Abstract
As odorant receptors (ORs) are thought to be critical determinants of olfactory sensory neuron (OSN) axon targeting and organization, we examined the spatiotemporal onset of mice ORs expression from the differentiation of OSNs in the olfactory placode to an aging olfactory epithelium. ORs were first detected in the placode at embryonic day 9 (E9), at the onset of OSN differentiation but before axon extension. By E13, 22 of 23 ORs were expressed. Onset of individual OR expression was diverse; levels and patterns of expression were unique for each OR. Regional distribution of ORs within zones of the olfactory epithelium appeared stable across development; adult-like patterns were observed by E13. Finally, analysis of OR expression and chromosomal location suggests that ORs are not stochastically expressed; they show evidence of coordinated expression. Collectively, these studies demonstrate that ORs are not equally represented in the “olfactome” across an animal's lifespan.
Introduction
Individual olfactory sensory neurons (OSNs) express only 1 of ∼1100 different odor receptors (ORs) and the axons from OSNs expressing the same OR converge into 2–3 glomeruli in each olfactory bulb (OB) (Chess et al., 1994; Ressler et al., 1994; Vassar et al., 1994; Mombaerts, 2006). Substitution of ORs disrupts or redirects the convergence of axons in the OB, changing the molecular/glomerular map, leading to the suggestion that ORs are critical determinants of OSN axon targeting and organization (Mombaerts et al., 1996; Wang et al., 1998; Feinstein and Mombaerts, 2004). Recent data further suggest that OR downstream signaling, via adenylyl cyclase III (ACIII) and cAMP is also involved, perhaps through the regulated expression of cell adhesion molecules (Imai et al., 2006; Chesler et al., 2007; Col et al., 2007; Zou et al., 2007). ORs are expressed in growth cones (Barnea et al., 2004; Strotmann et al., 2004) and recent data show OR mRNA translation can occur locally in growth cones (Dubacq et al., 2009). Moreover, odorants applied to OSN axon growth cones can trigger local cAMP synthesis and calcium influx (Maritan et al., 2009).
If ORs are implicated in OSN axon extension/coalescence, it seems reasonable that they would be expressed during early OSN differentiation and axon extension. Because of the implied importance of ORs in the initial extension of axons from developing OSNs, and later in their coalescence into glomeruli, we report here a systematic examination of OR expression across development. We focused on a selected subset of ORs that differ in regional localization in the olfactory epithelium (OE), chromosome location, and degree of homology. Embryonic day 9 (E9) was the earliest age that a subset of ORs (3/23) was detected, an age when OSN somata are first found in the olfactory placode but before the emergence of axons (Whitesides and LaMantia, 1996). Expression increased as development continued, with 22 of 23 ORs detected by E13, and all 23 recovered in every sample consistently by postnatal day 7 (PND7).
Initial expression of individual ORs was variable, extending across embryogenesis and into the earliest postnatal period. The number of OSNs expressing individual ORs across development unexpectedly revealed that several ORs exhibit transitory increases in frequency, followed by decreases, a sequence reminiscent of the early hypertrophy of OSN axon projections to the OB. Moreover, when we extended our analysis to the postnatal period using a custom mouse OR microarray, each OR studied presented a unique temporal pattern of expression.
At E13, the earliest age at which there were sufficient numbers of cells expressing an OR to observe zones, adult-like zonal patterns were observed. Finally, we also show for the first time that some OR genes located on the same chromosomes are overrepresented during early development, while others on different chromosomes are under represented, suggesting a differential regulation based on chromosomal location. These data suggest that OR expression may be regulated at many levels, and be partially dependent on the genes chromosomal location, the age of the animal and environmental influences.
Materials and Methods
Animals.
Pregnant, time-mated CD-1 mice (Charles River) were killed with CO2 before cesarean section. For in situ hybridization, the embryos, both male and female, were immersion fixed in 4% paraformaldehyde (PFA) in PBS [0.1 m phosphate buffer (PB) and 0.9% NaCl, pH 7.4] at 4°C overnight. Embryos were collected at E9, E9.5, E10, E10.5, E11, E11.5, E12, E13, and E17, where the day of conception is designated as E0. Because embryos of the same gestational age may differ in their stage of development, individual mouse embryos were staged using the Theiler criteria, described previously (Theiler, 1989). Mice at PND0 and PND2 were rapidly decapitated and immersion fixed in 4% PFA in PBS at 4°C overnight. For RNA extraction, pregnant, time-mated mice were killed with CO2, embryos harvested by cesarean section and dissected in cold PBS. The whole head (E9–E10), the frontonasal prominence (E10.5–E12), or the OE (E13–E17) was removed and dissolved in TRIzol (Invitrogen). Mice at PND0 were rapidly decapitated while those at PND7 and PND21 were killed with CO2 and the OE was removed and dissolved in TRIzol.
For microarray analysis, mouse tissue from C57BL/6 mice (The Jackson Laboratory) was collected at E15 and PND0, -10, -20, -60, -120, and -210. The C57BL/6 mice were used for this experiment so that aging mice (PND550, 18 months) could be obtained from the National Institute on Aging. For each time point, 3–5 biological replicates were collected each of which comprised OEs from 2 animals (adult and older postnatal) and a litter (embryonic and younger postnatal). For OE harvesting, pregnant, time-mated mice were killed with CO2, embryos harvested by cesarean section and dissected in cold PBS. The E15 OE was removed and placed in RNeasy (Qiagen) until use. Mice at PND0 were rapidly decapitated while those at PND10–PND550 were killed with a mixture of ketamine (100 mg/kg) xylazine (10 mg/kg) and the OE was removed and placed in RNeasy until use.
All procedures undertaken in this study were approved by Yale and/or Columbia University's Animal Care and Use Committee and conform to NIH guidelines.
Reverse transcriptase PCR.
Total RNA was extracted using TRIzol (Invitrogen) according to manufacturer instructions. Any traces of DNA were removed by treatment with Turbo DNase (Ambion), for 2 h. All samples were tested for DNA contamination by doing a PCR with actin primers (R&D Systems) in the absence of reverse transcriptase (RT). Specific OR primers sequences, degenerate primers sequences for ORs as well as RT-PCR protocols were previously reported (Malnic et al., 1999; Miyamichi et al., 2005) (see supplemental Table 1, available at www.jneurosci.org as supplemental material for primer details). After degenerate PCR, amplified fragments were cloned into pCRII-TOPO plasmid (Invitrogen) and sequenced. Over 400 colonies were screened, and corresponding sequences were subjected to BLAST analysis. To verify the specificity of the RT-PCR, selected bands of the expected OR product size were purified and sequenced. In all cases examined, the sequences corresponded to the targeted OR.
In situ hybridization.
Tissue was cryoprotected in 30% sucrose, frozen in O.C.T and serially sectioned on cryostat (20 μm). In situ hybridization was performed with digoxigenin-labeled RNA probes obtained by in vitro transcription. OR plasmids were gifts from Dr. Peter Mombaerts (Max Planck Institute of Biophysics, Frankfurt, Germany). Hybridization was done using established procedures (Ishii et al., 2004), and developed with NBT/BCIP (Roche). MOR28 antibody was a gift from Dr. Hitoshi Sakano (University of Tokyo, Tokyo, Japan). Double in situ hybridization-immunohistochemistry for MOR28 was done following an established protocol (Ishii et al., 2004).
Images were acquired with a Leica confocal microscope, using 40× or 63× oil-immersion objectives. Z-stacks were taken with 0.5 μm steps between images. Images displayed are maximum projections, generated with Leica confocal software. Digital images were color balanced using Adobe Photoshop CS2 (Adobe Systems). The composition of the images was not altered in any way. Plates were constructed using Corel Draw X3.
