Abstract
We report the application of single molecule-based sequencing technology for high-throughput profiling of histone modifications in mammalian cells. By obtaining over 4 billion bases of sequence from chromatin immunoprecipitated DNA, we generated genome-wide chromatin state maps of mouse embryonic stem cells, neural progenitor cells and embryonic fibroblasts. We find that lysine 4 and lysine 27 tri-methylation effectively discriminate genes that are expressed, poised for expression, or stably repressed, and therefore reflect cell state and lineage potential. Lysine 36 tri-methylation marks primary coding and non-coding transcripts, facilitating gene annotation. Lysine 9 and lysine 20 tri-methylation are detected at satellite, telomeric and active long-terminal repeats, and can spread into proximal unique sequences. Lysine 4 and lysine 9 tri-methylation mark imprinting control regions. Finally, we show that chromatin state can be read in an allele-specific manner by using single nucleotide polymorphisms. This study provides a framework for the application of comprehensive chromatin profiling towards characterization of diverse mammalian cell populations.
Introduction
One of the fundamental mysteries of biology is the basis of cellular state. Although their genomes are essentially identical, cell types in a multicellular organism maintain strikingly different behaviors that persist over extended periods. The most extreme case is lineage-commitment during development, where cells progress from totipotency to pluripotency to terminal differentiation; each step involves establishment of a stable state encoding specific developmental commitments that can be faithfully transmitted to daughter cells. Considerable evidence suggests that cellular state may be closely related to ‘chromatin state’ – that is, modifications to histones and other proteins that package the genome1–3. Accordingly, it would be desirable to construct ‘chromatin state maps’ for a wide variety of cell types, showing the genome-wide distribution of important chromatin modifications.
Chromatin state can be studied by chromatin immunoprecipitation (ChIP), in which an antibody is used to enrich DNA from genomic regions carrying a specific epitope. The major challenge to generating genome-wide chromatin state maps lies in characterizing these enriched regions in a scalable manner. Enrichment at individual loci is commonly assayed by PCR, but this method does not scale efficiently. A more recent approach has been ChIP-chip, in which enriched DNA is hybridized to a microarray4,5. This technique has been successfully used to study large genomic regions. However, ChIP-chip suffers from inherent technical limitations: (i) it requires large amounts (several micrograms) of DNA and thus involves extensive amplification, which introduces bias; (ii) it is subject to cross-hybridization which hinders study of repeated sequences and allelic variants; and (iii) it is currently expensive to study entire mammalian genomes. Given these issues, only a handful of whole-genome ChIP-chip studies in mammals have been reported.
In principle, chromatin could be readily mapped across the genome by sequencing ChIP DNA and identifying regions that are over-represented among these sequences. Importantly, sequence-based mapping could require relatively small quantities of DNA and provide nucleotide-level discrimination of similar sequences, thereby maximizing genome coverage. The major limitation has been that high-resolution mapping requires millions of sequences (Supplementary Note 1). This is cost-prohibitive with traditional technology, even with concatenation of multiple sequence tags6. However, recent advances in single molecule-based sequencing (SMS) technology promise to dramatically increase throughput and decrease costs7. In the approach developed by Illumina/Solexa, DNA molecules are arrayed across a surface, locally amplified, subjected to successive cycles of primer-mediated single-base extension (using fluorescently-labeled reversible terminators) and imaged after each cycle to determine the inserted base. The ‘read length’ is short (25–50 bases), but tens of millions of DNA fragments may be read simultaneously.
Here, we report the development of a method for mapping ChIP enrichment by sequencing (ChIP-Seq) and describe its application to create chromatin-state maps for pluripotent and lineage-committed mouse cells. The resulting data (1) define three broad categories of promoters based on their chromatin state in ES cells, including a larger than anticipated set of ‘bivalent’ promoters; (2) reveal that lineage commitment is accompanied by characteristic chromatin changes at bivalent promoters that parallel changes in gene expression and transcriptional competence; (3) demonstrate the potential for using ChIP for genome-wide annotation of novel promoters and primary transcripts, active transposable elements, imprinting control regions and allele-specific transcription. This study provides a technological framework for comprehensive characterization of chromatin-state across diverse mammalian cell populations.
Genome-wide chromatin state maps
We created genome-wide chromatin state maps for three mouse cell types: ES cells, neural progenitor cells (NPCs)8 and embryonic fibroblasts (MEFs). For each cell type, we prepared and sequenced ChIP DNA samples for some or all of the following features: pan-H3, H3K4me3, H3K9me3, H3K27me3, H3K36me3, H4K20me3 and RNA polymerase II (Supplementary Table 1).
In each case, we sequenced nanogram quantities of DNA fragments (~300 bp) on a Solexa 1GGA sequencer. We obtained an average of 10 million successful reads, consisting of the terminal 27–36 bases of each fragment. The reads were mapped to the genome and used to determine the number of ChIP fragments overlapping any given position (Fig. 1). Enriched intervals were defined as regions where this number exceeded a threshold defined by randomization (see Methods). The full data set consists of 18 chromatin-state maps, containing ~140 million uniquely aligned reads, representing over 4 billion bases of sequence.
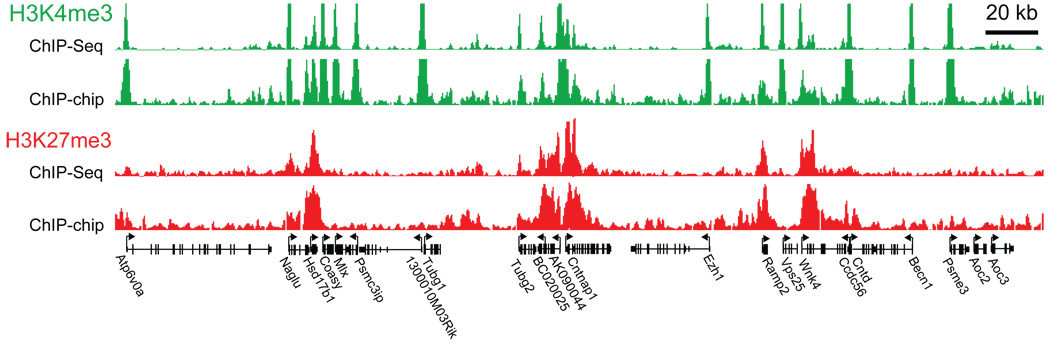
Figure 1. Comparison of ChIP-Seq and ChIP-chip data.
Direct comparison of H3K4me3 (green) and H3K27me3 (red) ChIP data across a 300 kb region in mouse ESCs from independent experiments assayed by SMS (absolute fragment counts) or tiling arrays (log p-values for enrichment relative to whole-cell extracts15).
We validated the chromatin state maps by computational analysis and by comparison to previous methods. ChIP-Seq maps of specific histone modifications show marked enrichment at specific locations in the genome, while the pan-H3 and unenriched samples show relatively uniform distributions (Supplementary Fig. 1–2). The maps show close agreement with our previously reported ChIP-chip data from ~2.5% of the mouse genome9 (Fig. 1). Also, ChIP-PCR assays of 50 sites chosen to represent a range of ChIP-Seq fragment counts showed 98% concordance and a strong, quantitative correlation (Supplementary Fig. 3; Supplementary Table 2).
Promoter state in ES and lineage-committed cells
We began our analysis by studying H3K4me3 and H3K27me3 patterns at known promoters. H3K4me3 is catalyzed by trithorax-group (trxG) proteins and associated with activation, while H3K27me3 is catalyzed by Polycomb-group (PcG) proteins and associated with silencing10,11. Recently, we and others observed that some promoters in ES cells carry both H3K4me3 and H3K27me39,12. We termed this novel combination a ‘bivalent’ chromatin mark and proposed that it serves to poise key developmental genes for lineage-specific activation or repression.
