The WASP family control formation of actin filaments through the Arp2/3 complex. Subfamiles include WASP, SCAR/WAVE, WASH, and WHAMM. We show that the family is unexpectedly ancient and that all subfamilies are now identified. This work also identifies a subfamily-specific control mechanism, and an emerging bias towards vesicular roles of actin.
Abstract
WASP family proteins control actin polymerization by activating the Arp2/3 complex. Several subfamilies exist, but their regulation and physiological roles are not well understood, nor is it even known if all subfamilies have been identified. Our extensive search reveals few novel WASP family proteins. The WASP, WASH, and SCAR/WAVE subfamilies are evolutionarily ancient, with WASH the most universally present, whereas WHAMM/JMY first appears in invertebrates. An unusual Dictyostelium WASP homologue that has lost the WH1 domain has retained its function in clathrin-mediated endocytosis, demonstrating that WASPs can function with a remarkably diverse domain topology. The WASH and SCAR/WAVE regulatory complexes are much more rigidly maintained; their domain topology is highly conserved, and all subunits are present or lost together, showing that the complexes are ancient and functionally interdependent. Finally, each subfamily has a distinctive C motif, indicating that this motif plays a specific role in each subfamily's function, unlike the generic V and A motifs. Our analysis identifies which features are universally conserved, and thus essential, and which are branch-specific modifications. It also shows the WASP family is more widespread and diverse than currently appreciated and unexpectedly biases the physiological role of the Arp2/3 complex toward vesicle traffic.
INTRODUCTION
The actin cytoskeleton plays a crucial role in the lives of all eukaryotes. Its dynamic nature stems from the continuous turnover of individual actin filaments. Initiation of new filaments (called “nucleation” because the rate-determining step is the formation of a three-monomer nucleus) has a relatively high energy threshold (Sept and McCammon, 2001) and is the cell's principal control mechanism over actin filament formation. The Arp2/3 complex is a seven-subunit assembly that constitutes one of the most important nucleators of new actin filaments (Pollard, 2007). It binds to the side of an existing filament and makes a branch using a stub of two actin-related proteins (Rouiller et al., 2008). The addition of the first actin monomer to Arp2/3 is catalyzed by WASP family proteins and leads to a new growing, branched actin filament.
WASP family proteins are defined by their catalytic VCA domain, which consists of a Verprolin homology and a Central and an Acidic region (Miki et al., 1998). A more accurate name for the verprolin homology domain is the WASP Homology 2 (WH2) motif, and it is this motif that binds the nascent actin monomer (Paunola et al., 2002; Dominguez, 2007). The C region was initially thought to be homologous to cofilin and is named accordingly. However the sequence similarity with cofilin is poor, and the X-ray structure of cofilin has a different secondary structure than that of the VCA domain (compare PDB:1TVJ and PDB:1EJ5). As a consequence, it is no longer believed that the C motif is functionally similar to the proposed corresponding region of cofilin, and the region in between the WH2 motif and acidic region is now usually referred to as the Connecting or Central region (Marchand et al., 2001; Boczkowska et al., 2008). The Central region has affinity for both actin and Arp2/3 and may play a role in the transfer of the actin monomer to the Arp2/3 complex (Marchand et al., 2001; Kelly et al., 2006). The acidic region is a stretch of low complexity and highly enriched in acidic residues. A characteristic feature of the region is a tryptophan residue that is present two positions from the C-terminus, which is essential for Arp2/3 binding and enhances Arp2/3 complex activation (Marchand et al., 2001).
WASP, the founding member of the family, was first discovered over 15 years ago as the gene mutated in Wiskott-Aldrich syndrome patients (Derry et al., 1994). A few years later the related gene SCAR (also called WAVE) was discovered in Dictyostelium (Bear et al., 1998; Miki et al., 1998). SCAR has a C-terminal VCA domain similar to that of WASP but the N-terminal domain is different. This difference is reflected by their physiological functions. WASP subfamily proteins are involved in podosome formation and endocytosis, whereas SCAR subfamily proteins organize pseudopodia and lamellipodia; see Campellone and Welch (2010), for an extensive review. Until recently WASP and SCAR/WAVE comprised the entire WASP family, but in the last two years a number of new family members have been identified in the human genome, specifically WASH, WHAMM, and JMY (Linardopoulou et al., 2007; Campellone et al., 2008; Zuchero et al., 2009).
The recent flurry of articles on new WASP family proteins raises the question of how extensive the family actually is. When did these proteins first emerge and how have their domain structures and their associated proteins evolved? Completed genome sequences are now available for organisms in four of the eukaryotic kingdoms, namely plants, chromalveolates, excavates, and unikonts (Baldauf, 2003; Keeling et al., 2005; Cavalier-Smith, 2010). The availability of this genomic data for the first time allows a eukaryote-wide analysis of the WASP protein family. We have searched 54 genomes across the eukaryotic tree and explored the common and distinct features of WASP family proteins, traced back the roots of family, and also investigated the coevolution of binding partners and of the Arp2/3 complex.
MATERIALS AND METHODS
Databases and Sequence Alignments
To identify WASP family homologues, the predicted proteins of each genome were first searched using BLASTP (National Center for Biotechnology Informatics; http://blast.ncbi.nlm.nih.gov/Blast.cgi; Altschul et al., 1990) with a WASP family protein of the most closely related organism. If this did not yield any results, the search was repeated in the DNA contigs and expressed sequence tags using TBLASTN. Used databases were those from GenBank (http://blast.ncbi.nlm.nih.gov), Joint Genome Institute (http://genome.jgi-psf.org), TIGR (http://blast.jcvi.org), the BROAD institute (http://www.broadinstitute.org), Tetrahymena Genome Database (http://www.ciliate.org), the ParameciumDB (http://paramecium.cgm.cnrs-gif.fr), and the Cyanidioschyzon merolae Genome Project (http://merolae.biol.s.u-tokyo.ac.jp). See Supplemental Table 1 for a complete list of all organisms that were searched.
To classify putative WASP family proteins that were thus found, their sequences were aligned with members of known WASP subfamilies. All sequence alignments were done with ClustalX using default parameters (Larkin et al., 2007). Phylogenetic trees were calculated in ClustalX using Neighborhood Joining and were bootstrapped (1000 trials) as necessary. All trees were drawn in TreeView (http://taxonomy.zoology.gla.ac.uk/rod/treeview.htm; Page, 2002). Sequences with unusually long branch lengths were subjected to further examination and were reclassified where needed. Domain borders shown in the domain topology figures were identified using either the SMART Web site (http://smart.embl-heidelberg.de) or by visual inspection of sequence alignments.
Dictyostelium Cell Strains, Constructs, and Culture Conditions
The axenic Dictyostelium discoideum strain AX3 was used as wild type. Cells were cultivated in Petri dishes under HL5 medium (Formedium). The open reading frames of WASP B [DictyBase:DDB_G0272811] and WASP C [DictyBase:DDB_G0283827] were amplified from cDNA using PCR. The full length genes were then cloned into green fluorescent protein (GFP) expression vector pDM448 (Veltman et al., 2009). The resulting plasmids were electroporated into Dictyostelium using standard conditions and transformants were selected with 50 μg/ml hygromycin. For double expression with clathrin, the clathrin light-chain gene [DictyBase:DDB_G0277403] was cloned into the monomeric red fluorescent protein (mRFP) shuttle vector pDM413 (Veltman et al., 2009). The mRFP-clathrin light-chain fusion cassette was then excised using NgoMIV and cloned into the unique NgoMIV site of the WASP B or WASP C expression vector. To reduce autofluorescence, cells were incubated under Lo-Flo medium (Formedium) for 18 h before imaging.
