Abstract
Alopecia areata (AA) is among the most highly prevalent human autoimmune diseases, leading to disfiguring hair loss due to the collapse of immune privilege of the hair follicle and subsequent autoimmune attack1,2. The genetic basis of AA is largely unknown. We undertook a genome-wide association study (GWAS) in a sample of 1,054 cases and 3,278 controls and identified 139 single nucleotide polymorphisms that are significantly associated with AA (P ≤ 5 × 10−7). Here we show an association with genomic regions containing several genes controlling the activation and proliferation of regulatory T cells (Treg cells), cytotoxic T lymphocyte-associated antigen 4 (CTLA4), interleukin (IL)-2/IL-21, IL-2 receptor A (IL-2RA; CD25) and Eos (also known as Ikaros family zinc finger 4; IKZF4), as well as the human leukocyte antigen (HLA) region. We also find association evidence for regions containing genes expressed in the hair follicle itself (PRDX5 and STX17). A region of strong association resides within the ULBP (cytomegalovirus UL16-binding protein) gene cluster on chromosome 6q25.1, encoding activating ligands of the natural killer cell receptor NKG2D that have not previously been implicated in an autoimmune disease. By probing the role of ULBP3 in disease pathogenesis, we also show that its expression in lesional scalp from patients with AA is markedly upregulated in the hair follicle dermal sheath during active disease. This study provides evidence for the involvement of both innate and acquired immunity in the pathogenesis of AA. We have defined the genetic underpinnings of AA, placing it within the context of shared pathways among autoimmune diseases, and implicating a novel disease mechanism, the upregulation of ULBP ligands, in triggering autoimmunity.
AA affects about 5.3 million people in the United States alone, including males and females across all ethnic groups, with a lifetime risk of 1.7% (refs 1, 2). Autoimmunity develops against the hair follicle, resulting in non-scarring hair loss that may begin as patches that can coalesce and progress to cover the entire scalp (alopecia totalis) or eventually the entire body (alopecia universalis) (Supplementary Fig. 1). The phenomenon of ‘sudden whitening of the hair’ is ascribed to the acute onset of AA at times of profound grief, stress or fear3, in which the pigmented hair is selectively shed while the white hair persists. AA spares the stem cell compartment and attacks only the base of the hair follicle, which is surrounded by infiltrating lymphocytes. Despite these marked perturbations in the hair follicle, there is no permanent organ destruction, and regrowth of the hair remains possible. The concept of an autoimmune mechanism as the basis for AA emerged during the twentieth century from multiple lines of evidence4. AA hair follicles are surrounded by an immune infiltrate with activated T-helper cells (TH cells), cytotoxic T cells (TC cells) and natural killer (NK) cells, characterized as a TH1-type inflammatory response5,6. The notion of a collapse of immune privilege is thought to be a key event in triggering AA4,7.
Evidence supporting a genetic basis for AA stems from multiple lines of research, including the observed heritability in first-degree relatives8,9, twin studies10 and, most recently, from our family-based linkage studies11. Although a number of candidate-gene association studies were performed over the past two decades, the informativeness of these studies was inherently limited by small sample sizes and preselection of candidate genes.
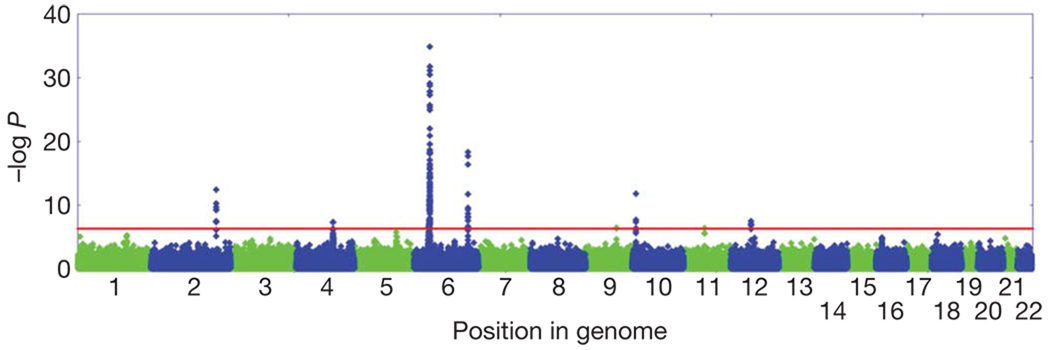
To determine the genetic architecture of AA, we genotyped or used publicly available data for up to 1,054 AA cases and 3,278 controls with a combination of Illumina 610K and 550K arrays. We performed association tests adjusted for residual population stratification (λ = 1.051) and found 139 single nucleotide polymorphisms (SNPs) significant at 5 × 10−7 (Fig. 1 and Supplementary Table 1).
