbid proband groups, at around 22–23%.
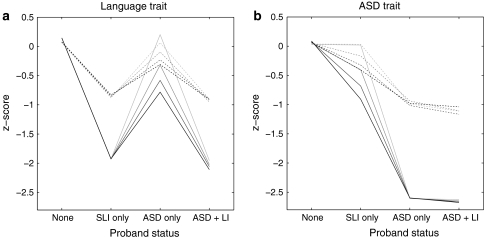
As well as looking at categorical outcomes, we can inspect mean scores on liability markers for language impairment, which may be seen as proxies for language test scores or a quantitative measure of autistic features. Figure 4 shows how the profiles seen in probands (unbroken lines) and their first degree relatives (dashed lines) change, as degree of genetic correlation increases. Note that relatives who themselves meet criteria for ASD are excluded, as is usually done in family studies (e.g., Lindgren et al. 2009). Relatives are not, however, usually screened for SLI, and so those who meet criteria for this condition (i.e., have language liability scores below −1.5) are retained in the sample. (As is evident from Fig. 3, the number of relatives with ASD was very small, and including them makes no visible difference to the plots). The plots of mean liability markers show that (nonautistic) relatives have a profile resembling that of probands, but less extreme. As the genetic correlation increases, probands with ‘pure’ disorders and their first degree relatives show a tendency to be impaired on the liability marker for the other condition, i.e., pure ASD probands and their relatives have depressed scores on the language liability marker, and pure SLI probands and their relatives have depressed scores on the ASD liability marker. The relatives of comorbid cases score as poorly as relatives of pure cases on the relevant liability marker (i.e., mean scores are comparable to pure SLI relatives on the language trait, and comparable to pure ASD relatives on the ASD trait).
Fig. 4.
Simulated data from correlated additive risks model showing mean scores for probands (unbroken lines) and their first degree relatives (dashed lines) in relation to proband diagnosis, for 10-gene model, with 0, 2, 4 or 6 pleiotropic genes, denoted by shading from pale grey (0 pleiotropic) to black (6 pleiotropic). Relatives who meet criteria for ASD are excluded from the means. SLI specific language impairment, ASD autism spectrum disorder, ASD+LI ‘comorbid’ cases
Comparison with observed data
As we have seen, the CAR model can explain why rates of comorbidity occur in excess of the chance rate that would be expected if the two disorders were independent. It also is compatible with molecular genetic evidence of common risk variants for ASD and SLI. However, it further predicts that relatives of those with comorbid ASD+LI should resemble relatives of those with pure ASD on ASD trait markers, and resemble relatives of those with pure SLI on language measures. As noted above, this is inconsistent with observed data from studies by Bishop et al. (2004), Lindgren et al. (2009) and Whitehouse et al. (2007). The CAR model therefore fails to provide a plausible account of etiological overlaps.
There are other models of overlapping risk factors that have been proposed in the literature, but in all cases they predict that relatives of those with comorbid ASD+LI should have an increased rate of language impairments. Thus, Tager-Flusberg and Joseph (2003) suggested that pure ASD and ASD+LI might be distinct subtypes, with a common neurocognitive phenotype for ASD+LI and SLI: however the subtype hypothesis predicts that ASD+LI will ‘breed true’, so relatives of affected individuals would show language difficulties.
Another model of overlapping risk factors is the dimensional account of ASD by Ronald et al. (2006). Rather than assuming two liability distributions, one for autism and one for SLI, they suggest that the different components of autism (social interaction, communication and behavioural repertoire) are independently heritable, and only when all dimensions were impaired would they qualify as cases of autism. A problem for this view is that the communication dimension is not well-specified. As noted above, while an autism diagnosis requires that the child have difficulties with communication, this does not necessarily mean problems with structural aspects of language of the kind seen in SLI. Thus, to represent the full range of observed phenotypes, we need at least four dimensions, corresponding to language structure, pragmatic aspects of communication, social interaction and behavioural repertoire. As noted by Bishop (2003), such a model has the advantage of being able to capture a wide range of clinical conditions, including classic SLI (only language structure impaired), pragmatic language impairment (communication impaired, with or without poor language structure), Asperger syndrome (impairment in all domains except language structure) and autism (impairment in pragmatics, social interaction and behavioural repertoire, with or without poor language structure). The model runs into difficulties, however, in accounting for patterns of trait markers in relatives, because, like the CAR model, it predicts that the different dimensions should ‘breed true’, so relatives of those with comorbid autism+LI should resemble relatives of those with SLI on language trait markers, with both being impaired. Furthermore, it is hard to specify thresholds on the four causal traits that can generate plausible prevalence rates for the different types of disorder. If the traits are semi-independent, then the more traits that are impaired, the rarer the disorder will be. Autism without structural language impairment should therefore be far more common than autism with language impairment, which is not what is found (Loucas et al. 2008).