Microscopy
Total Internal Reflection Fluorescence (TIRF) microscopy was performed on a Nikon Eclipse TE2000-U that was fitted with a custom condenser and a Nikon 1.45 NA 100× Plan Apo TIRF objective (Melville, NY). GFP was excited with the 473-nm line of an Omicron (South Bend, IN) diode laser, and mRFP was excited at 561 nm. Images were recorded on a backlit Photometrics cascade II camera (Tucson, AZ). MetaMorph software (Universal Imaging, West Chester, PA) was used to control the camera, shutters, and light sources.
RESULTS
WASP
The founding member of the WASP family was first identified in human (Derry et al., 1994; Miki et al., 1996), but several model organisms, such as yeast, Drosophila and Dictyostelium also express WASP orthologues (Naqvi et al., 1998; Bogdan and Klambt, 2003; Myers et al., 2005; Schafer et al., 2007). Although these organisms are separated by hundreds of millions of years of evolution, they all belong to the kingdom of unikonts and WASP proteins have thus far not been reported outside of this kingdom.
To determine the evolutionary range of WASP subfamily proteins, we searched 54 completed genomes for the presence of WASP genes (see Supplementary Table 1 for a complete list and abbreviations). At least one WASP homologue was found for each genome in the unikont kingdom, with the exception of the microsporidium Encephalitozoon, which has an unusually small genome. In contrast to the near-perfect of conservation of WASP in the unikont kingdom, WASP is found in only three of the 38 organisms of the remaining kingdoms, namely the chromalveolates Paramecium and Tetrahymena and the excavate Naegleria. The WASP-interacting protein (WIP) has been found to play a major role in controlling WASP function in unikonts (Ho et al., 2001, 2004). A WIP homologue is also present in Naegleria (JGI:77992), indicating that both have coevolved and that WASP function has been dependent on WIP for 2 billion years.
WASP Protein Features
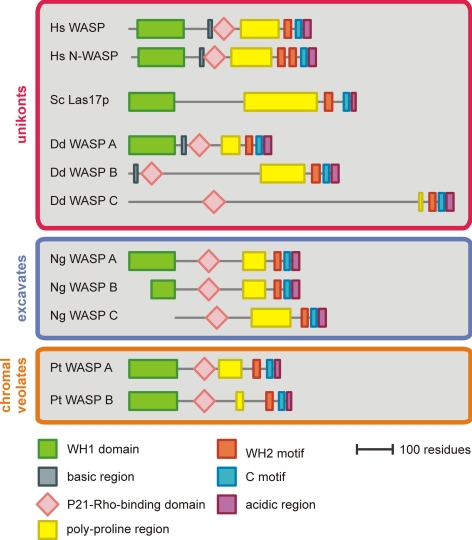
The common domain organization of WASP was determined on the basis of a sequence alignment (Figure 1 and Supplementary Figure 1). The domain structure of WASP in the evolutionarily distant Naegleria and Paramecium is very similar to that seen in mammals, indicating that the ancestral WASP protein must have closely resembled WASP as it is currently found in human. The sequence conservation of the WASP homology 1 domain (WH1 domain) and P21-Rho–binding domain (PBD; Burbelo et al., 1995) is very good across distant organisms, reflecting their functional importance. In contrast, the sequence in between these domains shows no regions of high homology.
Figure 1.
Domain topology diagrams of WASP orthologues from unikonts, excavates, and chromalveolates. WASP is not found in plants. The regions in between the indicated domains show no sequence similarity when comparing orthologues across different kingdoms.
One of the more prominent differences in evolutionarily distant WASPs compared with mammalian WASP regards the basic region. Residues in the basic region bind to negatively charged phospholipids, which is essential for optimal activation (Rohatgi et al., 2000; Hemsath et al., 2005). In vertebrate WASP this region is located immediately upstream of the PBD. A basic region can be found in most unikont WASP proteins, but the position of the region shifts away from the PBD in organisms that branched earlier from the tree (Supplementary Figure 1). Also, relatively more acidic residues are present in more primitive organisms. In Tetrahymena and Paramecium the number of acidic residues matches the number of basic residues, leaving a region that is very polar, but with no net charge. In Naegleria the basic region is totally absent. These findings indicate that the ancestral WASP may not have been regulated by negatively charged phospholipids, but that this level of regulation has evolved only later in metazoa.
Unconventional Dictyostelium WASP
Dictyostelium contains a single WASP homologue with the conventional domain topology (WASP A). However, two additional proteins are present that contain both a PBD and a VCA domain. The PBD aligns very well with PBD of human WASP (Supplementary Figure 1). In addition, the VCA domains of the proteins align best with the VCA domains of WASP subfamily proteins, further implying that these proteins are WASP homologues. We have consequently named these proteins WASP B [DictyBase:DDB_G0272811] and WASP C [DictyBase:DDB_G0283827]. A gene very similar to WASP B and C is found in the genome of Dictyostelium purpureum [JGI:81492], indicating these novel genes are not simply pseudogenes of the conventional WASP A. Notably, neither protein contains any sequence that is homologous to the WH1 domain. In fact, the regions flanking the PBD do not resemble any other protein in the GenBank nonredundant database.
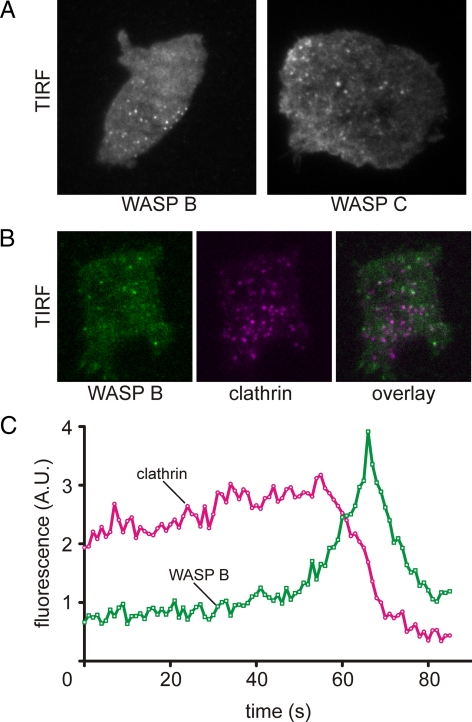
To investigate effect of the loss of the WH1 domain on the function of WASP B and C, we amplified the genes from cDNA, fused them to GFP and expressed the recombinant genes in wild-type cells. GFP-WASP B was visible as small transient puncta at the basal membrane (Figure 2A). No localization was observed at the leading edge during cell migration. Mammalian N-WASP, yeast Las17p, and Dictyostelium WASP A are involved in clathrin-mediated endocytosis (Naqvi et al., 1998; Merrifield et al., 2004). To investigate if WASP B is also involved in clathrin-mediated endocytosis, we coexpressed GFP-WASP B together with clathrin light chain fused to mRFP. The results closely match the observations of conventional WASP; puncta of clathrin are present at the basal membrane for ∼1–2 min and disappear shortly after the recruitment of WASP B (Figure 2, B and C, and Supplementary Movie 1). A quantification of several time-lapse movies reveals that >95% of all clathrin puncta show a recruitment of WASP before disappearing. A similar result was obtained with WASP C, indicating that both may have overlapping functions. The major difference between WASP B and C is that WASP C is also visible in long strands, similar to that of actin filament bundles, as seen with the F-actin probe LimEΔcoil (Bretschneider et al., 2004). WASP B and C thus contain all the regulatory sequences required for targeting to coated pits, meaning that at least in Dictyostelium, the WH1 domain is not essential for targeting and that other domains are responsible.