Figure 1. Manhattan plot of the joint analysis of the discovery GWAS and the replication GWAS.
Results are plotted as negative log-transformed P values from a genotypic association test controlled for residual population stratification as a function of the position in the genome. Odd chromosomes are in green and even chromosomes in blue. Eight genomic regions contain SNPs that exceed the genome-wide significance threshold of 5 × 10−7 (red line).
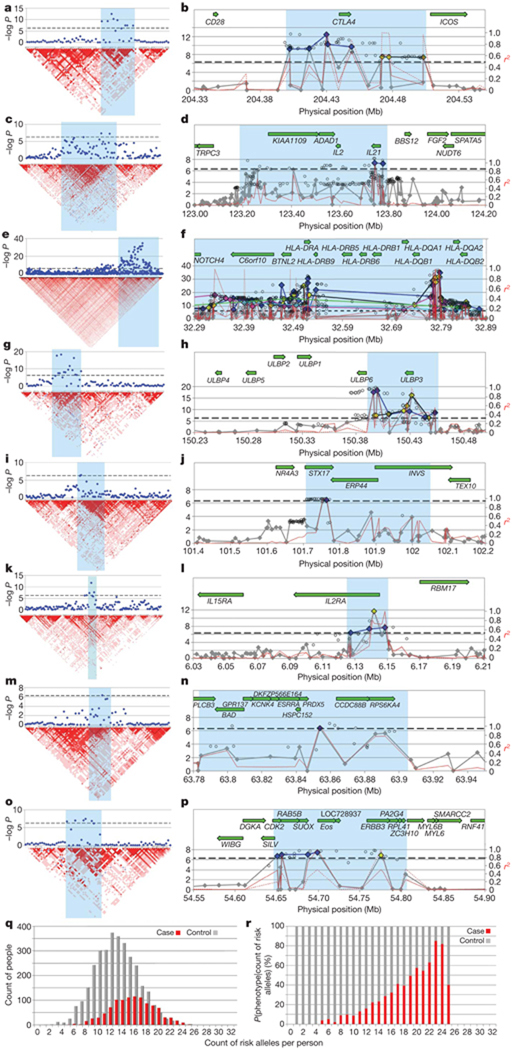
Our analysis identified several susceptibility loci for AA, most of which clustered into eight genomic regions and fell within discrete linkage disequilibrium (LD) blocks (Fig. 2 and Table 1). These include loci on chromosome 2q33.2 containing CTLA4, chromosome 4q27 containing IL-2/IL-21, chromosome 6p21.32 containing the HLA, chromosome 6q25.1 harbouring the ULBP genes, chromosome 10p15.1 containing IL-2RA (CD25), and chromosome 12q13 containing Eos (IKZF4) and ERBB3. One SNP resides on chromosome 9q31.1 within syntaxin 17 (STX17), and one resides on chromosome 11q13, upstream from peroxiredoxin 5 (PRDX5). Several of these LD blocks coincide with regions of linkage that we reported previously on chromosomes 6p (HLA), 6q (ULBPs), 10p (IL-2RA) and 18p (PTPN2)11. We also identified an additional 163 SNPs that were nominally significant (10−4 < P < 5 × 10−7), which include 12 regions containing genes involved in the immune response, notably IL-13, IL-6, IL-26, IFNG, SOCS1 and PTPN2 (Supplementary Tables 2 and 3). Finally, imputation analysis from HapMap release 2.2 identified additional statistically significant SNPs within each of the eight regions and one additional SNP in PTPN2 that raised it above statistical significance (Supplementary Tables 3 and 4).
Figure 2. LD structure and haplotype organization of the implicated regions from GWAS.