Phenomimicry
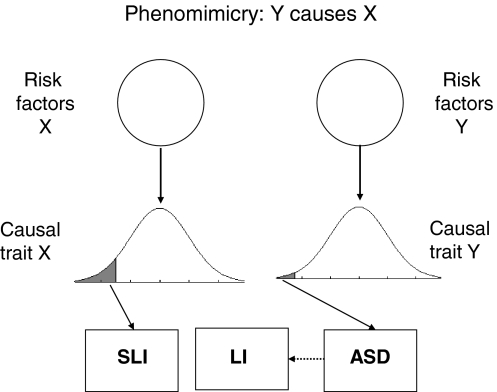
A very different way of accounting for similar language problems in ASD and SLI is to argue that phenotypic continuities between conditions are more apparent than real, resulting from ‘phenomimicry’. I use this term to refer to the situation when the causal route for one disorder can lead to an outcome resembling the other disorder. Unlike ‘phenocopy’, which refers to an environmentally caused disorder that resembles a genetically-determined disorder, the term ‘phenomimicry’ makes no assumptions about whether etiology is genetic or environmental, or, as in this case, complex and multifactorial. It corresponds to what Neale and Kendler (1995) termed ‘multiformity’. In the case of ASD and SLI, this would mean that having a causal trait for one disorder could lead to a clinical picture resembling the other disorder. Figure 5 shows one version of phenomimicry where a child with ASD develops language deficits similar to those in SLI. The converse is also possible: a child with SLI might develop a clinical picture resembling ASD, perhaps because social interaction is difficult and stressful. Note that a phenomimicry account essentially is a variant of the traditional model (Fig. 1), and treats any phenotypic overlap between disorders as superficial.
Fig. 5.
Phenomimicry model: the phenotype of autism spectrum disorder (ASD) can lead to language impairment (LI), but the resemblance with specific language impairment (SLI) is superficial and those with ASD+LI do not have risk factors for SLI
An account similar to that shown in Fig. 5 was proposed by Williams et al. (2008), who argued that apparently similar language deficits in SLI and ASD had different underlying causes. They argued that, although some children with ASD made errors in using inflectional endings on verbs, the types of errors differed from those seen in SLI. In a similar vein, Whitehouse et al. (2007) noted that the pattern of errors on a nonword repetition test was different in the two disorders, and argued that deficient phonological memory, indexed by disproportionate difficulty with long nonwords, may be implicated only in pure SLI.
Problems for a phenomimicry account
Although data on relatives appear compatible with a phenomimicry account, there are two problems for this explanation of comorbidity. First, it cannot readily explain the finding that CNTNAP2 is implicated as a risk factor for both SLI and ASD. Furthermore, we also have to explain why only a subset of individuals with ASD have SLI-like language problems, if such problems are a consequence of having ASD. An obvious possibility is that the likelihood of language problems increases with severity of ASD, as suggested by Whitehouse et al. (2007). However, this was not found in the larger study by Lindgren et al. (2009), nor in the epidemiologically-based sample studied by Loucas et al. (2008).
The evidence for etiological overlap between ASD and SLI is therefore somewhat inconsistent. On the one hand, phenotypic similarities between language deficits in the two disorders, and findings that CNTNAP2 variants confer risk for both ASD and SLI, suggest a common causal pathway. On the other hand, qualitative differences in language phenotypes, coupled with the relatively spared language abilities in relatives of those with ASD points to distinct etiologies.
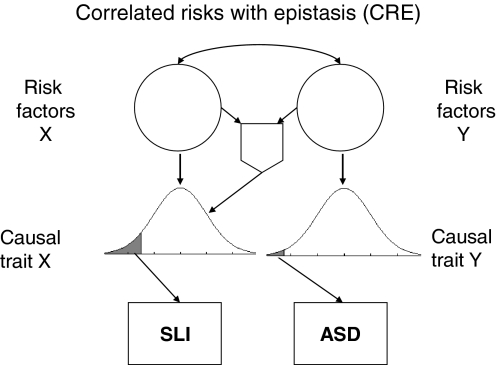
A modified model: correlated risks with epistasis (CRE)
A possible way of resolving the inconsistencies is to incorporate nonadditive interactions between genes in an etiological model. There are several lines of evidence that suggest that a genetic model of autism needs to include interactions between genes, rather than just additive effects. The first is an analysis by Pickles et al. (1995), who considered frequency of autism in relatives (twins and other family members) for probands with autism. If the etiology involves many genetic variants with additive effects, then the prediction would be that the rate of autism in DZ twins or sibs of those with autism should be around 50% of the rate seen in MZ twins. In fact, the rate in these first degree relatives is considerably less than that (with around 10% of siblings and DZ affected with ASD, compared to around 80% in MZ twins; Pickles et al. 1995). The authors concluded that nonadditive genetic influences must be implicated in the etiology. A similar conclusion was reached by Risch et al. (1999), who compared the proportion of alleles with a common identity by descent (IBD) in affected sib-pairs vs. discordant sib pairs. They found that there was a small increase in IBD-sharing for affected sib pairs across all 360 markers that they considered, rather than an effect confined to a few loci. They concluded that the etiology of autism involved a large number of loci, perhaps more than 15, and probably involves interactions between genes (i.e., epistasis) as well as additive effects. A third line of evidence comes from consideration of the functional networks in which genes are involved; Bill and Geschwind (2009) noted that many autism susceptibility genes are involved in the same pathways, suggesting possible interactions between proteins in signalling pathways.
