Abstract
The B cell antigen receptor (BCR) plays an essential role in all phases of B cell development. Here, we show the extracellular domains of murine and human Igβ form an I-set immunoglobulin-like structure with an inter-chain disulfide between cysteines on their G-strands. Structural and sequence analysis suggests that Igα displays similar fold as Igβ. An Igαβ heterodimer model was generated based on the unique disulfide bonded Igβ dimer. Solution binding studies showed that the extracellular domains of Igαβ preferentially recognized the constant region of BCR with the μ-chain specificity, suggesting a role for Igαβ to enhance the BCRμ-chain signaling. Cluster mutations on Igα, Igβ and mIgM based on the structural model identified distinct area of potential contacts involving charged residues on both subunits of the co-receptor and the Cμ4 domain of mIgM. These studies provide the first structural model for understanding BCR function.
Introduction
The B cell antigen receptor (BCR) plays a critical role in all stages of B cell development and function (Geisberger et al., 2006; Reth, 1992). It consists of two principal components: an antigen binding and a signaling subunit. The antigen binding subunit is a membrane bound form of immunoglobulin (mIg) with a short cytoplasmic tail lacking any signaling motifs. Through non-covalent interactions, mIg associates with a disulfide linked Igαβ (CD79a/CD79b) signaling heterodimer (Campbell et al., 1991; Hermanson et al., 1988; Kashiwamura et al., 1990; Venkitaraman et al., 1991) forming a complex with 1:1 stoichiometry (Schamel and Reth, 2000; Tolar et al., 2005). Both Igα and Igβ contain a single immunoreceptor tyrosine-based activation motif (ITAM) in their cytoplasmic domains (Cambier, 1995; Reth, 1989). Upon antigens binding, the ITAMs of Igα and Igβ are phosphorylated by the Src-family kinase, Lyn initiating a signaling cascade in B cells (Dal Porto et al., 2004; Gauld et al., 2002; Jumaa et al., 2005). Importantly, both positive and negative selection of developing B lymphocytes as well as the survival and activation of mature B cells depend critically on Igα and Igβ (Nemazee et al., 2000; Rajewsky, 1996). It was also established that mIgM is absolutely dependent on the association with Igαβ heterodimer for its cell surface expression, whereas mIgG1 is not (Venkitaraman et al., 1991).
A critical gap in our knowledge of how the BCRs transduce signals is the molecular architecture of mIg-Igαβ complex. It is well established that many multi-chain immune receptors, such as T cell receptors (TCRs) and activating natural killer cell receptors, associate with their signaling adaptor molecules though interactions between positively and negatively charged amino acid pairs in their transmembrane (TM) domains (Lanier, 2005). For the BCR, only Igα has a charged residue in its transmembrane domain and mIgM and Igβ contain only two polar residues in their TM regions (Campbell et al., 1991; Reth, 1992). The presence of a charged Glu residue in the TM portion of Igα led to the hypothesis that interactions between mIgM and the Igαβ heterodimer are primarily through Igα (Reth, 1992). However, recent studies utilizing fluorescence resonance energy transfer (FRET) have demonstrated that the cytoplasmic C-terminus of Igβ is positioned closer to mIg than Igα (Tolar et al., 2005; Wienands, 2005). Mutational studies confirmed a critical role of polar residues in transmembrane region of mIgM and revealed that YS to VV mutation in TM region of μ-chain abolishes its association with Igαβ heterodimer (Grupp et al., 1993). Later experiments proposed that there is an ordered association of BCR components during BCR assembly (Foy and Matsuuchi, 2001). An intriguing aspect of the BCR signaling subunit is its structural and functional similarity to CD3 molecules in TCR assembly. Unlike many signaling subunits in immune receptors that do not have appreciable extracellular domains, Igα and Igβ have sizable extracellular domains as do CD3δε or CD3γε heterodimers in TCRs (Call et al., 2002; Clevers et al., 1988; Kuhns et al., 2006; Wegener et al., 1995). However, unlike Igαβ, CD3δε and CD3γε are not disulfide-bonded heterodimers.
The exact role of extracellular domains of Igαβ in BCR assembly and signaling remains unresolved. It was reported that the extracellular domain of Igα affects the expression level of mIgM (Hombach et al., 1990). Moreover, Igα and Igβ that lack extracellular domains did not mediate transport of IgM to B cell surface (Alfarano et al., 1999; Indraccolo et al., 2002). Recently it was also reported that both extracellular and transmembrane regions of Igαβ must be properly associated for correct BCR assembly (Dylke et al., 2007). It is also interesting to note that an excess of Igβ have been observed in the endoplasmic reticulum of B cells as a disulfide bonded homodimer, although its functional relevance is uncertain (Brouns et al., 1995; Schamel et al., 2003). Based on amino acid sequences the extracellular domains of Igα and Igβ are predicted to have a C2- and a V-set immunoglobulin-like (Ig-like) fold, respectively (Hermanson et al., 1988; Kashiwamura et al., 1990). In addition to the classical Ig-fold intra chain bond disulfide, both Igα and Igβ contain additional cysteines that form an inter chain heterodimeric disulfide bond. Igβ also has an additional intra-molecular disulfide bond. At present, the cysteine assignment for the intra- and inter-molecular disulfide bonds in Igβ remains controversial (Campbell et al., 1991; Hermanson et al., 1988; Kashiwamura et al., 1990; Reth, 1992; Siegers et al., 2006). To further investigate the function of Igαβ and its association with BCR, we determined crystal structures of the extracellular domains of murine and human Igβ, generated a structural model for Igαβ, carried out solution binding studies between Igαβ and various isotypes of the B cell receptors, and identified, through mutational analysis, residues on the extracellular portion of Igαβ as well as in the Cμ4 domain of mIgM involved in the receptor and co-receptor association. Together, these studies provide the first structural model for our understanding of BCR architecture and activation.
Results and Discussion
The extracellular domains of Igα and Igβ preferentially recognized BCR μ-chain
In addition to mediating BCR signaling, Igαβ promotes the surface expression of mIg heavy chains, in particular the μ-chain of BCR (Venkitaraman et al., 1991). In the absence of Igαβ, μ-chain of BCR is retained in the endoplasmic reticulum (ER) through interactions of its TM domain with calnexin (Grupp et al., 1995). The association between Igαβ and mIg has been probed extensively through mutations in the TM region of mIg and pinpointed to Tyr-Ser pair of amino acids (Blum et al., 1993; Grupp et al., 1993; Pleiman et al., 1994; Venkitaraman et al., 1991; Williams et al., 1994). The results showed that Igαβ interact with mIg primarily through their TM regions and that the extracellular domains of Igαβ appear to be dispensable for BCR signaling. However, the interactions between Igαβ and mIg in TM region alone are insufficient to explain the ER retention of μ-chain but not γ-chain of mIg as they share 70% similar TM sequences with Tyr-Ser pair conserved for all mIg isotypes.
