Abstract
AIM: To study the germline mutation of hPMS2 gene in 26 unrelated Chinese hereditary nonpolyposis colorectal cancer (HNPCC) probands and to fulfill the screening strategy for HNPCC in Chinese.
METHODS: Genomic DNA was extracted from the peripheral blood. To avoid the interference of pseudogene in detection of the remaining 11 exons (exon 1-5, 9, 11-15), long-range polymerase chain reaction (PCR) was conducted to amplify the complete coding region of hPMS2 gene firstly. Then 1/8 of the PCR products were used as template to amplify the individual exon respectively and DNA sequencing was done. Direct DNA sequencing of the conventional PCR products of exon 6, 7, 8 and 10 of hPMS2 gene was performed. The same analysis was made in 130 healthy persons without family histories of HNPCC to further investigate the pathological effects of the detected missense mutation.
RESULTS: One HNPCC proband fulfilled Bethesda guidelines and was found to carry the germline mutation of hPMS2 gene, which has not been reported in Chinese HNPCC families. It was a missense mutation at c.1532C>T of exon 11. It was detected in three controls as well with an occurrence rate of 2.3% (3/130). Since it could not be found in the PMS2-single nucleotide polymorphism (SNP) database, this missense mutation is a new SNP unreported up to date. Meanwhile, 260 reported SNPs of hPMS2 gene were detected in the 26 HNPCC probands. The 2nd and 5th exons were probably the hot SNP regions of hPMS2 gene in Chinese HNPCC families involving 53.1% of all reported SNP.
CONCLUSION: The germline mutation of hPMS2 gene may be rare in Chinese HNPCC families. The 2nd and 5th exons are hot SNP regions of hPMS2 gene.
Keywords: Hereditary nonpolyposis colorectal cancer, hPMS2, Missense mutation, Single nucleotide polymorphism, Colorectal cancer
INTRODUCTION
Hereditary nonpolyposis colorectal cancer (HNPCC), or Lynch syndrome, is an autosomal dominantly inherited disease with cancer-susceptibility. Perhaps it is the most common cause of hereditary colorectal cancer, accounting for 5%-10% of the total colorectal cancers worldwide[1-3]. People inheriting this predisposition are at a particularly high risk of developing colorectal cancer with an early age of onset[3,4]. The affected patients always carry germline mutations in DNA mismatch repair (MMR) genes, mostly in hMLH1, hMSH2, and hMSH6[5,6]. Less commonly, mutations in other MMR genes are present. We analyzed the abnormalities of hMSH2/hMLH1/hMSH6 genes in a series of Chinese HNPCC families fulfilling different clinical criteria. We studied germline mutation, large genomic variations of the entire coding regions of the three genes and methylation of hMLH1 promoter in 58 Chinese HNPCC probands, in which 24 fulfilled Amsterdam criteria (AC)[7], 15 fulfilled Japanese criteria (JC)[8] and 19 met Bethesda guidelines (BG)[7]. The total detected gene abnormality rate was only 53.4% (31/58), including 29 cases of germline mutation and 2 cases of methylation of hMLH1 promoter[9-14]. So the aberrant MMR genes other than hMSH2/hMLH1/hMSH6 are suspected to be involved in Chinese HNPCC.
In order to accomplish our serial studies of Chinese HNPCC, we detected hPMS2 germline mutation in 26 Chinese HNPCC families by long-range polymerase chain reaction (LR-PCR)-based sequencing in this study, and evaluated this manner in the molecular genetics screening of Chinese HNPCC.
MATERIALS AND METHODS
Materials
Twenty-six unrelated HNPCC probands registered from January 1998 to October 2005 at the Department of Abdominal Surgery in Shanghai Cancer Center were retrieved. Five of them fulfilled AC, 10 fulfilled JC and the remaining 11 fulfilled BG. Germline abnormalities of MSH2/MLH1/MSH6 were excluded in all the 26 probands by PCR-based sequencing. Ten milliliter peripheral blood was collected from each proband for genomic DNA preparation. The peripheral blood samples of 130 healthy volunteers without any family history of hereditary disease or development of colon cancer in early age were obtained for control. The informed consents were signed by all the probands and volunteers before blood drawing. This study was approved by the Medical Ethical Committee of Shanghai Cancer Center, Fudan University. The whole procedures of the study were in accordance with the international rules and regulations.