Figure 2.
Dictyostelium WASP B and C characterization. (A) TIRF microscopy image of a Dictyostelium cell expressing GFP-tagged WASP B or C. (B) TIRF image of a Dictyostelium cell coexpressing GFP-WASP B and mRFP-clathrin light chain. (C) A time-lapse movie was recorded for a cell as shown in B. The average green and red fluorescence intensity of an area enclosing a single clathrin punctum was measured every second, and the results were plotted against time.
Although the similarity between WASPs from divergent organisms shows that the ancestral WASP must have closely resembled the current human WASP, there is a great diversity in domain composition. Indeed, it becomes hard to define WASP, as each of the individual domains except the C-terminal, catalytic VCA can be lost without loss of biological function. In Dictyostelium WASP B, the WH1 domain is lost, whereas in Saccharomyces the PBD is lost and in Naegleria the basic region is absent.
Origin of the Human WASP and N-WASP Paralogues
The human genome encodes two WASP paralogues, WASP and N-WASP, whereas invertebrates typically encode only 1 WASP protein. To determine the evolutionary roots of human WASP and N-WASP, we identified a large number of metazoan WASP proteins and constructed a phylogenetic tree (Figure 3). WASP homologues from invertebrate organisms group together on a branch that clearly separates them from their vertebrate counterparts. Surprisingly, all examined invertebrate organisms encode a WASP with a tandem WH2 motif. In contrast, nonmetazoan WASP orthologues have only a single WH2 motif. Apparently, the ancestral WASP only possessed a single WH2 motif, and during early metazoan evolution the WH2 motif of WASP was duplicated.
Figure 3.
Phylogenetic tree of metazoan WASP homologues. (A) Full-length WASP protein sequences from a large number of metazoa were aligned in ClustalX, and a distance tree was calculated from the results. Bootstrap values (1000 trials) are given for the branches that separate the different colored groups. A yellow dot indicates the presence of a tandem WH2 motif at the C-terminus. Alphabetical suffixes are added for paralogues that fall within the same colored group. (B) Schematic showing the most likely sequence of events that explains the distribution of WASPs with single and tandem WH2 motifs across metazoa.
Vertebrate WASP proteins form two separate branches. One branch contains human WASP, and the other branch contains human N-WASP, indicating that the split between WASP and N-WASP occurred at the onset of vertebrate evolution. Curiously, hematopoietic WASP has lost the extra WH2 motif that was acquired in early metazoa, whereas the ubiquitously expressed N-WASP remains unchanged. Thus, unlike its naming suggests, N-WASP with the tandem WH2 motif forms the direct link with the ancestral WASPs. This is consistent with the shared function of vertebrate N-WASP, Saccharomyces Las17p and Dictyostelium WASP in clathrin-mediated endocytosis and implies that the original function of the WASP subfamily was the organization of endocytosis and vesicle trafficking. Hematopoietic WASP branched from this line during the onset of vertebrates and acquired novel roles such as podosome formation (Linder et al., 1999).
SCAR/WAVE
The second member of the WASP family, SCAR/WAVE, drives actin nucleation in pseudopodia and lamellipodia and has so far been studied in metazoa, Dictyostelium and Arabidopsis; because the SCAR name was used before (Bear et al., 1998; Miki et al., 1998), we will use it hereafter (it is more common to use WAVE in mammalian cells and SCAR in other model organisms). Our search in 54 completed genomes reveals no fewer than 32 species with SCAR proteins. No yeast genome encodes SCAR, but SCAR is found in Batrachochytrium, which is a member of the Chytridiomycota, the most basal fungal phylum. SCAR is also found in heterokonts such as Phytophtora and excavates such as Naegleria and Trichomonas.
SCAR/WAVE Family Tree
In vivo, SCAR is part of a pentameric complex together with PIR121 (homologues in other organisms are also called CYFIP1, Sra1, GEX-2, or KLUNKER), Nap (also called Hem1, KETTE, or GEX-3), Abi and HSPC300 (also called Brick1; Eden et al., 2002; Derivery et al., 2009a). Essentially all unikonts, excavates, and plants that encode SCAR also encode the other complex members (Supplementary Figure 2). The only exception is found in chromalveolates, where Abi is consistently absent. However, Abi has the lowest sequence conservation of all SCAR complex members (compare metazoan Abi to plant Abi in Supplementary Figure 3), and without biochemical evidence it cannot be confirmed whether chromalveolates have lost Abi or whether their Abi orthologues are simply undetectable using BLAST searches. The sporadic inability to find all complex members, such as in Emiliania and Aureococcus, is probably due to incomplete genome sequence coverage. The other way around, organisms that lose SCAR also lose the remaining complex members. The excellent correlation between the presence of SCAR and the other complex members indicates that the complex is fundamental to SCAR function and that a role for individual subunits outside of the complex is not supported.
Conservation of SCAR Complex Protein Features
SCAR in plants differs substantially from its unikont homologues. Plant SCARs have a long, poorly defined region between the SCAR homology domain and the VCA domain, and their poly-proline region is much reduced compared with their unikont counterparts (Figure 4). To determine if the ancestral SCAR more closely resembled the current plant homologues or the current unikont homologues, we constructed domain topology diagrams of SCAR proteins from organisms across the eukaryotic tree. As can be seen in Figure 4, the excavate and chromalveolate SCAR look remarkably like their unikont homologues, and it is the plant SCAR that deviates from the common theme. Therefore, the ancestral SCAR must have resembled the current human SCARs and the long insertion after the SCAR homology domain and the decimation of the poly-proline region are specific to the plant lineage.
Figure 4.
Domain topology diagrams of SCAR and Abi orthologues from several different organisms in each kingdom. The SCAR and Abi homology domains are defined as the largest region on the N-terminus that shows sequence similarity when comparing orthologues from different kingdoms. The regions in between indicated domains show no sequence similarity when comparing orthologues across different kingdoms.
The story is different for the SCAR complex subunit Abi. An alignment of Abi homologues reveals that only the N-terminal 150 residues are conserved when comparing organisms across different kingdoms (Supplementary Figure 3). We have termed this region, which must have been present in the ancestral protein, the Abi homology domain (Figure 4). The Abi homology domain encompasses the full SCAR binding region (Echarri et al., 2004) and SNARE motif (Echarri et al., 2004), but only half of the proposed homeodomain homology region (Dai and Pendergast, 1995). However, the most notable finding is that only fungal and metazoan Abi homologues have a C-terminal SH3 domain. This domain is absent from Abi homologues in lower unikonts and the other kingdoms. It is thus most likely that the ancestral Abi had no SH3 domain and that this domain was added later during unikont evolution.
In contrast to SCAR and Abi, no subdomains have been identified in the SCAR complex subunits PIR121, Nap and HSPC300, and the function of these proteins is still largely unknown. To identify conserved features of PIR121, Nap, and HSPC300, we aligned sequences from various organisms across different kingdoms (not shown). In all three cases, the orthologous proteins show very high sequence similarity over their entire length. No large insertions or deletions have occurred, and none of the proteins have any recognizable domains. In addition, no other proteins contain a fragment of Nap, PIR121, or HSPC300, suggesting that they can only function as a whole. Thus, unlike with SCAR or Abi, phylogenetic analysis does not provide any clues on how the potential function of these proteins can be dissected.