To investigate the function of the extracellular region of Igαβ, we examined the binding of soluble Igαβ to different immunoglobulin (Ig) isotypes, including IgM, IgG1, IgD, IgA and IgE. Human Igαβ heterodimer was expressed in insect cells as a leucine zipper fusion protein (Igαβ-LZ). The constant portion of a human BCR μ-chain, corresponding to Cμ2-Cμ4 domains (Fcμ), was expressed in CHO cells as a disulfide-bonded dimer and reacted with an anti-human IgM antibody. The solution binding experiments were carried out using immobilized human Igαβ-LZ on CM5 sensor chips via primary amine attachment. The analytes consisted of serial dilutions of the recombinant human Fcμ, human IgM, IgA, IgD, IgE and IgG1. The solution binding showed that Igαβ bound Fcμ tightest with a dissociation constant of ~2μM (Figure 1, Table 1). Interestingly, the intact IgM bound Igαβ significantly less than Fcμ despite it is 10x larger than Fcμ(Table 1), suggesting that the presence of J-chain on IgM interferes with Igαβ binding. Similarly, IgA displayed weak binding to Igαβ at high level of the co-receptor immobilization and failed to bind the co-receptor at low immobilization levels (Table 1). No bindings were observed between IgG1, IgD, IgE and immobilized Igαβ under any surface densities. Thus, the solution binding results showed a preferential μ-chain association by the extracellular domains of Igαβ, suggesting a unique role for the co-receptor in enhancing the BCRμ signaling. This may contribute to lowering the signaling threshold of the receptor and compensate for the lower μ-chain antigen affinity during B cell development.
Figure 1.
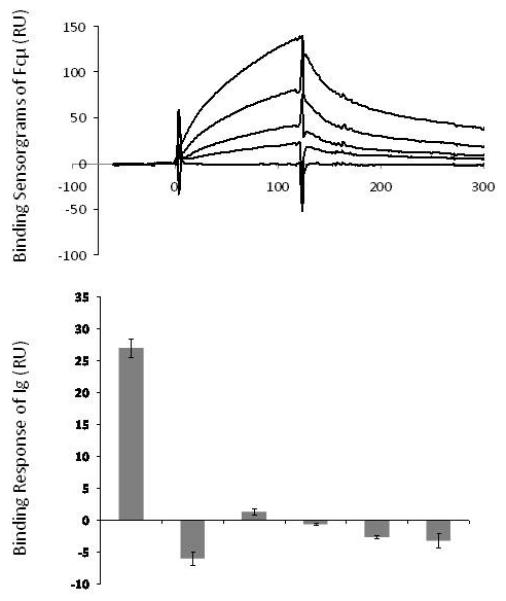
Solution binding of immunoglobulins to immobilized Igαβ at 540 RU of surface densities. (A) The binding sensorgrams of recombinant Fcμ at 10, 5, 2.5, 1.25 and 0.625 μM concentrations to immobilized Igαβ. The calculated dissociation constant KD is 2.3 ×10−6 ± 1.1 ×10−6 μM. (B) The binding responses of 3μM Fcμ and various human immunoglobulins to immobilized Igαβ. The responses were taken at the time point immediate before the dissociation phase.
Table 1.
Solution affinities of Fcμ and immunoglobulins binding to Igαβ
| Igαβ |
||||
|---|---|---|---|---|
| Immobilization (RU) | 250 | 540 | 990 | 2096 |
| Analyte (concentrations) | Dissociation constants, KD (μM) | |||
| Fcμ (0-20μM) | 2.4±2.9 | 2.3±1.1 | 2.2±2.2 | 0.9 |
| IgM (0-20μM) | ND | ND | ND | >10 |
| IgA(0-20μM) | ND | ND | ND | >10 |
| IgG1(0-6μM) | ND | ND | ND | ND |
| IgE(0-6μM) | ND | ND | ND | ND |
| IgD(0-6μM) | ND | ND | ND | ND |
ND – None Detectable binding.
Structure determination of murine and human Igβ
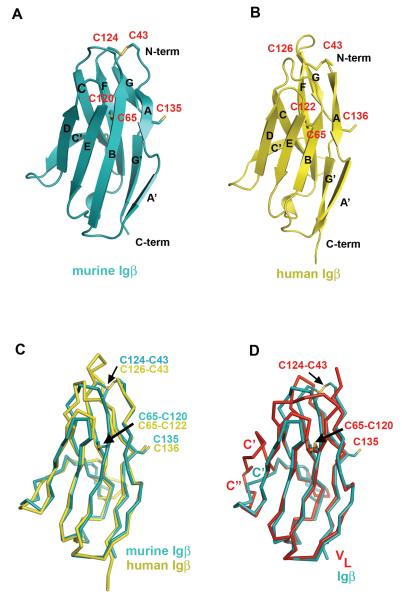
To gain structural insights to the signaling chain of BCR, we expressed the extracellular portions of human Igα (amino acids 33-143), Igβ (amino acids 26-159) as well as their murine homologs as recombinant proteins in E.coli. The refolded human Igα existed in mostly a monomeric form, while human and murine Igβ formed both monomers and dimers (Supplemental Figure 1). Both Igα and Igβ, when immobilized on a CM5 sensorchip, bound Fcμ as well as their respective antibodies suggesting that the refolded Igα and Igβ were functional. We crystallized the monomeric and dimeric forms of murine Igβ as well as the dimeric form of human Igβ. The crystals of the monomeric Igβ belonged to an orthorhombic space group P21212 and contained one Igβ monomer in each asymmetric unit. The structure was determined by molecular replacement method despite of less than 30% sequence identity between Igβ and the known structures of V-set Ig-like domains. The structure was refined to 1.7 Å resolution with crystallographic and free R-factors of Rcryst =18.7% and Rfree=19.7%, respectively (Table 2). The refined electron density map was contiguous from residue Cys 43 to Leu 142 except for the FG loop, between residues 127 and 133, which appeared disordered (Supplemental Figure 2). The refined monomeric Igβ structure was then used as the search model in molecular replacement to solve both murine and human dimeric Igβ structures (Table 2). The structure of the murine Igβ dimer was refined to 3.1 Å resolution with the final Rcryst and Rfree of 20.6% and 25.8%, respectively. The electron densities are continuous between Cys 43 and Leu 142 for the A-subunit except for the FG loop (residues 125-129), and between Ile 46 and Phe 144 for the B-subunit except for part of the C’D (residues 90-91) and FG (residues 124-132) loops. The dimeric human Igβ structure was refined to resolution with the final Rcryst and Rfree of 18.1% and 26.3%, respectively (Table 2, Figures 2 and 3). The refined (2Fo-Fc) map showed good electron density in both subunits throughout the whole structure between Cys 43-Phe 145.
Table 2.