DNA extraction
Genomic DNA was extracted from the peripheral blood using the QIAGEN (Hilden, Germany) DNA extraction kit and following the manufacturer’s instructions. Concentrations of the genomic DNA were determined by an ultraviolet spectrophotometer (Beckman, DU640 type).
PCR amplification and DNA sequencing
LR-PCR (exon 1-5, 9, and 11-15): Since exon 1-5, 9, and 11-15 of hPMS2 genes were severely hampered by the presence of multiple pseudogenes with highly similar sequences. LR-PCR was conducted to preferentially amplify hPMS2 gene and avoid the interference of the pseudogenes.
Four overlapping sets of primers were designed to amplify the complete coding region of hPMS2 gene by LR-PCR[15,16] (Table 1). The LR-PCR amplification profile is also shown in Table 1. Then 1/8 of the four LR-PCR products were used as template to amplify the 11 exons (exon 1-5, 9, 11-12 and 13-15) individually. The primer sequences are listed in Table 2.
Table 1.
Primer sequences of long-range polymerase chain reaction
| Primer name | Sequence (5’-3’) | Size (bp) | Exon |
| LRPCR1 | |||
| For | ACGTCGAAAGCAGCCAATGGGAGTT | 9964 | Exon 1-5 |
| Rev | CTTCCACCTGTGCATACCACAGGCT | ||
| LRPCR2 | |||
| For | GGTCCAGGTCTTACATGCATACTGT | 9440 | Exon 9 |
| Rev | CTGACTGACATTTAGCTTGTTGACA | ||
| LRPCR3 | |||
| For | GCGTTGATATCAATGTTACTCCAGA | 8812 | Exon 11, 12 |
| Rev | AGTAGTCAGGGTAAAACATTCCAGT | ||
| LRPCR4 | |||
| For | AAAATTAGTCAGACTTGATGGTGTG | 9804 | Exon 13-15 |
| Rev | CCTTCCATCTCCAAAACCAGCAAGA |
Table 2.
Primer sequences and polymerase chain reaction condition of individual exon of hPMS2 gene
| Exon | Primer sequence (5’-3’) | Size (bp) | AT (°C) | CN |
| 1 | M13F-ACGTCGAAAGCAGCCAATGGGAGTT | 475 | 66 | 28 |
| M13R-CAGGTAGAAAGGAAATGCATTCAGT | ||||
| 2 | M13F-ACAGTGTTGAGTCATTTCCCACAGT | 455 | 66 | 28 |
| M13R-TTCTTAGCATAACACCTGCCTGGCA | ||||
| 3 | M13F-TAGTCTGGGCTAGTAAATAGCCAGA | 705 | 68 | 35 |
| 4 | M13R-TATGACTTAGATTGGCAGCGAGACA | |||
| 5 | M13F-CTTGATTATCTCAGAGGGATCGTCA | 540 | 68 | 35 |
| M13R-TCTCACTGTGTTGCCCAGTCCTAAT | ||||
| 6 | M13F-TGCTTCCCTTGATTTGTGCGATGAT | 504 | 67 | 32 |
| M13R-TGAGGCAGGAGAATTGCTTGAATCT | ||||
| 7 | M13F-ACCCACGAGTTTGACATTGCAGTGA | 498 | 60 | 35 |
| M13R-GTAGAGGTTGCAGTGAGCCAAGATA | ||||
| 8 | M13F-AGATTTGGAGCACAGATACCCGTGA | 414 | 61 | 32 |
| M13R-TGCGGTAGACTTCTGTAAATGCACA | ||||
| 9 | M13F-CCTTCTAAGAACATGCTGGTTGGTT | 279 | 64 | 45 |
| M13R-ATCTCATTCCAGTCATAGCAGAGCT | ||||
| 10 | M13F-AGCCCTTCCGTATTTTGTCTATTCA | 719 | 61 | 32 |
| M13R-GCTTTAGAAGCTGTTTGTACACTGT | ||||
| 11 | M13F-TCACATAAGCACGTCCTCTCACCAT | 1021 | 64 | 45 |
| M13R-GCAACAGAGCAAGACTCTGTCTCAA | ||||
| 12 | M13F-GCCAAGATTGTGCCATTGCACTGTA | 493 | 64 | 25 |
| M13R-AGTAGATACAAGGTCTTGCTGTGTT | ||||
| 13 | M13F-GTGACACTTAGCTGAGTAGTGTTGT | 372 | 64 | 35 |
| M13R-ATGTTAGCCAGGCTGGTCTCAAACT | ||||
| 14 | M13F-GGTCTGTATCTCCTGACCTCATGAT | 473 | 64 | 35 |
| M13R-GCACGTAGCTCTCTGTGTAAAATGA | ||||
| 15 | M13F-GCTGAGATCTAGAACCTAGGCTTCT | 522 | 64 | 35 |
| M13R-ACACACGAGCGCATGCAAACATAGA |
AT: Anneal temperature; CN: Cycle number. The sequence of M13F was 5’-GTAAAACGACGGCCAGT-3’. The sequence of M13R was 5’-AACAGCTATGACCATG-3’.