Immunohistochemistry.
Immunostaining was performed as previously described (Treloar et al., 2009). Briefly, sections were blocked with 2% bovine serum albumin (Sigma) in TBS containing 0.3% Triton X-100 (TBST) for 30 min at room temperature then incubated with either mouse anti-β-tubulin III (Sigma; 1:1000) or rat anti-neuronal cell adhesion molecule (NCAM; Millipore; 1:1000) in block solution overnight at 4°C. Sections were rinsed three times in TBST, incubated in either Alexa 488-conjugated donkey anti-mouse IgG secondary antibodies (Invitrogen) or Alexa 555-conjugated goat anti-rat IgG secondary antibodies (Invitrogen) diluted 1:1000 together with fluorescent DNA dye Draq5 (1:1000, Biostatus) for 1 h at room temperature, rinsed three times in TBST, once in TBS, and coverslipped in NPG fluorescent mounting media (5% n-propyl gallate in glycerol and 25 mm NaHCO3).
Microarray.
Samples were prepared according to protocols described in the Expression Analysis Technical Manual (Affymetrix). We used DCHIP software (version 2004 and 2008) to do the normalization and model-based expression value calculation. Based on invariant difference selection algorithm, invariant probe sets were chosen for normalization between different samples. The PM-MM difference model was applied for expression value calculation. Significant analysis of microarrays, which is a supervised learning software for microarray data analysis, was used to estimate the false discovery rate of differential analysis between different samples. To analyze the time course data, only OE samples at different time points were grouped for normalization. Raw expression values were exported to EDGE software for differential analysis (Leek et al., 2006). Genes without significant change (p-value cutoff at 0.05) over time course were excluded. The mean ± SEM of expression values obtained from the MOR custom array are shown.
Results
Olfactory sensory neurons (OSNs) differentiate in the olfactory placodes, epithelial thickenings found bilaterally on the rostrolateral regions of the E8.5–E9.5 embryo. As OSNs differentiate, the placodal epithelium thickens (E10–E10.5), forming olfactory discs which develop indentations with distinct marginal lips. By E11 the discs invaginate to form nasal pits, lined by olfactory epithelium (Theiler, 1989). The adult-like organization of the nasal cavity, including the delineation of turbinates and recesses, is apparent by ∼E13. At this point it is prudent to note that inconsistencies in embryo staging protocols often make it difficult to compare the rapidly occurring events of early embryogenesis. To help overcome this problem, we used an established protocol in each study and normalized our embryos to the Theiler staging protocol (Theiler, 1989). We adopted an equivalent staging (ES) strategy for use with published data that report alternate methods to stage their embryos. By converting the day of positive plug to Theiler stage 1 (TS1), all studies can then be compared. As such, when we report an equivalent staging modification from an original manuscript, our reference will include an annotation of “ES” after the year to indicate that we have verified the staging.
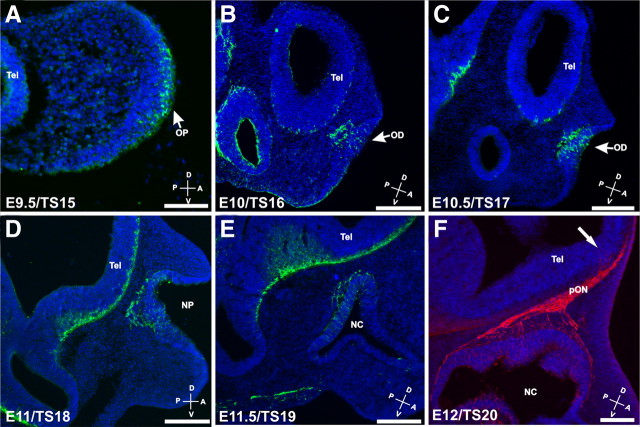
We examined the spatiotemporal onset of OSN differentiation in the developing olfactory system using β-tubulin-III as a marker of neuronal identity. Neurons were first identifiable in the placode at E9.5/TS15 (Fig. 1A). In the olfactory discs (E10–E10.5/TS16–TS17), differentiation of OSNs occurred in the middle of the discs, not in the periphery near the marginal lips (Fig. 1B,C). Interestingly, we found OSNs axons and migratory cells exiting the discs as early as E10/TS16, 12–24 h earlier than previously reported (Cuschieri and Bannister, 1975a,b).
Figure 1.
β-Tubulin III expression in the developing olfactory system. Sagittal sections through mice embryo heads from E9.5 (A) to E12 (F) were stained for β-tubulin III (green), a neuronal marker (A–E), and NCAM (red), a marker of olfactory sensory neurons (F). Neurons were observed in the olfactory placode as early as E9.5 and migrating out of the placode from E10 (B, C). OSN axons were observed in the underlying mesenchyme by E11 (D, E). The development of the OB starts after the first OSN axons reach the telencephalic vesicle (arrow, F). Scale bars: A, 100 μm; B--F, 200 μm. A, Anterior; D, dorsal; NC, nasal cavity; NP, nasal pit; OD, olfactory disc; OP, olfactory placode; P, posterior; pON, presumptive olfactory nerve; Tel, telencephalic vesicle; V, ventral.
OSN axons reached the telencephalic vesicle in the region of the presumptive OB (pOB) at E11.5, marking the initial formation of the olfactory nerve. The arrival of the OSN axons and formation of the nerve predated the evagination of the pOB by 12–24 h (Fig. 1F, arrow) (Treloar et al., 1996; Shay et al., 2008) consistent with reports that OSN axons stimulate OB formation (Gong and Shipley, 1995). Thus, OSNs are among the first sensory neurons to differentiate in the nervous system, with a corresponding precocious formation of the olfactory nerve.
Odor receptors have been repeatedly implicated as important determinants of OSN axon targeting, primarily because the substitution of OR coding regions redirects, or in some cases disrupts, coalescence in the OB (Mombaerts et al., 1996; Wang et al., 1998; Feinstein and Mombaerts, 2004). We reasoned that since OSN axons extend so early in development, with the nascent olfactory nerve present by E11.5, if ORs have a role in axon guidance and/or extension, they would most likely be expressed before, or coincident with, OSN axon extension. Previous in situ hybridization studies reported some ORs expressed in the OE as early as E11.5 in mice (Sullivan et al., 1995) and in the migratory mass as early as E11.5 (Schwarzenbacher et al., 2004) and perhaps as early as E10 (Nef et al., 1992).
These discrepancies in age of onset, particularly during a period in which profound developmental changes are occurring in the olfactory system, suggest some diversity may exist in onset of OR expression. Therefore, to establish OR expression profiles we adopted a multifaceted approach to study a large subset of ORs from the formation of the OE to mouse senescence. To screen for onset of expression of 23 OR genes we used an RT-PCR approach with previously published primers (Miyamichi et al., 2005). These 23 ORs were selected because their expression in the OE is restricted to either the dorsomedial (zone 1), intermediate (zone 2/3) or ventrolateral region (zone 4) and their expression patterns do not overlap in the adult (Miyamichi et al., 2005). Using this strategy, we found that OR genes had very different temporal onsets of expression, varying from E9 in the olfactory placode to PND0 (in >50% of samples; Figs. 2, 3). As seen in Figure 2A, onsets (yellow arrowheads) varied significantly for different ORs. Figure 2B summarizes the RT-PCR data, and includes information of the proportion of samples which showed a specific band in four repeat experiments. At E9, 3 OR genes were consistently detected in the samples; MOR144-1 (100%), MOR163-1 (≥50%), and MOR244-1 (100%). These 3 ORs are from each of the 3 adult OE regions, suggesting that there are no temporal preferences for onset of OR expression in one zone versus another.
Figure 2.