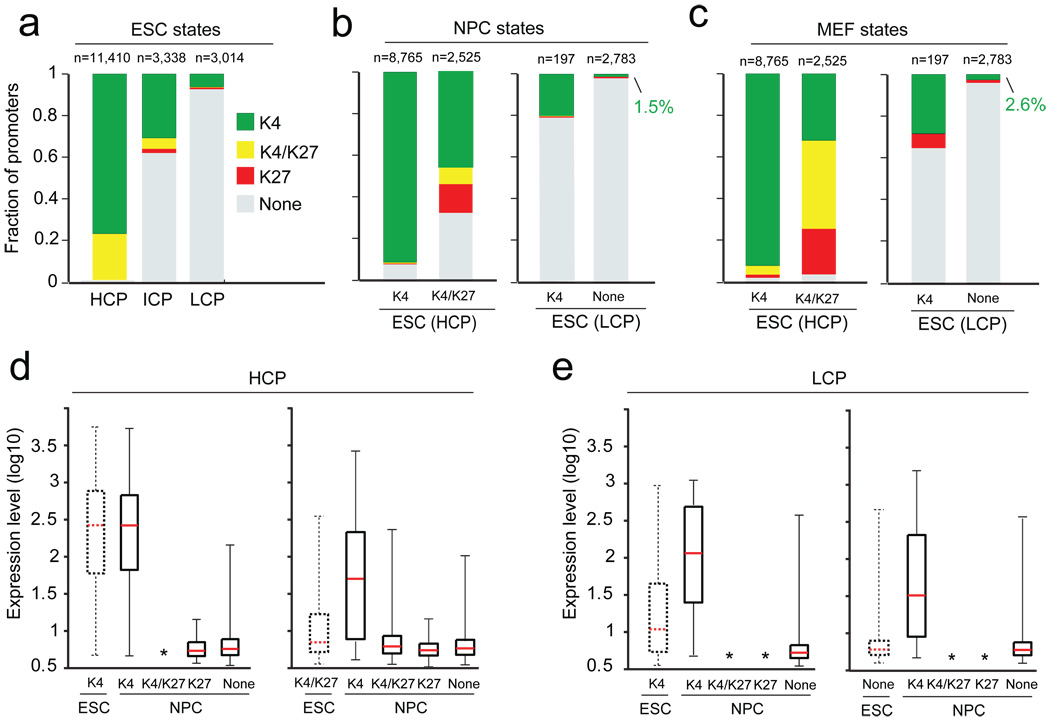
We studied 17,762 promoters inferred from full-length cDNAs (Supplementary Table 3). Mammalian RNA Polymerase II promoters are known to occur in at least two major forms13,14 (Supplementary Fig. 4). CpG-rich promoters are associated with both ubiquitously expressed ‘housekeeping’ genes, and genes with more complex expression patterns, particularly those expressed during embryonic development. CpG-poor promoters are generally associated with highly tissue-specific genes. Accordingly, we divided our analysis to focus on high CpG promoters (HCP; n=11,410) and low CpG promoters (LCP; n=3,014) separately. To ensure a clean separation, we defined a set of intermediate CpG content promoters (ICP; n=3,338); this class shows properties consistent with being a mixture of the two major classes.
High CpG promoters in ES cells
Virtually all HCPs (99%) are associated with intervals of significant H3K4me3 enrichment in ES cells (Fig. 2a). The modified histones are typically confined to a punctate interval of 1–2 kb (Supplementary Fig. 5). As observed previously15,16, there is a strong correlation between the intensity of H3K4me3 and the expression level of the associated genes (Spearman’s ρ=0.67). However, not all promoters associated with H3K4me3 are active.
Figure 2. Histone tri-methylation state predicts expression of HCP and LCP promoters.
(a) Mammalian promoters can be readily classified into sets with high (HCPs), intermediate (ICPs) or low (LCPs) CpG-content. In ES cells (ESCs), virtually all HCPs are marked by H3K4me3, either alone (green) or in combination with H3K27me3 (yellow). In contrast, most LCPs have neither mark (grey). Few promoters are only enriched for H3K27me3 (red). (b) Tri-methylation states of HCPs and LCPs in NPCs (indicated by colors), conditional on their ESC state (indicated below each bar). HCPs marked by H3K4me3 only in ESCs tend to retain this mark. HCPs marked by H3K4me3 and H3K27me3 tend to lose one or both marks, although some remain bivalent. Small, partially overlapping subsets of LCPs are marked by H3K4me3. (c) Tri-methylation states of HCPs and LCPs in MEFs. (d) Changes in expression levels of HCP genes with H3K4me3 alone (left) or also with H3K27me3 (right) upon differentiation to NPCs. Resolution of bivalent promoters to H3K4me3 is associated with increased expression. Boxplots show median (red bar), 25th and 75th percentile expression levels in ESCs. Whiskers show 2.5th and 97.5th percentiles. Asterisks indicate classes with less than 15 genes. (e) Changes in expression levels of LCP genes with H3K4me3 (left) or no mark (right) upon differentiation to NPCs. Gain of H3K4me3 is associated with increased expression.
The chromatin state maps reveal that ~22% of HCPs (n=2,525) are actually bivalent, exhibiting both H3K4me3 and H3K27me3 (Fig. 2a). A minority (n=564) are ‘wide’ bivalent sites in which H3K27me3 extends over a region of at least 5 kb and resemble those described previously9. The majority (n=1,961) are ‘narrow’ bivalent sites, with more punctate H3K27me3, that correspond to many additional PcG target promoters17–19. Bivalent promoters show low activity despite the presence of H3K4me3, suggesting that the repressive effect of PcG activity is generally dominant over the ubiquitous trxG activity (Supplementary Fig. 6; Supplementary Table 4).
The different types of chromatin marks at HCP promoters are closely related to the nature of the associated genes (Supplementary Table 5). Monovalent promoters (H3K4me3) generally regulate genes with ‘housekeeping’ functions including replication and basic metabolism. By contrast, bivalent promoters are associated with genes with more complex expression patterns, including key developmental transcription factors, morphogens and cell surface molecules. In addition, several bivalent promoters appear to regulate transcripts for lineage-specific microRNAs.
High CpG promoters in NPCs and MEFs
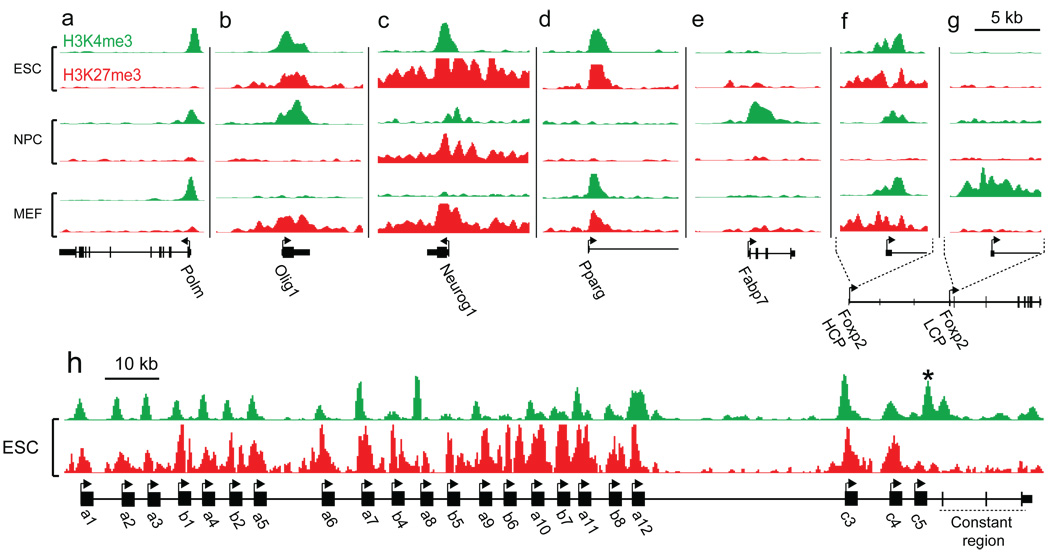
The vast majority of HCPs marked with H3K4me3 alone in ES cells retain this mark both in NPCs and MEFs (92% in each; Fig. 2b,2c,3a). This is consistent with the tendency for this sub-class of promoters to regulate ubiquitous housekeeping genes. A small proportion (~4%) of these promoters have H3K27me3 in MEFs, and are thus bivalent or marked by H3K27me3 alone. This correlates with lower expression levels and may reflect active recruitment of PcG proteins to new genes during differentiation20. An example is the transcription factor gene Sox2, where the promoter is marked by H3K4me3 alone in ES cells and NPCs, but H3K27me3 alone in MEFs. Notably, this locus is flanked by CpG islands with bivalent markings in ES cells (see below), suggesting the locus may be poised for repression upon differentiation.