PCR (exon 6, 7, 8 and 10): Conventional PCR was performed to detect the four exons (exon 6, 7, 8 and 10) which were seldom influenced by pseudogenes. Four sets of primers and PCR amplification profile are listed in Table 2.
DNA sequencing: The conventional PCR products were subjected to 2% agarose gel electrophoresis, while for LR-PCR products, 1% agarose was used with 9Kb as marker. After observation of clear and expected size bands, the products were purified and used as a template for sequencing reactions with BigDye terminator cycle sequencing kit (Applied Biosystems, Foster City, CA, USA). The sequencing primers were M13F or M13R. Automated fluorescence analysis was performed on a 3700 DNA sequence system (ABI, USA).
Bioinformatics analysis
Each result of sequencing was analyzed by DNAStar 5.08 bioanalysis software. The type of mutations and potential significance were determined by comparing the corresponding amino acids and proteins in the following databases (http://www.ncbi.nlm.nih.gov/; http://www.ensemble.org/homosapies; and http://www.insight-group. org).
RESULTS
Germline mutation of hPMS2 gene in HNPCC probands
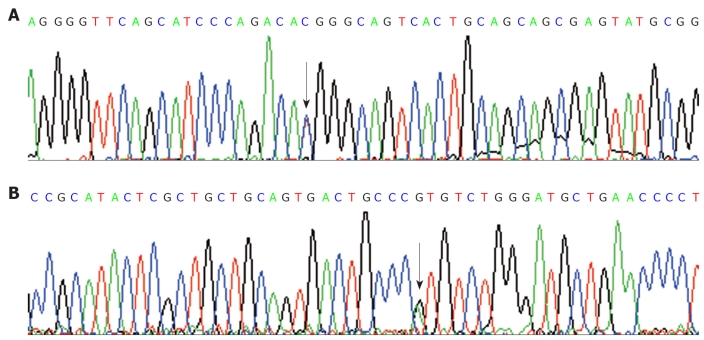
Among the 26 unrelated HNPCC probands, only one (H13) was found to carry the germline mutation of hPMS2 gene. She was a 30-year-old female BG patient. The mutation was a missense mutation at codon 511 (ACG>ATG, Thr>Met) (Figure 1). To further investigate the pathological effects of the missense mutation, we analyzed the related exon 11 in 130 controls by PCR-based sequencing. The results showed that the mutation of codon 511, consistent with the HNPCC case at c.1532C>T of exon 11 of hPSM2 gene, was also found in three healthy controls. The occurrence rate was approximately 2.3% (3/130). It could not be found in the PMS2-SNP database (http://www.nfdht.nl; http://www.insight-group.org; and http://www.ensembl.org). Thus, the mutation at c.1532C>T of hPSM2 gene which we detected in the HNPCC patient is an unreported new single nucleotide polymorphism (SNP).
Figure 1.