Data Collection and Refinement Statistics
| Data Collection | murine Igβ monomer |
murine Igβ homodimer |
human Igβ homodimer |
|---|---|---|---|
| Space group | P21212 | P41212 | P4132 |
| Unit cell (Å) | a=35.63, b=79.72, c=34.32 | a=b=87.97, c=75.29 | a=129.83 |
| Resolution limit (Å) | 1.7 | 3.1 | 3.2 |
| Unique reflections | 10471(898)a | 5597(541) | 6616(630) |
| Redundancy | 5.5(2.3) | 6.1(5.4) | 27.6(28.7) |
| Completeness (%) | 93.8(67.4) | 97.4(98.7) | 99.9(100.0) |
| Rsym (%)b | 6.5(31.7) | 13.5(50.6) | 7.0(33.9) |
| <I/σ(I)> | 21.5(2.6) | 14.1(3.2) | 55.6(13.1) |
|
| |||
| Refinement | |||
| Resolution (Å) | 40.-1.7 | 48.-3.1 | 46.-3.2 |
| No. reflections | 10071 | 5407 | 6407 |
| No. protein atoms | 757 | 1485 | 1678 |
| No. solvent atoms | 92 | 26 | 2 |
| Rcryst(%) | 18.7(27.0) | 20.6(27.8) | 18.1(20.7) |
| Rfree(%)c | 19.7(29.0) | 25.8(32.0) | 26.3(36.2) |
| Mean B-factor (Å2) | 31.7 | 52.1 | 77.2 |
| Wilson B-factor (Å2) | 20.6 | 59.7 | 75.8 |
| r.m.s.d. bond lengths (Å) | 0.006 | 0.007 | 0.010 |
| r.m.s.d. bond angles (°) | 1.18 | 1.39 | 1.39 |
Values in parentheses are for highest resolution shells: 1.7-1.74Å, 3.21-3.10Å, and 3.31-3.20Å for murine Igβ monomer, murine Igβ homodimer, and human Igβ homodimer, respectively.
Rsym=100×ΣǀIh-<Ih> ǀ/ΣIh, where <Ih> is the mean intensity of multiple measurements of symmetry equivalent reflections.
Rfree was calculated using test set of 5%.
Figure 2.
Ribbon drawing of a monomer from murine (A) and human (B) Igβ homodimers, respectively. All secondary structure elements on Igβs are marked in accordance with the sequence alignment in Figure 4. Cysteins (labeled in red) and disulfide bonds are shown in stick representation, N- and C-termini are marked. (C) Structural comparison between murine Igβ (cyan) and human Igβ (yellow) monomers. Positions of disulfide bonds are shown with arrows. Labels colored according to the color of corresponding molecule. (D) Structural comparison between murine Igβ (cyan) and VL domain of IgG1 (red, PDB code 1YQV). Cysteins and disulfide bonds are shown in stick representation, labeled according to Igβ sequence. C’- and C”-strands, that are different in the two structures are labeled and colored according to the color of corresponding molecule. This figure and all subsequent ribbon drawings are prepared using the program PyMol (DeLano, W.L. The PyMOL Molecular Graphics System (2002), http://www.pymol.org). See also Figure S2.
Figure 3.
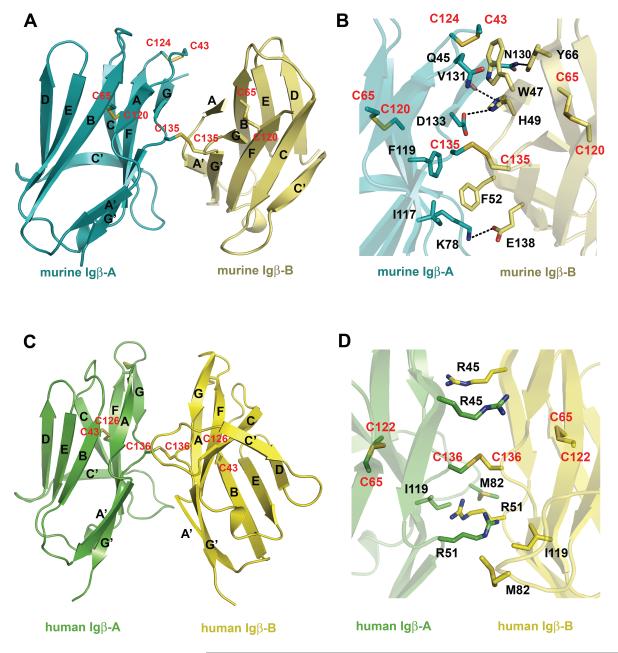
Murine and human Igβ homodimer formation. (A) Murine Igβ homodimer with secondary structure elements labeled. Monomers A and B are colored cyan and lemon, respectively. (B) Detailed view of the hydrophobic core and hydrogen bonds (black dashed lines) at the homodimer interface with major side chains involved in the interactions. (C) Human Igβ homodimer. Monomers A and B are colored green and yellow, respectively. (D) Detailed view of major side chains involved in the interactions at human Igβ interface. Cystine residues are shown in stick representation and labeled red. See also Figure S3.
The subunit structures of murine and human Igβ are very similar to each other with root mean square deviation (r.m.s.d.) of among 86 core Cα atoms (Figure 2C). The overall structure of the Igβ extracellular domain assumes an I-type Ig-fold with the two anti-parallel β-sheets formed by A-B-E-D and A’-G-F-C strands and a characteristic disulfide bond between Cys 65-Cys 120 (Cys 122 in human Igβ) from the B- and F-strands, respectively (Figure 2) (Harpaz and Chothia, 1994). Like a V-type fold, a conserved proline residue, Pro 50, breaks the first β-strand into two shorter strands, A and A’. However, unlike the classical V-type domain, the I-type Igβ does not have a C” β-strand leaving C’ strand to bridge the two β-sheets (Figure 2). As a consequence, the loop corresponding to the putative second complementarity determining region (CDR2) is absent in Igβ. The structural comparison between Igβ and several other members of Ig superfamily such as the VH and VL domains of an IgG1 (PDB entry 1YQV), the Vα and Vβ domains of a TCR (1AO7), the Vγ and Vδ of a γδTCR (1HXM), and CD8 (1CD8) resulted in root mean square deviations (r.m.s.d.) of 1.1-1.3 Å for 75-87 Cα atoms. In addition, Igβ contains a second intra-chain disulfide bond formed between Cys 43 and Cys 124 (Cys 126 in human Igβ), and an inter-chain disulfide bond between Cys 135 (Cys 136 in human Igβ) from both subunits (Figure 3). Our structural data is in accordance with the reported Igαβ heterodimer formation through Cys 135 of murine Igβ in Drosophila S2 cells (Siegers et al., 2006).