Figure 3. Cell type-specific chromatin marks at promoters.
(a) Multiple ‘housekeeping genes’, such as DNA Polymerase mu (Polm), are associated with HCPs marked by H3K4me3 in all cell types. (b) The neural transcription factor gene Olig1 (HCP) is bivalent in ESCs, but resolves to H3K4me3 in NPCs and H3K27me3 in MEFs. (c) The neurogenesis transcription factor gene Neurog1 (HCP) remains bivalent upon differentiation to NPCs, but resolves to H3K27me3 in MEFs. (d) The adipogenesis transcription factor gene Ppar-γ (HCP) remains bivalent in MEFs, but loses both marks in NPCs. (e) The neural progenitor marker gene Fabp7 (LCP) is marked by H3K4me3 in NPCs only. (f) The brain and lung expressed transcription factor gene Foxp2 is associated with an HCP that is bivalent in ES cells, but resolves to H3K4me3 in NPCs and remains bivalent in MEFs. (g) Foxp2 also has an LCP marked by H3K4me3 in MEFs only. (h) Multiple, distinct bivalent chromatin marks at the variable region promoters of Pcdh-γ. A promoter proximal to the constant region exons (*) is marked by H3K4me3 only.
The majority of HCPs with bivalent marks in ES cells resolve to a monovalent status in the committed cells. In NPCs, 46% resolve to H3K4me3 only and these genes show increased expression (Fig. 2b,2d,3b). Of the remaining promoters, 14% resolve to H3K27me3 alone and 32% lose both marks, with both outcomes being associated with low levels of expression. Importantly, 8% remain bivalent and these genes also continue to be repressed (Fig. 2b,2d,3c). A somewhat less resolved pattern is seen in MEFs, with 32% marked by H3K4me3 alone, 22% marked by H3K27me3 alone, 3% without both marks, and the remaining (43%) still bivalent (Fig. 2c). The relatively high number of bivalent promoters in MEFs may reflect a less differentiated state and/or heterogeneity in the population.
Distinct regulation of Low CpG Promoters
The LCPs show a strikingly different pattern than the HCPs. Only a small minority (6.5%, n=207) of LCPs have significant H3K4me3 in ES cells and virtually none have H3K27me3 (Fig. 2a). Most of these promoters have lost H3K4me3 in NPCs and MEFs, while a small number of other LCPs (1.5% and 2.6%, respectively) have gained the mark (Fig. 2b,2c,3e). In all three cell types, the expression levels of the associated genes strongly correlate with presence or absence of H3K4me3 (Fig. 2e; Supplementary Fig. 6).
The genes with LCPs marked by H3K4me3 are closely related to tissue-specific functions. In NPCs, they include genes encoding several known markers of neural progenitors in vivo (such as Fabp7, Cp, Gpr56). In MEFs, they include genes encoding extracellular matrix components and growth factors (such as Col3a1, Col6a1, Postn, Aspn, Hgf, Figf), consistent with the mesenchymal origin of these cells (see below).
We conclude that HCPs and LCPs are subject to distinct modes of regulation. In ES cells, all HCPs appear to be targets of trxG activity, and may therefore drive transcription unless actively repressed by PcG proteins. In committed cell types, a subset of HCPs appear to lose the capacity to recruit trxG activity (possibly due to other epigenetic modifications, such as DNA methylation21). In contrast, CpG-poor promoters appear to be inactive by default, independent of repression by PcG proteins, and may instead be selectively activated by cell type- or tissue-specific factors.
Alternative promoter use
We note that genes with alternative promoters may have multiple, distinct chromatin states. An ‘active’ state at any one of these may be sufficient to drive expression. A common situation involves genes with one major HCP and one or more alternative LCPs. An example is the transcription factor Foxp2, which is expressed at moderate levels in both NPCs and MEFs (Fig. 3f,g). The Foxp2 HCP is marked by H3K4me3 in NPCs, but is bivalent in MEFs. However, an alternative LCP is marked by H3K4me3 exclusively in MEFs. The protocadherin-γ (Pcdh-γ) locus is a more extreme case: the N-terminal variable regions of this gene are transcribed from at least 20 different HCPs in neurons22, all of which carry bivalent chromatin marks in ES cells. Pcdh-γ expression is nevertheless detected by microarrays, possibly due to a single promoter in front of the C-terminal constant region marked by H3K4me3 alone (Fig. 3h).
Although only ~10% of the genes analyzed here have more than one known promoter, recent ‘cap-trapping’ studies suggest that alternative promoter use may be substantially more common23. The ability of ChIP-Seq to assess chromatin state at known promoters, as well as to identify novel promoters (see below), should prove valuable in analysis of transcriptional networks.
Promoter state reflects lineage commitment and potential
Given their association with epigenetic memory, we next examined whether the patterns of H3K4me3 and H3K27me3 can reflect developmental potential. Both of the committed cell types studied here have been shown to be multipotent ex vivo. NPCs can be differentiated to glial and neuronal lineages8, while primary MEFs have been differentiated into adipocytes24, chondrocytes25 and osteoblast-like cells26.
Lineage-specific resolution and retention of bivalent marks
We first examined a set of genes involved in in vivo differentiation pathways known to be, at least partially, recapitulated by MEFs, NPCs or neither. These genes all have bivalent promoters in ES cells. We found that their resolution in lineage-committed cells is closely related to their demonstrated developmental potential (Supplementary Table 6):
Genes restricted to regulation or specialized functions in unrelated lineages, such as hematopoietic (Cdx4, PU.1), epithelial (Cncf, Krt2–4), endoderm (Gata6, Pdx1) or germ line (Tenr, Ctcfl), have generally resolved to monovalent H3K27me3 or carry neither mark in both NPCs and MEFs.
Genes related to adipogenesis and chondro/osteogenesis often remain bivalent in MEFs, but not in NPCs. Examples include Ppar-γ, which is a key regulator of apipogenesis, and Sp7, which promotes chondro/osteogenic pathways. Early mesenchymal markers, such as Runx1 and Sox9 resolved to H3K4me3 alone in MEFs.
Genes related to gliogenesis and neurogenesis often resolve to H3K4me3 alone or remain bivalent in NPCs, while resolving to H3K27me3 alone in the MEFs. Gliogenesis and neurogenesis are thought to be mutually opposing pathways27, and we find that genes promoting gliogenesis are more likely to resolve to H3K4me3 in NPCs. Examples include Bmp2 and the miRNA mir-9-3, which promotes glial but inhibits neuronal differentiation28. Several genes known to promote neuronal differentiation, such as Neurog1 and Neurog2, remain bivalent while others, such as Bmp6, appear to resolve to H3K27me3 alone. In our hands, the NPCs differentiate to astrocytes with significantly higher efficiency than to neurons (M. Wernig, unpublished data). The observed chromatin patterns may reflect this gliogenic bias.
Correlation with expression in adult tissues
We next analyzed gene expression in adult tissues with major contributions from neuroectodermal or mesenchymal lineages. We reasoned that if H3K4me3 is generally not restored once lost, then differential loss of H3K4me3 at promoters early in these lineages (as represented by NPCs and MEFs, respectively) might be reflected in differential gene expression patterns in related adult tissues.