a–p, In all graphs the genome-wide significance threshold (5 × 10−7) is indicated by a black dotted line. Results from the eight regions are aligned with LD maps (a, c, e, g, i, k,m, o) and transcript maps (b, d, f, h, j, l, n, p): chromosomes 2q33 (a, b), 4q26–27 (c, d), 6p21.3 (e, f), 6q25.1 (g, h), 9q31.1 (i, j), 10p15–p16 (k, l), 11q13 (m, n) and 12q13 (o, p). For the plots with the LD maps, red indicates high LD as measured by D′. For the plots with the transcript maps, results for imputed SNPs are indicated by open circles. Typed SNPs that do not reach significance are in grey; significantly associated typed SNPs are in colour, coded by the risk haplotypes. For example in b, conditioning on any of the blue SNPs will decrease evidence for association of the other blue SNPs but will not affect evidence of any of the yellow-coded SNPs. On chromosome 6p in the HLA, significantly associated SNPs can be organized into at least five distinct haplotypes (blue, green, yellow, pink and light blue). The red lines show pairwise LD as measured by r2, for the most significant SNP in each haplotype, and define the LD block that is demonstrating association. q, r, The cumulative effect of risk haplotypes is indicated by the distribution of the genetic liability index in cases and controls. We chose the most significantly associated SNP from each haplotype to serve as a proxy for the haplotype, and show in q that the distribution of independent genetic risk factors changes as a function of phenotype, with an average of 13 risk alleles found in controls (grey) and 16 found in cases (red). As the number of risk alleles in an individual increases, the proportion affected by AA increases. The conditional probability of phenotype given count of risk alleles is shown in r (AA in red, control in grey).
Table 1.
Genes with significant association to AA
| Region | Gene | Function | Strongest association (P value) | Maximum odds ratio | Involved in other autoimmune disease |
|---|---|---|---|---|---|
| 2q33.2 | CTLA4 | Co-stimulatory family | 3.55 × 10−13 | 1.44 | T1D, RA, CeD, MS, SLE, GD |
| ICOS | Co-stimulatory family | 4.33 × 10−8 | 1.32 | ||
| 4q27 | IL-21/IL-2 | T-, B- and NK-cell proliferation | 4.27 × 10−8 | 1.34 | T1D, RA, CeD, PS |
| 6q25.1 | ULBP6 | NKG2D activating ligand | 4.49 × 10−19 | 1.65 | None |
| ULBP3 | NKG2D activating ligand | 4.43 × 10−17 | 1.52 | None | |
| 9q31.1 | STX17 | Premature hair greying | 3.60 × 10−7 | 1.33 | None |
| 10p15.1 | IL-2RA | T-cell proliferation | 1.74 × 10−12 | 1.41 | T1D, MS, GD, GV |
| 11q13 | PRDX5 | Antioxidant enzyme | 4.14 × 10−7 | 1.33 | MS |
| 12q13 | Eos (IKZF4) | Treg transcription factor | 3.21 × 10−8 | 1.34 | T1D, SLE |
| ERBB3 | Epidermal growth factor receptor | 1.27 × 10−7 | 1.34 | T1D, SLE | |
| 6p21.32 | MICA | NKG2D activating ligand | 1.19 × 10−7 | 1.44 | T1D, RA, CeD, UC, PS, SLE |
| (HLA) | NOTCH4 | Haematopoietic differentiation | 1.03 × 10−8 | 1.61 | T1D, RA, MS |
| C6orf10 | Unknown | 1.45 × 10−16 | 2.36 | T1D, RA, PS, GV | |
| BTNL2 | Co-stimulatory family | 2.11 × 10−26 | 2.70 | T1D, RA, UC, CD, SLE, MS, GV | |
| HLA-DRA | Antigen presentation | 2.93 × 10−31 | 2.62 | T1D, RA, CeD, MS, GV | |
| HLA-DQA1 | Antigen presentation | 3.60 × 10−17 | 2.15 | T1D, RA, CeD, MS, SLE, PS, CD, UC, GD | |
| HLA-DQA2 | Antigen presentation | 1.38 × 10−35 | 5.43 | T1D, RA | |
| HLA-DQB2 | Antigen presentation | 1.73 × 10−13 | 1.60 | RA |
Each of the eight regions implicated in our study contains multiple significant SNPs, which are detailed in Supplementary Tables 1 and 2. Here we display candidate genes within the implicated regions, and include the P value of the most significant SNP, and the odds ratio for the SNP with the largest effect estimate. Diseases are listed for which a GWAS or previous candidate gene study identified the same region (http://www.genome.gov/gwastudies, http://www.cdc.gov/genomics/hugenet): Crohn’s disease (CD), celiac disease (CeD), Graves disease (GD), generalized vitiligo (GV), multiple sclerosis (MS), psoriasis (PS), rheumatoid arthritis (RA), system lupus erythematosus (SLE), type I diabetes (T1D), and ulcerative colitis (UC).