Igβ forms a distinct dimer
Although its functional relevance is unclear, excess of Igβ could be found in B cells as a disulfide bonded homodimer (Brouns et al., 1995; Schamel et al., 2003). The two subunits of Igβ are linked by a disulfide bond between Cys 135 (Cys 136 in human Igβ) from the opposing G-strands (Figure 3). Unlike many covalent dimers, such as CD8, NKG2D, and NKG2A/CD94 receptors, that usually form disulfide bonds in their membrane proximal stalk regions, Igβ inter-subunit disulfide bond is located in the middle of its Ig domain. Similar to the V-domain structures of antibodies and T cell receptors, the structure of Igβ contains a glycine residue, Gly 136 (Gly 137 in human Igβ), in the middle of the G-strand creating a β–buldge that splits the strand into G- and G’-strands (Chothia, et al 1998, Richardson,1981). Interestingly, Cys 135 (Cys 136 in human Igβ) is located in the middle of the buldge, whose conformation makes the cysteine readily accessible for the inter-chain disulfide bond formation (Figures 2 and 3). Although similar in overall dimer formation, the relative orientations of two Igβ subunits differ between the murine and human homodimers as a result of a rotation around the inter-chain disulfide bond (Figure 3A & C). The murine dimer is asymmetrical and stabilized by interactions between subunits A and B, including two salt bridges (Lys 78 (A)–Glu 138 (B) and Asp 133 (A)–His 49 (B)), one hydrogen bond ( Asn 130 (A) -Tyr 66 (B)), and hydrophobic interactions, involving Ile 117 (A), Phe 119 (A), Gln 45 (A), Lys 78 (A), and Trp 47 (B) and Phe 52 (B) (Figure 3A &B). In contrast, the human Igβ homodimer is symmetrical and primarily formed through hydrogen bonds from the side chains of Arg 45 and Arg 51 to the main chain atoms of the opposing subunit (Figure 3C&D). In addition to the current Igβ-type dimer, several types of immunoglobulin dimers have been reported to date, including those in the V-domains of antibodies, TCRs, CD8, CTLA-4, CD28, and TREM-1 (Cohen et al., 1996; Evans et al., 2005; Garboczi et al., 1996; Harris et al., 1992; Leahy et al., 1992; Ostrov et al., 2000; Radaev et al., 2003; Schwartz et al., 2001) (Supplemental Figure 3). However, all these reported dimers would position the inter-chain cysteines of Igβ at distances between 12-25 Å too far for disulfide bond formation, and thus are incompatible with the Igαβ structure.
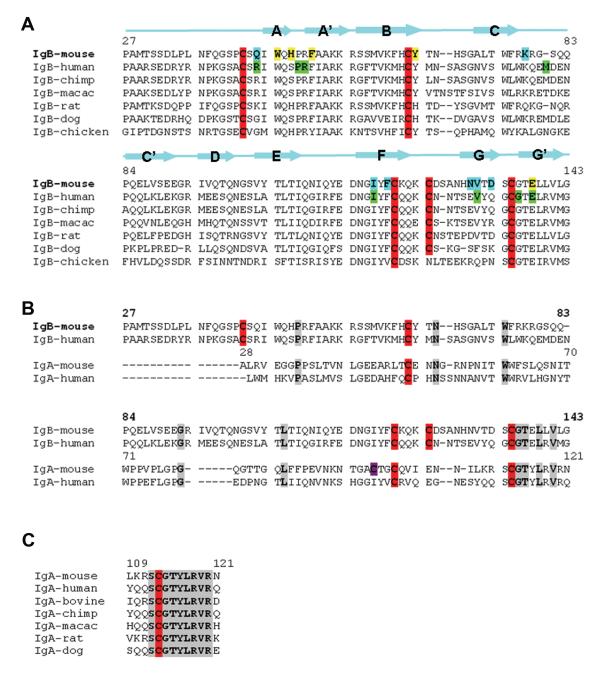
Based on initial sequence analysis Igα was proposed to have a C2-set Ig-like fold (Kashiwamura et al., 1990). The sequences of Igα and Igβ share less than 20% identity (Figure 4). However, several structural features of Igβ appear to be also present in the structure of Igα. First, a conserved Pro 35 in the first β-strand of Igα suggests its first strand is split into A- and A’-strands like those in the V-type and I-type Ig-fold. Second, the most conserved region between Igα and Igβ (~60% of sequence identity) corresponds to the GG’ β-buldge and the G’-strand of Igβ (Figure 4B). In particular, the four residues at β-buldge of Igβ (SCGT) are invariant in all Igα sequences (Figure 4C), suggesting that the β-buldge conformation of Igβ is also present in the structure of Igα. This, together with the conserved Pro 35 in Igα sequences, likely creates A’-and G’- strand pairing in Igα that is a similar to that of Igβ. The interchain disulfide forming cystine, Cys 113 of Igα, is aligned against the dimer forming cysteine 135 of murine Igβ (Figure 4B), suggesting that the heterodimeric disulfide bond is formed between Cys 113 of Igα and Cys 135 of Igβ (murine numbering). The sequence of Igα is also compatible with the hydrogen bonding observed in the structure of asymmetric murine Igβ dimer (Supplemental Figure 4). Since the dimerization mode observed in Igβ is the only one compatible with the inter-chain disulfide bond through the GG’ β-buldge cysteines, we modeled the structure of Igαβ heterodimer based on that of the asymmetric murine Igβ dimer (Supplemental Figure 4).
Figure 4.
Sequence alignments of Igβs and Igαs. (A) Sequence comparison of several mammalian and chicken Igβs. The numbering is consistent with the sequence of murine Igβ. The secondary structure elements of Igβ are illustrated as arrows for β-strands. Cysteins are highlighted red. Residues involved in interactions at the interface of the murine Igβ homodimer are highlighted according to the subunit color, i.e. cyan and yellow for A- and B-subunit, respectively. Residues involved in the interactions at the interface of human Igβ highlighted green. (B) Sequence alignment of murine and human Igβs and Igαs. The numbering is consistent with murine Igβ (bold font) and Igα (regular font). Cysteins are highlighted red. Unique cystein 98 in murine Igα is highlighted magenta. (C) Sequence alignment of several mammalian Igαs in the area of predicted G-, G’-strands. Conserved residues are highlighted gray. See also Figure S4.
