Abstract
The Escherichia coli signal recognition particle (SRP) system plays an important role in membrane protein biogenesis. Previous studies have suggested indirectly that in addition to its role during the targeting of ribosomes translating membrane proteins to translocons, the SRP might also have a quality control role in preventing premature synthesis of membrane proteins in the cytoplasm. This proposal was studied here using cells simultaneously overexpressing various membrane proteins and either SRP, the SRP protein Ffh, its 4.5S RNA, or the Ffh M domain. The results show that SRP, Ffh, and the M domain are all able to selectively inhibit the expression of membrane proteins. We observed no apparent changes in the steady-state mRNA levels or membrane protein stability, suggesting that inhibition may occur at the level of translation, possibly through the interaction between Ffh and ribosome-hydrophobic nascent chain complexes. Since E. coli SRP does not have a eukaryote-like translation arrest domain, we discuss other possible mechanisms by which this SRP might regulate membrane protein translation when overexpressed.
IMPORTANCE
The eukaryotic SRP slows down translation of SRP substrates by cytoplasmic ribosomes. This activity is important for preventing premature synthesis of secretory and membrane proteins in the cytoplasm. It is likely that an analogous quality control step would be required in all living cells. However, on the basis of its composition and domain structure and limited in vitro studies, it is believed that the E. coli SRP is unable to regulate ribosomes translating membrane proteins. Nevertheless, several in vivo studies have suggested otherwise. To address this issue further in vivo, we utilized unbalanced conditions under which E. coli simultaneously overexpresses SRP and each of several membrane or cytosolic proteins. Surprisingly, our results clearly show that the E. coli SRP is capable of regulating membrane protein synthesis and demonstrate that the M domain of Ffh mediates this activity. These results thus open the way for mechanistic characterization of this quality control process in bacteria.
INTRODUCTION
The Escherichia coli version of the signal recognition particle (SRP) system includes two essential proteins, Ffh and FtsY, homologues of the mammalian SRP54 protein and the SRP receptor α subunit (SRα), respectively (1, 2), and a small stable RNA (4.5S RNA) (3). The bacterial system plays an important role in expression, membrane targeting, and proper integration of inner membrane proteins (4, 5), as shown both by genetic studies (6–12) and by use of in vitro systems (e.g., references 13 and 14). In addition to its role in targeting ribosomes translating SRP substrates to the translocon, the eukaryotic SRP can slow their translation on cytoplasmic ribosomes. This activity is mediated through the physical interaction of SRP with cytoplasmic ribosome-hydrophobic nascent chain complexes (translation arrest, reviewed in reference 15). Recent studies have shown that the physiological role of the SRP-mediated translation arrest in eukaryotes can be demonstrated in vivo (16, 17). It is likely that in vivo, the nascent chain elongation pause would be more pronounced and noticeable under unbalanced conditions, such as limiting amounts of the SRP receptor (8, 18, 19), or in normally existing unbalanced situations (17). Interestingly, several studies have raised the possibility that SRP might be able to regulate membrane protein translation also in E. coli (12, 20), but through a different mechanism, because E. coli SRP does not contain a domain that is analogous to the arrest domain of the eukaryotic complex. Here, we examined this proposal by implementing unbalanced situations in vivo in E. coli, which enabled detection of changes in membrane protein expression in a reasonably quantitative manner. This was accomplished by overexpressing SRP, 4.5S RNA (encoded by the ffs gene), and Ffh or its separated domains NG and M.
Taken together, the results show that SRP, Ffh, and its M domain have the capacity to selectively limit the synthesis of membrane proteins in vivo. The extent of inhibition does not fully account for the phenotype observed previously in FtsY-depleted cells (12), suggesting additional, yet-unknown regulatory modes. We propose that the observations made under overexpression conditions demonstrate the capabilities of the system under less extreme conditions, such as those that might exist when membrane proteins are highly induced in response to physiological demands.
RESULTS
Effect of SRP overexpression on the expression of membrane proteins.
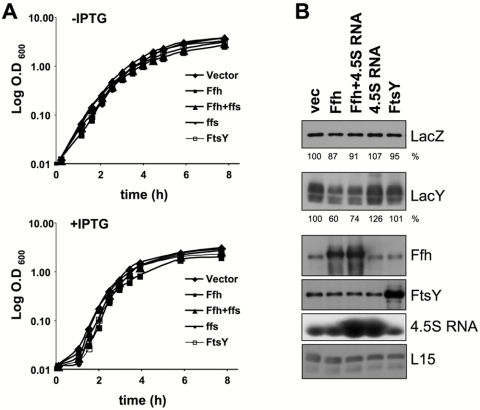
Binding of the eukaryotic SRP to cytosolic ribosomes translating secretory or membrane proteins causes a nascent chain elongation pause via its Alu domain (reviewed in reference 15), which reaches into the elongation factor-binding site of the ribosome (21). In E. coli, the SRP does not contain an Alu domain, and therefore, it was proposed that this SRP has no translation arrest activity (22). Nevertheless, since previous indirect studies suggested otherwise (12, 20), we decided to examine this question in vivo. Initially, we overexpressed Ffh and/or 4.5S RNA or FtsY as a control, together with the membrane protein LacY (an SRP substrate; see reference 6). Figure 1A shows the results for a typical growth experiment for the various transformants. Clearly, overexpressed Ffh inhibits growth, but only slightly, and we hypothesize that this is due to its effect on membrane protein expression (see below). As shown in Fig. 1B, LacY expression was largely inhibited only when Ffh or Ffh plus 4.5S RNA was overexpressed. In contrast, the expression of the cytosolic protein LacZ was hardly affected in all samples. Interestingly, when FtsY or 4.5S RNA alone was overexpressed, it had little or no affect on the level of LacY. The results indicate that overexpression of Ffh alone might be responsible for the decreased LacY expression, although a possible role for 4.5S RNA (together with Ffh) cannot be excluded.
FIG 1 .
Expression of LacY and LacZ under conditions of Ffh and/or 4.5S RNA or FtsY overexpression. E. coli cells harboring plasmid carrying arabinose-inducible ffh and/or ffs or ftsY were induced to overexpress the indicated proteins. (A) Growth was followed by measuring optical density at 600 nm, and representative semilog growth curves of the various transformants with and without IPTG induction are shown. (B) Expression of LacZ, LacY, Ffh, and FtsY was analyzed by Western blotting. The expression levels of LacZ and LacY were quantified and are shown as percentages of their expression levels in samples with empty vector. The experiments were repeated three times, and the results shown are representative, with standard deviations that did not exceed 10%. 4.5S RNA expression was analyzed by Northern blotting. As a loading control, antibodies against the ribosomal protein L15 were used.