We next assessed the extent to which these genetic risk factors contribute to AA. First, we decreased redundancy in our association evidence by using conditional analysis to determine which SNPs represent independent risk factors within each region (Supplementary Fig. 2), thus identifying a set of 16 risk haplotypes (Fig. 2 and Supplementary Table 1). The distribution of risk haplotypes was significantly different between cases and controls (P = 1.1 × 10−107) (Fig. 2q, r). To determine the relative contribution of different alleles to the genetic burden of AA, population-attributable risks were calculated for genotypes of individual SNPs and showed large contributions from individual alleles (ranging from 16 to 69%) (Supplementary Table 5). Together with the high concordance in siblings8,9, these findings demonstrate an overwhelming contribution of risk from genetic factors in AA, which awaits confirmation in a validation study.
Our GWAS study in AA implicates a new class of NKG2D ligands in autoimmune disease. The ULBP genes reside in a 180-kilobase MHC class I-related cluster on human chromosome 6q25.1 (Fig. 2g, h) that arose through duplication of the MHC locus12. Each of the ULBP genes has been shown to function as an NKG2D-activating ligand13,14. NKG2D ligands, including MICA/B and ULBPs, are stress-induced molecules that act as danger signals to alert NK, natural killer T, δγ T and CD8+ T lymphocytes through the engagement of the receptor NKG2D13.
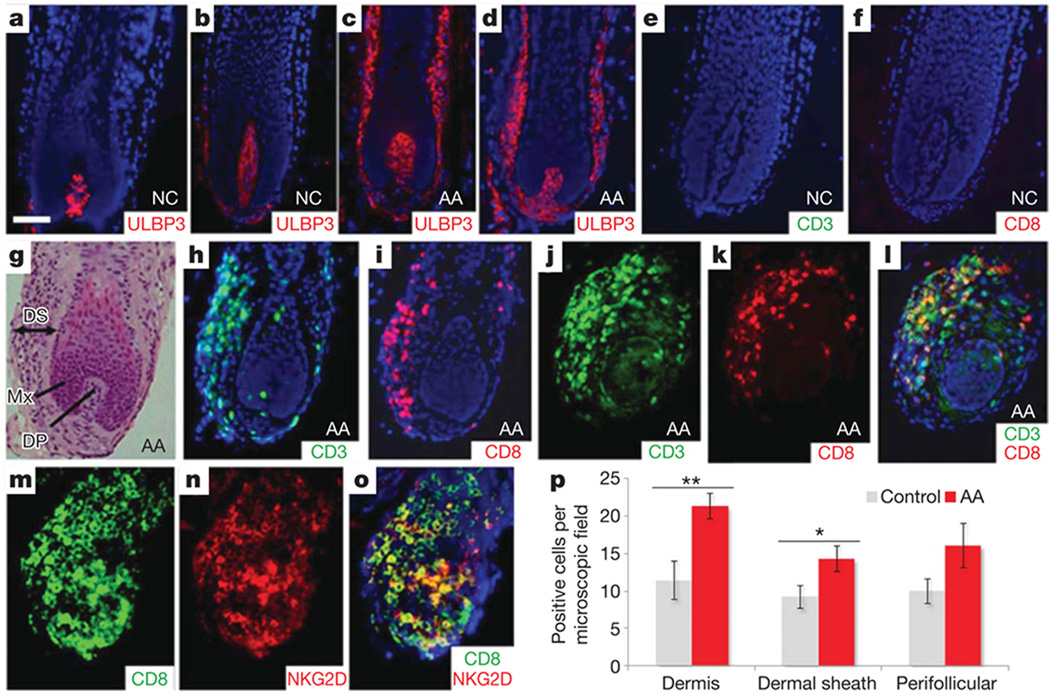
We next considered whether perturbations in the hair follicle microenvironment contribute to the initiation of AA. It has been postulated that NKG2D ligands, if overexpressed in genetically susceptible individuals, can trigger an autoimmune response against the tissue expressing the ligand15. To probe this hypothesis in the setting of AA, we examined ULBP3 expression within the hair follicle of unaffected scalp (Fig. 3a, b) and patients with AA (Fig. 3c, d). Whereas ULBP3 is expressed at low levels within the dermal papilla in normal hair follicle (Fig. 3a, b), in two different patients with early active AA lesions, we observed marked upregulation of ULBP3 expression in the dermal sheath as well as the dermal papilla (Fig. 3c, d), but not in control individuals or those with other inflammatory scalp disorders (data not shown). Quantitative immunohistochemistry corroborated a significantly increased number of ULBP3+ cells in 16 additional AA samples (Fig. 3p). We also noted a massive inflammatory cell infiltrate characterized by CD8+CD3+ T cells (Fig. 3g–l), but only rare NK cells (data not shown). Double immunostaining with an anti-CD8 and an anti-NKG2D antibody revealed that most NKG2D+ cells were CD8+ T cells (Fig. 3m–o). These results suggest that the autoimmune destruction in AA may be mediated in part by CD8+NKG2D+ cytotoxic T cells, whose activation may be induced by upregulation of ULBP3 in the dermal sheath of the hair follicle.