Identification of the contact regions between Igαβ and the μ-chain of BCR
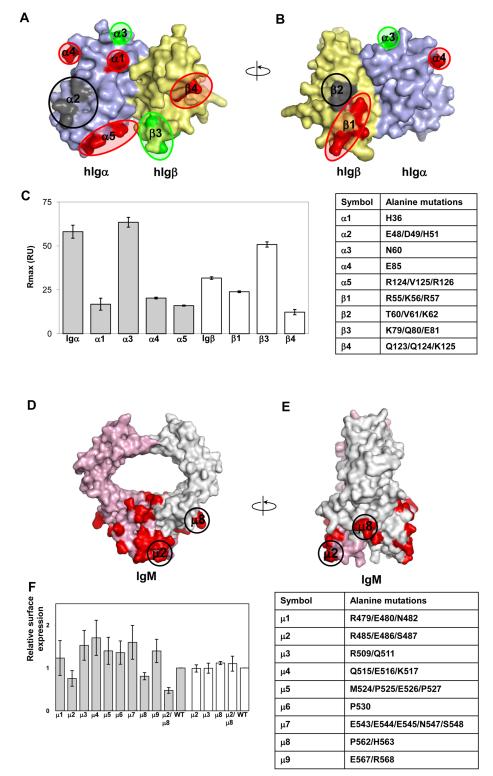
The structure model of Igαβ reveals approximately nine polar and charged patches on the surface of the heterodimer. To further define the contact region between μ-chain of BCR and Igαβ, we carried out a cluster mutagenesis on both subunits of Igαβ heterodimer as well as the μ-chain of BCR. Cluster mutations, in which clusters of amino acids are replaced with alanines, generally result in larger effects than single alanine mutations and can be effectively used to map a ligand binding site. However, a potential complication of cluster mutations is their higher tendency to disrupt native conformations. Thus, the approach requires conformational-sensitive validations, such as antibody binding. Based on the human Igαβ model, two surface-exposed Igα clusters, E48/D49/H51 (α2) and R124/V125/R126 (α5), together with three individual amino acid sites, H36 (α1), N60 (α3) and E85 (α4) were selected for alanine mutations (Figure 5). Four Igβ clusters, R55A/K56A/R57A (β1), T60A/V61A/K62A (β2), K79A/Q80A/E81A (β3) and Q123A/Q124A/K125A (β4), were also generated. The mutations were carried out on Igα and Igβ subunits to avoid mutations affecting the Igαβ heterodimer formation which would complicate the mapping of BCR binding site. Seven out of nine mutants were successfully expressed in E.coli and refolded in vitro. Two mutants, E48A/D49A/H51A (α2) of Igα and T60A/V61A/K62A (β2) of Igβ failed to express. The refolded Igα and Igβ mutants bound to their respective antibodies in solution under experimental conditions. The mutational effects on BCR association were analyzed by solution binding experiments on immobilized Fcμ sensorchips. The wild-type Igα and Igβ exhibited similar binding affinities to immobilized Fcμ with dissociation constants KD of ~1 μM, suggesting their equal contribution to μ-chain association (Supplemental Figure 5). The binding of Igα and Igβ mutants, reconstituted at 4uM concentration in PBS, to Fcμ varied significantly depending on the mutations. In particular, three of the Igα mutants, H36A (α1), E85A (α4) and R124A/V125A/R126A (α5) resulted in 5-10 fold reductions in their binding to Fcμ compared to the wild type. One mutation, N60A (α3) of Igα, exhibited a two-fold increase in Fcμ binding (Figure 5C). Among the three mutants of human Igβ, R55A/K56A/R57A (β1) and Q123A/Q124A/K125A (β4) each reduced their BCR binding by six-fold and two-fold, respectively, whereas K79A/Q80A/E81A (β3) increased binding moderately. First, the mutations further support the conclusion that both subunits of Igαβ are involved in BCR binding. Second, the mutations affected Fcμ binding are broadly distributed rather than clustered on the surface of Igαβ. Third, the loss of Fcμ binding involves both positively and negatively charged residues, such as arginine and glutamate, suggesting significant electrostatic interactions present at the BCR and Igαβ interface. In addition, the location of these mutations and the size of Igαβ extracellular domain suggest the membrane proximal Cμ4 of BCR as the most likely contact domain for the extracellular domains of Igαβ heterodimer.
Figure 5.
Binding of mutant human Igα and Igβ to recombinant human Cμ2-Cμ4. (A)Surface representation of human Igαβ heterodimer model. Point mutations resulting in reduction or increase of binding circuled red and green, respectively. Mutations causing absence of protein expression are circled black. Alpha and beta chains of Igαβ heterodimer are colored pale blue and yellow, respectively. (B) 180 degree rotation of the previous view of the Igαβ heterodimer model. (C) Binding of Igα (grey bars) and Igβ (white bars) mutants to Cμ2-Cμ4. Mutant concentration in the analyte was kept constant at 4μM. At least three sets of experiment were performed for each mutant. A table with mutant description is given for reference. (D) Structural model Cμ3-Cμ4 domains of IgM. IgM chains are painted pink and grey with groups of amino acid selected for mutations in red. Groups of mutations μ2 (R485, E486, S487) and μ8 (P562, H563) that affect IgM surface expression are circled. Arginine 485 and proline 562 are highly concerved in IgM. (E) 90 degree rotation of the previous view. (F) Relative surface expression of different IgM groups of mutants (grey bars); white bars represent relative surface expression for IgM YS/VV construct. The YS/VV mutation in transmembrane domains of IgM allows surface IgM expression without binding to the Igαβ heterodimer. That indicates that the effects of the mutations were likely through disrupting interactions with Igαβ. A table with mutant description is given for reference. See also Figure S5.
To investigate if the Cμ4 domain of mIgM is involved in the binding to Igαβ, we used the mIgM surface expression on J558L cells to measure Igαβ association as mIgM expression in this cell line is dependent on the assembly with Igαβ (Reth, 1992). A structural model of Cμ2-Cμ4 domains of mIgM was generated from the crystal structure of IgA to guide the selection of the mutational sites (Herr et al., 2003) (Figure 5). Based on this model, nine groups of amino acids (clusters μ1-μ9) were selected evenly across the surface of Cμ4 for cluster mutational analysis to probe the Igαβ contact site. Mutations were introduced into the B1-8 IgM heavy chain. Both the wild type and mutant mIgM were transfected into J558L cells together with an Igα-YFP plasmid as J558L cells express endogenous Igβ (Tolar et al., 2005). While several cluster mutations mildly enhanced mIgM expression, two mutations, R485/E486/S487 (μ2) and P562/H563 (μ8) reduced surface expression of mIgM compared to that of the wild type as determined by surface staining with anti-IgM antibodies (Figure 5F). Staining of permeabilized cells indicated that all mIgM constructs were present at similar levels intracellularly suggesting variations in mIgM surface expression are not the result of transfection efficiency (Supplemental Figure 5). Combined mutations of R485/E486/S487 (μ2) and P562/H563 (μ8) lead to a further reduction in IgM surface expression without affecting intracellular levels of the IgM. The constructs were expressed normally when we introduced a YS/VV mutation into their transmembrane domains, which allows surface IgM expression without binding to the Igαβ heterodimer (Figure 5F), indicating that the effects of the mutations were likely through disrupting interactions with Igαβ. Interestingly, residues in clusters μ2 and μ8 are conserved between human and mouse IgM but variable among other immunoglobulins. Importantly, residues affecting the association with Igαβ are spaced apart on the Cμ4 domain, which is in accordance with our mutational studies on Igα and Igβ. Moreover, the IgM mutants with reduced binding to Igαβ also involve charged residues like Arg, His and Glu that again points to the probable electrostatic nature of interactions between BCR and Igαβ.
In summary, we determined the crystal structures of human and murine Igβ. The structures adopt an IgV-like fold with a unique C’-strand positioned across the β-sandwich bridging the C- and D-strands. Both human and murine Igβ form disulfide-bonded dimers through cysteins from their G-strands that are distinct from the structures of existing Ig-like dimers. From the sequence conservation and hydrogen bonding patterns, we modeled the Igαβ heterodimer structure based on the structure of the murine Igβ dimer. Solution binding experiments using soluble Igαβ demonstrated a preferential recognition of μ-chain but not α-, γ-, δ-, and ε-chains of BCR, suggesting a role for Igαβ to enhance BCR μ-chain signaling during B cell development, and to compensate low antigen affinity of μ-chain. To define the contact region between μ-chain of BCR and extracellular domains of Igαβ, a series of cluster mutations were generated on Igα and Igβ as well as the μ-chain of BCR. The binding analysis based on these cluster mutations showed an extensive contact surface between Igαβ and BCR involving both subunits of Igαβ through multiple charged residues.