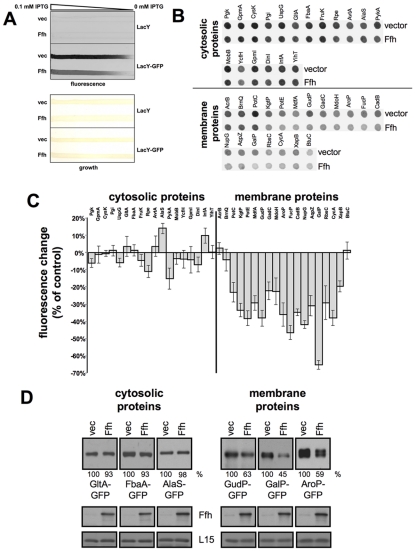
Next, we tested whether the observed inhibition of LacY expression by Ffh is representative of other integral, polytopic (6- to 12-transmembrane helices) membrane proteins, which have been selected randomly. In this experiment, we simultaneously overexpressed Ffh with each of several membrane proteins fused to green fluorescent protein (GFP) at their cytoplasmically oriented C termini and several cytoplasmic proteins also fused to GFP, and the levels of their expression were monitored by GFP fluorescence. Figure 2A shows the results for a control experiment in which we tested (i) the GFP fluorescence as a function of induction of LacY-GFP and (ii) the fluorescence of cells that do not express GFP (only LacY). The results clearly show that cells devoid of GFP do not fluoresce and that GFP fluorescence depends on the level of induction of LacY-GFP. Figure 2B and C show that Ffh overexpression inhibited the expression of 16 out of 19 membrane proteins by 15 to 60% but that the expression of cytosolic proteins was generally much less affected. We hypothesize that the extent of Ffh (SRP)-dependent inhibition of membrane protein expression might be inversely correlated with the efficiency of targeting to and insertion in the membrane. These processes depend on hydrophobicity and several other chemical and structural properties of the nascent chain (e.g., reference 23). In order to verify the results shown in Fig. 2B, we analyzed the expression of several hybrids by Western blotting (Fig. 2D). The results show a decrease in expression of the integral membrane hybrids GudP-GFP, GalP-GFP, and AroP-GFP, whereas no significant change was detected in the expression of the cytosolic hybrids GltA-GFP, FbaA-GFP, and AlaS-GFP. These results demonstrate that Ffh (SRP) overexpression leads to selective inhibition of expression of many membrane proteins.
FIG 2 .
Effect of Ffh overexpression on expression of cytosolic and membrane proteins. (A) For a control experiment, we used E. coli cells expressing chromosomally encoded LacY-GFP or LacY as indicated and harboring plasmids encoding arabinose-inducible Ffh or carrying empty vector. The transformants were grown overnight on a nylon membrane covering an LB agar plate containing a linear concentration gradient of IPTG (0 to 0.1 mM) for LacY or LacY-GFP induction and 0.02% arabinose for Ffh induction. The upper panel shows the fluorescence as recorded using Typhoon. The lower panel shows the bacterial growth on the nylon membrane. (B) E. coli cells harboring plasmids carrying arabinose-inducible ffh or empty vector were transformed with vectors encoding the indicated GFP hybrids. The cells were grown overnight on LB agar plates covered with nylon membranes and supplemented with 0.02% arabinose. The fluorescence levels of the colonies on the nylon membrane were recorded (B) and quantified (C). The experiment was repeated nine times, and the error bars represent the standard deviations. (D) Cells harboring a plasmid encoding arabinose-inducible Ffh or carrying an empty vector were transformed with a second plasmid encoding either of the indicated GFP hybrids and grown in LB broth with 0.2% arabinose. After disruption, equal amounts of total proteins from each extract were analyzed by Western blotting with anti-GFP and anti-Ffh antibodies. As a loading control, antibodies against the ribosomal protein L15 were used. The proteins’ expression levels were quantified and are shown as percentages of their expression levels in samples with empty vector. The experiments were repeated three times, and the results shown are representative, with standard deviations that did not exceed 10%.
Effect of the Ffh M domain on membrane protein expression.
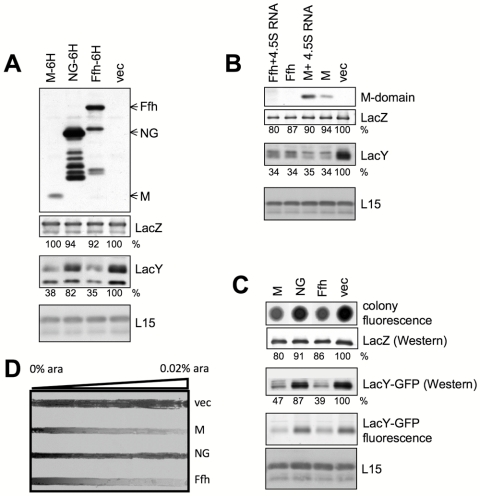
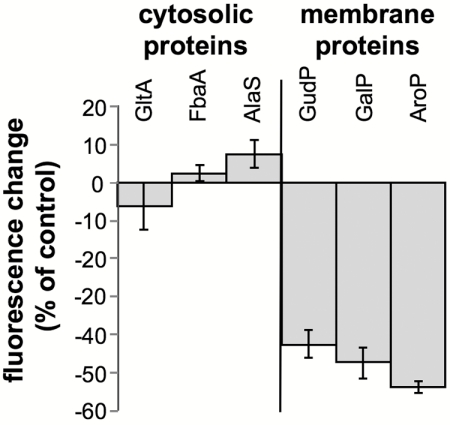
Ffh has two domains connected by a linker (reviewed in reference 24). The N-terminal NG domain is homologous to the NG domain of FtsY and harbors the GTP-binding and hydrolysis activity. The C-terminal M domain (methionine rich) is responsible for interaction with 4.5S RNA and hydrophobic nascent peptides that emerge from ribosomes (reviewed in reference 24). To identify which domain of Ffh is responsible for the observed inhibition of membrane protein expression, Ffh or its NG or M domain was expressed with or without a C-terminal 6-histidine tag in cells induced for expression of the membrane protein LacY or the cytosolic protein LacZ (as a control). Initially, the amounts of LacY and LacZ in each strain were examined by Western blotting, and the results show that the M domain has a selective inhibitory effect on the expression of LacY (Fig. 3A, panels below upper panel). Only a minor difference was observed in the expression of LacY in cells overexpressing the NG domain, even though the amount of NG was substantially larger than that of the M domain (Fig. 3A, upper panel). The results were essentially the same with M, NG, or Ffh lacking a 6-His tag (data not shown and Fig. 3B). Interestingly, although coexpression of 4.5S RNA had a putative stabilizing effect on the M domain (Fig. 3B), as shown previously for Ffh (25–27), this coexpression did not change the phenotype observed in cells expressing the M domain alone. As noted above, overexpression of 4.5S RNA alone did not decrease the expression of LacY (Fig. 1). The effect of the M domain was further evaluated by utilizing a LacY-GFP hybrid (Fig. 3C). The GFP moiety enabled detection of differential expression in vivo (Fig. 3C, top panel) and with the GFP fluorescence of LacY-GFP separated by SDS-PAGE (Fig. 3C, panel marked “LacY-GFP fluorescence”). Overall, the results observed with LacY-GFP were identical to those described for LacY (Fig. 3A). Cells expressing LacY-GFP were also utilized to examine if the effect of M domain expression is concentration dependent. Cells coexpressing LacY-GFP and the arabinose-regulated M domain, NG domain, or Ffh protein were plated on an arabinose concentration gradient. After growth, the plate was scanned for GFP fluorescence as a measure for LacY-GFP expression (Fig. 3D), and the results clearly show that the inhibitory effects of the M domain and Ffh are concentration dependent. In accordance with the results shown above (Fig. 3A and C), the expression of LacY-GFP in cells harboring empty vector or overexpressing NG remained much less affected by elevated arabinose concentrations (Fig. 3D). Finally, we tested the effect of the M domain on the expression of other membrane proteins (Fig. 4), and the results show that the M domain-dependent inhibition is not restricted to LacY and that cytoplasmic proteins are not affected. Together, these studies suggest that the inhibitory effect of Ffh on membrane protein expression seems to be mediated by its M domain.