Figure 3. ULBP3 expression and immune cell infiltration of AA hair follicles.
a, b, Low levels of expression of ULBP3 in the dermal papilla of hair follicles from two unrelated, unaffected individuals. c, d, Massive upregulation of ULBP3 expression in the dermal sheath of hair follicles from two unrelated patients with AA in the early stages of disease. e, f, Absence of immune infiltration in two control hair follicles. g, Haematoxylin and eosin staining of AA hair follicle. DS, dermal sheath; Mx, matrix; DP, dermal papilla. h, i, Immunofluorescence analysis using CD3 (h) and CD8 (i) cell-surface markers for T-cell lineages. There is a marked inflammatory infiltrate in the dermal sheath of two affected AA hair follicles. j–l, Double-immunofluorescence analysis with anti-CD3 (j) and anti-CD8 (k) antibodies. l, The merged image clearly shows infiltration of CD3+CD8+ T cells in the dermal sheath of the AA hair follicle. Panels d and g–l are serial sections of the same hair follicle of an affected individual. Counterstaining with 4′,6-diamidino-2-phenylindole is shown in blue (a–f, h, i, l). Scale bar, 50 µm. AA, alopecia areata patients; NC, normal control individuals. m–o, Double immunostainings with anti-CD8 (m) and anti-NKG2D (n) antibodies revealed that most NKG2D+ cells co-expressed CD8+; o, merged image. p, Quantification of immunohistochemical staining (positive cells per microscope field at a magnification of × 200) for ULBP3 in 16 patients with AA and in 7 controls showed a significantly increased number of ULBP3+ cells in the dermis and dermal sheath in patients with AA compared with control skin. In addition, positive cells were also upregulated in perifollicular regions in AA samples, although this was not statistically significant. Data were analysed by Mann–Whitney test for unpaired samples and are expressed as means ± s.e.m.; asterisk, P < 0.05; two asterisks, P < 0.01.
The localization of an NK-activating ligand in the outermost layer of the hair follicle places it in an ideal position to express a danger signal16 and engage NKG2D on immune cells in the local milieu. Inducible overexpression of the ULBP homologues Rae1ε and Rae1α in mouse epidermis and pulmonary epithelium were previously shown to markedly alter the immune microenvironment within the skin and lung, respectively17,18. We postulate that in genetically susceptible individuals, upregulation of ULBP3 may have a similar effect on initiating the immune response in AA, and/or may become induced as part of an inflammatory cascade. Consistent with these findings, upregulation of the NK ligand MICA has recently been demonstrated in the hair follicle of patients with AA19. Taken together with the increased numbers of perifollicular NKG2D+CD8+ cells that we and others observed in lesional skin of patients with AA (Fig. 3)19, these data implicate a mechanism involving the expression of NKG2D ligands, including ULBPs, and infiltration of NKG2D-expressing cells in the aetiology of AA.
In addition to ULBP3/ULBP6, we identified other genes that are expressed in the hair follicle and may provide insight into the initiating events (Supplementary Figs 3 and 4). For example, STX17 (rs10760706, P = 3.60 × 10−7) is expressed in the hair follicle20 and is associated with the grey hair phenotype in horses, which is of interest because AA preferentially attacks pigmented hairs21. PRDX5 (rs694739, P = 4.14 × 10−7) is an antioxidant enzyme involved in the cellular response to oxidative stress, a process which is dysregulated in AA scalp22. PRDX5 has been implicated in the degeneration of target cells in several autoimmune disorders23–25, and other PRDX family members can serve as autoantigens26 (Supplementary Table 7). We found evidence for several genes whose expression in the hair follicle may contribute to a disruption in the local milieu, resulting in the onset of autoimmunity.