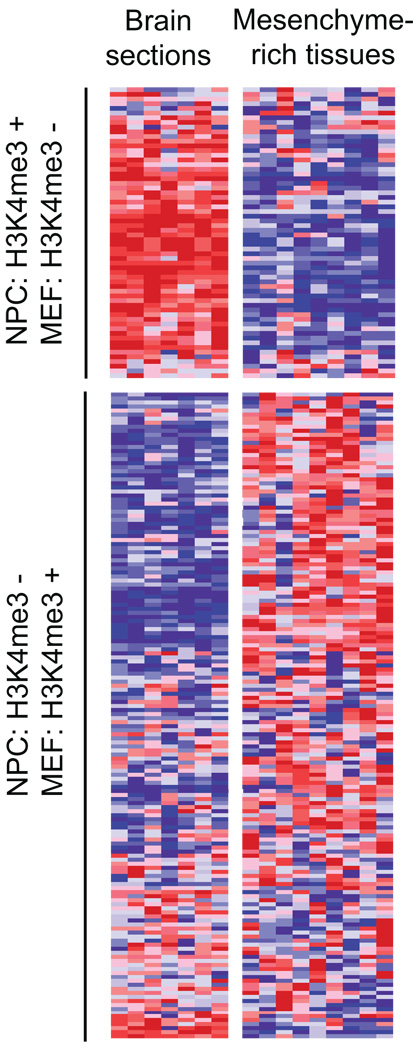
Strikingly, we observed a clear bias in relative expression levels between relevant adult tissues for genes that retain H3K4me3 in NPCs only versus genes that retain H3K4me3 in MEFs only. The former are strongly biased toward higher expression in various brain sections, while the latter are biased towards higher expression in bone, adipose and other mesenchyme-rich tissues (Fig. 4).
Figure 4. Correlation between chromatin state changes and lineage expression.
Relative expression levels across adult mouse brain (frontal and cerebral cortex, substantia nigra, cerebellu, amygdale, hypothalamus, hippocampus) and relatively mesenchyme-rich tissues (bone, white fat, brown fat, trachea, digits, lung, bladder, uterus, umbilical cord) are shown for genes with bivalent chromatin marks in ES cells that retain H3K4me3 in NPCs but lose this mark in MEFs (n=62) or vice versa (n=160). Red, white and blue indicates higher, equal and lower relative expression, respectively.
These analyses are of course limited by alternative promoter usage, the cell models used, and the heterogeneity of the adult tissues. Nonetheless, the data show clear trends that support an important role for retention and resolution of bivalent chromatin in the regulation of hierarchical lineage commitment.
Genome-wide annotation of promoters and primary transcripts
We next considered genome-wide maps of H3K36me3. This mark has been linked to transcriptional elongation and may serve to prevent aberrant initiation within gene bodies29–33. Our chromatin maps reveal a global pattern of H3K36me3 in mammals similar to that previously observed in yeast29.
In all three cell types, H3K36me3 is strongly enriched across the transcribed regions of active genes (Fig. 5a), beginning immediately after the promoter H3K4me3 signal. The level of H3K36me3 is strongly correlated with the level of gene expression (Spearman’s ρ=0.77), although the dynamic range is compressed (1–2 orders of magnitude for H3K36me3 vs 3–4 for expression levels; Supplementary Fig. 7). Genes with bivalent promoters rarely show H3K36me3, consistent with their low expression. Notably, there is essentially no overlap between intervals significantly enriched for H3K36me3 and for H3K27me3, consistent with a role for PcG complexes in the exclusion of polymerases11.
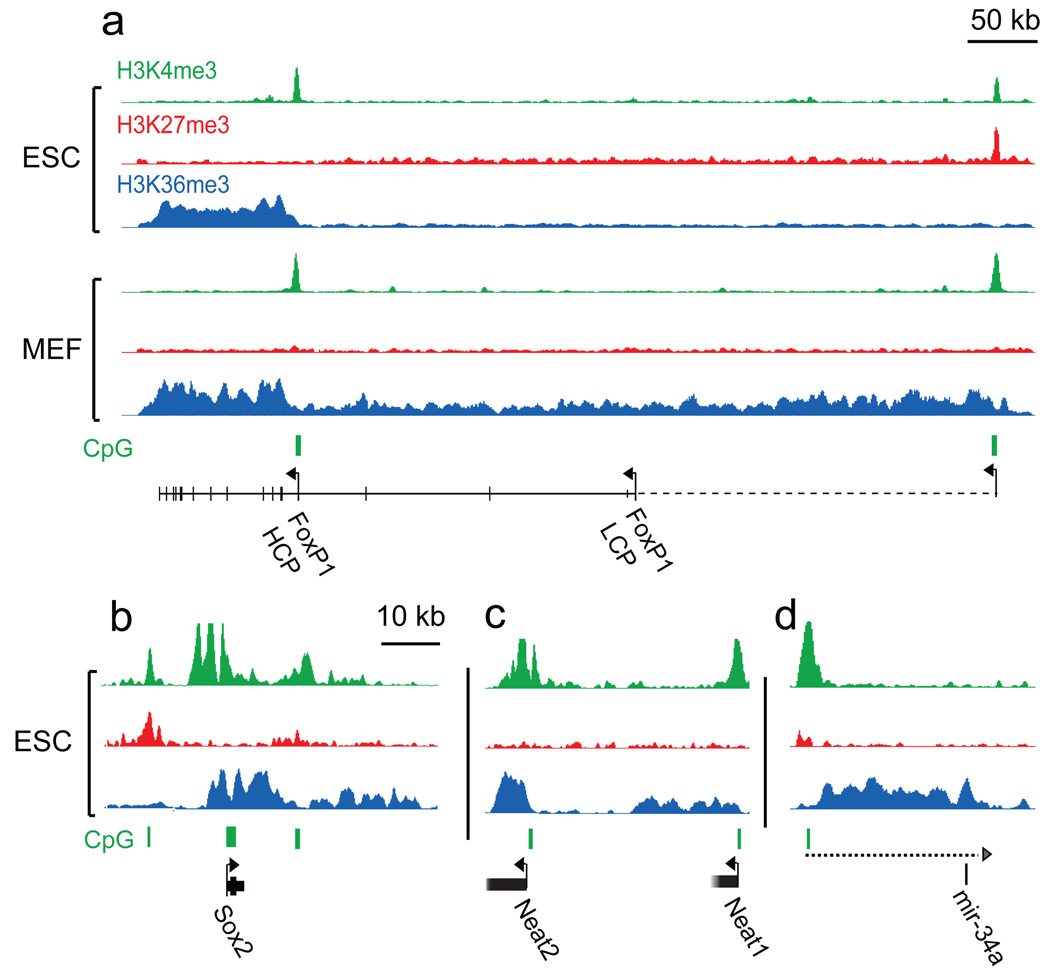
Figure 5. H3K4me3 and H3K36me3 annotate genes and non-coding RNA transcripts.
(a) Foxp1 has two annotated promoters (based on RefSeq and UCSC Known Genes), only one of which shows H3K4me3 in ES cells. The corresponding transcriptional unit is marked by H3K36me3. In MEFs, H3K36me3 extends an additional 500 kb upstream to an H3K4me3 site that appears to reflect an alternate promoter (this site is bivalent in ES cells). (b) H3K36me3 enrichment extends significantly downstream of Sox2. Though highly active in ES cells, Sox2 is flanked by two bivalent CpG islands that may poise it for repression. (c) H3K4me3 and H3K36me3 indicate two highly expressed non-coding RNAs, and (d) the putative primary transcript (dashed line) for a single annotated microRNA.
The vast majority of intervals significantly enriched for H3K36me3 is associated with known genes (~92% in ESCs), but there are at least ~500 additional regions across the genome (median size ~2 kb), with most being adjacent to sites of H3K4me3. Inspection revealed a number of interesting cases, falling into three categories.
The first category corresponds to H3K36me3 that extends significantly upstream from the annotated start of a known gene, often until an H3K4me3 site. These appear to reflect the presence of unannotated alternate promoters. A notable example is the Foxp1 locus. In ES cells, one annotated Foxp1 promoter is marked by H3K4me3 and another CpG-rich region located ~500 kb upstream carries a bivalent mark. In MEFs, this CpG island is marked by H3K4me3 only, and H3K36me3 extends from this site to the 3’ end of Foxp1 (Fig. 5a). Although no transcript extending across this entire region has been reported in mouse, the orthologous position in human has been shown to act as a promoter for the orthologous gene. The ChIP-Seq data contain many other examples where the combination of H3K36me3 and H3K4me3 appear to reveal novel promoters.