FIG 3 .
Effect of the Ffh M domain on expression of membrane proteins. E. coli cells harboring a plasmid encoding arabinose-inducible M, NG, and/or Ffh were induced to overexpress the indicated proteins. The expression levels of the indicated proteins were analyzed by Western blotting (panels A, B and C, using anti-Ffh, LacY, LacY-GFP, LacZ, or anti-His tag antibodies) or by “in-gel” fluorescence (C). The expression levels of LacZ and LacY were quantified and are shown as a percentages of their expression levels in samples with empty vector. The experiments were repeated three times, and the results shown are representative, with standard deviations that did not exceed 10%. As a loading control, antibodies against the ribosomal protein L15 were used. (D) E. coli IY228 (expressing chromosomally encoded LacY-GFP) harboring plasmids encoding arabinose-inducible M domain, NG domain, or Ffh or carrying empty vector were grown overnight on nylon membranes covering an LB agar plate containing a linear concentration gradient of arabinose (0 to 0.02%). The fluorescence was recorded using Typhoon.
FIG 4 .
Effect of the Ffh M domain on expression of membrane and cytosolic proteins. E. coli cells harboring plasmids encoding arabinose-inducible M domain or carrying empty vector (as a control) were transformed with plasmids encoding the indicated GFP hybrids. The cells were grown overnight on LB agar plates supplemented with 0.02% arabinose and covered with nylon membranes. The fluorescence was recorded and quantified. The experiment was repeated three times, and the error bars represent the standard deviations.
Expression of the M domain does not affect membrane protein stability or mRNA levels.
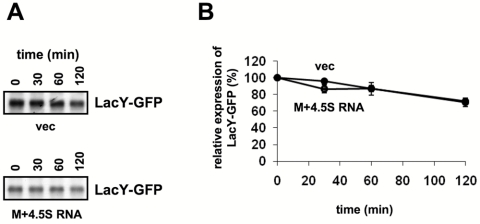
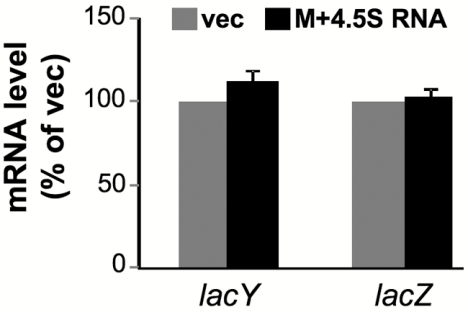
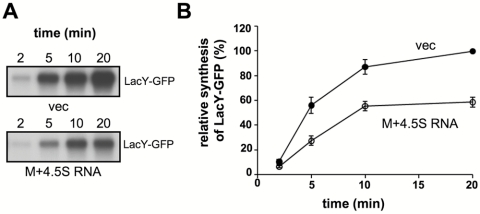
How does the M domain regulate the expression of membrane proteins? To answer this question, we examined the effect of M domain overexpression on the stability and rate of synthesis of a membrane protein and the fate of its coding mRNA in cells harboring a chromosomally encoded LacY-GFP (Fig. 5; see also Fig. 7) or LacY (Fig. 6). Because of the stabilizing effect of 4.5S RNA on the M domain (Fig. 3B), we included 4.5S RNA coexpression in all the following experiments. The stability of LacY-GFP was investigated by pulse-chase experiments, and as expected, the results show that the level of LacY-GFP expression is substantially lower in cells expressing the M domain (Fig. 5A). Importantly, however, no difference in LacY-GFP stability was observed between cells overexpressing the M domain and control cells (Fig. 5B). Therefore, posttranslational degradation does not explain the low expression level of LacY in M domain-expressing cells. Next, quantitative real-time PCR studies were conducted with total RNA samples prepared from control cells and cells overexpressing the M domain. Figure 6 shows that the amounts of the LacY-encoding mRNA are similar in both cell types. We therefore suggest that the decreased expression of membrane proteins in Ffh- or M domain-overexpressing cells might be due to inhibition of translation. This notion is consistent with the results of a pulse experiment, where the kinetics of expression of LacY-GFP in cells overexpressing the M domain was measured (Fig. 7A and B). The results of this experiment clearly show that LacY synthesis is substantially more rapid in control cells than in those coexpressing the M domain.
FIG 5 .
Effect of the Ffh M domain on the stability of LacY-GFP. (A) E. coli IY228 harboring plasmids encoding arabinose-inducible M and carrying ffs or empty vector were induced with arabinose, and then IPTG was added to induce expression of LacY-GFP. The cells were pulse-labeled for 3 min with [35S]methionine and [35S]cysteine and chased with an excess of methionine and cysteine. Equal samples were withdrawn at the indicated time points, and LacY-GFP was immunoprecipitated using anti-GFP antibodies. The precipitates were solubilized in 50 µl SDS sample buffer, and 25 µl was loaded on SDS-PAGE gel (12%), which was then dried and subjected to autoradiography. (B) The amount of labeled LacY-GFP was quantified by densitometry, and the data shown are averages of results from three independent experiments.
FIG 7 .
Effect of the Ffh M domain on the rate of synthesis of LacY-GFP in vivo. E. coli IY228 harboring plasmids encoding arabinose-inducible M and carrying ffs or empty vector were induced with arabinose, and then IPTG was added to induce expression of LacY-GFP. The cells were pulse-labeled with [35S]methionine and [35S]cysteine, and equal samples were withdrawn at the indicated time points. LacY-GFP was immunoprecipitated using anti-GFP antibodies. The precipitates were solubilized in 50 µl SDS sample buffer, and 25 µl was loaded on SDS-PAGE gel (12%), which was then dried and subjected to autoradiography. (B) The amount of labeled LacY-GFP was quantified by densitometry, and the averages of results from three independent assays are shown, with error bars representing standard deviations.