Our data implicate several factors that may act together to induce and promote immune dysregulation in the pathogenesis of AA. We found strong evidence for genes involved in the differentiation and maintenance of both immunosuppressive Treg cells, as well as their functional antagonists, pro-inflammatory T helper cells (TH17). Treg cells have a critical function in preventing immune responses against autoantigens, and their differentiation depends on the early expression of IL-2RA (CD25) (rs3118470, P = 1.74 × 10−12), as well as a key lineage-determining transcription factor, Foxp3. Foxp3-mediated gene silencing is critical in determining that Treg cells effectively suppress immune responses. Both IL-2 (rs7682241, P = 4.27 × 10−8) and its high-affinity receptor IL-2RA (rs3118470, P = 1.74 × 10−12) are central in controlling the survival and proliferation of Treg cells. It was recently found that Eos (IKZF4) (rs1701704, P = 3.21 × 10−8), a member of the Ikaros family of transcription factors, is a key co-regulator of FoxP3-directed gene silencing during Treg differentiation27. Although Treg cells probably use several different mechanisms to suppress immune responses, the high expression of CTLA4 (rs1024161, P = 3.55 × 10−13) has been proposed as a major determinant of their suppressive activity, particularly because CTLA4 is essential for the inhibitory activity of Treg cells on antigen-presenting cells28. The IL-2 locus is tightly linked with IL-21 (rs7682241, P = 4.27 × 10−8), which has pleiotropic effects on multiple cell lineages, including CD8+ T cells, B cells, NK cells and dendritic cells. IL-21 is a major product of proinflammatory TH17 (IL-17-producing CD4+ TH cells) and has been shown to have a key role in both promoting the differentiation of TH17 cells and limiting the differentiation of Treg cells29. Collectively, the constellation of immunoregulatory genes implicated in AA clearly point to Treg cells and TH17 cells as avenues for future studies and novel therapies.
The common-cause hypothesis of autoimmune diseases has received robust validation from GWAS in recent years30. This hypothesis evolved initially from epidemiological studies that demonstrated the aggregation of autoimmune diseases within families, and was further supported by findings of shared susceptibility regions in linkage studies. Our GWAS upheld the previously reported associations of HLA genes in AA and other autoimmune disorders (reviewed in ref. 4), whereas we did not find strong evidence for the other loci previously tested in AA using the candidate gene approach (Supplementary Table 6). In accordance with the common-cause hypothesis, our GWAS revealed several risk loci in common with other forms of autoimmunity, such as rheumatoid arthritis, type I diabetes, coeliac disease, systemic lupus erythematosus, multiple sclerosis and psoriasis: in particular, CTLA4, IL-2/IL-21, IL-2RA and genes critical to Treg maintenance (Table 1 and Supplementary Table 1). The commonality with rheumatoid arthritis, type I diabetes and coeliac disease is particularly noteworthy in view of the significance of the NKG2D receptor in the pathogenesis of each of these three diseases15.
Our GWAS establishes the genetic basis of AA, revealing several loci that contribute to disease susceptibility. These findings open new avenues of exploration for therapy based on the underlying mechanisms of AA with a focus not only on T-cell subsets and mechanisms common to other forms of autoimmunity, but also on unique mechanisms that involve signalling pathways downstream of the NKG2D receptor.
METHODS SUMMARY
Cases were ascertained through the National Alopecia Areata Registry (NAAR) with approval from institutional review boards and genotyped on the Illumina HumanHap 610 chip. Three sets of previously published control data sets were used for comparison of allele frequencies. These had been genotyped on the Illumina HumanHap 550v2. All samples were confirmed to be of European ancestry by principal component analysis with ancestry informative markers. Stringent quality control measures were used to remove samples and markers that did not exceed predefined thresholds. Tests of association were run with and without measures to control for residual population stratification. Tissue specimens and RNA from human scalp biopsies were obtained with approval from institutional review boards. All experiments were performed according to the Helsinki guidelines.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Material