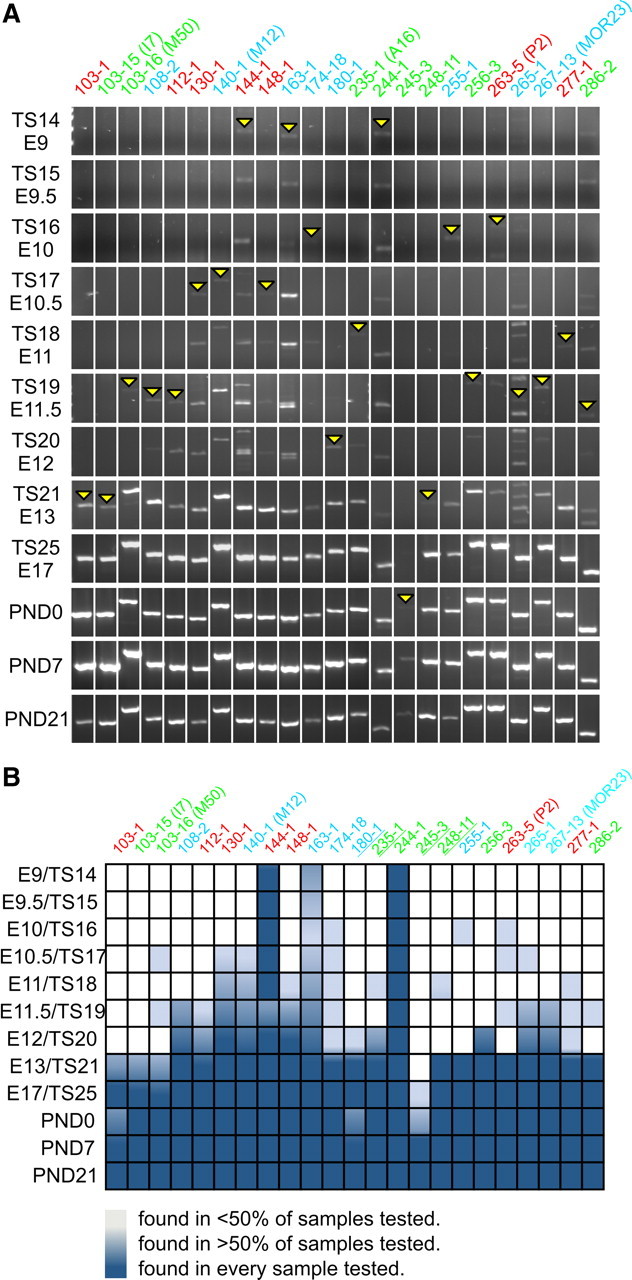
Onset of expression for 23 different ORs during OE development. A, Specific OR primers were selected such as dorsomedial/zone 1 (MORs 108-2, 140-1, 163-1, 174-18, 180-1, 255-1, 265-1, 267-13), intermediate/zones 2–3 (MORs 103-1, 112-1, 130-1, 144-1, 148-1, 263-5, 277-1) and ventrolateral/zone 4 (MORs 103-15, 103-16, 235-1, 244-1, 245-3, 248-11, 256-3, 286-2) of the adult OE were represented. Analysis demonstrates the presence of at least 3 OR mRNAs by E9 (MORs144-1, 163-1, and 244-1). As expected, the number of OR mRNAs expressed increases during development and by E13 not only are almost all of the ORs studied expressed, but PCR products show robust expression of the mRNA. MOR245-3 was the last one to be turned on and it was only consistently detected after birth. B, Summary of OR onset during OE development. The results of 4 different RT-PCRs from 2 independent tissue sample preparations are shown. Dark blue, the specific expected band was detected in all samples; mid blue: at least half of the samples produced a band of the predicted size; light blue, less than half of the samples produced a band of the predicted size; white, the expected band was never observed. Earlier stages of development show nonspecific bands for some ORs (144-1, 163-1, 265-1, 267-13, and 286-2) that were not detected at later stages. Underlined ORs reside on chromosome 2. Note that in A and B the ORs are color coded to represent zonal organization, such that dorsomedial is blue, intermediate is red, and ventrolateral is green.
Figure 3.
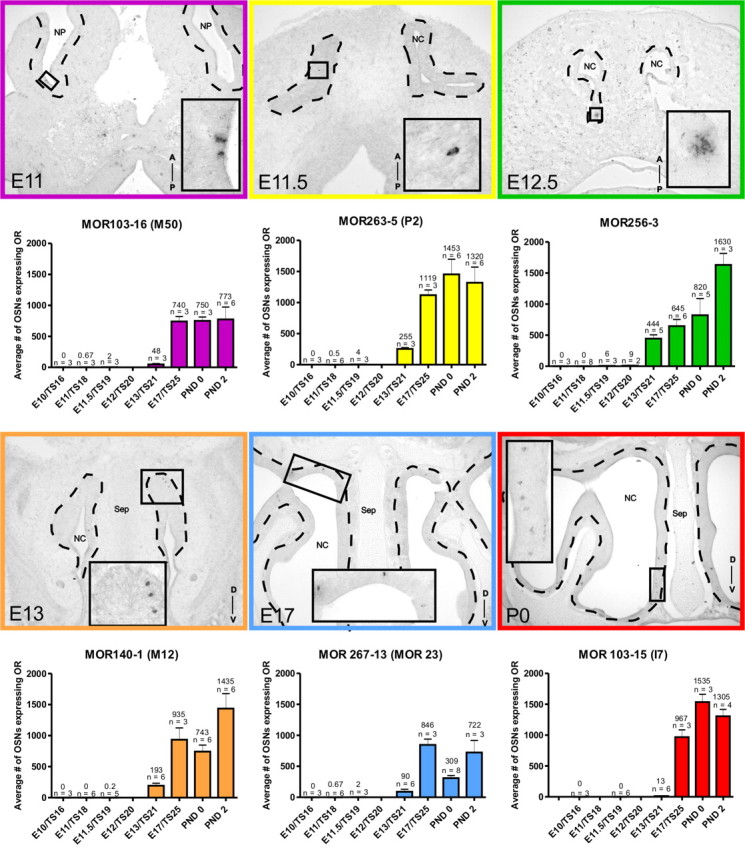
In situ hybridization and quantification of OSNs expressing specific ORs during OE development. In situ positive cells in embryonic (horizontal E10–E12; coronal E13–E17) and postnatal (coronal) sections. High-magnification images of positive cells are shown in the inserts. Quantification involved counting every cell in serial sections from E10 to E12 and counting positive cells in every fifth section at E13–E17 and every tenth section at PND0–PND2 followed by Abercrombie's extrapolation. Histograms show mean ± SEM and number of animals analyzed are indicated above each bar. Dashed line delineates the olfactory epithelium. A, Anterior; D, dorsal; NP, nasal pit; NC,: nasal cavity; P, posterior; Sep, septum; V, ventral.
While most ORs were first detected between E9 and E12 (20/23), expression was variable. In some cases, ORs were detected at an early stage but not at the subsequent stage [e.g., 103-16 (M50) and 263-5 (P2)]. This may reflect very low levels of expression, or a limited number of animals in the sample expressing the OR. Robust expression, defined as expression in a minimum of 50% of the samples, was not found until E13 (mid blue, Fig. 2B). At E12 only 12 of 23 (∼52%) of ORs tested were robustly expressed, but by E13, 22 of 23 (∼96%) of ORs had robust expression. Only one OR, MOR245-3, did not show evidence of robust expression until PND0 (Figs. 2, 3). As is evident in Figure 2B by PND7, every OR studied was detected in all samples.