The second category corresponds to H3K36me3 that extends significantly downstream of a known gene. An example is the Sox2 locus, which encodes a pluripotency-associated transcription factor that also functions during neural development. In ES cells, Sox2 has an unusually large region of H3K4me3 (>20 kb) accompanied by H3K36me3 extending far beyond the annotated 3’-end (>15 kb); non-coding transcription throughout the locus has been noted previously34 and may serve a regulatory role (Fig. 5b).
The third category appears to reflect transcription of non-coding RNA genes. For example, two regions with H3K36me3 and adjacent H3K4me3 correspond to recently discovered nuclear transcripts with possible functions in mRNA processing35 (Fig. 5c). In addition, a number of these presumptive transcriptional units overlap microRNAs (Fig. 5d). A striking example is a >200 kb interval within the Dlk1-Dio3 imprinted locus (Fig. 6a). This region harbors over 40 non-coding RNAs, including clusters of microRNAs and small nucleolar RNAs36. The ChIP-Seq data suggest that the entire region is transcribed as a single unit that initiates at an H3K4me3 marked HCP.
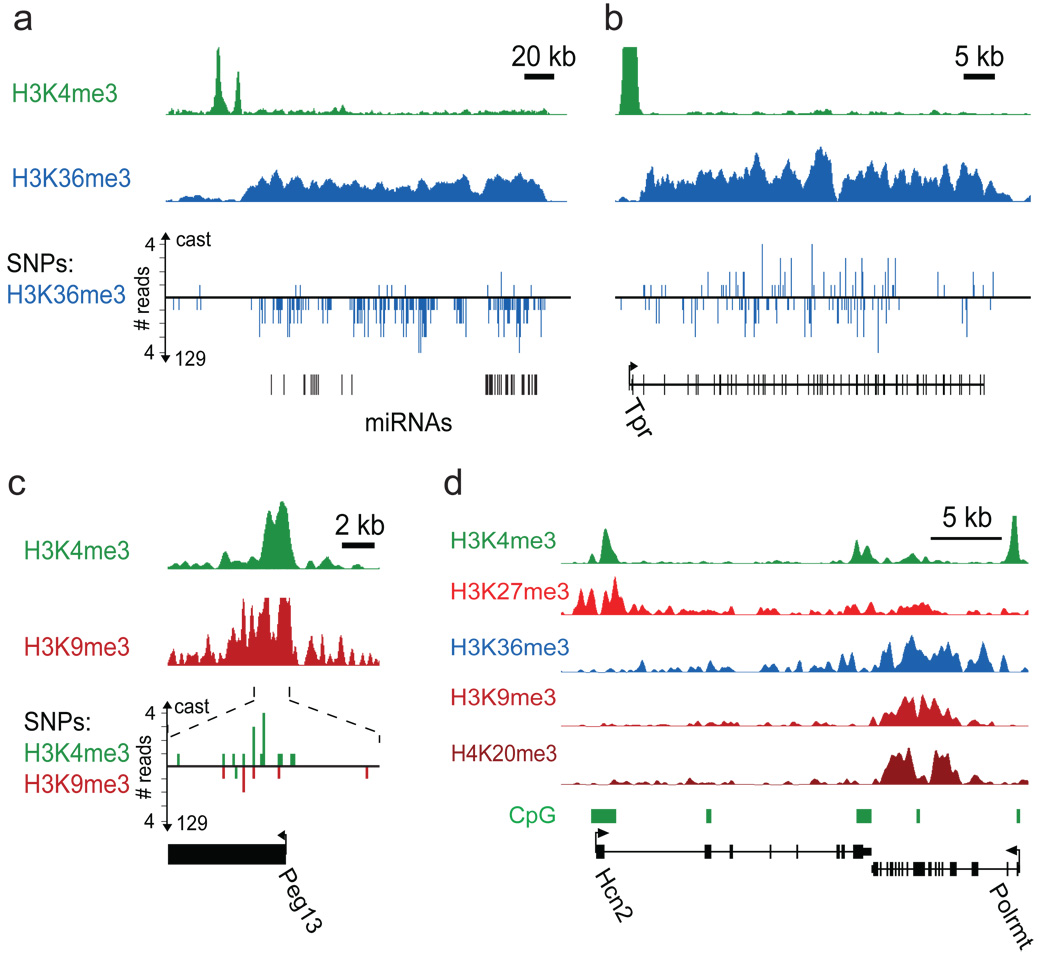
Figure 6. Allele-specific histone methylation and genic H3K9me3/H4K20me3.
(a) H3K4me3 and H3K36me3 indicate a primary microRNA transcript in the Dlk1-Dio3 locus. The allele-specificity of this transcript is read out using ChIP-Seq data for hybrid ES cells and a SNP catalogue. The H3K36me3 reads overwhelmingly correspond to maternal 129 alleles, consistent with the known maternal expression of these microRNAs36. (b) In contrast, a non-imprinted transcript shows roughly equal proportions of reads assigned to 129 and castaneus alleles. (c) Peg13 is marked by H3K4me3 and H3K9me3 in ES cells; 19 of 21 H3K4me3 reads correspond to the paternal castaneus allele, while 6 of 6 H3K9me3 reads correspond to the maternal 129 allele, consistent with paternal expression of this gene. (d) H3K9 me3 and H4K20me3 enrichment evident at the Polrmt gene may reflect transcriptional interference due to antisense transcription from the 3’ UTR CpG island of Hcn2 (see text).
These findings suggest that genome-wide maps of H3K4me3 and H3K36me3 may provide a general tool for defining novel transcription units. The capacity to define the origins and extents of primary transcripts will be of particular value for characterizing the regulation of microRNAs and other non-coding RNAs that are rapidly processed from long precursors37. Finally, the relatively narrow dynamic range of H3K36me3 may offer advantages over RNA-based approaches in assessing gene expression and defining cellular states.
H3K9 and H4K20 tri-methylation associated with specific repetitive elements
We next studied H3K9me3 and H4K20me3, both of which have been associated with silencing of centromeres, transposons and tandem repeats38–40. We sought first to assess the relative enrichments of H3K9me3 and H4K20me3 across different types of repetitive elements by aligning ChIP-Seq reads directly to consensus sequences for various repeat families (~40 million reads could be aligned this way).
H3K9me3 and H4K20me3 show nearly identical patterns of enrichment in ES cells. The strongest enrichments are observed for telomeric, satellite, and long terminal repeats (LTRs). The LTR signal primarily reflects enrichment of intracisternal A-particles (IAP), early transposon (ETn) elements, and the LTRIS sub-family (Supplementary Fig. 8).
IAP and ETn elements are active in murine ES cells and produce double-stranded RNAs41,42. RNA has also been implicated in maintaining satellite and telomeric heterochromatin38. Hence, these enrichment data are consistent with a global role for RNA in targeting repressive chromatin marks in mammalian ES cells, analogous to that observed in lower eukaryotes38,39.
We next examined the distributions of H3K9me3 and H4K20me3 across unique sequence in the mouse genome. We identified ~1800 H3K9me3 sites (median size ~300 bp) in ES cells, with the vast majority also showing H4K20me3. Fully 78% of the sites lie within two kb of a satellite repeat or LTR (primarily IAP and ETn elements). This suggests that repressive marks are capable of spreading from repeat insertions and could potentially regulate proximal unique sequence.