FIG 6 .
Effect of the Ffh M domain on the amount of LacY-encoding mRNA. RNA preparations from wild-type E. coli harboring empty vector (vec) or plasmid encoding arabinose-inducible M and carrying ffs (M+4.5S RNA) were used for real-time PCR analysis. The expression levels of lacY and lacZ mRNAs were normalized to that of ribosomal 16S rRNA, and the vector-derived results were set to 100%. The averages of results from three separate assays are shown, with error bars representing standard deviations.
The Ffh M domain comigrates with cytosolic ribosomes.
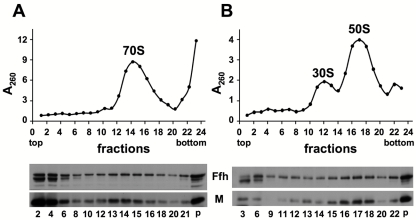
The interaction between SRP and ribosomes in E. coli is well studied and characterized (21). Since translation inhibition might be mediated through direct interaction with the ribosome, we asked whether the M domain interacts with cytosolic ribosomes in our experimental system as shown previously (20). Cells transformed with a plasmid encoding the M domain were disrupted, and the ultracentrifuged pellets (including membranes and ribosomes) were separated by sucrose gradient centrifugation. Fractions were collected and examined for rRNA (A260) and M domain (Western blot) content. Figure 8A shows that free 70S ribosomes migrate in fractions 13 to 15 and that large portions of both the M domain and chromosomally expressed Ffh migrate in the same fractions. To distinguish between the possibility that the migration of the M domain at high sucrose density is due to aggregation and the possibility that this migration is due to association with the ribosomes, we utilized conditions under which ribosomes dissociate into their subunits, and the results show that both the M domain and Ffh now migrate in a lower density, primarily with the large (50s) subunit (Fig. 8B). These results suggest that Ffh might inhibit membrane protein expression via the direct interaction of its M domain (alone or with 4.5S RNA) with the large subunits of cytosolic ribosomes.
FIG 8 .
The Ffh M domain comigrates with the cytoplasmic ribosomes in a sucrose gradient. E. coli cells harboring a plasmid encoding arabinose-inducible M and carrying ffs were induced and harvested, and cell extracts were ultracentrifuged. The pellet fractions were loaded on top of a 10% to 30% (wt/vol) sucrose gradient containing either 10.5 mM MgCl2 (A) or 1 mM MgCl2 (B) and separated by ultracentrifugation for 70 min (A) or 105 min (B). The ODs (260 nm) of the fractions were measured (upper panels), and fractions were analyzed by Western blotting with antibodies against Ffh (and its M domain) (lower panels).
DISCUSSION
In our previous study, we asked how FtsY or Ffh depletion influences the synthesis, localization, and functional assembly of several test membrane proteins (12). The results indicated that FtsY depletion drastically inhibited the expression of membrane proteins. In the present study, we asked whether SRP might be responsible for the inhibitory effect observed under FtsY depletion conditions. This was addressed by overexpressing SRP, Ffh, the Ffh M domain, or 4.5S RNA and analyzing membrane protein expression. The results show that of all the test cases, only cells overexpressing SRP, Ffh, or its M domain exhibited considerable degrees of inhibition of synthesis of many membrane proteins but that cytosolic proteins usually remained unaffected. The finding that the M domain alone has the same effect as SRP or Ffh suggests that the observed phenotype is FtsY independent, since there is no evidence for FtsY-M domain interaction (20, 28). By studying various stages of membrane protein expression under these conditions, we showed that the inhibition might occur at the level of translation. However, although significant, the extent of inhibition does not fully account for the phenotype observed in FtsY-depleted cells (12).
Our results suggest that SRP might play a selective role in translation inhibition of membrane proteins by cytoplasmic ribosomes, since soluble proteins were generally unaffected. The inherent association of SRP with ribosomes, as also shown here with Ffh and its M domain (Fig. 8), raises the possibility that cytosolic ribosomes translating membrane proteins might be inhibited by direct interaction with SRP. Whereas these surprising results shed light on the potential capacity of SRP to regulate membrane protein synthesis, the physiological implication is not that trivial. Previous studies have suggested that the amount of Ffh in E. coli cells is small (~500) (25) compared to the number of ribosomes (~30,000) (29). In exponentially growing E. coli cells, ~6% of all ribosomes are membrane bound (~2,000) (19), and these ribosomes presumably represent the population of ribosomes that are involved in membrane protein translation. Theoretically, therefore, a significant effect on translation can be achieved if every SRP particle is able to regulate, by transient engagement, several ribosomes that translate membrane proteins. Notably, the affinity of SRP to ribosome-hydrophobic nascent chain complexes is substantially higher (~100-fold) than its affinity to nontranslating ribosomes (30). The results presented in Fig. 7, which show that SRP slows down only synthesis, support the possibility that this scenario is plausible.
Eukaryotic in vitro protein-targeting assays showed that SRP binding to ribosome signal peptide harboring nascent chain complexes affects the rate of nascent-chain elongation from transient pausing in homologous mammalian in vitro assays (31) to prolonged inhibition of translation in heterologous in vitro systems (32). Although in vitro the nascent-chain-elongation pause is not essential for proper targeting and translocation (33, 34), this pause plays an important regulatory role in vivo (16, 17). In this regard, a similar physiological role for SRP in preventing premature synthesis of membrane proteins in the cytoplasm might also exist in bacteria. Previously, it was shown by utilizing an in vitro translation system that there is no SRP-mediated translation inhibition in E. coli (35). However, other indirect observations supported the notion that such activity might exist also in this bacterium (20). Since our results also demonstrate selective translation inhibition of membrane proteins in vivo, it would be interesting to identify which factors, if any, in addition to Ffh (and possibly 4.5S RNA), might be required for the reconstitution of this phenomenon in vitro.
As proposed previously, evading aggregation of membrane proteins in the cytosol is possibly one of the main evolutionary driving forces for the preservation of a cotranslational targeting mechanism for membrane proteins (36). Therefore, it would be reasonable to suggest that a quality control mechanism exists, which takes care of nontargeted ribosomes that translate membrane proteins in the cytosol. There could be several mechanisms for avoiding this (as discussed in reference 37). Our results suggest that the bacterial SRP protein might also be involved in such a quality control pathway.