Six ORs were selected for further analysis via in situ hybridization (ISH). We specifically selected 6 well characterized ORs so comparisons could be made with previous reports. MOR103-16 (M50), MOR263-5 (P2), MOR267-13 (MOR23), MOR140-1 (M12), MOR103-15 (I7), and MOR256-3 were selected to comprise a set spanning all zones of the epithelium. Our aim was to characterize the population dynamics of expression onset and to address the following questions: 1) do all populations follow a similar temporal pattern of expression regardless of when onset occurs (i.e., did the population expand at a similar rate); 2) Are cell numbers comparable between populations at equivalent time points after onset of expression; and 3) Is there evidence of progressive OSN neurogenesis, or is there overproduction followed by attrition of the population over time?
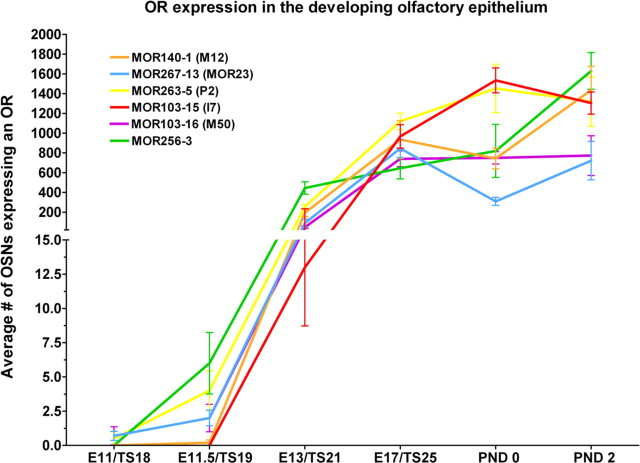
The 6 ORs displayed distinct temporal onsets and population profiles over the embryonic and postnatal periods examined (Fig. 3). No two profiles were the same, varying in onset date, total number of cells and the population dynamics over time. For example, MOR103-16 (M50), MOR263-5 (P2), and MOR267-13 (MOR23) all had onset dates of E11. However, MOR103-16 (M50) reached a plateau before birth (E17), MOR263-5 (P2) reached a plateau after birth (PND0), and MOR267-13 (MOR23) showed a decrease at birth which recovered by PND2 (Fig. 3). To ensure some of the transient changes were not artifacts we increased the numbers of embryos analyzed and confirmed the changes observed. We also observed that while MOR263-5 (P2), MOR140-1 (M12), and MOR103-15 (I7) all reach similar population sizes by PND2, their onset of expression varied from E11 for MOR140-1 (M12), E11.5 for MOR263-5 (P2) and E13 for MOR103-15 (I7). These changes in population dynamics can be best compared by collapsing the data onto a single graph (Fig. 4; raw data shown in supplemental Table 2, available at www.jneurosci.org as supplemental material). As is evident, each OR has a unique distribution curve, all of which show a general trend of increasing with age, but some with transient decreases [MOR140-1 (M12) and MOR267-13 (MOR23)], some with plateaus [MOR103-16 (M50)] and some with decreases after birth [MOR103-15 (I7) and MOR263-5 (P2)].
Figure 4.
OR expression in the developing olfactory system. This combined graph, obtained from the in situ hybridization data, shows that expression profiles are unique for each OR. Each data point is the mean ± SEM. The graph has been plotted with 2 different scales on the y-axis so that low numbers of cells at early ages can be visualized.
To further analyze the expression patterns observed during development, we extended our studies from E15/TS23 through PND550 (18 months) with a custom mouse odorant OR microarray (Zhang et al., 2004b). We focused our analyses on the six ORs previously analyzed via ISH as well as a further six well known ORs: MOR171-3p (M72), MOR174-9 (mOR-EG), MOR187-1 (MOR10), MOR235-1 (A16), MOR244-1 (MOR28), and MOR244-3 (MOR83). The MOR array detects expression in all positive cells; consequently signal is a reflection both of the number of neurons expressing the gene as well as the expression level in single cells. Consistent with the embryonic data, the overall expression was unique for each OR analyzed (Fig. 5A,B). ORs fell into 2 broad classes; high-expressing (Fig. 5A) and low-expressing (Fig. 5B). To examine whether high expression was due to more cells expressing a gene or just a higher copy number per cell, we correlated the ISH and array data at PND0 (Fig. 5C). There is a significant correlation (p = 0.0151) between OR-expressing cells (ISH) and RNA level detected on the MOR microarray. This suggests that most of the variation in microarray expression is accounted for by changes in number of cells expressing a given OR rather than changes in transcript copy number within cells.
Figure 5.
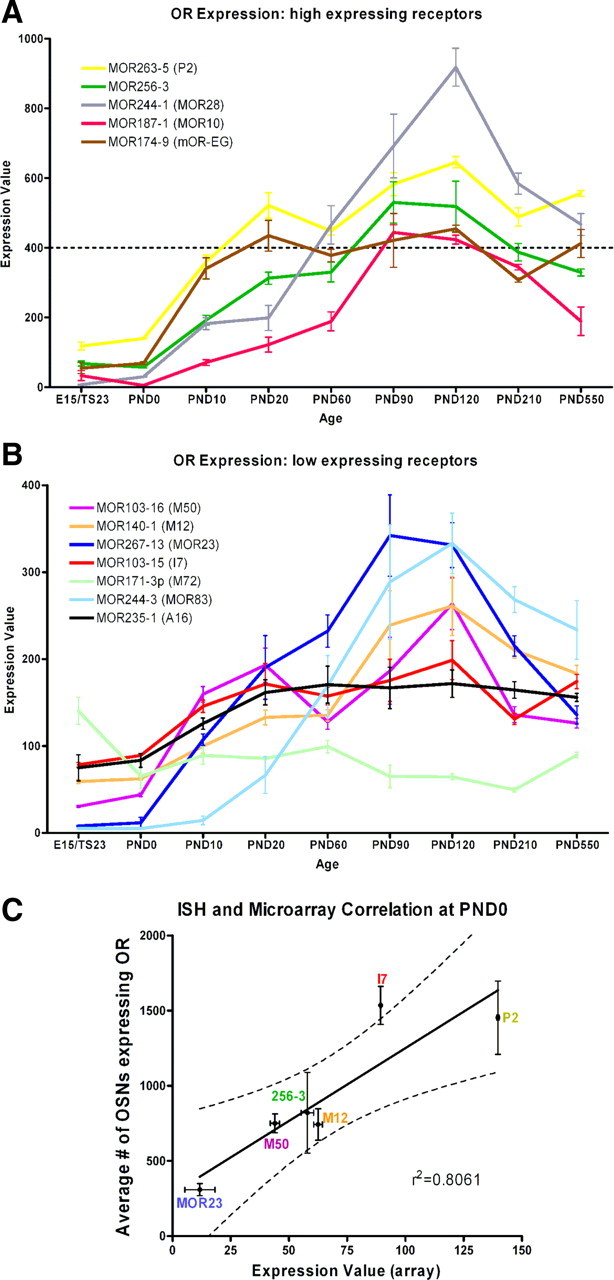
Temporal expression profile of 12 selected ORs. The mean raw expression value ± SEM for each OR obtained with the array from E15 to PND550 is shown. ORs fell into 2 categories, high expressers (A) and low expressers (B). High expression was defined as having a peak expression level above 400 (dashed line in A) while low was having a peak below 400. As was seen with the in situ data, each OR has a unique expression profile. C, Correlation between in situ data and array data at PND0. Correlation was significant (p = 0.0151); r2 = 0.8061.