Materials and Methods
Protein expressions and purifications
The extracellular portions of human Igα (residues 33-143) and Igβ (residues 26-159) and murine Igβ (residues 27-159) were subcloned into a pET-30a vector using NdeI and XhoI restriction sites. Human Igα and Igβ mutants for binding studies were generated using standard protocol with “Quick Change” kit (Stratagene). The recombinant proteins were expressed in Escherichia coli BL21 (DE3) cells as inclusion bodies and then reconstituted in vitro similar to previously described (Radaev et al., 2003). Human and mouse Igβ showed high tendency in forming disulfide bonded homodimers, however, during murine Igβ refolding, monomers with free cysteine blocked by gluthatione were also observed. The renaturated proteins were purified through a Ni-NTA affinity column, followed by a size exclusion column (Superdex 200, GE Healthcare). Purified proteins were dialyzed against following buffers: murine Igβ against 10mM Na Acetate, pH 5.2; human Igβ against water; human Igα against 50mM NaCl, 5mM Tris pH 9.0. The identity of the refolded proteins was confirmed by N-terminal amino acid sequencing that showed some N-terminal degradation for murine Igβ.
The extracellular domains of human and murine Igαβ fused with a leucine zipper (Igαβ-LZ) were expressed in insect cells using similar procedure for both proteins. In brief, the extracellular portion of Igα followed by a basic leucine zipper and a six histidine tag was inserted into the pBACpl0p (Kozono et al., 1994) between Xho I and Mro I sites. The extracellular part of Igβ followed by an acidic leucine zipper and a FLAG tag was inserted into the same vector between EcoR I and Sph I sites. A thrombin site was engineered between Igα or Igβ and the leucine zippers. The vector was co-trasfected with BaculoGold Linearized Baculovirus DNA (Pharmingen) into sf9 cells. Recombinant virus was amplified and proteins were expressed in High Five cells cultured in Express Five SFM medium infected at a MOI of 6. Igαβ-LZ was purified using a Ni-NTA affinity column followed by a size exclusion Superdex 200 column. The Fc portion of a human BCR μ-chain (Fcμ, amino acid residues 258-586), corresponding to Cμ2-Cμ4 regions, was expressed as a disulfide-bonded dimer in CHO cells. Total RNA of a human B cell line, Daudi (ATCC, Manassas, VA), was isolated using TRIZOL Reagent (Invitrogen). The cDNA synthesis of human membrane bound form IgM was performed utilizing SuperScript First-Strand Synthesis System for RT-PCR kit (Invitrogen). Human Fcμ was PCR-amplified from the cDNA using primers 5′-GGTTCTGCCTTTTCTCACCACCATCACCACCACCATCATATTGCTGAGCTGCCTCCC-3′ and 5′-CGGGATCCTCAGGTGGACTTGTCCACGGTCC-3′. A second PCR was performed with primers 5′-AAAGGGGCTAGCGCCACCATGAAGTGGGTAACCTTTCTCCTCCTC-3′ and 5′-AGAAAAGGCAGAACCGGAGATGAAGAGGAGGAGGAGAAAGGTTAC-3′ to insert a rat serum albumin leader sequence at the 5′ prime followed by an 8 × His-tag sequence. The segment was cloned into the pIRES-neo3.0 plasmid (Clontech Laboratories, Inc.), which was then stably transfected into CHO-lec3.2.8.1 cells (kindly provided by Dr. Pamela Stanley, Albert Einstein College of Medicine, NY) using Lipofectamine 2000 (Invitrogen). G418 resisting clones were screened for Fcμ expression by ELISA and the clone with the highest expression level was selected for protein preparation. Conditioned CHO-S-SFM II spent medium containing Igαβ-LZ was collected using a Ni-NTA column and further purified on a Superdex 200 column.
Crystallization and Structure Determination
Single crystals of monomeric form of murine Igβ were obtained by vapor diffusion in hanging drops at room temperature using reservoir solution containing 20% Peg 750MME, 0.2M MgCl2 and 0.1M sodium citrate, pH 5.0. Crystals of murine Igβ homodimer grew up in hanging drops with reservoir solution containing 0.72-1.0M ammonium sulfate and 0.1M Tris, pH 8.0-9.0, while the crystals of human Igβ homodimer were obtained in 0.6M Na formate and Na acetate, pH 4.0. The crystals of momomeric form of murine Igβ diffracted to , and contained one molecule per asymmetric unit. X-ray datasets were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions are listed at http://www.ser-cat.org. The data were processed and scaled with HKL2000 (Otwinowski, 1997). The structure was solved by molecular replacement using CNS v1.1 (Brunger et al., 1998). The polyalanine-based search model consisted of 70 core amino acids from the structure of TREM-1 (PDB code 1Q8M) that encompass portions of the B, C, D, E, F and G β-strands conserved among the V-type immunoglobulin structures. The program “O” was used for model building and adjustments (Kleywegt and Jones, 1997). Positional and individual B-factor refinement was carried out using the maximum likelihood target function of CNS v1.1 (Brunger et al., 1998), followed by REFMAC5 of CCP4 program suite (1994; Murshudov et al., 1997). A well-defined glutathione molecule forming a disulfide bond to Cys 124 was built into the electron density. The structure of the murine Igβ homodimer was determined to 3.1 Å resolution by molecular replacement using the Igβ monomer structure as a search model. The refinement and model building were done using programs CNS v1.1 and “O”, respectively. Two well defined sulfate ions were located in the electron density map. The structure of human Igβ homodimer was determined to resolution using murine Igβ monomer as the search model and refined using programs CNS v1.1 (Brunger et al., 1998), PHENIX.REFINE (Afonine et al., 2005), “O” (Kleywegt and Jones, 1997) and COOT (Emsley and Cowtan, 2004).
Surface plasmon resonance
Human IgD (Calbiochem), human IgE, human IgA (Athens Research & Technology), human IgM, human IgG1 kappa, and human IgG1 lambda (SIGMA) were purchased from commercial sources as indicated. Binding studies were performed with a BIAcore 3000 instrument and analyzed with BIAevaluation 4.1 software (Biacore AB). Recombinant Igαβ leucine zipper fusion proteins were immobilized individually onto a carboxymethylated dextran (CM5) chip using primary amine-coupling in 10mM sodium acetate pH4.5-5.5 at flow rate of 10ml/min to levels between 250-2000 response units (RU). The binding experiments were performed using serial dilutions of analytes in PBS buffer at a flow rate of 10 or 20 μl/min. Only the binding experiments with Fcμ as analyte displayed good binding kinetics. Following antibodies were used as controls in binding studies: anti-human CD79a (clone JCB117, Lab Vision), anti-human CD79b (clone CB3-1, BD Biosciences), and anti-human IgM (SIGMA). All recombinant proteins showed specific interactions with their corresponding antibodies. Surfaces were regenerated by brief injection of either 10mM NaOH or 10mM Glycine at pH 3.0. Dissociation constants (KD) were determined from either kinetic or steady state fittings with BIAevaluation software. Binding studies on human Igα and Igβ mutants were performed in reverse orientation, where recombinant Fcμ was immobilized onto a CM5 chip using primary amine-coupling in 10mM sodium acetate pH5.0. Anti-human CD79a (clone JCB117, Lab Vision) and anti-human CD79b (clone CB3-1, BD Biosciences) antibodies were immobilized in separate flow cells on the same sensorchip to monitor the conformational integrity of mutant Igα and Igβ. The analytes consist of individual mutants dissolved in PBS buffer at a constant concentration of 4μM. Similar binding affinities were obtained for the wildtype Igα and Igβ in this reverse immobilization. Surfaces were regenerated by brief injection of 5mM NaOH and 5mM glycine pH 2.0. At least three sets of experiments were performed for each mutant.