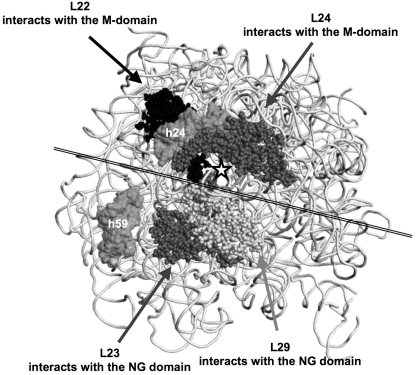
In mammalian cells, SRP binds to the elongation-factors’ binding site(s) of the ribosome through the SRP-RNA Alu domain and the accompanied SRP9/14 proteins, as observed by cryoelectron microscopy (38), and thereby inhibits translation. In contrast, the E. coli SRP does not have an Alu domain, and therefore, the mechanism by which E. coli utilizes the SRP to regulate translation of membrane proteins in the cytosol must be different from that proposed for the eukaryotic system. One possibility is that the SRP imposes an inhibitory conformational effect upon binding at the ribosome region flanking the nascent peptide exit site. Cryoelectron microscopy and single-particle analysis (39) revealed that the E. coli SRP interacts with a translation-arrested 70S ribosome via both its NG and its M domains (depicted in Fig. 9). NG interacts through the N domain with the ribosomal proteins L23/L29 in the large subunit, and the M domain interacts with several rRNA helices and possibly with the ribosomal proteins L22 and L24. The pairs L22/L24 and L23/L29 are located on opposite sides of the ribosome exit tunnel (Fig. 9). In the context of our results, this may suggest that the conformational response of L22/L24 upon the binding to the M domain might lead to regulation of translation by slowing down the movement of the nascent chain through the tunnel. Particularly interesting is the postulated interaction with L22, because this protein was shown to be involved in translation regulation under various conditions (40). Elucidation at high resolution of the precise contacts of the M domain with the ribosome large subunit is crucial for understanding the mechanism of the postulated translation regulation by the SRP and its further evaluation.
FIG 9.
A model for the interaction of Ffh domains with the E. coli 50S ribosomal subunit (Protein Data Bank [PDB] code 2AWB). The large-subunit rRNA (23S) is shown as a view from the ribosome exit tunnel (☆). Two of its helices are shown, h24 and h59. Large-subunit proteins are shown as a sphere presentation. A double line across the figure approximately separates the M and NG domain binding sites on the ribosome. This figure was created by the software program PyMol (DeLano Scientific LLC, San Carlos, CA [http://www.pymol.org/]).
MATERIALS AND METHODS
E. coli strains and growth conditions.
E. coli BW25113 (41) and IY52 (see below) were used in overexpression experiments. E. coli IY228 was used in the LacY-GFP florescence experiments. E. coli IY52 (harboring an intact lac operon) was constructed by P1 transduction using BW25113 (41) and P1 lysates of DY378 (42). Positive transductants were selected on an M9 plate containing lactose as the only carbon source and verified by PCR and growth on MacConkey agar plates. E. coli IY228 was constructed by inserting gfp in frame with lacY (regulated by the native lac promoter) using homologue recombination, followed by excision of the kanamycin resistance gene (41). Plasmid p(EGFP-kan) was used as a template for PCR amplification of gfp-kan, using forward and reverse primers 5′-CCCGCTTTCCCTGCTGCGTCGTCAGGTGAATGAAGTCGCTGTGAGCAAGGGCGAGGAGCTGTTC-3′ and 5′-ATAGCTTGCCTGCTCTTATTCTTTCGGTCATTGGCATGTTCCCGGATCCCATATGAATATCCTCC-3′, respectively. The PCR fragment was transformed and recombined into BW25113/pKD46 (41), thus inserting gfp in-frame with lacY. The resulting strain, IY228, was verified by sequencing. In overexpression experiments, overnight cultures were diluted to an optical density at 600 nm (OD600) of 0.01 and grown at 37°C. After 1 h, arabinose was added (0.02%) to induce expression of SRP components (see below), and the cells were grown for an additional 30 min. Then, IPTG (isopropyl-β-d-thiogalactopyranoside) was added (0.5 mM) to induce expression of LacY and LacZ, and the growth continued for 2.5 h, after which cells were chilled on ice. Expression of the tested cytosolic or membrane proteins (ASKA clones) was carried out without addition of IPTG. After 2 h, arabinose was added (0.02%) to induce SRP components, and the growth continued for 4 h.
Plasmids.
For constructing plasmids expressing inducible SRP components, we utilized the kanamycin resistance gene amplified from plasmid pKD4 (41) by PCR utilizing forward (5′-AATTGAGCTCGTGTAGGCTGGAGCTGCTTCG-3′) and reverse (5′-TTAAGAGCTCATATGAATATCCTCCTTAGTTCC-3′) primers. The PCR product was digested with SacI and ligated to SacI-linearized plasmid pT7-5(araP-mdfA-6H) (R. Edgar and E. Bibi, unpublished), and the kanamycin resistance gene, together with the arabinose promoter, was amplified by PCR using primers 5′-CACCGAAACGCGCGAGGCCCA-3′ and 5′-ATTGCGCAGCGTGCGCGACAAACGATCGGTTAAATTATCAAACATGGTCTTACTCCATCCAGAAAAACAG-3′. The PCR product was used for homologous recombination with plasmid pT7-5(ffh-6H+ffs) (A. Seluanov and E. Bibi, unpublished) to yield pT7-5(Kan-araP-ffh-6H+ffs). A fragment containing only ffh under the arabinose promoter was cloned into the pACYC184 derivative pCV3 (43), utilizing the enzyme XbaI and the Klenow fragment, yielding pCV3(araP-ffh-6H). The ffs gene under its native promoter was PCR amplified using forward (5′-AAAAGGAATTCCTTTTTCCATCTTTTCTTCC-3′) and reverse (5′-TTTTCTGCAGCACGCCGCACAGCCCGTCACG-3′) primers and cloned into pCV3(araP-ffh-6H), utilizing enzymes PstI and EcoRI, to yield pCV3(araP-ffh-6H-ffs). pCV3(araP-ftsY) was prepared by digestion of plasmid pT7-5(ftsY) (44) with NcoI and HindIII and ligation into pBAD24. The ftsY gene under the arabinose promoter was then amplified by PCR using forward primer 5′-ACCCTATGCTACTCCGTCAAGC-3′ and reverse primer 5′-GGGTTATTGTCTCATGAGCGGA-3′ and cloned into PvuII-digested pCV3, yielding pCV3(araP-ftsY). The plasmid expressing ffs under the arabinose promoter was constructed as follows. The kanamycin resistance gene was amplified using forward (5′-GAGAATTCGTGTAGGCTGGAGCTGCTTC-3′) and reverse (5′-CCGGATCCCATATGAATATCCTCCTTA-3′) primers and cloned into AfeI-linearized pBAD24 or StuI-digested p(EGFP) (Clontech) to yield pBAD24(RV-kan) or p(EGFP-kan), respectively. The new plasmid, pBAD24(RV-kan), was