Expression patterns fell into a variety of classes. For example, while most ORs analyzed showed a peak of expression after birth, MOR171-3p (M72) showed the highest level of expression at E15. Interestingly, MOR244-1 (MOR28) which by RT-PCR was detected as early as E9, displayed the highest levels of expression of the ORs analyzed. Despite different expression levels, half of the ORs showed a trend of increasing expression up to PND90–PND120 followed by a decrease in expression at subsequent ages [MOR140-1 (M12), MOR187-1 (MOR10), MOR244-1 (MOR28), MOR244-3 (MOR83), MOR256-3, MOR267-13 (MOR23); Fig. 5A,B]. Lee et al. (2009) recently reported data on the expression of ORs in aging mice. Our aging data are consistent, particularly in showing a significant decrease in MOR267-13 (MOR23) expression beyond 3 months of age.
We also investigated the coincidence of onset of mRNA and protein expression of one OR, MOR28. At E13, the earliest age we could reliably detect MOR28 protein expression, most cells were colabeled using in situ hybridization and immunohistochemistry. However, a few cells were observed which exhibited only ISH signals (supplemental Fig. 1, available at www.jneurosci.org as supplemental material, open arrow) indicating onset of protein expression had yet to occur in this cell.
The ORs analyzed by ISH can be grouped according to zonal expression in either the dorsomedial [MOR267-13 (MOR23) and MOR140-1 (M12)], intermediate [MOR263-5 (P2)], or ventrolateral [MOR103-16 (M50), MOR 103-15 (I7), and MOR 256-3] regions of the OE. This enabled us to analyze when a topographical distribution is first observed in the developing OE. The total number of OR-expressing cells, for any of the characterized ORs is <10 at stages younger than E13, making any attempt at studying a topographic distribution implausible (cf. Figs. 4 and 5). However, by E13 the number of OR-expressing cells increases significantly making a topographic analysis feasible. As shown in Figure 6, at E13, even with relatively few OR-expressing cells in each section, MOR267-13 (MOR23) and MOR 140-1 (M12) are restricted to the dorsal recess of the nasal cavity (Fig. 6, red tones), MOR 103-16 (M50), MOR 103-15 (I7) and MOR 256-3 are restricted to the ventral nasal cavity (Fig. 6, green tones), and MOR 263-5 (P2) resides in an intermediate position (Fig. 6, yellow). These patterns are more elaborated at PND0 (Fig. 6), and are consistent with the reports of expression in adult OE (Miyamichi et al., 2005).
Figure 6.
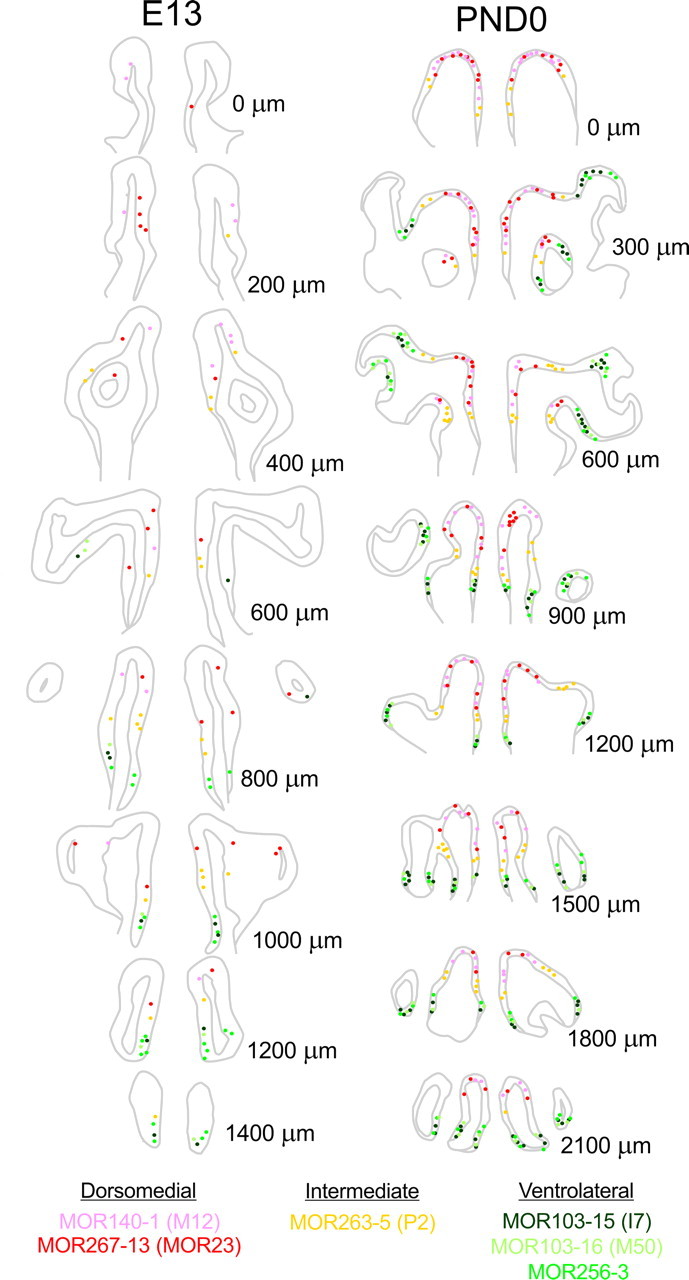
Spatially restricted expression of ORs along the rostral-caudal extension of the nasal cavity. The position of specific OR-expressing cells is shown on nasal cavity schemes every 200 μm (E13) and 300 μm (PND0) over the length of the rostral (top) to caudal (bottom) extension of the nasal cavity. As early as E13 the spatially restricted expression of ORs into dorsomedial (MOR140-1 and MOR267-13), intermediate (MOR263-5) or ventrolateral (MOR103-15, MOR103-16 and MOR256-3) could be observed.
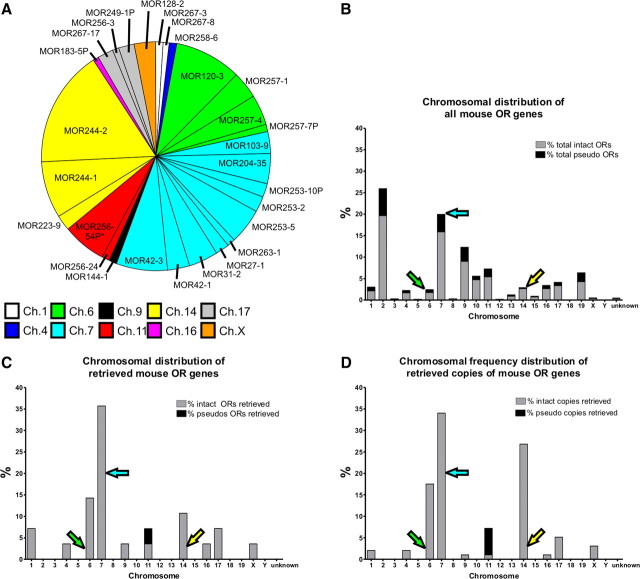
Because positive and negative regulatory elements have been shown to affect OR expression at the chromosomal level, we next asked whether OR onset was related to chromosomal organization (for review, see Imai et al., 2006). We reasoned that if chromosome location was important, we would see a disproportionate representation of genes from some chromosomes. Using degenerate primers to the conserved transmembrane domains 3 and 6, as reported by Malnic et al. (1999), we used a PCR-based screen to retrieve clones from E11 OE mRNA. We selected 400 transformants; DNA was isolated, and from these, 98 viable sequences were retrieved. Figure 7A details the ORs that were retrieved and their relative frequency, color coded by chromosome. We retrieved both class I (MOR27-1, MOR31-2, MOR42-1, MOR42-3) as well as many class II ORs. A pseudogene, MOR256-54P was retrieved, although it should be noted that its status as a pseudogene was current at time of manuscript preparation but may have changed as updates to the mouse genome continue. Many OR pseudogenes have been upgraded to intact genes as sequence data improves in subsequent genome rebuilds. This is evident by four genes identified (MOR183-5P, MOR249-1P, MOR253-10P, and MOR257-7P) which were originally classified as pseudogenes but have been reclassified as intact genes.