Missense germline mutation of exon 11 of hPMS2 gene in the proband of H13 hereditary nonpolyposis colorectal cancer kindreds. A: The forward sequence; B: The reverse sequence. Arrow indicates the mutation site, the single basyl substitution was transversed from C to T (C>T) at the codon 511, the codon from ACG to ATG, causing the amiod acid changes from Thr>Met, the change was identified as a new single nucleotide polymorphism.
SNP detection and analysis of hPMS2 gene
By DNA sequencing, 27 loci on the exons of hPMS2 gene including 260 reported SNP (http://www.ensembl.org/homo_sapies) were detected in the 26 HNPCC probands. Among them, 30% (78/260) were located in the 2nd exon, 23.1% (60/260) in the 5th exon, 13.8% (36/260) in the 15th exon, 10% (26/260) in the 7th exon, and 9.2% (24/260) in the 11th exon. However, none variant was detected in the remaining exons of the 1st, 3rd, 6th, 8th, 9th and 10th. The 2nd and 5th exons were probably the hot SNP regions of hPMS2 gene because 53.1% of the reported SNP were located in them. Distribution of the SNP of hPMS2 gene is shown in Table 3.
Table 3.
Distribution of single nucleotide polymorphism of hPMS2 gene in 26 probands
| Exon | Nucleotide change | Amino acid change | n | SNP (%) |
| 2 | c.24-4C>T | - | 14 | 78 (30) |
| c.89A>C | Gln30Pro | 15 | ||
| c.117A>G | Val39Val | 15 | ||
| c.120G>A | Lys40Ly | 4 | ||
| c.121G>A | Glu41Lys | 15 | ||
| c.124T>A | Leu42Ile | 8 | ||
| 4 | c.288C>T | Ala96Ala | 8 | 18 (6.9) |
| c.295A>C | Thr99Pro | 10 | ||
| 5 | c.406A>G | Met136Val | 10 | 60 (23.1) |
| c.418A>G | Asn140Asp | 10 | ||
| c.429T>C | Ile143Ile | 10 | ||
| c.452G>A | Arg151His | 10 | ||
| c.478C>A | Gln160Lys | 10 | ||
| c.492C>T | Ser164Ser | 10 | ||
| 11 | c.1408C>T | Pro470Ser | 7 | 24 (9.2) |
| c.1454C>A | Thr485Lys | 11 | ||
| c.2006+6G>A | - | 7 | ||
| 12 | c.2007-4G>A | - | 11 | 12 (4.6) |
| c.2007-7C>T | - | 1 | ||
| 13 | c.2253T>C | Phe751Phe | 1 | 1 (0.4) |
| 14 | c.2324A>G | Asn775Ser | 3 | 5 (1.9) |
| c.2340C>T | Pro780Pro | 2 | ||
| 15 | c.2466T>C | Leu822Leu | 12 | 36 (13.8) |
| c.2570G>C | Gly857Ala | 2 | ||
| c.92dupA | - | 17 | ||
| c.17G>C | - | 5 |
SNP: Single nucleotide polymorphism.
DISCUSSION
HNPCC, also called Lynch syndrome, is one of the most common autosomal dominantly inherited cancer syndromes with a high risk of colorectal cancer as well as other tumors occurring in endometrium, stomach, ovary, urinary tract, pancreas, small intestine, brain and skin. People with HNPCC take about 80% risk to develop colorectal cancer in their lifetime. It accounts for 2%-15% of all colorectal cancers. Compared to sporadic colorectal cancer, HNPCC possesses its own characteristics in clinical presentations, treatment, genetic features and management of kindred[17,18]. Many countries have established the clinical diagnostic criteria for HNPCC, such as AC, JC and BG. Defects in MMR genes, mainly in hMLH1, hMSH2 and hMSH6 were considered to be closely related to the genetic mechanism of HNPCC. The defection would consequently lead to the dysfunction of MMR system, ultimately resulting in the development of neoplasm. So, detection of MMR gene mutation is the only gold criteria to make a diagnosis of HNPCC.