Recent studies have described a handful of active genes with H3K9me3 and H4K20me3, raising the possibility that these ‘repressive’ marks also function in transcriptional activation31,32. One-third of the ~1800 H3K9me3 enriched sites reside within an annotated gene, which is roughly the proportion expected by chance. However, H3K9me3 sites that are larger and/or more distant from LTRs are more likely to occur within genes (Supplementary Fig. 9). The largest genic site in ES cells (~6 kb) coincides with the Polrmt gene (Fig. 6d). This case is notable because the downstream gene (Hcn2) is convergent and contains a CpG island at its 3’ end. Transcription from 3’ promoters has been proposed as a potential mechanism of transcriptional interference by producing antisense transcripts23. This example may therefore reflect a link between transcriptional interference and H3K9me3, as has been suggested for a few other mammalian loci43,44. Our results thus confirm the presence of H3K9me3 within a subset of genes, although the functional implications remain to be elucidated.
Imprinting control regions show overlapping H3K4 and H3K9 tri-methylation
We next studied chromatin marks associated with imprinting. This epigenetic process typically involves allele-specific DNA methylation of CpG-rich imprinting control regions (ICRs)45. Several reports have also described allele-specific chromatin modification at a handful of ICRs, with H3K9me3 and H4K20me3 on the DNA methylated allele and H3K4me3 on the opposite allele46,47.
We searched for regions showing overlapping H3K9me3 and H3K4me3 in ES cells. Strikingly, 13 of the top 20 sites, as ranked by enrichment of the two marks, are located within known imprinted regions, coincident with ICRs or imprinted gene promoters. An example is the Peg13 promoter (Fig. 6c). Conversely, of the ~20 known and putative autosomal imprinted loci that contain ICRs, 17 have at least one with the overlapping chromatin marks (Supplementary Table 5). We conclude that overlapping H3K9me3 and H3K4me3 is a common signature of ICRs in ES cells.
Allele-specific histone methylation
To explore the feasibility of inferring allele-specific chromatin states, we constructed chromatin-state maps in male ES cells derived from a more distant cross (129 (maternal) × M. castaneus (paternal)), and used a catalog of ~3.5 million SNPs to assign ChIP-Seq reads to one of the two parental alleles.
As a positive control, we first compared results for chromosome X and the autosomes for reads derived by H3K4me3 ChIP. Virtually all (97%) of ~3700 informative reads on chromosome X, and roughly half (57%) of the 178,000 informative reads on the autosomes, were assigned to the 129 strain. These proportions correspond roughly to the expected 100% and 50%.
We then examined the allelic distribution at overlapping H3K4me3 and H3K9me3 sites coincident with putative ICRs (see above). Six of the ICRs had enough reads (≥10) containing SNPs to assess allelic bias. In every case, the SNPs showed significant bias in the expected direction (p<0.02; Fig. 6c; Supplementary Table 7).
We applied the same approach to search for allelic imbalance in intervals with significant H3K36me3 enrichment, which would predict differential transcription of the two alleles. A striking interval corresponds to a microRNA cluster within the Dlk1-Dio3 locus known to be imprinted in the embryo proper36 (Fig. 6a–b). Of the additional imprinted genes with H3K36me3 enrichment, four (Snrpn, Grb10, Impact, Peg3) had enough reads containing SNPs to assess allelic bias. In every case, the data showed significant bias in the expected direction (p<0.02). The data also revealed novel instances of allele-specific transcription. For example, a transcript of unknown function (BC054101), first identified in trophoblast stem cells48, showed highly significant maternal bias for H3K36me3, as well as H3K4me3 (p<10−15; Supplementary Fig. 10).
The results suggest that, with sufficiently deep coverage and dense SNP maps, ChIP-Seq will provide a powerful means for identifying allele-specific chromatin modifications. With data from reciprocal crosses, it should be possible to discriminate novel cases of imprinting from strain-specific differences.
Conclusion
Genome-wide chromatin-state maps provide a rich source of information about cellular state, yielding insights beyond what is typically obtained by RNA expression profiling. Analysis of H3K4me3 and H3K36me3 allows recognition of promoters together with their complete transcription units. This should help define alternative promoters and their usage in specific cell types; identify the structure of genes encoding non-coding RNAs; detect gene expression (given the narrower dynamic range); and detect detecting allele-specific transcription. In addition, analysis of H3K9me3 and H4K20me3 should facilitate study of heterochromatin, spreading and imprinting mechanisms.
Most interestingly, analysis of H3K4me3 and H3K27me3 provides a rich description of cellular state. Our results suggest that promoters may be classified as active, repressed or poised for alternative developmental fates. Conceivably, chromatin state at key regulatory genes may suffice to describe developmental commitment and potential.
Given the technical features of ChIP-Seq (high throughput, low cost and input requirement), it is now appropriate to contemplate projects to generate catalogs of chromatin-state maps representing a wide range of human and mouse cell types. These should include varied developmental stages and lineages, from totipotent to terminally differentiated, with the aim of precisely defining cellular states at the epigenetic level and observing how they change over the course of normal development. Chromatin-state maps should also be systematically cataloged from situations of abnormal development. Cancer cells are the most obvious targets, as they are frequently associated with epigenetic defects and many appear to have acquired characteristics of earlier developmental stages. A comprehensive public database of chromatin-state maps would be a valuable resource for the scientific community.
Methods Summary
Murine V6.5 ES cells (129SvJae x C57BL/6; male), hybrid ES cells (129SvJae x M. castaneus F1; male) and NPCs were cultured as described8,9. Primary MEFs (129SvJae x C57BL/6; male) were obtained at d13.5.
ChIP experiments were carried out as described15. Sequencing libraries were generated from 1–10 ng of ChIP DNA by adapter ligation, gel purification and 18 cycles of PCR. Sequencing was carried out using the Illumina/Solexa system according to the manufacturer’s specifications.
Reads were aligned to the reference genome and the fragment count at any given position (25 bp resolution) was estimated as the number of uniquely aligned reads oriented towards it and within 300 bp. Enriched intervals were identified by comparison of the mean fragment count in 1kb windows against a sample-specific expected distribution estimated by randomization (H3K4me3, H3K27me3), or using a supervised Hidden Markov Model (H3K36me3, H3K9me3, H4K20me3).
Promoters were inferred from full-length mouse RefSeqs. HCPs contain a 500 bp interval within −0.5 kb to +2 kb with a (G+C)-fraction ≥ 0.55 and a CpG observed to expected ratio (O/E) ≥ 0.6. LCPs contain no 500 bp interval with CpG O/E ≥ 0.4. Chromatin states of promoters were determined by overlap with H3K4me3 and H3K27me3 enriched intervals. Correlations with expression levels were calculated from the mean fragment count over each promoter or transcript.
ESC, NPC and MEF expression data were generated using GeneChip arrays (Affymetrix) and GenePattern [www.broad.mit.edu/cancer/software/]. Expression data for adult tissues were downloaded from Novartis [expression.gnf.org].
Repeat class enrichments were determined by aligning reads to consensus sequences [www.girinst.org]. Mouse SNP maps were obtained from Perlegen [mouse.perlegen.com]. Allele specific bias was evaluated by a binomial test of the null hypothesis that ChIP fragments were drawn uniformly from both alleles.
Methods
Cell Culture
V6.5 murine ES cells (genotype 129SvJae x C57BL/6; male; passages 10–15) and hybrid murine ES cells (genotype 129SvJae x M. castaneus F1; male; passages 4–6) were cultivated in 5% CO2 at 37° on irradiated MEFs in DMEM containing 15% FCS, leukemia-inhibiting factor, penicillin/streptomycin, L-glutamine, nonessential amino acids and 2-mercaptoethanol. Cells were subject to at least two to three passages on 0.2% gelatin under feeder-free conditions to exclude feeder contamination. V6.5 ES cells were differentiated into neural precursor cells (NPCs) through embryoid body formation for 4 days and selection in ITSFn media for 5–7 days, and maintained in FGF2 and EGF2 (R&D Systems) as described8. The cells uniformly express nestin and Sox2 and can differentiate into neurons, astrocytes and oligodendrocytes. Mouse embryonic fibroblasts (genotype 129SvJae x C57BL/6; male; d13.5; passages 4–6), were grown in DMEM with 10% fetal bovine serum and penicillin/streptomycin at 37°, 5% CO2.