The models in Figs. 1 and 2 are derived from the classic ACE model, which assumes only additive influences. The question then arises as to whether modifying the model to allow for nonadditive effects in the form of gene–gene (G × G) interaction might accommodate findings such as those of Lindgren et al. (2009), given that one signature of epistasis is reduced similarity between probands and their first degree relatives. The simulation was therefore modified to incorporate interaction between risk genes for SLI and ASD. Figure 6 shows one specific version of the model in which presence of autism risk factors enhances the impact of a risk factor for LI, but it would be logically possible to have the opposite situation (impact of autistic risk factor increased if LI risk factors present), or epistatic interaction affecting both language and ASD traits. All these options can be explored using the simulation; the one shown in Fig. 6 is selected here because it gives a reasonable account of observed data.
Fig. 6.
Correlated risks with epistasis model. The shield-shaped symbol depicts an AND gate, whereby there is an extra impact on the language trait only if both risk factors X and Y are present
Methods
The Matlab script in the Appendix includes the option of specifying G × G interactions. Each gene is identified by number, and the user specifies a list of genes involved in each interaction. The first gene in the list has the effect of its risk genotype increased by a specified amount if and only if all the subsequent genes in the list have the risk genotype. For instance, if a G × G term is specified as [1 7 8] this means that an individual with a homozygous risk genotype (aa) for gene 1 will have the effect of that genotype amplified if a homozygous risk genotype is also present for genes 7 and 8. This means that even if two relatives have the risk genotype for language impairment, they may differ in terms of the effect of that genotype, because the added effect depends on the presence of a constellation of genotypes on other (ASD) genes. The probability of relatives sharing such a constellation decreases with the number of genes involved in epistasis.
Results
Table 1 shows the categorical results for diagnoses in relatives in relation to proband diagnosis for a model where there are 10 genes in total, one of which is pleiotropic, and has its impact doubled when its homozygous risk genotype (aa) occurs in combination with risk genotypes of any one of two pairs of ASD genes. It can be seen that relatives of comorbid ASD+LI probands now have a reduced rate of language impairment compared with the CAR model, and mean scores of relatives on the language liability measure are also less impaired. The pattern of results is thus more compatible with empirical data than was seen with the CAR model. The relatives of comorbid cases still show more language impairment than relatives of unaffected individuals; however, the relatively small increase in rates of LI over population prevalence, and more modest deficit on the language trait would only be detectable with a large sample. Although the overall pattern of affectedness in relatives is similar to that of Lindgren et al. (2009), the rates of language impairment in relatives are lower in the CRE model. Thus, the percentages of relatives with pure LI are 21%, 6% and 14% for the SLI, ASD and ASD+LI probands in the CRE model, compared with 52%, 12% and 27% for the sample of Lindgren et al. To some extent the discrepancy may reflect the fact that the criterion of LI used by Lindgren et al. was a language score of 1 SD below average on one of two measures, whereas the model cutoff was at 1.5 SD. Re-running the CRE model with cutoff for LI of -1 SD improved the fit, giving rates of affected relatives in SLI, ASD and ASD+LI of 34%, 10% and 25%, but the observed rate in SLI relatives was still higher than predicted by the model. Note, however, that the Lindgren et al. sample included several relatives from each family, and so rates could be inflated if there were clustering of affectedness within families, either because of assortative mating, or shared genes. In addition, affectedness in siblings was determined on the basis of a low score on either one of two tests, whereas in parents, a single test (nonword repetition) was used. This complicates comparisons of absolute frequencies with simulated data.
Table 1.