Origins of Mammalian SCAR Complex Protein Paralogues
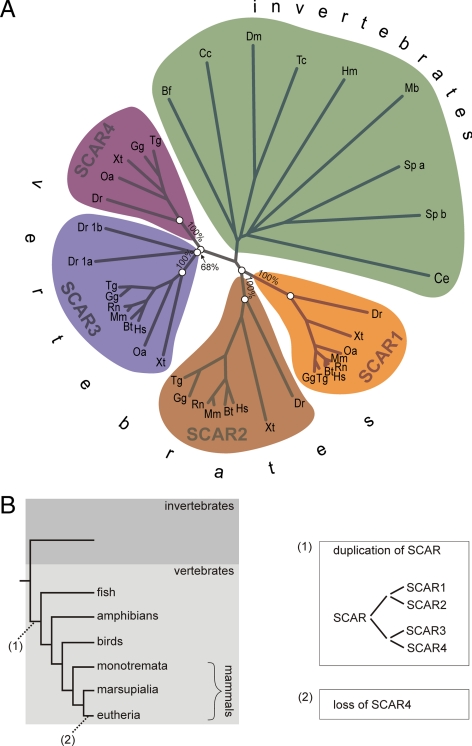
Similar to that of WASP, the number of SCAR paralogues in metazoa is variable. The human genome encodes 3 paralogues, whereas Danio has five paralogues and most invertebrates have only one copy of the SCAR gene. Paralogues generally have distinct, specialized functions. This property can be helpful to interpret experimental data from different model organisms, but it is currently not clear what the relation between the different vertebrate SCAR homologues is. To discriminate which genes are orthologues and which are species-specific gene duplications, we aligned all SCAR proteins from several metazoa and calculated a phylogenetic tree from the result (Figure 5A). Human SCAR1, SCAR2, and SCAR3 are located on different branches that we have named accordingly. All other vertebrates also have a SCAR homologue on the SCAR1, SCAR2, and SCAR3 branch, whereas all invertebrate SCAR homologues group together on a different branch, clearly indicating that SCAR1, SCAR2, and SCAR3 diverged at the onset of vertebrate evolution. Surprisingly, a number of vertebrates contain an additional SCAR that does fall within any of the established groups. Instead, these SCARs group together on a separate branch, indicating that they are orthologues and not species-specific duplications. We have called this extra group SCAR4. Inspection reveals that all vertebrates, except eutheria (placental mammals), encode such a SCAR4 orthologue. It should be noted here that the SCAR4 that is currently annotated in the human genome is a species-specific duplication of SCAR2 that is most likely a pseudogene and that is not a member of the vertebrate SCAR4 group that we define here.
Figure 5.
The evolutionary origins of SCAR1, 2, 3, and 4. (A) Full-length SCAR protein sequences from a large number of metazoa were aligned in ClustalX, and a distance tree was calculated from the results. Bootstrap values (1000 trials) are given for the branches that separate the different colored groups. Alphabetical suffixes are added for paralogues that fall within the same colored group. (B) Schematic showing the most likely split and loss of SCAR homologues that has led to the current distribution of SCAR paralogues in vertebrates.
When combining the SCAR gene tree with the species tree, it is possible to reconstruct a sequence of events that has led to the current distribution of SCAR genes in vertebrates (Figure 5B). At the split of vertebrates, a point where the genome is known to have duplicated twice (Dehal and Boore, 2005), the single ancestral SCAR duplicated into four paralogues. Most vertebrates have kept all four copies of SCAR, but in eutheria SCAR4 was lost.
A similar alignment and gene tree was made for the other SCAR complex subunits (Supplementary Figure 4). The gene trees of the different subunits are remarkably similar. The paralogues PIR121/Sra1, Nap1/Hem1, and Abi1/Abi2 all split at the onset of vertebrates. Only the time point of splitting of Abi3 (NESH) remains ambiguous, as these sequences are very divergent and they do not form a separate branch. Nonetheless, the results clearly indicate that the SCAR complex greatly diversified at a single point in evolution, at the onset of vertebrates.
WASH
The WASP protein family expanded in 2007 with the identification of WASH (WASP and SCAR homologue; Linardopoulou et al., 2007). The gene is present in the subtelomeric region of the human genome, which accounts for the relatively late discovery of this protein. Several recent articles find that WASH has a role in vesicle fission during retrograde traffic (Derivery et al., 2009b; Gomez and Billadeau, 2009; Liu et al., 2009).
WASH Family Tree
WASH homologues have thus far been identified only in unikonts such as metazoa, Dictyostelium, and Entamoeba. However, a more complete search uncovers homologues in the other kingdoms as well. In excavates, WASH is found in four of five organisms with completed genomes. Many chromalveolates and plants also encode WASH homologues. For example, WASH is found in the green algae Ostreococcus. Divergent WASH homologues can even be found in the moss Physcomitrella, the nonflowering plant Selaginella and as high as the monocot group of flowering plants such as Sorghum. The most likely explanation why the presence of WASH in the plant kingdom has thus far been overlooked is the loss of WASH in Arabidopsis.
Conservation of WASH Complex Protein Features
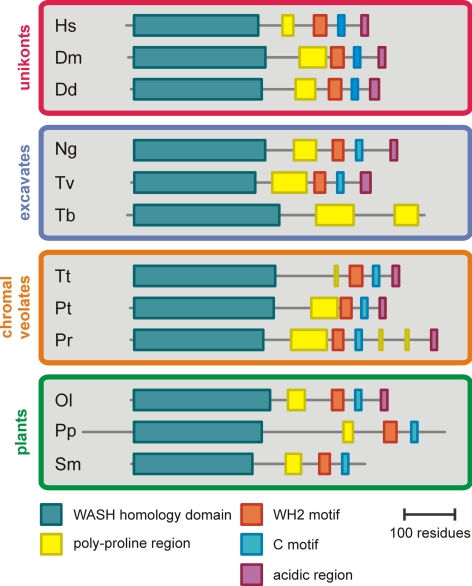
A domain structure diagram of WASH that was drawn on the basis of a sequence alignment (Supplementary Figure 5) shows that the overall domain topology of WASH homologues from organisms out of all four kingdoms is remarkably similar (Figure 6). The most notable variation on the common theme is the replacement of the VCA domain by a poly-proline region in Trypanosoma. Curiously, Trypanosoma does encode all Arp2/3 complex subunits, but no WASP family proteins other than WASH, which poses the interesting question of how Trypanosoma WASH regulates Arp2/3. Another notable variation on the common theme is the loss of the acidic region in the VCA domains of WASH in land plants such as Physcomitrella and Selaginella. This loss is specific for WASH as all SCAR homologues in land plants have well conserved VCA domains that include the acidic region.
Figure 6.
Domain topology diagrams of WASH orthologues from several organisms in each kingdom. The WASH homology domain is the largest region on the N-terminus that shows sequence similarity when comparing orthologues from different kingdoms. The regions in between domains show no sequence similarity when comparing orthologues across different kingdoms.