Figure 7.
OR expression at E11 sorted by chromosome location. A, Retrieved ORs were grouped according to chromosomal location. Each slice represents the frequency with which each OR was observed. Asterisk indicates that this OR is classified as a pseudogene (MOR256-54P). B, All ORs were grouped according to their chromosomal location, and represented as a percentage of the total ORs. Intact ORs are represented as gray bars while pseudogenes are represented as black bars. C, Retrieved ORs were grouped according to chromosomal location, and represented as a percentage of the total retrieved ORs. D, Retrieved ORs were grouped according to chromosomal location, and represented as a percentage of the total retrieved ORs accounting for the number of times each OR was retrieved in the screening.
Chromosomal distribution of all known mouse OR genes is shown in Figure 7B, while Figure 7C plots the distribution of our retrieved genes. It is immediately apparent that some chromosomes, including chromosome 6 (Ch.6), Ch.7, and Ch.14 are overrepresented in our retrieved dataset, while others are not present. For example, we did not retrieve any genes from Ch.2, despite the fact that Ch.2 has the largest number of OR genes in the genome. This is consistent with our initial screen using specific OR primers; MOR180-1, MOR 235-1, MOR245-3, and MOR248-11—all of them encoded in Ch.2—showed an onset of expression later than E12 (Fig. 2B). Figure 7D is a weighted distribution plot, which takes into account the number of times a gene was retrieved, making the overrepresentation even more considerable. These data support the notion that ORs located on different chromosomes have a differential onset of expression. That is, chromosomal location may be a determinant of the onset of OR expression.
Discussion
The first putative OSNs, recognized as β-tubulin III-expressing cells, appear around E9.5 (Fig. 1) and continue to proliferate as the surface area and thickness of the OE increases. Axons first extend from OSNs at E10/TS16, 1 d earlier than previous reports (A. Miller and C. Greer, unpublished observations). The first OSN axons reach the nascent OB by ∼E11.5, but the first synapses are not apparent in appreciable numbers until E15 (Hinds and Hinds, 1976) (TS23 ES). In this context, we asked when OR gene expression occurs. Here, we used PCR primers for 23 different OR genes, selected for their restricted OE zonal expression pattern, and the lack of overlapping expression between zones (Miyamichi et al., 2005). As early as E9, mRNA from one OR from each of the three representative zones was expressed. Likewise, the subsequent onset of other ORs did not showed any preferential zonal expression. Importantly, onset of OR expression was asynchronous, with some ORs retrievable as soon as the first OSNs differentiate, and others not appearing until the late embryonic/early postnatal period. Prior analyses of onset of OR expression reported results varying from E10 to E12.5 for the main olfactory epithelium (Nef et al., 1992; Sullivan et al., 1995; Saito et al., 1998; Royal and Key, 1999; Conzelmann et al., 2002; Schwarzenbacher et al., 2004). The data presented here extend onset to an earlier time, but nevertheless support the prior studies in showing that different ORs have different onsets of expression and expression profiles. Studying multiple ORs enabled us to recognize the variability not evident when studying just one or a few ORs.
OR receptor gene choice has been proposed to be stochastic, but like the zebrafish (Barth et al., 1996; Kratz et al., 2002) OR expression in mammals does not appear to correspond to a stochastic model. Our data, and that of others, suggests there are multiple mechanisms regulating OR expression (Kratz et al., 2002; Serizawa et al., 2004; Imai and Sakano, 2009). First, temporally regulated cues are likely acting as onset of ORs is asynchronous. This is consistent with reports in zebrafish where OR onset is also asynchronous (Barth et al., 1996). Second, the restricted topographic expression of OR genes in the adult in the OE suggests that topographically regulated cues may restrict/control expression (Iwema et al., 2004; Miyamichi et al., 2005). We show here that as early as E13 the restriction of ORs to specific OE zones is established, suggesting that by E13 progenitors are topographically and lineage restricted.
Finally, our data suggest that some regulatory elements may influence expression based on chromosomal position. We show that it is not until late in embryonic development that Ch.2 OR genes begin to be expressed, even though Ch.2 contains the largest number of OR genes (26%; (Zhang et al., 2004a). No Ch.2 genes were detected in our degenerate PCR screen at E11 (Fig. 7) and the 4 ORs from Ch.2 in our specific PCR analysis (Fig. 2A) did not show robust expression until at least E13 (Fig. 2B). Consistent with the notion that there may be regulatory elements affecting chromosomal expression, at E11 three chromosomes showed overrepresentation compared with expected expression levels in our degenerate PCR screen (using total ORs on a chromosome as the expected value). Of the retrieved clones, 35.7% were on Ch.7, which contains the second largest number of OR genes (20%). Ch.6, which contains one of the lower number of OR genes (2.5%), accounted for 14.3% of the retrieved clones while 10.7% were on Ch.14, which also contains one of the lower number of OR genes (3%).
It is interesting to note that MOR244-1 and MOR244-2 [the highest frequency of retrieval, and both from Ch.14 (Nagawa et al., 2002)] are under the control of the H-element (Serizawa et al., 2003; Fuss et al., 2007). While other enhancers have not yet been described, it is tempting to speculate that similar enhancers may exist on chromosomes 7 and 6 accounting for the enhanced expression of OR genes on these chromosomes at E11.
Key olfactory signaling molecules, show different expression patterns; for example ACIII is present by E13 (Treloar et al., 2005), but Golf and OCNC1, present in the odor transduction cascade of mature OSNs, do not appear robustly until ∼E15 (Menco et al., 1994). Before E15 immature OSNs appear to express Gs in lieu of Golf (Chesler et al., 2007). PCR data showed that even though OR expression turns on early in development, it is not until E13 that most of the characterized ORs show consistent and robust expression. Many more cells begin to express ORs ∼E13, which is evident from our ISH data showing an average 100-fold increase in OR expression from E11.5 to E13 (Fig. 4). Additionally, increased expression may also be accounted for by the onset of ciliogenesis, which occurs ∼E13 (Schwarzenbacher et al., 2005; Treloar et al., 2005). The increase in the number and length of OSN cilia would likely increase expression of the gene (i.e., more copies per cell) due to the increased protein insertion into the membrane.
Expression profiles were unique for each OR studied (Fig. 4, 5). Interestingly, no correlation was found between day of onset and total number of cells expressing an OR. For example, MOR103-15 (I7) turned on relatively late at E13 but had one of the higher expressions (via in situ) at PND2. This trend was not continued however as I7 was among the lower-expressing ORs during maturity (via microarray), reaching a plateau ∼PND20. This result of unique expression profiles is in agreement with the developmental profile of nine ORs developmentally characterized in the septal organ (Tian and Ma, 2008).
One of the most striking results is the transient perinatal decrease in the number of OSNs expressing one specific OR (Fig. 3, 4). Elsewhere in the CNS developmental overproduction of cells is adjusted by programmed cell death. Similarly, OSN axon projections to the olfactory bulb undergo significant refinement and remodeling, including the loss of supernumerary glomeruli, but the timeframe of remodeling appears to be unique for different ORs (Zou et al., 2004; Kerr and Belluscio, 2006). It is likely that the loss of supernumerary glomeruli in the OB may account for the reduction in the number of OSNs expressing a specific OR.