Results from correlated risks with epistasis simulation, with 10 genes, one of which is pleiotropic. The effect of the pleiotropic gene is magnified by risk genotypes from ASD genes (see text)
| Proband diagnosis | ||||
|---|---|---|---|---|
| None | SLI | ASD | ASD+LI | |
| Prevalence (%) | 92.14 | 6.20 | 0.98 | 0.67 |
| Percentage relatives with pure LI | 5.11 | 21.59 | 6.01 | 13.80 |
| Percentage relatives with pure ASD | 0.89 | 0.74 | 8.35 | 5.19 |
| Percentage relatives with ASD+LI | 0.52 | 1.53 | 4.58 | 8.75 |
| LI trait mean in probands | 0.15 | −1.93 | −0.53 | −2.37 |
| ASD trait mean in probands | 0.07 | −0.31 | −2.74 | −3.09 |
| LI trait mean in relativesa | 0.08 | −0.81 | 0.05 | −0.53 |
| ASD trait mean in relativesa | 0.07 | −0.07 | −0.83 | −0.84 |
aExcluding relatives with ASD liability score below cutoff
SLI specific language impairment, ASD autism spectrum disorder, ASD+LI ‘comorbid’ cases, LI language impairment
Another inconsistency with data from Lindgren et al. (2009) is that their ASD+LI and SLI probands had similar scores on language measures, whereas Table 1 indicates poorer performance in the ASD+LI probands. This simulation results are, however, compatible with some other empirical studies, which report poorer language test scores in ASD+LI cases than SLI for some receptive language measures (Loucas et al. 2008; Rapin and Dunn 2003).
The simulation was re-run with different values specified for G × G interactions, number of genes, and the size of epistatic effect. Examples of outputs are shown in Supplementary Material. Including more than two ASD genes in interaction with a LI gene makes outcomes of relatives less similar to the proband, but also makes it less likely that conditions for epistasis will be met, and so has little impact unless the size of epistatic effect is large. Altering the number of genes did not, in general, affect outcomes, unless the proportion of genes involved in epistasis was too low to exert much influence on overall trait scores. Increasing the size of epistatic effect decreased similarity between relatives, but large values induced strong skew, and sometimes bimodality, in the data.
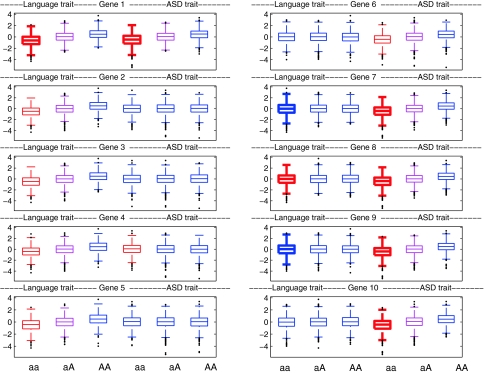
The skew in liability trait markers induced by epistasis is of potential interest for those who aim to detect epistasis using association analysis. Figure 7 shows boxplots of phenotypic scores on SLI and ASD liability markers for aa, aA and AA genotypes for each gene in the simulation described above, based on 5000 probands (to give a sample size that is in the range of realism). A plot shown in red denotes that the mean on the trait for aa genotype is significantly lower than for other genotypes on t-test, and magenta indicates that the mean for the aA genotype is lower than for AA. This kind of comparison relates to the statistical testing for association for a quantitative trait that is performed in conventional molecular genetic analyses. As expected, the left-hand set of genes (1–5) show differences between genotypes for the language trait, and the right-hand set (6–10) as well as the pleiotropic gene (1) show differences between genotypes for the ASD trait. Gene 4 gives a false positive result on the ASD trait, which disappears if a larger sample is taken. Overall, the t-test comparisons show that if we simply compare mean values of language trait scores for different genotypes for genes 6–10, which interact with gene 1, we are unlikely to detect an effect of an ASD gene on a language phenotype, with gene 8 being the only one to show a significant (though very small) effect. This confirms the low power of means comparisons for detecting epistasis.
Fig. 7.
Simulated data from correlated risks with epistasis model showing distributions of scores for 5000 probands on language trait (three leftmost plots) and ASD trait (three rightmost plots) for aa, aA and AA genotypes of 10 genes. For genes 1–5, the a allele confers risk for language impairment, and for genes 1 and 6–10 the a allele confers risk for ASD. G × G interaction is specified so that when the aa genotype is present for gene 1 in the context of either aa genotypes for genes 7 and 8, or for genes 9 and 10, its effect is doubled. Red denotes that the mean on a trait for aa genotype is significantly lower than for other genotypes on t-test, and magenta that the mean for the aA genotype is lower than for AA. Bold lines denote cases where the variance on a trait for the aa genotype is significantly greater than for other genotypes on F-test
It is striking, however, that the distributions of phenotype scores are more skewed for risk genotypes that enter into G × G interactions. Bold lines denote cases where the variance on a trait for the aa genotype is significantly greater than for other genotypes on F-test. It is evident from inspection that this is a feature that characterises just those genes that are implicated in epistasis. This is because the genotype usually has no effect on language ability, but occasionally exerts a large effect, when occurring in the context of a set of other risk genotypes. This skew is distinctive compared with that seen for the other genotypes of this gene, and for distributions of genotypes of other genes. If this kind of G × G interaction is in play, then significant differences in variances of phenotypic trait scores between genotypes could provide evidence that a gene may be implicated in epistasis.