Chromatin Immunoprecipitation (ChIP)
ChIP experiments were carried out as described in Bernstein et al., 2005 and at www.upstate.com. Briefly, chromatin from fixed cells was fragmented to a size range of 200 to 700 bases with a Branson 250 Sonifier or a Diagenode Bioruptor. Solubilized chromatin was immunoprecipitated with antibody against H3K4me3 (Abcam #8580), H3K9me3 (Abcam #8898), H3K27me3 (Upstate #07-449), H3K36me3 (Abcam #9050), H4K20me3 (Upstate #07-463), pan-H3 (Abcam #1791) or RNA polymerase II (Covance MMS-126R). Antibody-chromatin complexes were pulled-down using Protein A-sepharose (or anti-IgM conjugated agarose for RNA polymerase II), washed and then eluted. After cross-link reversal and Proteinase K treatment, immunoprecipitated DNA was extracted with phenol-chloroform, ethanol precipitated, and treated with RNase. ChIP DNA was quantified using PicoGreen.
Library Preparation and Solexa sequencing
One to ten nanograms of ChIP DNA (or unenriched whole cell extract) were prepared for Solexa sequencing as follows: DNA fragments were repaired to blunt ends by T4 DNA polymerase and phosphorylated with T4 Polynucleotide kinase using the END-IT kit (Epicentre). Then, a single ‘A’ base was added to 3’ ends with Klenow (3’→5’ exo−, 0.3 U/µl). Double-stranded Solexa adaptors (75 bp with a ‘T’ overhang) were ligated to the fragments with DNA ligase (0.05 U/µl). Ligation products between 275 and 700 base pairs were gel purified on 2% agarose to remove unligated adaptors, and subjected to 18 PCR cycles. Completed libraries were quantified with PicoGreen.
DNA sequencing was carried out using Illumina’s Solexa sequencing system. Cluster amplification, linearization, blocking and sequencing primer reagents were provided in the Solexa Cluster Amplification kits and were used according to the manufacturer’s specifications as described here. To obtain single strand templates, the sample prep was first denatured in NaOH (0.1N final concentration) and diluted in Solexa hybridization buffer (4°C) to a final concentration of either 2 or 4pM. Sample loading was carried out as follows. A template sample was loaded into each lane of a Solexa flowcell mounted on a Solexa cluster station on which all subsequent steps were performed. The temperature was increased to 95°C for 1min and slowly decreased to 40°C to allow for annealing onto complementary adapter oligos on the flowcell surface. Cluster formation was then carried out as follows. The template strands were extended with Taq polymerase (0.25U/ul) to generate a fixed copy of the template on the flowcell. The samples were then denatured with formamide (Sigma-Aldrich, F-5786, >99.5% (GC)) and washed (Solexa Wash buffer) to remove the original captured template leaving behind a single stranded template ready for amplification. Clusters were then amplified under isothermal conditions (60°C) for 30 cycles using Solexa Amplification mix containing Bst I DNA polymerase (0.08U/ul). After each amplification cycle, the templates were denatured with formamide (as above). Fresh amplification mix was added after each denaturation step. Following amplification, the clusters were linearized with Solexa Linearization mix, and any unextended flowcell surface capture oligos were blocked with ddNTPs (2.4uM mix in the presence of 0.25U/ul terminal transferase). The linearized clusters were then denatured (0.1N NaOH) to remove and wash away the linearized strands. The single-stranded templates in the cluster were then annealed with the Solexa sequencing primer (10uM). The flowcells were removed from the cluster station and then transferred onto the 1G Genetic Analyzer which performed the sequencing according to its own standard protocols. We followed the protocol without any modifications.
Read alignment and generation of density maps and modified intervals
Sequence reads from each ChIP library are compiled, post-processed and aligned to the reference genome sequence using a general purpose computational pipeline. We first pre-compute a table that associates each possible 12-mer with all of its occurrences in the reference genome. Then, for each k-bp read, we scan both it and its reverse complement, and for each of its constituent 12-mers, we find each potential start point on the reference genome, and then compute the number of mismatches in the corresponding alignment. These computations are dynamically terminated so that only "unique" alignments are reported, according to the following rule: if an alignment A has only x mismatches, and if there is no alternative alignment having ≤ x + 2 mismatches, then we call A unique. To minimize the risk of amplification bias, only one read was kept if multiple reads aligned to the same start point.
For each ChIP (or control) experiment, we next estimate the number of end-sequenced ChIP fragments that overlap any given nucleotide position in the reference genome (here, at 25-bp resolution). For each position, we count the number of aligned reads that are oriented towards it and closer than the average length of a library fragment (~300 bp).
To identify the portion of the mouse genome that can be interrogated with SMS reads of a given length (k) and alignment stringency, we aligned every k-mer that occurs in the reference sequence (mm8) using the same pipeline as for SMS reads. Nucleotide positions in the reference genome where less than 50% of the 200 flanking k-mers on each side had “unique” alignments, were masked as repetitive and disregarded from further analysis (<28% of the genome). Although we analyzed reads spanning 27–36 bp, all data were conservatively masked at k=27.
We identified genomic intervals enriched with a specific chromatin mark from the mean fragment count in 1kb sliding windows. To account for varying read numbers and lengths, we generated sample-specific expected distributions of fragment counts under the null hypothesis of no enrichment by moving each aligned read to a randomly chosen, “unique” position on the same chromosome. Nominal p-values for enrichment at a particular position were obtained by comparison to a randomized version of the same dataset (due to the large number of reads, multiple randomizations gave identical results). Genome-wide maps of enriched sites were created by identification of windows where the nominal p-value fell below 10−5, and merging any enriched windows that were less than 1 kb apart, into continuous intervals. To improve sensitivity to the more diffuse enrichment observed from H3K9me3 and H4K20me3 near repetitive regions and from H3K36me3 across large transcripts, we also developed a Hidden Markov Model (HMM) to segment the reference genome into ‘enriched’ and ‘unenriched’ intervals (Koche, R. in preparation). The observed fragment densities were discretized to four categories, in a sample dependent manner (‘masked, ‘sub-threshold’, ‘near-threshold’ and ‘above threshold’). Emission and transition probabilities were fitted using supervised learning on limited intervals (~10 Mb total) chosen to reflect diverse chromatin landscapes, and the resultant models were applied genome-wide.
Validation of ChIP-Seq by comparison to ChIP-chip and real-time PCR
ChIP-Seq data for H3K4me3 and H3K27me3 in ES cells were compared to published ChIP-chip profiles across ~2% of the mouse genome9. Significantly enriched sites in the ChIP-chip data were defined using a previously validated p-value threshold of 10−4, and compared to the ChIP-Seq sites. In addition, a set of 50 PCR primer pairs (Supplementary Table 2) was designed to amplify 100–140 bp fragments from genomic regions showing a wide range of signal for H3K4me3 and H3K27me3 by ChIP-Seq. Real-time PCR was carried out using Quantitect SYBR green PCR mix (Qiagen) on a 7000 ABI detection system, using 0.25 ng ChIP or WCE DNA as template. Fold-enrichments reflect two independent ChIP assays, each evaluated in duplicate by real-time PCR.
Promoter classification and definition of gene and transcript intervals
The analyzed promoters were based on transcription start sites inferred from full-length mouse RefSeqs (downloaded from the UCSC Genome Brower April 02, 2007). Promoters containing a 500 bp interval within −0.5 kb to +2 kb with a (G+C)-fraction ≥ 0.55 and a CpG observed to expected ratio (O/E) ≥ 0.6 were classified as HCPs. Promoters containing no 500 bp interval with CpG O/E ≥ 0.4 were classified as LCPs. The remainder were classified as ICPs. The chromatin states of promoters were determined by overlap with cell type specific H3K4me3 and H3K27me3 intervals. For comparison with expression levels, the chromatin states of genes with more than one known promoter were classified according to the most ‘active’ mark (i. e. a gene with an H3K4me3 marked promoter and a bivalent promoter, would be classified as ‘H3K4me3’). Correlation between H3K4me3 enrichment and expression levels was calculated from the mean fragment density over promoter from −0.5 kb to +1 kb. Correlation between H3K36 me3 and expression levels was calculated from the mean fragment density over each RefSeq transcript.