Within the family of MMR genes, germline mutations in the coding region of hMSH2 and hMLH1 could be detected in up to 45%-64% of all HNPCC families, while hMSH6 about 10%. Previously we analyzed germline mutations and large genomic variations of the entire coding regions of hMSH2/hMLH1/hMSH6 genes and the methylation of hMLH1 promoter in 58 Chinese HNPCC probands, resulting in 29 germline mutations and 2 exhaustive inherited methylations of hMLH1 promoter (excluding 3 part-methylations of hMLH1 promoter). The total gene abnormality rate was only 53.4% (31/58). We suspected that the other MMR gene mutations might be associated with the remaining probands without hMSH2, hMLH1 or hMSH6 gene abnormalities.
The hPMS2 gene is a member of a set of human mismatch repair genes, located on chromosome 7. It encodes the protein that plays an essential role in repairing DNA by forming an active protein complex with the MLH1 protein which interacts with MSH2 bound to mismatched bases. In 1994, Nicolaides et al[19] firstly found the germline mutation of hPMS2 gene in a HNPCC patient. Since then, more and more data have proved that hPMS2 germline mutation is involved in the development of HNPCC. In some reports, it could be detected in as high as 62% of HNPCC probands[20]. The hPMS2 gene was suggested as the first candidate gene for testing germline mutations in HNPCC families in which hMSH2, hMLH1and hMSH6 aberrant was excluded. However, genetic testing for germline mutation of hPMS2 gene was technically challenging because they were severely hampered by a large family of highly homologous pseudogenes located on the same chromosome as the true hPMS2, such as PMS2CL. They shared similar sequences to hPMS2 but had no functions. Data from literature indicated that the exon 6 to 8 and exon 10 of hPMS2 could be easily screened by direct sequencing of genomic DNA without interference of pseudogenes. But detection of exon 1-5, 9 and exon 11-15 was complicated due to the interference of PMS2CL. LR-PCR was recommended as a useful method to preferentially identify hPMS2 but not the pseudogenes. In this study, we used LR-PCR to investigate the germline mutation of hPMS2 gene in those HNPCC probands who did not carry hMLH1/hMSH2/hMSH6 germline mutations investigated by the previous studies. Four overlapping sets of primers were designed to amplify the complete coding region of hPMS2 gene by LR-PCR firstly. Then, exon-specific amplifications from the LR-PCR products were performed to obtain a clear sequence with no evidence of pseudogene contamination. We only found one missense mutation in 26 probands, which has not been reported in Chinese HNPCC families. This mutation could also be detected in the 130 control persons with an occurrence rate of about 2.3%. Since it could not be found in the PMS2-SNP database (http://www.nfdht.nl; http://www.insight-group.org; and http://www.ensembl.org), the mutation at c.1532C>T of hPMS2 gene in our HNPCC case was an unreported new single nucleotide polymorphism (SNP). Our results showed that the germline mutation of hPMS2 gene was probably a rare event in Chinese HNPCC, even in those probands without hMLH1/hMSH2/hMSH6 mutations. It was consistent with the results of some other studies[21]. Interestingly, another mutation was found in the same nucleotide, c1532_1533 delCGinsAC, causing the amiod changes from Thr to Asn (http://www.insight-group.org). So, the exon 11 may be a hot SNP or mutation region of hPMS2 gene.
The frequency of germline mutation in hPMS2 gene was reported to be up to 62% if patients whose tumor tissues lacked protein expression of hPMS2 or had MSI-H features, were selected[22]. Among the HNPCC families with monoallelic mutation in hPMS2, 65.5% were complied with BG. Recently, Niessen et al[23] identified 4 patients with pathogenic mutation of hPMS2 among 97 patients with suspected Lynch syndrome who carried no germline mutation in hMLH1, hMSH2 or hMSH6. All these 4 patients fulfilled BG and their corresponding tumor cells showed MSI-H and loss of expression of hPMS2. Clendenning et al[24] reported that a kind of frame-shift mutation of hPMS2 occurred in 12 ostensibly unrelated Lynch syndrome patients with 20% being the deleterious mutation. However, those families with pathogenic mutation did not have significantly high incidence of Lynch syndrome associated malignant tumors, indicating that the germline mutation of hPMS2 and occurrence of HNPCC were not concurrent sometimes. The patient with hPMS2 gene mutation in our group also met the requirements of BG. By reviewing the family history of our mutation positive patient, we found that in her first-degree relatives, three suffered from colorectal cancer but diagnosed at age over 60 years, not in accordance with the typical feature of HNPCC. Although we are not so certain about this, the non-classical presentation of her family history, to some extent, represents the phenomenon of separation of HNPCC occurrence and hPMS2 gene mutation.