General discussion
Although ASD and SLI have traditionally been regarded as distinct disorders, they often involve similar language deficits, raising the question of whether this is merely a superficial resemblance, or indicative of a deeper similarity, with overlap in etiology. Three models of etiology were considered. The first, of CAR, was simulated to test its predictions. The simulation confirmed that a model of correlated additive genetic risks can explain the relatively high rate of comorbid ASD+LI cases, but it does not predict observed data showing that relatives of people with ASD+LI tend to do better than relatives of those with pure SLI on language measures. The second model, in terms of ‘phenomimicry’, could account both for comorbidity and the patterns of deficit in relatives, but is unable to explain why the CNTNAP2 gene has been found, in independent samples, to be associated with both ASD and SLI. It also leaves unexplained why only a subset of those with ASD have language difficulties resembling SLI.
The final model was a modified version of the CAR model that incorporated G × G interaction. With one pleiotropic gene, whose impact was enhanced when a risk genotype occurred in the context of ASD risk genotypes, the model gave a pattern of results more in line with observed findings. In particular, the model could account for (a) comorbidity of ASD+LI at above chance levels, (b) similar or more severe levels of language impairment in ASD+LI as in SLI probands, while at the same time predicting (c) higher rates of language impairment in relatives of SLI cases than in relatives of ASD+LI cases.
Of course, the fact that a simulation can fit a pattern seen in observed data does not mean that the model is correct. Phenomimicry could also be implicated: we need more studies of qualitative aspects of language phenotypes in ASD and SLI to test this hypothesis convincingly. Other mechanisms, such as gene–environment interaction or assortative mating, could also be involved. Nevertheless, the simulation program used here showed that incorporating epistasis allows us to retain a model that postulates overlapping genetic etiology for ASD and SLI, in line with the molecular genetic findings on CNTNAP2. The G × G interaction reduces the correlation between probands and first degree relatives, and so can accommodate the result observed by Lindgren et al. (2009) whereby relatives of those with comorbid ASD+LI were less impaired on the language trait than relatives of pure LI cases, even though the comorbid probands themselves were at least as impaired as SLI cases on language measures. Although a model with G × G is less parsimonious than the basic CAR model, it is compatible with evidence from other sources that point to epistasis being implicated in the etiology of complex disorders in general (Carlborg and Haley 2004) and in ASD in particular (Bill and Geschwind 2009; Pickles et al. 1995; Risch et al. 1999).
The model not only provides a better fit to the observed data on relatives; it also suggests a way to identify genes that are involved in epistatic interactions. The CRE model included a single gene whose effect was magnified when its risk genotype co-occurred with risk genotypes on other genes. When this condition was met, this gene had a substantial impact on the pattern of observed data, introducing a skew in the tail of the distribution of liability markers. Nevertheless, when effect size is measured simply by comparing overall liability marker scores for those with the risk and nonrisk versions of the gene, the effect sizes of the interacting genes were small. Importantly, as shown in Fig. 7, the genes involved in epistatic interaction could, however, be distinguished from other genes in terms of the shape of the distributions of liability marker scores for risk and nonrisk genotypes. It follows that if such mechanisms do operate, then one could identify genes likely to be involved in epistasis by considering the distribution of liability marker scores associated with different allelic variants. In practice, non-normal distributions are often regarded as a nuisance to be corrected by removal of outliers, transformation or use of nonparametric tests (see also Bishop 2005). The sheer scale of information now available about the human genome means that one needs a strategy for prioritising which genes to analyse in order for association studies to be both statistically and methodologically tractable (Tabor et al. 2002). These simulations suggest that increased skew in phenotypic distributions for one genotype vs. others may be an indicator that a gene is likely to be involved in epistatic interactions.
Acknowledgments
The author is supported by a Principal Research Fellowship from the Wellcome Trust (ref 082498/Z/07/Z). The ideas in this paper were stimulated by discussions at a meeting organised by Gina Conti-Ramsden and funded by the Economic and Social Research Council, ‘Language and Social Understanding in Developmental Disorders’. The author likes to thank Andrew Whitehouse and Courtenay Frazier Norbury for their insightful comments on an earlier draft of this paper.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Edited by Gitta Lubke.