Expression data
RNA expression data for ES cells, NPCs and MEFs were generated from polyA RNA using GeneChip Mouse Genome 430 2.0 Arrays (Affymetrix). Expression data for adult tissues were downloaded from the Novartis Gene Expression Atlas at expression.gnf.org. Pre-processing, normalization (GC-RMA) and hierarchical clustering (Pearson, log-transformed, row-centered values) were performed using GenePattern [www.broad.mit.edu/cancer/software/].
Analysis of repetitive elements
Chromatin state at repetitive elements was evaluated by aligning SMS reads directly to a library of repetitive element consensus sequences [http://www.girinst.org]. The proportion of reads aligning to each class was calculated for H3K9me3 and H4K20me3, and enrichment determined by comparison to WCE and pan-H3. We also applied an orthogonal approach based on HMM intervals of H3K9me3 in unique sequence (see above). For each repetitive element type or class, we calculated the number of occurrences within 1 kb of a unique H3K9me3 site, controlling against a set of randomly placed sites of the same length distribution.
Allele-specific histone methylation
Mouse SNP between the 129 and M. castaneus strains were obtained from Perlegen at mouse.perlegen.com. Allele specific bias was evaluated by a binomial test of the null hypothesis that ChIP fragments were drawn uniformly from both alleles.(H3K4me3 and H3K9me3 reads were pooled prior to the test, see Supplementary Table 7). We note that the 129 strain is closer to the B6-derived reference genome, and this may cause a slight bias towards assigning aligned reads to this strain. To minimize this bias, aligned reads were kept for analysis if no alternative alignment had the same number of mismatches to the reference sequence.
Supplementary Material
Acknowledgements
We thank S. Fisher, M. Kellis, B. Birren and M. Zody for technical assistance and constructive discussions. We acknowledge L. Zagachin in the MGH Nucleic Acid Quantitation core for assistance with real-time PCR. E.M. was supported by an institutional training grant from NIH (T32CA09216). M.W. was supported by fellowships from the Human Frontiers Science Organization Program and the Ellison Foundation. This research was supported by funds from the National Human Genome Research Institute, the National Cancer Institute, the Burroughs Wellcome Fund, Massachusetts General Hospital, and the Broad Institute of MIT and Harvard.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
Author Information
All analyzed data sets can be obtained from www.broad.mit.edu/seq_platform/chip/. Microarray data have been submitted to the GEO repository under accession number GSE8024. Reprints and permissions information is available from www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 3.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Buck MJ, Lieb JD. ChIP-chip: considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics. 2004;83:349–360. doi: 10.1016/j.ygeno.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Mockler TC, et al. Applications of DNA tiling arrays for whole-genome analysis. Genomics. 2005;85:1–15. doi: 10.1016/j.ygeno.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Roh TY, Cuddapah S, Zhao K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 2005;19:542–552. doi: 10.1101/gad.1272505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Service RF. Gene sequencing. The race for the $1000 genome. Science. 2006;311:1544–1546. doi: 10.1126/science.311.5767.1544. [DOI] [PubMed] [Google Scholar]
- 8.Conti L, et al. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005;3:e283. doi: 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 10.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 11.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 12.Azuara V, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 13.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weber M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 15.Bernstein BE, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Kim TH, et al. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006 doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 18.Lee TI, et al. Control of developmental regulators by polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Squazzo SL, et al. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res. 2006;16:890–900. doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The Polycomb Group protein Suz12 is required for Embryonic Stem Cell differentiation. Mol Cell Biol. 2007 doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 22.Wang X, Su H, Bradley A. Molecular mechanisms governing Pcdh-gamma gene expression: evidence for a multiple promoter and cis-alternative splicing model. Genes Dev. 2002;16:1890–1905. doi: 10.1101/gad.1004802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 24.Alexander DL, Ganem LG, Fernandez-Salguero P, Gonzalez F, Jefcoate CR. Aryl-hydrocarbon receptor is an inhibitory regulator of lipid synthesis and of commitment to adipogenesis. J Cell Sci. 1998;111(Pt 22):3311–3322. doi: 10.1242/jcs.111.22.3311. [DOI] [PubMed] [Google Scholar]
- 25.Lengner CJ, et al. Primary mouse embryonic fibroblasts: a model of mesenchymal cartilage formation. J Cell Physiol. 2004;200:327–333. doi: 10.1002/jcp.20118. [DOI] [PubMed] [Google Scholar]
- 26.Garreta E, Genove E, Borros S, Semino CE. Osteogenic differentiation of mouse embryonic stem cells and mouse embryonic fibroblasts in a three-dimensional self-assembling peptide scaffold. Tissue Eng. 2006;12:2215–2227. doi: 10.1089/ten.2006.12.2215. [DOI] [PubMed] [Google Scholar]
- 27.Doetsch F. The glial identity of neural stem cells. Nat Neurosci. 2003;6:1127–1134. doi: 10.1038/nn1144. [DOI] [PubMed] [Google Scholar]
- 28.Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24:857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rao B, Shibata Y, Strahl BD, Lieb JD. Dimethylation of histone H3 at lysine 36 demarcates regulatory and nonregulatory chromatin genome-wide. Mol Cell Biol. 2005;25:9447–9459. doi: 10.1128/MCB.25.21.9447-9459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bannister AJ, et al. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem. 2005;280:17732–17736. doi: 10.1074/jbc.M500796200. [DOI] [PubMed] [Google Scholar]
- 31.Kim A, Kiefer CM, Dean A. Distinctive signatures of histone methylation in transcribed coding and noncoding human beta-globin sequences. Mol Cell Biol. 2007;27:1271–1279. doi: 10.1128/MCB.01684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vakoc CR, Sachdeva MM, Wang H, Blobel GA. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol. 2006;26:9185–9195. doi: 10.1128/MCB.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 34.Fantes J, et al. Mutations in SOX2 cause anophthalmia. Nat Genet. 2003;33:461–463. doi: 10.1038/ng1120. [DOI] [PubMed] [Google Scholar]
- 35.Hutchinson JN, et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seitz H, et al. A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res. 2004;14:1741–1748. doi: 10.1101/gr.2743304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cullen BR. Transcription and processing of human microRNA precursors. Mol Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 38.Zaratiegui M, Irvine DV, Martienssen RA. Noncoding RNAs and gene silencing. Cell. 2007;128:763–776. doi: 10.1016/j.cell.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 39.Verdel A, Moazed D. RNAi-directed assembly of heterochromatin in fission yeast. FEBS Lett. 2005;579:5872–5878. doi: 10.1016/j.febslet.2005.08.083. [DOI] [PubMed] [Google Scholar]
- 40.Martens JH, et al. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. Embo J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baust C, et al. Structure and expression of mobile ETnII retroelements and their coding-competent MusD relatives in the mouse. J Virol. 2003;77:11448–11458. doi: 10.1128/JVI.77.21.11448-11458.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Svoboda P, et al. RNAi and expression of retrotransposons MuERV-L and IAP in preimplantation mouse embryos. Dev Biol. 2004;269:276–285. doi: 10.1016/j.ydbio.2004.01.028. [DOI] [PubMed] [Google Scholar]
- 43.Cho DH, et al. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol Cell. 2005;20:483–489. doi: 10.1016/j.molcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 44.Feng YQ, et al. The human beta-globin locus control region can silence as well as activate gene expression. Mol Cell Biol. 2005;25:3864–3874. doi: 10.1128/MCB.25.10.3864-3874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol. 2007 doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 46.Delaval K, et al. Differential histone modifications mark mouse imprinting control regions during spermatogenesis. Embo J. 2007;26:720–729. doi: 10.1038/sj.emboj.7601513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feil R, Berger F. Convergent evolution of genomic imprinting in plants and mammals. Trends Genet. 2007;23:192–199. doi: 10.1016/j.tig.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 48.Strausberg RL, et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci U S A. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.