ORs, and members of their transduction cascade, are implicated in the glomerular coalescence of OSN axons that express the same OR (Mombaerts et al., 1996; Wang et al., 1998; Feinstein and Mombaerts, 2004; Imai et al., 2006; Chesler et al., 2007; Col et al., 2007; Zou et al., 2007). To establish whether the temporal expression OR pattern correlates with the extension and navigation of axons from the OE toward the OB, we examined onset at a time point before axon extension. We show that some ORs are expressed at E9, at least 1 d earlier that the first appearance of OSN axons in the OE, and ∼2 d before the initial contact of these axons with the developing OB (A. Miller and C. Greer, unpublished observations). This expression pattern strongly supports the notion that ORs have an instructive role in axon coalescence and the formation of glomeruli.
There is also evidence from both rodents and humans that embryos detect odorants in the amniotic fluid and that this can affect later postnatal behavior. For example, odor cues used to initiate rodent suckling during the early postnatal period are altered when a novel odor is introduced into the amniotic fluid before birth (Pedersen and Blass, 1982). Moreover, we previously showed that odorants introduced into the amniotic fluid result in an increase in functional activity in the OBs of late-stage embryos, suggesting that nascent connectivity between the OSN axons and their OB targets is established before birth (Pedersen et al., 1983). How these indices of embryonic olfactory function may relate to the variable onset of ORs is not yet known, but does pose the interesting hypothesis that OR onset may relate to developmentally specific functions or behaviors.
Together, our data provide compelling evidence of a much earlier onset of OR expression than previously reported. The earlier onset of OR expression is consistent with the proposed role of ORs in axon outgrowth and glomerular coalescence. Our data also highlight the diverse patterns of expression between different ORs over development and maturity. Some ORs are high expressers while others are low, some plateau early, some peak embryonically while others show a steady increase over life. No one rule appears to apply to all ORs supporting the notion of multiple levels of regulation of OR expression.
Footnotes
This work was supported in part by National Institutes of Health–National Institute on Deafness and Other Communication Disorders Grants DC00210, DC006291, and DC006972 to C.A.G., DC007600 to H.B.T., and DC003159 to S.J.F. A.M.M. was supported by the Yale Medical Scientist Training Program (GM0720) and F30 DC010324. A.T. was supported by the generosity of the Yale University School of Medicine Office of Student Research. We express their appreciation to all members of the Greer laboratory for helpful discussions and critical reading of the manuscript.
References
- Barnea G, O'Donnell S, Mancia F, Sun X, Nemes A, Mendelsohn M, Axel R. Odorant receptors on axon termini in the brain. Science. 2004;304:1468. doi: 10.1126/science.1096146. [DOI] [PubMed] [Google Scholar]
- Barth AL, Justice NJ, Ngai J. Asynchronous onset of odorant receptor expression in the developing zebrafish olfactory system. Neuron. 1996;16:23–34. doi: 10.1016/s0896-6273(00)80020-3. [DOI] [PubMed] [Google Scholar]
- Chesler AT, Zou DJ, Le Pichon CE, Peterlin ZA, Matthews GA, Pei X, Miller MC, Firestein S. A G protein/cAMP signal cascade is required for axonal convergence into olfactory glomeruli. Proc Natl Acad Sci U S A. 2007;104:1039–1044. doi: 10.1073/pnas.0609215104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- Col JA, Matsuo T, Storm DR, Rodriguez I. Adenylyl cyclase-dependent axonal targeting in the olfactory system. Development. 2007;134:2481–2489. doi: 10.1242/dev.006346. [DOI] [PubMed] [Google Scholar]
- Conzelmann S, Levai O, Breer H, Strotmann J. Extraepithelial cells expressing distinct olfactory receptors are associated with axons of sensory cells with the same receptor type. Cell Tissue Res. 2002;307:293–301. doi: 10.1007/s00441-001-0507-6. [DOI] [PubMed] [Google Scholar]
- Cuschieri A, Bannister LH. The development of the olfactory mucosa in the mouse: light microscopy. J Anat. 1975a;119:277–286. [PMC free article] [PubMed] [Google Scholar]
- Cuschieri A, Bannister LH. The development of the olfactory mucosa in the mouse: electron microscopy. J Anat. 1975b;119:471–498. [PMC free article] [PubMed] [Google Scholar]
- Dubacq C, Jamet S, Trembleau A. Evidence for developmentally regulated local translation of odorant receptor mRNAs in the axons of olfactory sensory neurons. J Neurosci. 2009;29:10184–10190. doi: 10.1523/JNEUROSCI.2443-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein P, Mombaerts P. A contextual model for axonal sorting into glomeruli in the mouse olfactory system. Cell. 2004;117:817–831. doi: 10.1016/j.cell.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Fuss SH, Omura M, Mombaerts P. Local and cis effects of the H element on expression of odorant receptor genes in mouse. Cell. 2007;130:373–384. doi: 10.1016/j.cell.2007.06.023. [DOI] [PubMed] [Google Scholar]
- Gong Q, Shipley MT. Evidence that pioneer olfactory axons regulate telencephalon cell cycle kinetics to induce the formation of the olfactory bulb. Neuron. 1995;14:91–101. doi: 10.1016/0896-6273(95)90243-0. [DOI] [PubMed] [Google Scholar]
- Hinds JW, Hinds PL. Synapse formation in the mouse olfactory bulb. I. Quantitative studies. J Comp Neurol. 1976;169:15–40. doi: 10.1002/cne.901690103. [DOI] [PubMed] [Google Scholar]
- Imai T, Sakano H. Odorant receptor gene choice and axonal projection in the mouse olfactory system. Results Probl Cell Differ. 2009;47:57–75. doi: 10.1007/400_2008_3. [DOI] [PubMed] [Google Scholar]
- Imai T, Suzuki M, Sakano H. Odorant receptor-derived cAMP signals direct axonal targeting. Science. 2006;314:657–661. doi: 10.1126/science.1131794. [DOI] [PubMed] [Google Scholar]
- Ishii T, Omura M, Mombaerts P. Protocols for two- and three-color fluorescent RNA in situ hybridization of the main and accessory olfactory epithelia in mouse. J Neurocytol. 2004;33:657–669. doi: 10.1007/s11068-005-3334-y. [DOI] [PubMed] [Google Scholar]
- Iwema CL, Fang H, Kurtz DB, Youngentob SL, Schwob JE. Odorant receptor expression patterns are restored in lesion-recovered rat olfactory epithelium. J Neurosci. 2004;24:356–369. doi: 10.1523/JNEUROSCI.1219-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr MA, Belluscio L. Olfactory experience accelerates glomerular refinement in the mammalian olfactory bulb. Nat Neurosci. 2006;9:484–486. doi: 10.1038/nn1673. [DOI] [PubMed] [Google Scholar]
- Kratz E, Dugas JC, Ngai J. Odorant receptor gene regulation: implications from genomic organization. Trends Genet. 2002;18:29–34. doi: 10.1016/s0168-9525(01)02579-3. [DOI] [PubMed] [Google Scholar]
- Lee AC, Tian H, Grosmaitre X, Ma M. Expression patterns of odorant receptors and response properties of olfactory sensory neurons in aged mice. Chem Senses. 2009;34:695–703. doi: 10.1093/chemse/bjp056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Monsen E, Dabney AR, Storey JD. EDGE: extraction and analysis of differential gene expression. Bioinformatics. 2006;22:507–508. doi: 10.1093/bioinformatics/btk005. [DOI] [PubMed] [Google Scholar]