Acknowledgements
We thank the many patients and their family members who participated in the National Alopecia Areata Registry from which the patient cohort was derived; S. Schwartz, D. A. Greenberg, S. E. Hodge, R. Ottman, K. Kiryluk, J. Lee, J. D. Terwilliger and R. Plenge for discussions about statistical methodology; A. Bowcock, M. Girardi, R. Clark, J. Trowsdale, R. Clynes, S. Ghosh and R. Bernstein for critical insights and perspectives on genetics, hair and immunobiology; C. Higgins, M. Kurban, M. Kiuru, H. Lam and M. Zhang for expert assistance in the laboratory; A. Martinez-Mir, M. Peacocke, A. Zlotogorski, M. Grossman, P. Schneiderman, D. Gordon and J. Ott for their critical input in the early phases of this study. We are grateful to the National Alopecia Areata Foundation (NAAF) for support of funding the initial studies, and to V. Kalabokes and her staff at NAAF for their efforts on our behalf. The patient cohort was collected and maintained by the National Alopecia Areata Registry (N01AR62279) (to M.D.). This work was supported in part by the DFG Cluster of Excellence, Inflammation at Interfaces (to R.P.) and by the National Institutes of Health grants R01AR44422 (to C.I.A. and P.K.G.), R01CA133996 and P30CA016772 (to C.I.A.) and R01AR52579 and R01AR56016 (to A.M.C.).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions L.P. performed technical aspects in preparation of samples for genotyping, the statistical analysis and preparation of the manuscript. M.D., V.P., M.H. and D.N. participated in phenotyping, diagnosis, and access to patient samples from the National Alopecia Areata Registry. Y.S., P.S. and H.K. provided expertise in RT–PCR and immunofluorescence. K.C.M. and R.P. provided expertise in immunhistochemistry. A.L. and P.K.G. provided control samples and performed genotyping as well as insight into autoimmune diseases. W.V.C. and C.I.A. provided additional statistical analysis and control samples from a distinct cohort. C.A.B.J. performed hair follicle microdissection and provided indispensable scientific expertise on the dermal sheath. A.M.C. provided oversight and conceptual guidance to the project, input into the functional significance of candidate genes, supervision of laboratory personnel, management of collaborations, preparation of the manuscript and all reporting requirements for granting agencies.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Cooper GS, Bynum ML, Somers EC. Recent insights in the epidemiology of autoimmune diseases: improved prevalence estimates and understanding of clustering of diseases. J. Autoimmun. 2009;33:197–207. doi: 10.1016/j.jaut.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Safavi KH, Muller SA, Suman VJ, Moshell AN, Melton LJ., III Incidence of alopecia areata in Olmsted County, Minnesota, 1975 through 1989. Mayo Clin. Proc. 1995;70:628–633. doi: 10.4065/70.7.628. [DOI] [PubMed] [Google Scholar]
- 3.Jelinek JE. Sudden whitening of the hair. Bull. N. Y. Acad. Med. 1972;48:1003–1013. [PMC free article] [PubMed] [Google Scholar]
- 4.Gilhar A, Paus R, Kalish RS. Lymphocytes, neuropeptides, and genes involved in alopecia areata. J. Clin. Invest. 2007;117:2019–2027. doi: 10.1172/JCI31942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilhar A, et al. Transfer of alopecia areata in the human scalp graft/Prkdc(scid) (SCID) mouse system is characterized by a TH1 response. Clin. Immunol. 2003;106:181–187. doi: 10.1016/s1521-6616(02)00042-6. [DOI] [PubMed] [Google Scholar]
- 6.Gilhar A, Shalaginov R, Assy B, Serafimovich S, Kalish RS. Alopecia areata is a T-lymphocyte mediated autoimmune disease: lesional human T-lymphocytes transfer alopecia areata to human skin grafts on SCID mice. J. Investig. Dermatol. Symp. Proc. 1999;4:207–210. doi: 10.1038/sj.jidsp.5640212. [DOI] [PubMed] [Google Scholar]
- 7.Ito T, Meyer KC, Ito N, Paus R. Immune privilege and the skin. Curr. Dir. Autoimmun. 2008;10:27–52. doi: 10.1159/000131412. [DOI] [PubMed] [Google Scholar]
- 8.McDonagh AJ, Tazi-Ahnini R. Epidemiology and genetics of alopecia areata. Clin. Exp. Dermatol. 2002;27:405–409. doi: 10.1046/j.1365-2230.2002.01077.x. [DOI] [PubMed] [Google Scholar]
- 9.Van der Steen P, et al. The genetic risk for alopecia areata in first degree relatives of severely affected patients. An estimate. Acta Derm. Venereol. 1992;72:373–375. [PubMed] [Google Scholar]