At the same time, we detected the reported SNP in these 26 probands and found some interesting results. Most of the SNP (21/27) were in the exons and 12 were non-synonymous coding SNP(cSNP). Since these non-synonymous cSNP can induce the change of amino acid and the relationship between cSNP and pathogenesis of HNPCC still remains unclear, whether they are involved in the development of HNPCC and HNPCC related tumors needs to be further investigated.
In conclusion, the germline mutation of hPMS2 gene is rare in the probands of Chinese HNPCC families. Since the testing of hPMS2 gene mutation is costly and complicated, it may be not reasonable to be included in the screening strategy of Chinese HNPCC. However, the frequency of SNP of hPMS2 gene is high and further studies are needed to identify its relationship with HNPCC.
COMMENTS
Background
Germline mutations in mismatched repair genes can lead to hereditary nonpolyposis colorectal cancer (HNPCC). Previously, the authors had analyzed the abnormalities of hMSH2/hMLH1/hMSH6 genes in a serious of Chinese HNPCC families and the total abnormality rate was only 53.4% (31/58). So the aberrant MMR genes such as hPMS2 were suspected to be involved in Chinese HNPCC.
Research frontiers
HNPCC or Lynch syndrome, is an autosomal dominantly inherited disease with cancer-susceptibility. The testing of hPMS2 gene mutation is costly and complicated, it may be not reasonable to be included in the screening strategy of Chinese HNPCC. However, the frequency of single nucleotide polymorphism (SNP) of hPMS2 gene is high and further studies are needed to identify its relationship with HNPCC.
Innovations and breakthroughs
One HNPCC proband was found to carry the germline mutation of hPMS2 gene. It was a new unreported coding SNP, which could also be detected in the control with an occurrence rate of 2.3% (3/130).
Applications
Germline mutations in genes can be used to diagnose early HNPCC and enrich the databases about HNPCC and SNP.
Peer review
This is an interesting article which deals with a remarkably rare germline mutation, namely PMS2, which is important in the etiology of Lynch syndrome (HNPCC). Their science appears to be sound.
Acknowledgments
The authors are grateful to the patients who took part in this study and to the Cancer Hospital for providing blood and tumor specimens. The authors also appreciate the help from Professor Sun MH, Wang CF, Cai Q and Yan SY for the detection of germline mutations in hMSH2, hMLH1 and hMSH6 genes of the probands of the Chinese HNPCC cases, and Professor Mo SJ for supplying the Chinese HNPCC cases.
Footnotes
Supported by The Initiation Fund for Prominent Young Researchers of Shanghai Medical College, Fudan University
Peer reviewers: Henry Thomson Lynch, MD, Department of Preventive Medicine, Creighton University School of Medicine, 2500 California Plaza, Omaha, NE 68178, United States; Minna Nyström, PhD, Department of Biological and Environmental Sciences, PO Box 56 (Viikinkaari 5 D), University of Helsinki, FI-00014 Helsinki, Finland
S- Editor Wang YR L- Editor Ma JY E- Editor Lin YP
References
- 1.Stephenson BM, Finan PJ, Gascoyne J, Garbett F, Murday VA, Bishop DT. Frequency of familial colorectal cancer. Br J Surg. 1991;78:1162–1166. doi: 10.1002/bjs.1800781005. [DOI] [PubMed] [Google Scholar]