References
- Alarcón M, Abrahams BS, Stone JL, Duvall JA, Perederiy JV, Bomar JM, Sebat J, Wigler M, Martin CL, Ledbetter DH, Nelson SF, Cantor RM, Geschwind DH. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008;82(1):150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association . Diagnostic and statistical manual of mental disorders. 4. Washington, DC: American Psychiatric Association; 1994. [Google Scholar]
- Arking DE, Cutler DJ, Brune CW, Teslovich TM, West K, Ikeda M, Rea A, Guy M, Lin S, Cook EH, Chakravarti A. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet. 2008;82(1):160–164. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey A, Le Couteur A, Gottestman I, Bolton P, Simonoff E, Yuzda E, Rutter M. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med. 1995;25:63–77. doi: 10.1017/S0033291700028099. [DOI] [PubMed] [Google Scholar]
- Bailey A, Palferman S, Heavey L, Le Couteur A. Autism: the phenotype in relatives. J Autism Dev Disord. 1998;28:369–392. doi: 10.1023/A:1026048320785. [DOI] [PubMed] [Google Scholar]
- Baird G, Simonoff E, Pickles A, Chandler S, Loucas T, Meldrum D, Charman T. Prevalence of disorders of the autism spectrum in a population cohort of children in South Thames: the Special Needs and Autism Project (SNAP) Lancet. 2006;368(9531):210–215. doi: 10.1016/S0140-6736(06)69041-7. [DOI] [PubMed] [Google Scholar]
- Barry JG, Yasin I, Bishop DVM. Heritable risk factors associated with language impairments. Genes Brain Behav. 2007;6:66–76. doi: 10.1111/j.1601-183X.2006.00232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartak L, Rutter M, Cox A. A comparative study of infantile autism and specific developmental receptive language disorder: I. The children. Br J Psychiatry. 1975;126:127–145. doi: 10.1192/bjp.126.2.127. [DOI] [PubMed] [Google Scholar]
- Bill BR, Geschwind DH. Genetic advances in autism: heterogeneity and convergence on shared pathways. Curr Opin Genet Dev. 2009;19:271–278. doi: 10.1016/j.gde.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DVM. Pragmatic language impairment: a correlate of SLI, a distinct subgroup, or part of the autistic continuum? In: Bishop DVM, Leonard LB, editors. Speech and language impairments in children: causes, characteristics, intervention and outcome. Hove, UK: Psychology Press; 2000. pp. 99–113. [Google Scholar]
- Bishop DVM. Autism and specific language impairment: categorical distinction or continuum? In: Bock G, Goode J, editors. Autism: neural basis and treatment possibilities Novartis Foundation Symposium 251. Chichester: John Wiley; 2003. pp. 213–226. [PubMed] [Google Scholar]
- Bishop DVM. DeFries-Fulker analysis of twin data with skewed distributions: cautions and recommendations from a study of children’s use of verb inflections. Behav Genet. 2005;35:479–490. doi: 10.1007/s10519-004-1834-7. [DOI] [PubMed] [Google Scholar]
- Bishop DVM. What causes specific language impairment in children? Curr Dir Psychol Sci. 2006;15:217–221. doi: 10.1111/j.1467-8721.2006.00439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DVM. Genes, cognition and communication: insights from neurodevelopmental disorders. Year Cognitive Neurosci: Ann NY Acad Sci. 2009;1156:1–18. doi: 10.1111/j.1749-6632.2009.04419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DVM, Hayiou-Thomas ME. Heritability of specific language impairment depends on diagnostic criteria. Genes Brain Behav. 2008;7:365–372. doi: 10.1111/j.1601-183X.2007.00360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DVM, Norbury CF. Speech and language disorders. In: Rutter M, Bishop DVM, Pine D, Scott S, Stevenson J, Taylor E, Thapar A, editors. Rutter’s child and adolescent psychiatry. Oxford: Blackwell; 2008. pp. 782–801. [Google Scholar]
- Bishop DVM, North T, Donlan C. Genetic basis of specific language impairment: evidence from a twin study. Dev Med Child Neurol. 1995;37:56–71. doi: 10.1111/j.1469-8749.1995.tb11932.x. [DOI] [PubMed] [Google Scholar]
- Bishop DVM, North T, Donlan C. Nonword repetition as a behavioural marker for inherited language impairment: evidence from a twin study. J Child Psychol Psychiatry. 1996;37:391–403. doi: 10.1111/j.1469-7610.1996.tb01420.x. [DOI] [PubMed] [Google Scholar]