used as a template for PCR amplification of ara-C, araP, and kan, using forward and reverse primers 5′-CCCGAGTGAAGTCGCATTGCGCAAGAAACCAGCATCTGGCACGCGATGGGCATATGAATATCCTCCTTAGTTCC-3′ and 5′-GTTGCGGGAGAACCAACAGAGCCCCCATTGAGAGCGTTGAGAACCAACGCATGGAGAAACAGTAGAGAGTTGCG-3′, respectively. The PCR fragment was transformed and recombined into BW25113/pKD46 (41), thus replacing the native ffs promoter with the arabinose promoter. Next, the genomic DNA of that strain was used as a template for PCR amplification of the arabinose-controlled ffs gene, using forward (5′-ACCCTATGCTACTCCGTCAAGC-3′) and reverse (5′-CCGAAGCGTACTGCGCAGCCA-3′) primers. The PCR product was cloned into the PvuII site of pCV3 to yield pCV3(araP-ffs). Plasmids expressing 6-His-tagged Ffh domains M-6H and NG-6H were constructed by deletions in pT7-5(ffh-6H) [constructed by NdeI deletion of ffs from pT7-5(ffh-6H+ffs)], using forward primer 5′-GTACTGTCGCTGATCGAAGATATCG-3′ and reverse primer 5′-CATGGTCTTACTCCATCCAGAAAAAC-3′, yielding pIY1044 (encoding M-6H), and forward primer 5′-CATCATCATCATCATCATTAAAAGCT-3′ and reverse primer 5′-GTCGCCCATGCCGAGAATA-3′, yielding pIY1024 (encoding NG-6H). Expression plasmids for Ffh, NG, and M were constructed by PCR amplifying each DNA fragment, using the forward primers 5′-GGAACATGTTTGATAATTTAACCGATCGTTTGTCGCGC-3′ (for ffh), 5′-GGCAAGCTTAGTCGCCCATGCCGAGAATA-3′ (for NG), and 5′-GCCCCATGGTACTGTCGCTGATCGAAGATA-3′ (for M), together with the same reverse primer, 5′-ATTAAGCTTAGCGACCAGGGAAGCCTGGGGG-3′. The PCR products were digested and ligated into NcoI- and HindIII-digested pIY1023 to yield pIY1030 (for Ffh), pIY1045 (for NG), and pIY1043 (for M). Plasmids encoding membrane and cytosolic proteins as GFP hybrids were obtained from the ASKA collection, constructed and maintained by Hirotada Mori, Nara Institute of Science and Technology, Japan (45).
Cell fractionation.
Cell extracts were prepared as described previously (19), with some modifications. Cell pellets (OD600, ~35) were kept on ice and suspended in 1 ml of 5% sucrose solution in buffer C (30 mM HEPES, pH 7.5, 10.5 mM MgCl2, 100 mM NH4CI, 0.1 mM EDTA, 0.1 mM dithiothreitol [DTT], 5 mM β-mercaptoethanol, 0.01% Igepal CA-630, and 1 mM phenylmethylsulfonyl fluoride [PMSF]). Extracts were prepared by three cycles of brief sonication (10 s) at 2-min intervals on ice, followed by a low-speed centrifugation (13,000 rpm for 10 min) to remove cell debris. Ribosomes were collected by ultracentrifugation (90 min at 70,000 rpm and 4°C; TLA-100.2 rotor) in a tabletop Optima TLX ultracentrifuge (Beckman). Pellets were resuspended in 120 µl of ice-cold 5% sucrose solution in buffer C or D (same as buffer C but with 1 mM MgCl2). Samples containing 6 (high Mg) or 2 (low Mg) A260 units were loaded on top of a preformed sucrose density gradient (1.4 ml containing 0.27-ml layers of 30% 25%, 20%, 15%, and 10%, wt/vol). Following ultracentrifugation (70 to 105 min at 54,000 rpm and 4°C; TLS-55 rotor), fractions (65 µl) were collected from the top. A260 was measured for each fraction using a NanoDrop sprectrophotometer.
Western blotting.
Protein concentration was measured using a Bradford assay or a modified Lowry procedure (46) in the presence of 2.5% (wt/vol) SDS, with BSA as a standard. Protein samples were incubated for 10 min at 75°C or 25 min at 37°C in experiments with membrane proteins. SDS-PAGE was conducted according to reference 47. Western blotting was performed as described previously (44), using rabbit antibodies to a C-terminal peptide of LacY (kindly provided by H. R. Kaback, UCLA), goat antibodies against L15, or HisProbe-horseradish peroxidase (HisProbe-HRP) (Pierce). Rabbit anti-FtsY and -Ffh antibodies were prepared in the course of this study, using nitrilotriacetic acid (NTA)-purified 6-His-tagged proteins. Monoclonal anti-β-galactosidase and anti-GFP antibodies were obtained from Boehringer Mannheim and BAbCO (Richmond, CA), respectively. Goat anti-rabbit, mouse anti-rabbit, or donkey anti-goat antibodies conjugated to horseradish peroxidase were used as secondary antibodies (Jackson Immunoresearch). Scanning densitometry was performed with a Bio-Rad imaging densitometer (Model GS-690). Each panel in the figures represents experiments done with the same samples, and therefore, the loading control is for all the samples in each panel.
Northern blotting.
RNA samples (10 µg) were denatured with formamide and electrophoresed on a 2% (wt/vol) agarose gel containing formaldehyde. The RNA was transferred and fixed using UV light to nylon (Hybond-N+; Amersham Pharmacia). The 4.5S RNA ffs gene product was visualized using digoxigenin (DIG) reagents, kits for nonradioactive nucleic acid labeling, and a detection system (Roche) according to the procedure specified by the manufacturer. The probe against 4.5S RNA was prepared by using a PCR DIG probe synthesis kit (Roche), with 5′-CTAGTCTAGAGGGGGCTCTGTTGGTTC-3′ and 5′-TCGCGGATCCGGGTGGGGGCCCTGCCAGC-3′ as primers.
Pulse and pulse-chase experiments.
E. coli IY228 harboring empty vector or plasmid encoding arabinose-inducible M+ 4.5S RNA was grown overnight at 37°C on an M9 plate containing 0.4% glycerol, 1 mM MgSO4, thiamine (1 µg/ml), ampicillin (200 µg/ml), and all amino acids (15 µg/ml) except for methionine and cysteine. Cells were diluted (1:40), grown for ~2.5 h, and induced by arabinose (0.02%) for expression of the M domain and 30 min later by IPTG (0.5 mM) for LacY-GFP induction. After 20 min of IPTG induction, the cultures were diluted to 0.5 OD420 units in 3 ml and transferred to a 34°C water bath. For pulse-labeling, aliquots (0.5 ml) were labeled with 20 µCi/ml of [35S]methionine and [35S]cysteine (1,000 Ci/mmol [Isolabel; Izotop]) for various amounts of time. For the pulse-chase experiment, aliquots were labeled for 3 min and an excess of unlabeled methionine and cysteine was added (final concentration, 2 mM). Immunoprecipitation was carried out using antibodies to GFP and protein A Sepharose, as described previously (48). Immunoprecipitated material was separated by 12% SDS-PAGE and subjected to autoradiography.