- 2.Lagerstedt Robinson K, Liu T, Vandrovcova J, Halvarsson B, Clendenning M, Frebourg T, Papadopoulos N, Kinzler KW, Vogelstein B, Peltomäki P, et al. Lynch syndrome (hereditary nonpolyposis colorectal cancer) diagnostics. J Natl Cancer Inst. 2007;99:291–299. doi: 10.1093/jnci/djk051. [DOI] [PubMed] [Google Scholar]
- 3.Wagner A, Tops C, Wijnen JT, Zwinderman K, van der Meer C, Kets M, Niermeijer MF, Klijn JG, Tibben A, Vasen HF, et al. Genetic testing in hereditary non-polyposis colorectal cancer families with a MSH2, MLH1, or MSH6 mutation. J Med Genet. 2002;39:833–837. doi: 10.1136/jmg.39.11.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu SR, Zhao B, Wang ZJ, Wan YL, Huang YT. Clinical features and mismatch repair gene mutation screening in Chinese patients with hereditary nonpolyposis colorectal carcinoma. World J Gastroenterol. 2004;10:2647–2651. doi: 10.3748/wjg.v10.i18.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park JG, Kim DW, Hong CW, Nam BH, Shin YK, Hong SH, Kim IJ, Lim SB, Aronson M, Bisgaard ML, et al. Germ line mutations of mismatch repair genes in hereditary nonpolyposis colorectal cancer patients with small bowel cancer: International Society for Gastrointestinal Hereditary Tumours Collaborative Study. Clin Cancer Res. 2006;12:3389–3393. doi: 10.1158/1078-0432.CCR-05-2452. [DOI] [PubMed] [Google Scholar]
- 6.Peltomäki P, Vasen HF. Mutations predisposing to hereditary nonpolyposis colorectal cancer: database and results of a collaborative study. The International Collaborative Group on Hereditary Nonpolyposis Colorectal Cancer. Gastroenterology. 1997;113:1146–1158. doi: 10.1053/gast.1997.v113.pm9322509. [DOI] [PubMed] [Google Scholar]
- 7.Lipton LR, Johnson V, Cummings C, Fisher S, Risby P, Eftekhar Sadat AT, Cranston T, Izatt L, Sasieni P, Hodgson SV, et al. Refining the Amsterdam Criteria and Bethesda Guidelines: testing algorithms for the prediction of mismatch repair mutation status in the familial cancer clinic. J Clin Oncol. 2004;22:4934–4943. doi: 10.1200/JCO.2004.11.084. [DOI] [PubMed] [Google Scholar]
- 8.Fujita S, Moriya Y, Sugihara K, Akasu T, Ushio K. Prognosis of hereditary nonpolyposis colorectal cancer (HNPCC) and the role of Japanese criteria for HNPCC. Jpn J Clin Oncol. 1996;26:351–355. doi: 10.1093/oxfordjournals.jjco.a023244. [DOI] [PubMed] [Google Scholar]
- 9.Yan SY, Zhou XY, Du X, Zhang TM, Lu YM, Cai SJ, Xu XL, Yu BH, Zhou HH, Shi DR. Three novel missense germline mutations in different exons of MSH6 gene in Chinese hereditary non-polyposis colorectal cancer families. World J Gastroenterol. 2007;13:5021–5024. doi: 10.3748/wjg.v13.i37.5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou HH, Yan SY, Zhou XY, Du X, Zhang TM, Cai X, Lu YM, Cai SJ, Shi DR. MLH1 promoter germline-methylation in selected probands of Chinese hereditary non-polyposis colorectal cancer families. World J Gastroenterol. 2008;14:7329–7334. doi: 10.3748/wjg.14.7329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai Q, Sun MH, Fu G, Ding CW, Mo SJ, Cai SJ, Ren SX, Min DL, Xu XL, Zhu WP, et al. [Mutation analysis of hMSH2 and hMLH1 genes in Chinese hereditary nonpolyposis colorectal cancer families] Zhonghua Binglixue Zazhi. 2003;32:323–328. [PubMed] [Google Scholar]
- 12.Wang CF, Zhou XY, Zhang TM, Sun MH, Xu Y, Shi DR. [The analysis for mRNA mutation of MLH1, MSH2 genes and the gene diagnosis for hereditary nonpolyposis colorectal cancer] Zhonghua Yixue Yichuanxue Zazhi. 2006;23:32–36. [PubMed] [Google Scholar]