A recent article describes that WASH, just like SCAR, is part of a multiprotein complex (Derivery et al., 2009b). The identified complex members are KIAA1033, KIAA0592, Strumpellin, and ccdc53 (Derivery et al., 2009b). We have searched for the WASH complex members in our set of organisms with completed genomes. There is a high degree of coconservation of WASH and the proposed complex members (Supplementary Figure 6). Thalassiosira is the only exception, because this organism encodes KIAA0592, KIAA1033, Strumpellin, and ccdc53 but not WASH. Interestingly, Thalassiosira also does not encode any subunits of the Arp2/3 complex and the incomplete complex thus has a function other than Arp2/3 activation. The inability to detect every subunit in some other species is likely due to incomplete genome sequence coverage. Nonetheless, out of the 33 organisms with WASH, 30 have ccdc43, 31 have KIAA0592, 32 have KIAA1033, and all have Strumpellin. The high degree of correlation between the presence of WASH and the other complex members strongly suggests that the function of the proteins is fully dependent on their presence in the complex.
Somewhat surprisingly, heterodimeric capping protein was also found to be associated with the WASH complex (Derivery et al., 2009b). The correlation between the presence of WASH and heterodimeric capping protein is conspicuously lower compared with the other complex members (Supplementary Figure 6). This suggests that capping protein is not centrally tied to WASH function, in agreement with capping protein's independent role in terminating actin polymerization.
WHAMM and JMY
WHAMM and JMY are the most recently discovered VCA proteins. WHAMM binds to membranes and microtubules and localizes to the perinuclear region (Campellone et al., 2008). JMY appears to have dual roles in transcriptional regulation and cell motility (Coutts et al., 2007; Zuchero et al., 2009). The WHAMM and JMY proteins show high sequence similarity, and it is clear that the genes have diverged relatively recently. Thus far homologues of WHAMM and JMY have only been described in vertebrates.
WHAMM/JMY Family Tree
We have searched for WHAMM/JMY homologues across the eukaryotic tree. In addition to the known vertebrate copies, we also found WHAMM/JMY homologues in the invertebrate organisms Strongylocentrotus, Branchiostoma, Lottia, and Capitella. Outside of bilateralia, no trace of either WHAMM or JMY could be found. The organisms in which invertebrate WHAMM/JMY homologues are found share a common ancestor with insects, but the gene has been lost in the insect lineage. The loss of WHAMM/JMY in Drosophila may be the reason why their presence in invertebrates has thus far been overlooked.
An alignment and a phylogenetic tree were generated from the identified WHAMM/JMY sequences (Figure 7). All invertebrate homologues group together on a separate branch, distinct from vertebrate WHAMM and JMY, indicating that the ancestral gene first evolved in invertebrates and that the split between the present day WHAMM and JMY occurred later at the onset of vertebrates. Because the invertebrate homologues are equally similar to WHAMM and JMY, we term this group WHAMY.
Figure 7.
The evolutionary origins of WHAMM and JMY proteins. (A) Full-length WHAMM/JMY protein sequences from indicated organisms were aligned in ClustalX, and a distance tree was calculated from the results. Bootstrap values (1000 trials) are given for the branches that separate the different colored groups. (B) Schematic showing the most likely chronological order of the mutation of the C-terminal phenylalanine and the gene duplication of WHAMY, based on the phylogenetic tree in A. The F indicates the presence of a phenylalanine near the C-terminus, and W indicates the presence of a tryptophan at this position.
Conservation of WHAMM/JMY Protein Features
It has been proposed that WHAMM and JMY use different mechanisms for actin nucleation. WHAMM has two WH2 motifs and is dependent on Arp2/3 for filament nucleation. Instead, JMY has three WH2 motifs and can also nucleate actin filaments by aligning several monomers in a similar manner to Spire/Cordon-bleu (Quinlan et al., 2005; Zuchero et al., 2009). Examination of the domain topology shows that the invertebrate WHAMY homologues only have one WH2 motif (Figure 8 and Supplementary Figure 7). Ancestral WHAMY is thus fully dependent on Arp2/3 for filament nucleation. Apart from the different number of WH2 motifs, invertebrate WHAMYs generally have the same domain topology as WHAMM and JMY. The only exception is Strongylocentrotus WHAMY that curiously contains a full repeat of the N-terminal and coiled coil domain.
Figure 8.
Domain topology diagram of metazoan WHAMM/JMY homologues. The borders of the N-terminal domain and coiled coil domain were determined by sequence alignment with WHAMM/JMY homologues where the domains have been defined.
A closer inspection of the invertebrate WHAMY VCA domains reveals another interesting difference with vertebrate WHAMM/JMY VCA domains. The acidic region of the VCA domain invariably contains a tryptophan residue close to the C-terminus that is involved in Arp2/3 binding. However, WHAMY in Strongylocentrotus, Lottia and Capitella, but not Branchiostoma, have a phenylalanine at this position (Supplementary Figure 7). Strongylocentrotus, Lottia and Capitella share a common ancestor that is older than that of Branchiostoma and vertebrates. Thus, the ancestral WHAMY protein first evolved with a phenylalanine in the acidic region. Only later during evolution this residue was changed to a tryptophan (Figure 7B). The implications of this remain unclear. All WASP, SCAR, and WASH homologues in Strongylocentrotus, Lottia, and Capitella have a tryptophan at their C-terminus, so preservation of the phenylalanine is specific for WHAMY. It is generally assumed that WASP family proteins require a C-terminal tryptophan to function, but the genomic data prove either that this is not necessarily the case or that ancestral WHAMY does not function as a conventional Arp2/3 activator.
Novel WASP Protein Families
The recent discovery of WASH and WHAMM/JMY raises the question whether all WASP protein families have now been identified or if there are still uncharacterized genes to be discovered. The defining property of WASP family proteins is the VCA domain, and we used this domain to search for novel WASP protein families. Typically, the predicted proteins of each genome were searched with the VCA domain of a known WASP family protein of that organism or with a VCA domain of a closely related organism. In two of the 54 searched genomes this search gave surprising results.
Entamoeba WASP Family Proteins
In Entamoeba, our search yielded four novel proteins with a C-terminal VCA domain. The proteins are highly homologous to each other over their entire sequence, clearly identifying them as paralogues. Their general domain topology is comparable to that of known WASP family members (Figure 9A), having an N-terminal homology region, followed by a poorly conserved linker region, a poly-proline region, and a C-terminal VCA domain. The VCA domain is very similar to that of known WASP family members; the WH2 motif, the C motif, and acidic region are all present, and the spacing between the regions is similar compared with the VCA domains of WASP or SCAR (Figure 9B). However, the N-terminal domain of these proteins has no sequence similarity with any of the known WASP family members. Instead, the N-terminal domain is most similar to the I-BAR domain (Habermann, 2004). In fact, these novel WASP family proteins are also identified by BLAST searching with the I-BAR domain of Dictyostelium IRSp53/IRTKS [DictyBase:DDB_G0274805]. I-BAR domain–containing adapter proteins such as IRSp53 and IRTKS couple membrane dynamics to actin polymerization by means of a C-terminal SH3 domain (Scita et al., 2008). These novel Entamoeba WASP family proteins may have cut such a pathway short by directly coupling the I-BAR domain to a VCA domain.
Figure 9.
Evolution of novel WASP family proteins. (A) Domain structures of four paralogues of a novel Entamoeba WASP family protein. (B) Sequence alignment of the VCA domain of the novel Entamoeba WASP family proteins. (C) Domain topology of Rattus SCAR2, SCAR3, and LOC302367. Numbers in the shaded areas indicate the percentage identity between the sequences. (D) Sequence alignment of the VCA domain of Rattus SCAR proteins and LOC302367. (E) Domain topology comparison of the divergent Rattus and Mus WASP family proteins. Numbers in the shaded areas indicate the percentage identity between the sequences.