Cell surface expression analysis of IgM mutants
The structure of Cμ3-Cμ4 domains of mouse IgM was modeled based on the structure of IgA (Herr et al., 2003). The nine clusters of surface residues of Cμ4 identified for alanine mutations were: R479/E480/N483 (μ1), R485/ E486/S487 (μ2), R509/Q511 (μ3), Q515/E516/K517 (μ4), M524/P525/E526/P527 (μ5), P530 (μ6), E543/E544/E545/N547/S548 (μ7), P562/H563 (μ8), and E567/R568 (μ9). Mutations were introduced into B1-8 IgM heavy chain in pcDNA6 plasmid using Quickchange kit (Stratagene). J558L cells were transiently co-transfected with the IgM constructs and Igα-YFP using Amaxa electroporation as described (Tolar et al., 2005). After 36 hours, cells were fixed in paraformaldehyde, permeabilized or not with 0.1 % Triton and stained with anti-IgM-Cy5 (Jackson Immunoresearch) antibodies. Surface expression was determined as mean fluorescence intensity of the IgM-positive cells normalized on the signal from IgM wild-type transfected cells.
Supplementary Material
Acknowledgements
We would like to thank Marina Zhuravleva and M.Gordon Joyce for their help in data collection. The use of the Advanced Photon Source was supported by the U. S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract No. W-31-109-Eng-38. This work is supported by the intramural research funding from the National Institute of Allergy and Infectious Diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Afonine PV, Grosse-Kunstleve RW, Adams PD. A robust bulk-solvent correction and anisotropic scaling procedure. Acta Crystallogr D Biol Crystallogr. 2005;61:850–855. doi: 10.1107/S0907444905007894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfarano A, Indraccolo S, Circosta P, Minuzzo S, Vallario A, Zamarchi R, Fregonese A, Calderazzo F, Faldella A, Aragno M, et al. An alternatively spliced form of CD79b gene may account for altered B-cell receptor expression in B-chronic lymphocytic leukemia. Blood. 1999;93:2327–2335. [PubMed] [Google Scholar]
- Blum JH, Stevens TL, DeFranco AL. Role of the mu immunoglobulin heavy chain transmembrane and cytoplasmic domains in B cell antigen receptor expression and signal transduction. J Biol Chem. 1993;268:27236–27245. [PubMed] [Google Scholar]
- Brouns GS, de Vries E, Borst J. Assembly and intracellular transport of the human B cell antigen receptor complex. Int Immunol. 1995;7:359–368. doi: 10.1093/intimm/7.3.359. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Call ME, Pyrdol J, Wiedmann M, Wucherpfennig KW. The organizing principle in the formation of the T cell receptor-CD3 complex. Cell. 2002;111:967–979. doi: 10.1016/s0092-8674(02)01194-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambier JC. Antigen and Fc receptor signaling. The awesome power of the immunoreceptor tyrosine-based activation motif (ITAM) J Immunol. 1995;155:3281–3285. [PubMed] [Google Scholar]
- Campbell KS, Hager EJ, Friedrich RJ, Cambier JC. IgM antigen receptor complex contains phosphoprotein products of B29 and mb-1 genes. Proc Natl Acad Sci U S A. 1991;88:3982–3986. doi: 10.1073/pnas.88.9.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C, Gelfand I, Kister A. Structural Determinants in the Sequences of Immunoglobulin Variable Domain. J. Mol. Biol. 1998;278:457–479. doi: 10.1006/jmbi.1998.1653. [DOI] [PubMed] [Google Scholar]
- Clevers H, Alarcon B, Wileman T, Terhorst C. The T cell receptor/CD3 complex: a dynamic protein ensemble. Annu Rev Immunol. 1988;6:629–662. doi: 10.1146/annurev.iy.06.040188.003213. [DOI] [PubMed] [Google Scholar]
- Cohen GH, Sheriff S, Davies DR. Refined structure of the monoclonal antibody HyHEL-5 with its antigen hen egg-white lysozyme. Acta Crystallogr D Biol Crystallogr. 1996;52:315–326. doi: 10.1107/S0907444995014855. [DOI] [PubMed] [Google Scholar]
- Dal Porto JM, Gauld SB, Merrell KT, Mills D, Pugh-Bernard AE, Cambier J. B cell antigen receptor signaling 101. Mol Immunol. 2004;41:599–613. doi: 10.1016/j.molimm.2004.04.008. [DOI] [PubMed] [Google Scholar]
- Dylke J, Lopes J, Dang-Lawson M, Machtaler S, Matsuuchi L. Role of the extracellular and transmembrane domain of Ig-alpha/beta in assembly of the B cell antigen receptor (BCR) Immunol Lett. 2007;112:47–57. doi: 10.1016/j.imlet.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Evans EJ, Esnouf RM, Manso-Sancho R, Gilbert RJ, James JR, Yu C, Fennelly JA, Vowles C, Hanke T, Walse B, et al. Crystal structure of a soluble CD28-Fab complex. Nat Immunol. 2005;6:271–279. doi: 10.1038/ni1170. [DOI] [PubMed] [Google Scholar]
- Harpaz Y, Chothia C. Many of the Immunoglobulin Superfamily Domains in Cell Adhesion Molecules and Surface Receptors Belong to a New Structural Set Which is Close to that Containing Variable Domains. J. Mol. Biol. 1994;238:528–539. doi: 10.1006/jmbi.1994.1312. [DOI] [PubMed] [Google Scholar]
- Foy SP, Matsuuchi L. Association of B lymphocyte antigen receptor polypeptides with multiple chaperone proteins. Immunol Lett. 2001;78:149–160. doi: 10.1016/s0165-2478(01)00256-5. [DOI] [PubMed] [Google Scholar]
- Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE, Wiley DC. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- Gauld SB, Dal Porto JM, Cambier JC. B cell antigen receptor signaling: roles in cell development and disease. Science. 2002;296:1641–1642. doi: 10.1126/science.1071546. [DOI] [PubMed] [Google Scholar]
- Geisberger R, Lamers M, Achatz G. The riddle of the dual expression of IgM and IgD. Immunology. 2006;118:429–437. doi: 10.1111/j.1365-2567.2006.02386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grupp SA, Campbell K, Mitchell RN, Cambier JC, Abbas AK. Signaling-defective mutants of the B lymphocyte antigen receptor fail to associate with Ig-alpha and Ig-beta/gamma. J Biol Chem. 1993;268:25776–25779. [PubMed] [Google Scholar]
- Grupp SA, Mitchell RN, Schreiber KL, McKean DJ, Abbas AK. Molecular mechanisms that control expression of the B lymphocyte antigen receptor complex. J Exp Med. 1995;181:161–168. doi: 10.1084/jem.181.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris LJ, Larson SB, Hasel KW, Day J, Greenwood A, McPherson A. The three-dimensional structure of an intact monoclonal antibody for canine lymphoma. Nature. 1992;360:369–372. doi: 10.1038/360369a0. [DOI] [PubMed] [Google Scholar]