RNA extraction and quantitative RT-PCR (qRT-PCR) amplifications.
For extraction of total RNA, 1 ml of culture was mixed with 0.1 ml of ice-cold phenol-ethanol stop solution (5% phenol in ethanol) and cells were collected by centrifugation. RNA was extracted by using a YRB50 kit (RBC Bioscience) according to the manufacturer’s protocol. Following elution, nucleic acid concentrations were determined by spectrophotometry (NanoDrop). Total RNA (1 µg) was reverse transcribed to cDNA using an ImProm-II reverse transcription (RT) kit (Promega, Madison, WI) with random hexamer primers according to the manufacturer’s instructions. Real-time PCR was done with an ABI 7300 machine (Applied Biosystems, Foster City, CA) with Sybr green PCR master mix (Applied Biosystems) in 15-μl volumes containing 0.75 ng cDNA. For amplification, we used the following primers: for lacY, 5′-GGTTGCCAATGCGGTAGGT-3′ (forward) and 5′-GCCAGCTTAAGGCTAAATGCC-3′ (reverse); for lacZ, 5′-ATCTTCCTGAGGCCGATACTGTC-3′ (forward) and 5′-CGTGCATCTGCCAGTTTGAG-3′ (reverse); and for 16S, 5′-CCTGGTAGTCCACGCCGTAA-3′ (forward) and 5′-CTCAAGGGCACAACCTCCAA-3′ (reverse). The expression levels of lacY and lacZ were normalized to the expression level of 16S rRNA.
Analysis of expression of GFP hybrids by colony and gel fluorescence measurements.
Cells were grown overnight in LB supplemented with appropriate antibiotics at 30°C in 96-well plates. Aliquots were spotted on top of nylon membrane (Schleicher and Schuell)-covered LB agar plates supplemented with 0.02% arabinose and incubated overnight at 30°C. The colony-containing membranes were removed from the plate, and their florescence was recorded using Typhoon 9400 (Molecular Dynamics) and analyzed using ImageJ. For in-gel fluorescence measurements, GFP-protein hybrids were separated by SDS-PAGE and the gels were washed (5 times) with an excess of water and scanned using Typhoon. Both nylon membranes and gels were scanned using a fluorescence mode (excitation wavelength, 488 nm; emission wavelength, 520 nm). For the concentration-dependent experiments (Fig. 3D), a linear gradient of arabinose (0 to 0.02%) was constructed as follows. LB agar supplemented with 0.5 mM IPTG and 0.02% arabinose was poured into a tilted plate. After the agar was solidified, the plate was moved to a horizontal plane and LB agar with 0.5 mM IPTG but no arabinose was added.
ACKNOWLEDGMENTS
This work was supported by GIF, the German-Israeli Foundation for Scientific Research and Development, and by the Israel Science Foundation.
Footnotes
Citation Yosef, I., E. S. Bochkareva, and E. Bibi. 2010. Escherichia coli SRP, its protein subunit Ffh, and the Ffh M domain are able to selectively limit membrane protein expression when overexpressed. mBio 1(2):e00020-10. doi:10.1128/mBio.00020-10.
REFERENCES
- 1. Bernstein H. D., Poritz M. A., Strub K., Hoben P. J., Brenner S., Walter P. 1989. Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature 340:482–486 [DOI] [PubMed] [Google Scholar]
- 2. Romisch K., Webb J., Herz J., Prehn S., Frank R., Vingron M., Dobberstein B. 1989. Homology of 54K protein of signal-recognition particle, docking protein and two E. coli proteins with putative GTP-binding domains. Nature 340:478–482 [DOI] [PubMed] [Google Scholar]
- 3. Brown S. 1991. 4.5S RNA: does form predict function? New Biol. 3:430–438 [PubMed] [Google Scholar]
- 4. Herskovits A. A., Bochkareva E. S., Bibi E. 2000. New prospects in studying the bacterial signal recognition particle pathway. Mol. Microbiol. 38:927–939 [DOI] [PubMed] [Google Scholar]
- 5. de Gier J. W., Luirink J. 2001. Biogenesis of inner membrane proteins in Escherichia coli. Mol. Microbiol. 40:314–322 [DOI] [PubMed] [Google Scholar]
- 6. Macfarlane J., Muller M. 1995. The functional integration of a polytopic membrane protein of Escherichia coli is dependent on the bacterial signal-recognition particle. Eur. J. Biochem. 233:766–771 [DOI] [PubMed] [Google Scholar]
- 7. de Gier J. W., Mansournia P., Valent Q. A., Phillips G. J., Luirink J., von Heijne G. 1996. Assembly of a cytoplasmic membrane protein in Escherichia coli is dependent on the signal recognition particle. FEBS Lett. 399:307–309 [DOI] [PubMed] [Google Scholar]
- 8. Seluanov A., Bibi E. 1997. FtsY, the prokaryotic signal recognition particle receptor homologue, is essential for biogenesis of membrane proteins. J. Biol. Chem. 272:2053–2055 [DOI] [PubMed] [Google Scholar]
- 9. Ulbrandt N. D., Newitt J. A., Bernstein H. D. 1997. The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell 88:187–196 [DOI] [PubMed] [Google Scholar]
- 10. Tian H., Boyd D., Beckwith J. 2000. A mutant hunt for defects in membrane protein assembly yields mutations affecting the bacterial signal recognition particle and Sec machinery. Proc. Natl. Acad. Sci. U. S. A. 97:4730–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Park S. K., Jiang F., Dalbey R. E., Phillips G. J. 2002. Functional analysis of the signal recognition particle in Escherichia coli by characterization of a temperature-sensitive ffh mutant. J. Bacteriol. 184:2642–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yosef I., Bochkareva E. S., Adler J., Bibi E. 2010. Membrane protein biogenesis in Ffh- or FtsY-depleted Escherichia coli. PLoS One 5:e9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Scotti P. A., Valent Q. A., Manting E. H., Urbanus M. L., Driessen A. J., Oudega B., Luirink J. 1999. SecA is not required for signal recognition particle-mediated targeting and initial membrane insertion of a nascent inner membrane protein. J. Biol. Chem. 274:29883–29888 [DOI] [PubMed] [Google Scholar]
- 14. Koch H. G., Hengelage T., Neumann-Haefelin C., MacFarlane J., Hoffschulte H. K., Schimz K. L., Mechler B., Müller M. 1999. In vitro studies with purified components reveal signal recognition particle (SRP) and SecA/SecB as constituents of two independent protein-targeting pathways of Escherichia coli. Mol. Biol. Cell 10:2163–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bui N., Strub K. 1999. New insights into signal recognition and elongation arrest activities of the signal recognition particle. Biol. Chem. 380:135–145 [DOI] [PubMed] [Google Scholar]
- 16. Mason N., Ciufo L. F., Brown J. D. 2000. Elongation arrest is a physiologically important function of signal recognition particle. EMBO J. 19:4164–4174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lakkaraju A. K., Mary C., Scherrer A., Johnson A. E., Strub K. 2008. SRP keeps polypeptides translocation-competent by slowing translation to match limiting ER-targeting sites. Cell 133:440–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rapoport T. A., Heinrich R., Walter P., Schulmeister T. 1987. Mathematical modeling of the effects of the signal recognition particle on translation and translocation of proteins across the endoplasmic reticulum membrane. J. Mol. Biol. 195:621–636 [DOI] [PubMed] [Google Scholar]