- Bishop DVM, Maybery M, Wong D, Maley A, Hill W, Hallmayer J. Are phonological processing deficits part of the broad autism phenotype? Am J Med Genet. 2004;128B:54–60. doi: 10.1002/ajmg.b.30039. [DOI] [PubMed] [Google Scholar]
- Cantwell D, Baker L, Rutter M. A comparative study of infantile autism and specific developmental language disorder. IV. Analysis of syntax and language function. J Child Psychol Psychiatry. 1978;19:351–362. doi: 10.1111/j.1469-7610.1978.tb00481.x. [DOI] [PubMed] [Google Scholar]
- Carlborg O, Haley CS. Epistasis: too often neglected in complex trait studies? Nat Rev Genet. 2004;5(8):618–625. doi: 10.1038/nrg1407. [DOI] [PubMed] [Google Scholar]
- Dover CJ, Le Couteur A. How to diagnose autism. Arch Dis Child. 2007;92(6):540–545. doi: 10.1136/adc.2005.086280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer DS. The inheritance of liability to certain diseases, estimated from the incidence among relatives. Ann Hum Genet. 1965;29:51–71. doi: 10.1111/j.1469-1809.1965.tb00500.x. [DOI] [Google Scholar]
- Fisher SE. Molecular windows into speech and language disorders. Folia Phoniatr Logop. 2007;59(3):130–140. doi: 10.1159/000101771. [DOI] [PubMed] [Google Scholar]
- Folstein S, Rutter M. Infantile autism: a genetic study of 21 twin pairs. J Child Psychol Psychiatry. 1977;18:297–321. doi: 10.1111/j.1469-7610.1977.tb00443.x. [DOI] [PubMed] [Google Scholar]
- Gottesman, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiat. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- Hallmayer J, Glasson EJ, Bower C, Petterson B, Croen L, Grether J, Risch N. On the twin risk in autism. Am J Hum Genet. 2002;71:941–946. doi: 10.1086/342990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller MC, Miller G. Resolving the paradox of common, harmful, heritable mental disorders: which evolutionary genetic models work best? Beh Brain Sci. 2006;29:385–452. doi: 10.1017/S0140525X06009095. [DOI] [PubMed] [Google Scholar]
- King M, Bearman P. Diagnostic change and the increased prevalence of autism. Int J Epidemiol. 2009;38(5):1224–1234. doi: 10.1093/ije/dyp261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjelgaard MM, Tager-Flusberg H. An investigation of language impairment in autism: implications for genetic subgroups. Lang Cog Proc. 2001;16:287–308. doi: 10.1080/01690960042000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka G, Bomar JM, Winden K, Coppola G, Jonsson ZO, Gao F, Peng S, Preuss TM, Wohlschlegel JA, Geschwind DH. Human-specific transcriptional regulation of CNS development genes by FOXP2. Nature. 2009;462(7270):213–217. doi: 10.1038/nature08549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BA, Thompson LA. A study of developmental speech and language disorders in twins. J Sp Hear Res. 1992;35:1086–1094. doi: 10.1044/jshr.3505.1086. [DOI] [PubMed] [Google Scholar]
- Lewis BA, Cox NJ, Byard PJ. Segregation analysis of speech and language disorders. Behav Genet. 1993;23:291–297. doi: 10.1007/BF01082469. [DOI] [PubMed] [Google Scholar]
- Lindgren KA, Folstein SE, Tomblin JB, Tager-Flusberg H. Language and reading abilities of children with autism spectrum disorders and specific language impairment and their first-degree relatives. Autism Res. 2009;2(1):22–38. doi: 10.1002/aur.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loucas T, Charman T, Pickles A, Simonoff E, Chandler S, Meldrum D, Baird G. Autistic symptomatology and language ability in autism spectrum disorder and specific language impairment. J Child Psychol Psychiatry. 2008;49:1184–1192. doi: 10.1111/j.1469-7610.2008.01951.x. [DOI] [PubMed] [Google Scholar]
- Neale MC, Kendler KS. Models of comorbidity of multifactorial disorders. Am J Hum Genet. 1995;57:935–953. [PMC free article] [PubMed] [Google Scholar]
- Newbury DF, Bonora E, Lamb JA, Fisher SE, Lai CSL, Baird G, Jannoun L, Slonims V, Stott CM, Merricks MJ, Bolton PF, Bailey AJ, Monaco AP, International Molecular Genetic Study of Autism Consortium FOXP2 is not a major susceptibility gene for autism or specific language impairment. Am J Hum Genet. 2002;70:1318–1327. doi: 10.1086/339931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newschaffer CJ, Fallin D, Lee NL. Heritable and nonheritable risk factors for autism spectrum disorders. Epidem Rev. 2002;24:137–153. doi: 10.1093/epirev/mxf010. [DOI] [PubMed] [Google Scholar]