- 10.Jackow C, et al. Alopecia areata and cytomegalovirus infection in twins: genes versus environment? J. Am. Acad. Dermatol. 1998;38:418–425. doi: 10.1016/s0190-9622(98)70499-2. [DOI] [PubMed] [Google Scholar]
- 11.Martinez-Mir A, et al. Genomewide scan for linkage reveals evidence of several susceptibility loci for alopecia areata. Am. J. Hum. Genet. 2007;80:316–328. doi: 10.1086/511442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Radosavljevic M, et al. A cluster of ten novel MHC class I related genes on human chromosome 6q24.2–q25.3. Genomics. 2002;79:114–123. doi: 10.1006/geno.2001.6673. [DOI] [PubMed] [Google Scholar]
- 13.Eagle RA, Trowsdale J. Promiscuity and the single receptor: NKG2D. Nature Rev. Immunol. 2007;7:737–744. doi: 10.1038/nri2144. [DOI] [PubMed] [Google Scholar]
- 14.Eagle RA, Traherne AJ, Hair JR, Jafferji I, Trowsdale J. ULBP6/RAET1L is an additional human NKG2D ligand. Eur. J. Immunol. 2009;39:3207–3216. doi: 10.1002/eji.200939502. [DOI] [PubMed] [Google Scholar]
- 15.Caillat-Zucman S. HowNKG2D ligands trigger autoimmunity? Hum. Immunol. 2006;67:204–207. doi: 10.1016/j.humimm.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 16.Matzinger P. The danger model: a renewed sense of self. Science. 2002;296:301–305. doi: 10.1126/science.1071059. [DOI] [PubMed] [Google Scholar]
- 17.Strid J, et al. Acute upregulation of an NKG2D ligand promotes rapid reorganization of a local immune compartment with pleiotropic effects on carcinogenesis. Nature Immunol. 2008;9:146–154. doi: 10.1038/ni1556. [DOI] [PubMed] [Google Scholar]
- 18.Borchers MT, et al. Sustained CTL activation by murine pulmonary epithelial cells promotes the development of COPD-like disease. J. Clin. Invest. 2009;119:636–649. doi: 10.1172/JCI34462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito T, et al. Maintenance of hair follicle immune privilege is linked to prevention of NK cell attack. J. Invest. Dermatol. 2008;128:1196–1206. doi: 10.1038/sj.jid.5701183. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Q, Li J, Deavers M, Abbruzzese JL, Ho L. The subcellular localization of syntaxin 17 varies among different cell types and is altered in some malignant cells. J. Histochem. Cytochem. 2005;53:1371–1382. doi: 10.1369/jhc.4A6508.2005. [DOI] [PubMed] [Google Scholar]
- 21.Rosengren Pielberg G, et al. A cis-acting regulatory mutation causes premature hair graying and susceptibility to melanoma in the horse. Nature Genet. 2008;40:1004–1009. doi: 10.1038/ng.185. [DOI] [PubMed] [Google Scholar]
- 22.Akar A, et al. Antioxidant enzymes and lipid peroxidation in the scalp of patients with alopecia areata. J. Dermatol. Sci. 2002;29:85–90. doi: 10.1016/s0923-1811(02)00015-4. [DOI] [PubMed] [Google Scholar]
- 23.Holley JE, Newcombe J, Winyard PG, Gutowski NJ. Peroxiredoxin V in multiple sclerosis lesions: predominant expression by astrocytes. Mult. Scler. 2007;13:955–961. doi: 10.1177/1352458507078064. [DOI] [PubMed] [Google Scholar]
- 24.Gerard AC, Many MC, Daumerie C, Knoops B, Colin IM. Peroxiredoxin 5 expression in the human thyroid gland. Thyroid. 2005;15:205–209. doi: 10.1089/thy.2005.15.205. [DOI] [PubMed] [Google Scholar]
- 25.Wang MX, et al. Expression and regulation of peroxiredoxin 5 in human osteoarthritis. FEBS Lett. 2002;531:359–362. doi: 10.1016/s0014-5793(02)03511-1. [DOI] [PubMed] [Google Scholar]
- 26.Karasawa R, Ozaki S, Nishioka K, Kato T. Autoantibodies to peroxiredoxin I and IV in patients with systemic autoimmune diseases. Microbiol. Immunol. 2005;49:57–65. doi: 10.1111/j.1348-0421.2005.tb03640.x. [DOI] [PubMed] [Google Scholar]
- 27.Pan F, et al. Eos mediates Foxp3-dependent gene silencing in CD4+ regulatory T cells. Science. 2009;325:1142–1146. doi: 10.1126/science.1176077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wing K, et al. CTLA-4 control over Foxp31 regulatory T cell function. Science. 2008;322:271–275. doi: 10.1126/science.1160062. [DOI] [PubMed] [Google Scholar]
- 29.Monteleone G, Pallone F, Macdonald TT. Interleukin-21 as a new therapeutic target for immune-mediated diseases. Trends Pharmacol. Sci. 2009;30:441–447. doi: 10.1016/j.tips.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 30.Gregersen PK, Olsson LM. Recent advances in the genetics of autoimmune disease. Annu. Rev. Immunol. 2009;27:363–391. doi: 10.1146/annurev.immunol.021908.132653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.