- 13.Wang CF, Zhou XY, Zhang TM, Xu Y, Cai SJ, Shi DR. Two novel germline mutations of MLH1 and investigation of their pathobiology in hereditary non-polyposis colorectal cancer families in China. World J Gastroenterol. 2007;13:6254–6258. doi: 10.3748/wjg.v13.i46.6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang CF, Zhou XY, Zhang TM, Sun MH, Shi DR. Detection of germline mutations of hMLH1 and hMSH2 based on cDNA sequencing in China. World J Gastroenterol. 2005;11:6620–6623. doi: 10.3748/wjg.v11.i42.6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Etzler J, Peyrl A, Zatkova A, Schildhaus HU, Ficek A, Merkelbach-Bruse S, Kratz CP, Attarbaschi A, Hainfellner JA, Yao S, et al. RNA-based mutation analysis identifies an unusual MSH6 splicing defect and circumvents PMS2 pseudogene interference. Hum Mutat. 2008;29:299–305. doi: 10.1002/humu.20657. [DOI] [PubMed] [Google Scholar]
- 16.Clendenning M, Hampel H, LaJeunesse J, Lindblom A, Lockman J, Nilbert M, Senter L, Sotamaa K, de la Chapelle A. Long-range PCR facilitates the identification of PMS2-specific mutations. Hum Mutat. 2006;27:490–495. doi: 10.1002/humu.20318. [DOI] [PubMed] [Google Scholar]
- 17.Luo DC, Cai Q, Sun MH, Ni YZ, Ni SC, Chen ZJ, Li XY, Tao CW, Zhang XM, Shi DR. Clinicopathological and molecular genetic analysis of HNPCC in China. World J Gastroenterol. 2005;11:1673–1679. doi: 10.3748/wjg.v11.i11.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai Q, Sun MH, Lu HF, Zhang TM, Mo SJ, Xu Y, Cai SJ, Zhu XZ, Shi DR. Clinicopathological and molecular genetic analysis of 4 typical Chinese HNPCC families. World J Gastroenterol. 2001;7:805–810. doi: 10.3748/wjg.v7.i6.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicolaides NC, Papadopoulos N, Liu B, Wei YF, Carter KC, Ruben SM, Rosen CA, Haseltine WA, Fleischmann RD, Fraser CM. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature. 1994;371:75–80. doi: 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- 20.Senter L, Clendenning M, Sotamaa K, Hampel H, Green J, Potter JD, Lindblom A, Lagerstedt K, Thibodeau SN, Lindor NM, et al. The clinical phenotype of Lynch syndrome due to germ-line PMS2 mutations. Gastroenterology. 2008;135:419–428. doi: 10.1053/j.gastro.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson E, Meldrum CJ, Crooks R, McPhillips M, Thomas L, Spigelman AD, Scott RJ. Hereditary non-polyposis colorectal cancer and the role of hPMS2 and hEXO1 mutations. Clin Genet. 2004;65:215–225. doi: 10.1111/j.1399-0004.2004.00214.x. [DOI] [PubMed] [Google Scholar]
- 22.Nakagawa H, Lockman JC, Frankel WL, Hampel H, Steenblock K, Burgart LJ, Thibodeau SN, de la Chapelle A. Mismatch repair gene PMS2: disease-causing germline mutations are frequent in patients whose tumors stain negative for PMS2 protein, but paralogous genes obscure mutation detection and interpretation. Cancer Res. 2004;64:4721–4727. doi: 10.1158/0008-5472.CAN-03-2879. [DOI] [PubMed] [Google Scholar]
- 23.Niessen RC, Kleibeuker JH, Westers H, Jager PO, Rozeveld D, Bos KK, Boersma-van Ek W, Hollema H, Sijmons RH, Hofstra RM. PMS2 involvement in patients suspected of Lynch syndrome. Genes Chromosomes Cancer. 2009;48:322–329. doi: 10.1002/gcc.20642. [DOI] [PubMed] [Google Scholar]
- 24.Clendenning M, Senter L, Hampel H, Robinson KL, Sun S, Buchanan D, Walsh MD, Nilbert M, Green J, Potter J, et al. A frame-shift mutation of PMS2 is a widespread cause of Lynch syndrome. J Med Genet. 2008;45:340–345. doi: 10.1136/jmg.2007.056150. [DOI] [PMC free article] [PubMed] [Google Scholar]