- Pickles A, Bolton P, Macdonald H. Latent class analysis of recurrence risks for complex phenotypes with selection and measurement error: a twin and family history study of autism. Am J Hum Genet. 1995;57:717–726. [PMC free article] [PubMed] [Google Scholar]
- Rapin I, Dunn M. Update on the language disorders of individuals on the autistic spectrum. Brain Dev. 2003;2003:166–172. doi: 10.1016/S0387-7604(02)00191-2. [DOI] [PubMed] [Google Scholar]
- Risch N, Spiker D, Lotspeich L, Nouri N, Hinds D, Hallmayer J, Kalaydjieva L, McCague P, Dimiceli S, Pitts T, Nguyen L, Yang J, Harper C, Thorpe D, Vermeer S, Young H, Hebert J, Lin A, Ferguson J, Chiotti C, Wiese-Slater S, Rogers T, Salmon B, Nicholas P, Petersen PB, Pingree C, McMahon W, Wong DL, Cavalli-Sforza LL, Kraemer HC, Myers RM. A genomic screen of autism: evidence for a multilocus etiology. Am J Hum Genet. 1999;65(2):493–507. doi: 10.1086/302497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald A, Happe F, Bolton P, Butcher LM, Price TS, Wheelwright S, Baron-Cohen S, Plomin R. Genetic heterogeneity between the three components of the autism spectrum: a twin study. J Am Acad Ch Adol Psychiat. 2006;45(6):691–699. doi: 10.1097/01.chi.0000215325.13058.9d. [DOI] [PubMed] [Google Scholar]
- Rutter M. Genetic influences and autism. In: Volkmar FR, Paul R, Klin A, Cohen D, editors. Handbook of autism and pervasive developmental disorders. 3. Hoboken, NJ: John Wiley and Sons; 2005. pp. 425–452. [Google Scholar]
- Rutter M. Incidence of autism spectrum disorders: changes over time and their meaning. Acta Paed. 2005;94:2–15. doi: 10.1080/08035250410023124. [DOI] [PubMed] [Google Scholar]
- Sing CF, Reilly SL. Genetics of common diseases that aggregate, but do not segregate in families. In: Sing CF, Hanis CL, editors. Genetics of cellular, individual, family and population variability. New York: Oxford University Press; 1993. pp. 140–161. [Google Scholar]
- Steffenburg S, Gillberg C, Hellgren L, Andersson L, Gillberg IC, Jakobsson G, et al. A twin study of autism in Denmark, Finland, Iceland, Norway and Sweden. J Child Psychol Psychiatry. 1989;30(3):405–416. doi: 10.1111/j.1469-7610.1989.tb00254.x. [DOI] [PubMed] [Google Scholar]
- Tabor HK, Risch NJ, Myers RM. Candidate-gene approaches for studying complex genetic traits: practical considerations. Nat Rev Genet. 2002;3(5):391–397. doi: 10.1038/nrg796. [DOI] [PubMed] [Google Scholar]
- Tager-Flusberg H, Joseph RM. Identifying neurocognitive phenotypes in autism. Philos Trans Roy Soc Lond B. 2003;358:303–314. doi: 10.1098/rstb.2002.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomblin JB, Buckwalter PR. Heritability of poor language achievement among twins. J Speech Lang Hear Res. 1998;41:188–199. doi: 10.1044/jslhr.4101.188. [DOI] [PubMed] [Google Scholar]
- Tomblin JB, Records NL, Buckwalter P, Zhang X, Smith E, O’Brien M. Prevalence of specific language impairment in kindergarten children. J Sp Hear Res. 1997;40(6):1245–1260. doi: 10.1044/jslhr.4006.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernes SC, Newbury DF, Abrahams B, Winchester L, Nicod J, Groszer M, Alarcón M, Oliver P, Davies K, Geschwind DH, Monaco AP, Fisher S. A functional genetic link between distinct developmental language disorders. New Eng J Med. 2008;359:2337–2345. doi: 10.1056/NEJMoa0802828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse A, Barry JG, Bishop DVM. The broader language phenotype of autism: a comparison with specific language impairment. J Child Psychol Psychiatry. 2007;48:822–830. doi: 10.1111/j.1469-7610.2007.01765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D, Botting N, Boucher J. Language in autism and specific language impairment: where are the links? Psychol Bull. 2008;134(6):944–963. doi: 10.1037/a0013743. [DOI] [PubMed] [Google Scholar]
- World Health Organization . The ICD-10 classification for mental and behavioural disorders: diagnostic criteria for research. Geneva: WHO; 1993. [Google Scholar]