Unusual Murine WASP Family Protein
Surprisingly, an unusual WASP family gene is also found in the genome and cDNA libraries of the Murinae Mus and Rattus, but not in the incomplete genomes of the rodents Cuniculus and Cavia. The predicted proteins show strong sequence identity with each other and are clear orthologues (Figure 9). Both proteins have a well-conserved VCA domain; the WH2 motif is confidently recognized by SMART (http://www.smart.embl-heidelberg.de), the C-terminal region is rich in acidic residues, and a tryptophan is present at the third-last position. Furthermore, a typical α-helix (discussed later) is found in the region in between the WH2 motif and the acidic region.
The N-terminal region of the Rattus orthologue, LOC302367, is similar to the SCAR homology domain, indicating that the gene originated from SCAR (Figure 9). Curiously, the gene is encoded by only a single exon, whereas the three Rattus SCAR genes each contain at least eight introns. The most plausible explanation for this is that a processed SCAR mRNA was reverse-transcribed and reintroduced into the genome. Ever since that event, which must have taken place in a common ancestor of Rattus and Mus, the gene has undergone a rapid evolution. The overall sequence identity of the gene compared with its closest relative, Rattus SCAR3, varies between 15 and 34% in different domains, whereas Rattus SCAR2 and SCAR3 (which have been separated for much longer) vary between 32 and 81% (Figure 9C). The typical basic region and the poly-proline region that are present in SCAR homologues across the eukaryotic tree are no longer present in LOC302367. However, the observations that the sequence conservation is highest in the critical VCA domain and that no stop codons were introduced in the gene despite the poor sequence conservation (at least eight gaps are introduced in the alignment between LOC302367 and Rattus SCAR3), indicate that the gene is still under selective pressure and that it is not merely a pseudogene.
The Evolutionary Origins of the WASP Protein Family
A complete overview of the WASP protein family can now be constructed by combining the WASP family gene tree with the species tree of eukaryotes. The exact form of the eukaryotic tree remains a controversial topic. The location of the root of the tree, in particular, is debated. We have based the tree in Figure 10 on the one from Keeling et al. (2005), who reviewed recent progress in the assembly of the eukaryotic tree. This consensus tree places the root of eukaryotes between the split of excavates, unikonts, and chromalveolates/plants. The distribution of WASP, SCAR, and WASH in this tree leads to the conclusion that these proteins were all present in the eukaryotic last common ancestor. This conclusion does not change when the root is placed basal to unikonts (Baldauf, 2003; Stechmann and Cavalier-Smith, 2003). Nonetheless, some alternative scenarios exist. A recent article places the root of eukaryotes basal to Euglenozoa (Cavalier-Smith, 2010), leaving the possibility open that WASH emerged first and SCAR and WASP evolved later. WASP is also rare enough in the more basal branches that horizontal gene transfer, though unlikely, cannot be ruled out.
Figure 10.
Distribution of WASP family proteins across the eukaryotic tree. An evolutionary tree is drawn of the 54 organisms with completed genomes that were searched for WASP family proteins (based on Keeling et al., 2005). Branch lengths do not reflect evolutionary distances. For each organism, the presence of formins, the Arp2/3 complex and WASP family proteins is indicated with colored disks. The Arp2/3 complex was considered to be complete if subunits ArpC1-ArpC5 were present. The number of missing pie slices in some disks indicates the number of missing complex subunits. Completeness of the SCAR complex (five subunits) and the WASH complex (seven subunits) is represented in a similar manner. Footnotes for the Arp2/3 complex: (a) Incomplete ArpC4 homologue found, all other subunits are absent (b) ArpC2 and ArpC5 are absent; ArpC1 is incomplete. (c) ArpC1 and ArpC5 are absent; ArpC2, ArpC3, and ArpC4 are incomplete. The same result is obtained in the closely related Ostreococcus taurus. (d) ArpC1 and ArpC2 have substantially diverged. ArpC3 is absent.
The presence of formins and the Arp2/3 complex is also indicated in the tree. Formins can be found in all but one of the investigated genomes, whereas the Arp2/3 complex is frequently and independently lost. Despite a comparative genomic analysis to determine the specific differences of actin-related proteins, it remains difficult to unambiguously discriminate Arp2 and Arp3 from other actin-related proteins in evolutionary distant species (Muller et al., 2005). We therefore confirmed the presence of the Arp2/3 complex by searching for the complex members ArpC1 to ArpC5 instead. All organisms that encode ArpC1–5 also encode at least one WASP family member, indicating that in principle there is no need to postulate unknown Arp2/3 activators. Conversely, all organisms with no Arp2/3 complex members do not encode any WASP family members, confirming the intimate relationship between these protein complexes.
Curiously, several organisms encode some, but not all Arp2/3 subunits. Although this can sometimes be attributed to incomplete genomic sequence data, this does not always appear to be the case. For example, the genomes of the closely related Volvox and Chlamydomonas encode only ArpC3 and ArpC4 and the first 200 amino acid residues of ArpC1. Even a close inspection of the genomic scaffold reveals no trace of the remainder of ArpC1 in the intergenic regions. The subunits ArpC2 and ArpC5 are completely absent from both organisms. The fact that the same truncated subset of Arp2/3 complex members is present in two distinct organisms indicates that this is not due to incomplete sequence information, but that instead the complex has substantially diverged. Neither Volvox nor Chlamydomonas encode any WASP family proteins and the partial Arp2/3 complex in these organisms is thus no longer activated in the conventional way.
The Physiological Functions of the VCA Domain
It is widely accepted that the C-terminal VCA domain of WASP family proteins brings the actin monomer into proximity with the Arp2/3 complex, but the exact mechanism is not understood, and neither is it known how VCA domains are activated. We aligned VCA domains from WASP subfamilies of several distantly related organisms in order to identify universally conserved features of the VCA domain.
The sequence alignment in Figure 11 starts with the WH2 motif that binds the nascent actin monomer. The high sequence conservation of actin across various species is reflected by the conservation of key binding residues in the WH2 motif. The α-helix with the LLxxIR consensus sequence that forms the N-terminal half of the WH2 motif and that binds in the groove between actin subdomain 1 and 3 is present in all examined VCA domains (Hertzog et al., 2004; Chereau et al., 2005). The WH2 motif of proteins such as thymosin β, MIM, and WIP is extended with a second α-helix (Chereau et al., 2005), but WH2 motif homology in all examined VCA domains ends after ∼20 residues. The region that follows shows very low sequence conservation, even among closely related organisms. The region is rich in proline residues that are known to break secondary structure. This suggests that it functions as a spacer and it also shows that short WH2 motifs are a universal feature of WASP family proteins.
Figure 11.
VCA domain sequence alignment. VCA domain sequences from the respective WASP subfamilies of indicated organisms were aligned in ClustalX using default parameters. The shown alignment starts with the most C-terminal WH2 motif, which location is drawn in the diagram underneath the alignment. A region with high sequence similarity that is predicted to fold into an α-helix is also indicated. We define this helix as the C motif.