- Hermanson GG, Eisenberg D, Kincade PW, Wall R. B29: a member of the immunoglobulin gene superfamily exclusively expressed on beta-lineage cells. Proc Natl Acad Sci U S A. 1988;85:6890–6894. doi: 10.1073/pnas.85.18.6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr AB, Ballister ER, Bjorkman PJ. Insights into IgA-mediated immune responses from the crystal structures of human FcalphaRI and its complex with IgA1-Fc. Nature. 2003;423:614–620. doi: 10.1038/nature01685. [DOI] [PubMed] [Google Scholar]
- Hombach J, Tsubata T, Leclercq L, Stappert H, Reth M. Molecular components of the B-cell antigen receptor complex of the IgM class. Nature. 1990;343:760–762. doi: 10.1038/343760a0. [DOI] [PubMed] [Google Scholar]
- Indraccolo S, Minuzzo S, Zamarchi R, Calderazzo F, Piovan E, Amadori A. Alternatively spliced forms of Igalpha and Igbeta prevent B cell receptor expression on the cell surface. Eur J Immunol. 2002;32:1530–1540. doi: 10.1002/1521-4141(200206)32:6<1530::AID-IMMU1530>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Jumaa H, Hendriks RW, Reth M. B cell signaling and tumorigenesis. Annu Rev Immunol. 2005;23:415–445. doi: 10.1146/annurev.immunol.23.021704.115606. [DOI] [PubMed] [Google Scholar]
- Kashiwamura S, Koyama T, Matsuo T, Steinmetz M, Kimoto M, Sakaguchi N. Structure of the murine mb-1 gene encoding a putative sIgM-associated molecule. J Immunol. 1990;145:337–343. [PubMed] [Google Scholar]
- Kleywegt GJ, Jones TA. Model building and refinement practice. Methods Enzymol. 1997;277:208–230. doi: 10.1016/s0076-6879(97)77013-7. [DOI] [PubMed] [Google Scholar]
- Kozono H, White J, Clements J, Marrack P, Kappler J. Production of soluble MHC class II proteins with covalently bound single peptides. Nature. 1994;369:151–154. doi: 10.1038/369151a0. [DOI] [PubMed] [Google Scholar]
- Kuhns MS, Davis MM, Garcia KC. Deconstructing the form and function of the TCR/CD3 complex. Immunity. 2006;24:133–139. doi: 10.1016/j.immuni.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- Leahy DJ, Axel R, Hendrickson WA. Crystal structure of a soluble form of the human T cell coreceptor CD8 at 2.6 A resolution. Cell. 1992;68:1145–1162. doi: 10.1016/0092-8674(92)90085-q. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nemazee D, Kouskoff V, Hertz M, Lang J, Melamed D, Pape K, Retter M. B-cell-receptor-dependent positive and negative selection in immature B cells. Curr Top Microbiol Immunol. 2000;245:57–71. doi: 10.1007/978-3-642-59641-4_3. [DOI] [PubMed] [Google Scholar]
- Ostrov DA, Shi W, Schwartz JC, Almo SC, Nathenson SG. Structure of murine CTLA-4 and its role in modulating T cell responsiveness. Science. 2000;290:816–819. doi: 10.1126/science.290.5492.816. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pleiman CM, Chien NC, Cambier JC. Point mutations define a mIgM transmembrane region motif that determines intersubunit signal transduction in the antigen receptor. J Immunol. 1994;152:2837–2844. [PubMed] [Google Scholar]
- Radaev S, Kattah M, Rostro B, Colonna M, Sun PD. Crystal structure of the human myeloid cell activating receptor TREM-1. Structure. 2003;11:1527–1535. doi: 10.1016/j.str.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- Reth M. Antigen receptor tail clue. Nature. 1989;338:383–384. [PubMed] [Google Scholar]
- Reth M. Antigen receptors on B lymphocytes. Annu Rev Immunol. 1992;10:97–121. doi: 10.1146/annurev.iy.10.040192.000525. [DOI] [PubMed] [Google Scholar]
- Richardson J. The Anatomy and Taxonomy of Protein Structure. Advances in Protein Chemistry. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- Schamel WW, Kuppig S, Becker B, Gimborn K, Hauri HP, Reth M. A high-molecular-weight complex of membrane proteins BAP29/BAP31 is involved in the retention of membrane-bound IgD in the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2003;100:9861–9866. doi: 10.1073/pnas.1633363100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schamel WW, Reth M. Monomeric and oligomeric complexes of the B cell antigen receptor. Immunity. 2000;13:5–14. doi: 10.1016/s1074-7613(00)00003-0. [DOI] [PubMed] [Google Scholar]
- Schwartz JC, Zhang X, Fedorov AA, Nathenson SG, Almo SC. Structural basis for co-stimulation by the human CTLA-4/B7-2 complex. Nature. 2001;410:604–608. doi: 10.1038/35069112. [DOI] [PubMed] [Google Scholar]
- Siegers GM, Yang J, Duerr CU, Nielsen PJ, Reth M, Schamel WW. Identification of disulfide bonds in the Ig-alpha/Ig-beta component of the B cell antigen receptor using the Drosophila S2 cell reconstitution system. Int Immunol. 2006;18:1385–1396. doi: 10.1093/intimm/dxl072. [DOI] [PubMed] [Google Scholar]
- Tolar P, Sohn HW, Pierce SK. The initiation of antigen-induced B cell antigen receptor signaling viewed in living cells by fluorescence resonance energy transfer. Nat Immunol. 2005;6:1168–1176. doi: 10.1038/ni1262. [DOI] [PubMed] [Google Scholar]
- Venkitaraman AR, Williams GT, Dariavach P, Neuberger MS. The B-cell antigen receptor of the five immunoglobulin classes. Nature. 1991;352:777–781. doi: 10.1038/352777a0. [DOI] [PubMed] [Google Scholar]
- Wegener AM, Hou X, Dietrich J, Geisler C. Distinct domains of the CD3-gamma chain are involved in surface expression and function of the T cell antigen receptor. J Biol Chem. 1995;270:4675–4680. doi: 10.1074/jbc.270.9.4675. [DOI] [PubMed] [Google Scholar]
- Wienands J. Unraveling B cell receptor mechanics. Nat Immunol. 2005;6:1072–1074. doi: 10.1038/ni1105-1072. [DOI] [PubMed] [Google Scholar]
- Williams GT, Peaker CJ, Patel KJ, Neuberger MS. The alpha/beta sheath and its cytoplasmic tyrosines are required for signaling by the B-cell antigen receptor but not for capping or for serine/threonine-kinase recruitment. Proc Natl Acad Sci U S A. 1994;91:474–478. doi: 10.1073/pnas.91.2.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.