- 19. Herskovits A. A., Bibi E. 2000. Association of Escherichia coli ribosomes with the inner membrane requires the signal recognition particle receptor but is independent of the signal recognition particle. Proc. Natl. Acad. Sci. U. S. A. 97:4621–4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Avdeeva O. N., Myasnikov A. G., Sergiev P. V., Bogdanov A. A., Brimacombe R., Dontsova O. A. 2002. Construction of the “minimal” SRP that interacts with the translating ribosome but not with specific membrane receptors in Escherichia coli. FEBS Lett. 514:70–73 [DOI] [PubMed] [Google Scholar]
- 21. Wild K., Halic M., Sinning I., Beckmann R. 2004. SRP meets the ribosome. Nat. Struct. Mol. Biol. 11:1049–1053 [DOI] [PubMed] [Google Scholar]
- 22. Luirink J., Sinning I. 2004. SRP-mediated protein targeting: structure and function revisited. Biochim. Biophys. Acta 1694:17–35 . [DOI] [PubMed] [Google Scholar]
- 23. Ullers R. S., Houben E. N., Brunner J., Oudega B., Harms N., Luirink J. 2006. Sequence-specific interactions of nascent Escherichia coli polypeptides with trigger factor and signal recognition particle. J. Biol. Chem. 281:13999–14005 [DOI] [PubMed] [Google Scholar]
- 24. Wild K., Rosendal K. R., Sinning I. 2004. A structural step into the SRP cycle. Mol. Microbiol. 53:357–363 [DOI] [PubMed] [Google Scholar]
- 25. Jensen C. G., Pedersen S. 1994. Concentrations of 4.5S RNA and Ffh protein in Escherichia coli: the stability of Ffh protein is dependent on the concentration of 4.5S RNA. J. Bacteriol. 176:7148–7154 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zheng N., Gierasch L. M. 1997. Domain interactions in E. coli SRP: stabilization of M domain by RNA is required for effective signal sequence modulation of NG domain. Mol. Cell 1:79–87 [DOI] [PubMed] [Google Scholar]
- 27. Batey R. T., Doudna J. A. 2002. Structural and energetic analysis of metal ions essential to SRP signal recognition domain assembly. Biochemistry 41:11703–11710 [DOI] [PubMed] [Google Scholar]
- 28. Grudnik P., Bange G., Sinning I. 2009. Protein targeting by the signal recognition particle. Biol. Chem. 390:775–782 [DOI] [PubMed] [Google Scholar]
- 29. Drew D., Fröderberg L., Baars L., de Gier J. W. 2003. Assembly and overexpression of membrane proteins in Escherichia coli. Biochim. Biophys. Acta 1610:3–10 [DOI] [PubMed] [Google Scholar]
- 30. Bornemann T., Jöckel J., Rodnina M. V., Wintermeyer W. 2008. Signal sequence independent membrane targeting of ribosomes containing short nascent peptides within the exit tunnel. Nat. Struct. Mol. Biol. 15:494–499 [DOI] [PubMed] [Google Scholar]
- 31. Wolin S. L., Walter P. 1989. Signal recognition particle mediates a transient elongation arrest of preprolactin in reticulocyte lysate. J. Cell Biol. 109:2617–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Walter P., Blobel G. 1981. Translocation of proteins across the endoplasmic reticulum. III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J. Cell Biol. 91:557–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Siegel V., Walter P. 1985. Elongation arrest is not a prerequisite for secretory protein translocation across the microsomal membrane. J. Cell Biol. 100:1913–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thomas Y., Bui N., Strub K. 1997. A truncation in the 14 kDa protein of the signal recognition particle leads to tertiary structure changes in the RNA and abolishes the elongation arrest activity of the particle. Nucleic Acids Res. 25:1920–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Raine A., Ullers R., Pavlov M., Luirink J., Wikberg J. E., Ehrenberg M. 2003. Targeting and insertion of heterologous membrane proteins in E. coli. Biochimie 85:659–668 [DOI] [PubMed] [Google Scholar]
- 36. Bernstein H. D., Hyndman J. B. 2001. Physiological basis for conservation of the signal recognition particle targeting pathway in Escherichia coli. J. Bacteriol. 183:2187–2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Koch H. G., Moser M., Muller M. 2003. Signal recognition particle-dependent protein targeting, universal to all kingdoms of life. Rev. Physiol. Biochem. Pharmacol. 146:55–94 [DOI] [PubMed] [Google Scholar]
- 38. Halic M., Becker T., Pool M. R., Spahn C. M., Grassucci R. A., Frank J., Beckmann R. 2004. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature 427:808–814 [DOI] [PubMed] [Google Scholar]
- 39. Halic M., Blau M., Becker T., Mielke T., Pool M. R., Wild K., Sinning I., Beckmann R. 2006. Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature 444:507–511 [DOI] [PubMed] [Google Scholar]
- 40. Berisio R., Schluenzen F., Harms J., Bashan A., Auerbach T., Baram D., Yonath A. 2003. Structural insight into the role of the ribosomal tunnel in cellular regulation. Nat. Struct. Biol. 10:366–370 [DOI] [PubMed] [Google Scholar]
- 41. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yu D., Ellis H. M., Lee E. C., Jenkins N. A., Copeland N. G., Court D. L. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97:5978–5983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Edgar R., Bibi E. 1999. A single membrane-embedded negative charge is critical for recognizing positively charged drugs by the Escherichia coli multidrug resistance protein MdfA. EMBO J. 18:822–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zelazny A., Seluanov A., Cooper A., Bibi E. 1997. The NG domain of the prokaryotic signal recognition particle receptor, FtsY, is fully functional when fused to an unrelated integral membrane polypeptide. Proc. Natl. Acad. Sci. U. S. A. 94:6025–6029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kitagawa M., Ara T., Arifuzzaman M., Ioka-Nakamichi T., Inamoto E., Toyonaga H., Mori H. 2005. A complete set of Escherichia coli open reading frames in mobile plasmids facilitating genetic studies. DNA Res. 12:291–299 [DOI] [PubMed] [Google Scholar]
- 46. Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265–275 [PubMed] [Google Scholar]
- 47. Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 48. Adler J., Bibi E. 2002. Membrane topology of the multidrug transporter MdfA: complementary gene fusion studies reveal a nonessential C-terminal domain. J. Bacteriol. 184:3313–3320 [DOI] [PMC free article] [PubMed] [Google Scholar]