The region that follows lacks a formal definition, but is usually referred to as the Connecting or Central region. The physiological function of this region is still poorly understood. In mammalian WASP it is known to fold into an α-helix and has a dual role; it binds both intramolecularly to the PBD, which causes its autoinhibition, (Kim et al., 2000) and to the Arp2/3 complex (Marchand et al., 2001). Interestingly, an α-helix is confidently predicted at this position for all WASP subfamilies, even for Dictyostelium WASP B and C, for the unique Entamoeba WASP family proteins and for the divergent Rattus LOC302367. The observation that the high degree of sequence conservation is confined to the region that is predicted to fold into an α-helix indicates that only the helix is of physiological importance. Close inspection learns that the helix motif is different for each of the WASP subfamilies. This observation is highly remarkable as some of the different subfamilies have been separated for 2 billion years, and it strongly suggests that the physiological function of the predicted α-helix is subfamily-specific. To reflect this, we propose to redefine this helix as the Coil motif, which is more specific than “central” or “connecting,” but preserves the abbreviation (C motif) that is currently used to describe this region.
In WASP, SCAR, and WHAMM/JMY the C motif is immediately followed by a stretch of mostly acidic residues. In WASH, however, the C motif is well separated from the acidic region by up to 100 residues. The sequence of this region is generally of low complexity and is scattered with prolines, again giving the impression that it serves as a spacer. The function of this spacer in WASH is a mystery. The prevailing idea is that the VCA domain brings the actin monomer that is bound to the WH2 motif into proximity of the Arp2/3 complex that is bound to the acidic region. However because of the wide separation between the WH2 motif and the acidic region in WASH, its VCA domain is clearly not optimized for this.
DISCUSSION
Our analysis shows that WASP, SCAR, and WASH are the only WASP family members that are found across multiple eukaryotic kingdoms. In the human genome, no WASP protein families were found in addition to WASP, SCAR, WASH, and WHAMM/JMY. However, the presence of unique proteins with a clear VCA domain in small branches of the eukaryotic tree, such as Murinae or Entamoeba, demonstrates that new WASP family proteins are still evolving. These findings indicate that there may be many more pockets of unique WASP protein subfamilies on other branches of the eukaryotic tree. As a result, we may never be able to call the WASP family complete.
The high degree of conservation of WASP, SCAR, and WASH and their complex members signifies that results from studies of these proteins can confidently be used across different organisms. However, our phylogenetic analysis does point out some exceptions to this rule. For example, Drosophila Abi has been found to regulate WASP activity via its SH3 domain, which leads to wonder whether the fundamental role of Abi is tied to WASP regulation (Bogdan et al., 2005). Several other lines of evidence do not support a role for Abi outside the SCAR complex, such as the lack of correlation between the presence of WASP and Abi and the previously obtained genetic and biochemical evidence (Ibarra et al., 2006; Derivery et al., 2009a). Our finding that the addition of C-terminal SH3 domain of Abi occurred relatively late during evolution now points out that regulation of WASP is not a fundamental role of Abi, but an extra level of control, possibly to coordinate the activity of SCAR and WASP in more complex organisms such as metazoa. This is particularly unexpected because most literature focuses on the SH3 domain as a major hub for signals that regulate SCAR activity (Dai and Pendergast, 1995; Innocenti et al., 2004; Ismail et al., 2009).
The increased complexity of multicellular animals has also led to an expansion of the WASP protein family. Invertebrates, which already have a complicated body plan and hematopoietic and nervous systems, utilize the additional function of WHAMY. The duplication of the SCAR complex and the WASP and the WHAMM/JMY paralogues did not occur until the onset of vertebrate evolution. The respective paralogues have been well conserved in all examined organisms, implying that their specialized functions have also remained the same. The most obvious gain from invertebrates to vertebrates is the development of bone. Thus the additional WASP family members may have key functions in myeloid cells, which we already known to be the case for WASP and Hem1. In a similar manner, the newly identified vertebrate SCAR4 group, which is lost only in placental mammals, is most likely to have a specialized function in placental-independent embryonic development.
The evolutionary history of WASP family proteins also offers a new perspective on the physiological role of Arp2/3-mediated actin nucleation. Loss of all WASP family proteins invariably leads to the loss of the Arp2/3 complex. This dependence puts the physiological function of WASP family proteins at the center stage. The massive recruitment of Arp2/3 to pseudopodia and lamellipodia in response to SCAR complex activation quickly established a role for the Arp2/3 complex in cell migration. However, the ancestral WASP, but also the novel WHAMM and JMY are involved in vesicle trafficking. WASH, which regulates vesicle fission, now emerges as the most widely conserved WASP family protein. The relative prevalence of these protein families now indicates that regulation of vesicle homeostasis is at least an equally important function of Arp2/3-mediated actin nucleation.
Detailed functioning of WASP family proteins is less well understood than is sometimes believed. The C motif that does not yet have a well-defined function now emerges as the most conserved feature of the VCA domain and discovery of the role of the C motif may be key to understanding how WASP family proteins are activated. On the other hand, some of the simplifying assumptions that are currently being held are not well supported. For example, WASP proteins can function without their N-terminal control domains, WHAMM/JMY proteins first evolved with an unconventional phenylalanine in their C-terminus and the WASH VCA domain is incompatible with simple apposition of actin monomers to Arp2 and Arp3. Taken together, our work shows that there is a great deal of Arp2/3 physiology yet to be discovered.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Laura Machesky and Dr. Peter Thomason for critical reading of the manuscript. The research was funded by Cancer Research UK.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-04-0372) on June 23, 2010.
REFERENCES
- Dominguez R. The beta-thymosin/WH2 fold: multifunctionality and structure. Ann. NY Acad. Sci. 2007;1112:86–94. doi: 10.1196/annals.1415.011. [DOI] [PubMed] [Google Scholar]
- Echarri A., Lai M. J., Robinson M. R., Pendergast A. M. Abl interactor 1 (Abi-1) wave-binding and SNARE domains regulate its nucleocytoplasmic shuttling, lamellipodium localization, and wave-1 levels. Mol. Cell. Biol. 2004;24:4979–4993. doi: 10.1128/MCB.24.11.4979-4993.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden S., Rohatgi R., Podtelejnikov A. V., Mann M., Kirschner M. W. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature. 2002;418:790–793. doi: 10.1038/nature00859. [DOI] [PubMed] [Google Scholar]
- Gomez T. S., Billadeau D. D. A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev. Cell. 2009;17:699–711. doi: 10.1016/j.devcel.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermann B. The BAR-domain family of proteins: a case of bending and binding? EMBO Rep. 2004;5:250–255. doi: 10.1038/sj.embor.7400105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemsath L., Dvorsky R., Fiegen D., Carlier M. F., Ahmadian M. R. An electrostatic steering mechanism of Cdc42 recognition by Wiskott-Aldrich syndrome proteins. Mol. Cell. 2005;20:313–324. doi: 10.1016/j.molcel.2005.08.036. [DOI] [PubMed] [Google Scholar]
- Hertzog M., et al. The beta-thymosin/WH2 domain; structural basis for the switch from inhibition to promotion of actin assembly. Cell. 2004;117:611–623. doi: 10.1016/s0092-8674(04)00403-9. [DOI] [PubMed] [Google Scholar]
- Ho H. Y., Rohatgi R., Lebensohn A. M., Le M., Li J., Gygi S. P., Kirschner M. W. Toca-1 mediates Cdc42-dependent actin nucleation by activating the N-WASP-WIP complex. Cell. 2004;118:203–216. doi: 10.1016/j.cell.2004.06.027. [DOI] [PubMed] [Google Scholar]
- Ho H. Y., Rohatgi R., Ma L., Kirschner M. W. CR16 forms a complex with N-WASP in brain and is a novel member of a conserved proline-rich actin-binding protein family. Proc. Natl. Acad. Sci. USA. 2001;98:11306–11311. doi: 10.1073/pnas.211420498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra N., Blagg S. L., Vazquez F., Insall R. H. Nap1 regulates Dictyostelium cell motility and adhesion through SCAR-dependent and -independent pathways. Curr. Biol. 2006;16:717–722. doi: 10.1016/j.cub.2006.02.068. [DOI] [PubMed] [Google Scholar]
- Innocenti M., Zucconi A., Disanza A., Frittoli E., Areces L. B., Steffen A., Stradal T. E., Di Fiore P. P., Carlier M. F., Scita G. Abi1 is essential for the formation and activation of a WAVE2 signalling complex. Nat. Cell Biol. 2004;6:319–327. doi: 10.1038/ncb1105. [DOI] [PubMed] [Google Scholar]
- Ismail A. M., Padrick S. B., Chen B., Umetani J., Rosen M. K. The WAVE regulatory complex is inhibited. Nat. Struct. Mol. Biol. 2009;16:561–563. doi: 10.1038/nsmb.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling P. J., Burger G., Durnford D. G., Lang B. F., Lee R. W., Pearlman R. E., Roger A. J., Gray M. W. The tree of eukaryotes. Trends Ecol. Evol. (Personal edition) 2005;20:670–676. doi: 10.1016/j.tree.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Kelly A. E., Kranitz H., Dotsch V., Mullins R. D. Actin binding to the central domain of WASP/Scar proteins plays a critical role in the activation of the Arp2/3 complex. J. Biol. Chem. 2006;281:10589–10597. doi: 10.1074/jbc.M507470200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A. S., Kakalis L. T., Abdul-Manan N., Liu G. A., Rosen M. K. Autoinhibition and activation mechanisms of the Wiskott-Aldrich syndrome protein. Nature. 2000;404:151–158. doi: 10.1038/35004513. [DOI] [PubMed] [Google Scholar]
- Larkin M.A., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Linardopoulou E. V., Parghi S. S., Friedman C., Osborn G. E., Parkhurst S. M., Trask B. J. Human subtelomeric WASH genes encode a new subclass of the WASP family. PLoS Genet. 2007;3:e237. doi: 10.1371/journal.pgen.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder S., Nelson D., Weiss M., Aepfelbacher M. Wiskott-Aldrich syndrome protein regulates podosomes in primary human macrophages. Proc. Natl. Acad. Sci. USA. 1999;96:9648–9653. doi: 10.1073/pnas.96.17.9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Abreu-Blanco M. T., Barry K. C., Linardopoulou E. V., Osborn G. E., Parkhurst S. M. Wash functions downstream of Rho and links linear and branched actin nucleation factors. Development. 2009;136:2849–2860. doi: 10.1242/dev.035246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand J. B., Kaiser D. A., Pollard T. D., Higgs H. N. Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat. Cell Biol. 2001;3:76–82. doi: 10.1038/35050590. [DOI] [PubMed] [Google Scholar]
- Merrifield C. J., Qualmann B., Kessels M. M., Almers W. Neural Wiskott Aldrich Syndrome Protein (N-WASP) and the Arp2/3 complex are recruited to sites of clathrin-mediated endocytosis in cultured fibroblasts. Eur. J. Cell Biol. 2004;83:13–18. doi: 10.1078/0171-9335-00356. [DOI] [PubMed] [Google Scholar]
- Miki H., Miura K., Takenawa T. N-WASP, a novel actin-depolymerizing protein, regulates the cortical cytoskeletal rearrangement in a PIP2-dependent manner downstream of tyrosine kinases. EMBO J. 1996;15:5326–5335. [PMC free article] [PubMed] [Google Scholar]
- Miki H., Suetsugu S., Takenawa T. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J. 1998;17:6932–6941. doi: 10.1093/emboj/17.23.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J., Oma Y., Vallar L., Friederich E., Poch O., Winsor B. Sequence and comparative genomic analysis of actin-related proteins. Mol. Biol. Cell. 2005;16:5736–5748. doi: 10.1091/mbc.E05-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S. A., Han J. W., Lee Y., Firtel R. A., Chung C. Y. A Dictyostelium homologue of WASP is required for polarized F-actin assembly during chemotaxis. Mol. Biol. Cell. 2005;16:2191–2206. doi: 10.1091/mbc.E04-09-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi S. N., Zahn R., Mitchell D. A., Stevenson B. J., Munn A. L. The WASp homologue Las17p functions with the WIP homologue End5p/verprolin and is essential for endocytosis in yeast. Curr. Biol. 1998;8:959–962. doi: 10.1016/s0960-9822(98)70396-3. [DOI] [PubMed] [Google Scholar]
- Page R. D. Visualizing phylogenetic trees using TreeView. Curr. Protocol. Bioinform. 2002 doi: 10.1002/0471250953.bi0602s01. Chapter 6, Unit 6 2. [DOI] [PubMed] [Google Scholar]
- Paunola E., Mattila P. K., Lappalainen P. WH2 domain: a small, versatile adapter for actin monomers. FEBS Lett. 2002;513:92–97. doi: 10.1016/s0014-5793(01)03242-2. [DOI] [PubMed] [Google Scholar]
- Pollard T. D. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu. Rev.Biophys. Biomol. Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- Quinlan M. E., Heuser J. E., Kerkhoff E., Mullins R. D. Drosophila Spire is an actin nucleation factor. Nature. 2005;433:382–388. doi: 10.1038/nature03241. [DOI] [PubMed] [Google Scholar]
- Rohatgi R., Ho H. Y., Kirschner M. W. Mechanism of N-WASP activation by CDC42 and phosphatidylinositol 4, 5-bisphosphate. J. Cell Biol. 2000;150:1299–1310. doi: 10.1083/jcb.150.6.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouiller I., Xu X. P., Amann K. J., Egile C., Nickell S., Nicastro D., Li R., Pollard T. D., Volkmann N., Hanein D. The structural basis of actin filament branching by the Arp2/3 complex. J. Cell Biol. 2008;180:887–895. doi: 10.1083/jcb.200709092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer G., Weber S., Holz A., Bogdan S., Schumacher S., Muller A., Renkawitz-Pohl R., Onel S. F. The Wiskott-Aldrich syndrome protein (WASP) is essential for myoblast fusion in Drosophila. Dev. Biol. 2007;304:664–674. doi: 10.1016/j.ydbio.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Scita G., Confalonieri S., Lappalainen P., Suetsugu S. IRSp53: crossing the road of membrane and actin dynamics in the formation of membrane protrusions. Trends Cell Biol. 2008;18:52–60. doi: 10.1016/j.tcb.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Sept D., McCammon J. A. Thermodynamics and kinetics of actin filament nucleation. Biophys. J. 2001;81:667–674. doi: 10.1016/S0006-3495(01)75731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stechmann A., Cavalier-Smith T. The root of the eukaryote tree pinpointed. Curr. Biol. 2003;13:R665–R666. doi: 10.1016/s0960-9822(03)00602-x. [DOI] [PubMed] [Google Scholar]
- Veltman D. M., Akar G., Bosgraaf L., Van Haastert P. J. A new set of small, extrachromosomal expression vectors for Dictyostelium discoideum. Plasmid. 2009;61:110–118. doi: 10.1016/j.plasmid.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Zuchero J. B., Coutts A. S., Quinlan M. E., Thangue N. B., Mullins R. D. p53-cofactor JMY is a multifunctional actin nucleation factor. Nat. Cell Biol. 2009;11:451–459. doi: 10.1038/ncb1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.