- Malnic B, Hirono J, Sato T, Buck LB. Combinatorial receptor codes for odors. Cell. 1999;96:713–723. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- Maritan M, Monaco G, Zamparo I, Zaccolo M, Pozzan T, Lodovichi C. Odorant receptors at the growth cone are coupled to localized cAMP and Ca2+ increases. Proc Natl Acad Sci U S A. 2009;106:3537–3542. doi: 10.1073/pnas.0813224106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menco BP, Tekula FD, Farbman AI, Danho W. Developmental expression of G-proteins and adenylyl cyclase in peripheral olfactory systems. Light microscopic and freeze-substitution electron microscopic immunocytochemistry. J Neurocytol. 1994;23:708–727. doi: 10.1007/BF01181645. [DOI] [PubMed] [Google Scholar]
- Miyamichi K, Serizawa S, Kimura HM, Sakano H. Continuous and overlapping expression domains of odorant receptor genes in the olfactory epithelium determine the dorsal/ventral positioning of glomeruli in the olfactory bulb. J Neurosci. 2005;25:3586–3592. doi: 10.1523/JNEUROSCI.0324-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P. Axonal wiring in the mouse olfactory system. Annu Rev Cell Dev Biol. 2006;22:713–737. doi: 10.1146/annurev.cellbio.21.012804.093915. [DOI] [PubMed] [Google Scholar]
- Mombaerts P, Wang F, Dulac C, Chao SK, Nemes A, Mendelsohn M, Edmondson J, Axel R. Visualizing an olfactory sensory map. Cell. 1996;87:675–686. doi: 10.1016/s0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- Nagawa F, Yoshihara S, Tsuboi A, Serizawa S, Itoh K, Sakano H. Genomic analysis of the murine odorant receptor MOR28 cluster: a possible role of gene conversion in maintaining the olfactory map. Gene. 2002;292:73–80. doi: 10.1016/s0378-1119(02)00670-4. [DOI] [PubMed] [Google Scholar]
- Nef P, Hermans-Borgmeyer I, Artières-Pin H, Beasley L, Dionne VE, Heinemann SF. Spatial pattern of receptor expression in the olfactory epithelium. Proc Natl Acad Sci U S A. 1992;89:8948–8952. doi: 10.1073/pnas.89.19.8948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen PE, Blass EM. Prenatal and postnatal determinants of the 1st suckling episode in albino rats. Dev Psychobiol. 1982;15:349–355. doi: 10.1002/dev.420150407. [DOI] [PubMed] [Google Scholar]
- Pedersen PE, Stewart WB, Greer CA, Shepherd GM. Evidence for olfactory function in utero. Science. 1983;221:478–480. doi: 10.1126/science.6867725. [DOI] [PubMed] [Google Scholar]
- Ressler KJ, Sullivan SL, Buck LB. Information coding in the olfactory system: evidence for a stereotyped and highly organized epitope map in the olfactory bulb. Cell. 1994;79:1245–1255. doi: 10.1016/0092-8674(94)90015-9. [DOI] [PubMed] [Google Scholar]
- Royal SJ, Key B. Development of P2 olfactory glomeruli in P2-internal ribosome entry site-tau-LacZ transgenic mice. J Neurosci. 1999;19:9856–9864. doi: 10.1523/JNEUROSCI.19-22-09856.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H, Mimmack M, Kishimoto J, Keverne EB, Emson PC. Expression of olfactory receptors, G-proteins and AxCAMs during the development and maturation of olfactory sensory neurons in the mouse. Brain Res Dev Brain Res. 1998;110:69–81. doi: 10.1016/s0165-3806(98)00096-0. [DOI] [PubMed] [Google Scholar]
- Schwarzenbacher K, Fleischer J, Breer H, Conzelmann S. Expression of olfactory receptors in the cribriform mesenchyme during prenatal development. Gene Exp Patterns. 2004;4:543–552. doi: 10.1016/j.modgep.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Schwarzenbacher K, Fleischer J, Breer H. Formation and maturation of olfactory cilia monitored by odorant receptor-specific antibodies. Histochem Cell Biol. 2005;123:419–428. doi: 10.1007/s00418-005-0790-5. [DOI] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, Yoshihara Y, Sakano H. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science. 2003;302:2088–2094. doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Sakano H. One neuron-one receptor rule in the mouse olfactory system. Trends Genet. 2004;20:648–653. doi: 10.1016/j.tig.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Shay EL, Greer CA, Treloar HB. Dynamic expression patterns of ECM molecules in the developing mouse olfactory pathway. Dev Dyn. 2008;237:1837–1850. doi: 10.1002/dvdy.21595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strotmann J, Levai O, Fleischer J, Schwarzenbacher K, Breer H. Olfactory receptor proteins in axonal processes of chemosensory neurons. J Neurosci. 2004;24:7754–7761. doi: 10.1523/JNEUROSCI.2588-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan SL, Bohm S, Ressler KJ, Horowitz LF, Buck LB. Target-independent pattern specification in the olfactory epithelium. Neuron. 1995;15:779–789. doi: 10.1016/0896-6273(95)90170-1. [DOI] [PubMed] [Google Scholar]
- Theiler K. The house mouse: atlas of mouse development. New York: Springer; 1989. [Google Scholar]
- Tian H, Ma M. Differential development of odorant receptor expression patterns in the olfactory epithelium: a quantitative analysis in the mouse septal organ. Dev Neurobiol. 2008;68:476–486. doi: 10.1002/dneu.20612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treloar HB, Nurcombe V, Key B. Expression of extracellular matrix molecules in the embryonic rat olfactory pathway. J Neurobiol. 1996;31:41–55. doi: 10.1002/(SICI)1097-4695(199609)31:1<41::AID-NEU4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Treloar HB, Uboha U, Jeromin A, Greer CA. Expression of the neuronal calcium sensor protein NCS-1 in the developing mouse olfactory pathway. J Comp Neurol. 2005;482:201–216. doi: 10.1002/cne.20431. [DOI] [PubMed] [Google Scholar]
- Treloar HB, Ray A, Dinglasan LA, Schachner M, Greer CA. Tenascin-C is an inhibitory boundary molecule in the developing olfactory bulb. J Neurosci. 2009;29:9405–9416. doi: 10.1523/JNEUROSCI.2356-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar R, Chao SK, Sitcheran R, Nuñez JM, osshall LB, Axel R. Topographic organization of sensory projections to the olfactory bulb. Cell. 1994;79:981–991. doi: 10.1016/0092-8674(94)90029-9. [DOI] [PubMed] [Google Scholar]
- Wang F, Nemes A, Mendelsohn M, Axel R. Odorant receptors govern the formation of a precise topographic map. Cell. 1998;93:47–60. doi: 10.1016/s0092-8674(00)81145-9. [DOI] [PubMed] [Google Scholar]
- Whitesides JG, 3rd, LaMantia AS. Differential adhesion and the initial assembly of the mammalian olfactory nerve. J Comp Neurol. 1996;373:240–254. doi: 10.1002/(SICI)1096-9861(19960916)373:2<240::AID-CNE7>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Zhang X, Rodriguez I, Mombaerts P, Firestein S. Odorant and vomeronasal receptor genes in two mouse genome assemblies. Genomics. 2004a;83:802–811. doi: 10.1016/j.ygeno.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Zhang X, Rogers M, Tian H, Zou DJ, Liu J, Ma M, Shepherd GM, Firestein SJ. High-throughput microarray detection of olfactory receptor gene expression in the mouse. Proc Natl Acad Sci U S A. 2004b;101:14168–14173. doi: 10.1073/pnas.0405350101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou DJ, Feinstein P, Rivers AL, Mathews GA, Kim A, Greer CA, Mombaerts P, Firestein S. Postnatal refinement of peripheral olfactory projections. Science. 2004;304:1976–1979. doi: 10.1126/science.1093468. [DOI] [PubMed] [Google Scholar]
- Zou DJ, Chesler AT, Le Pichon CE, Kuznetsov A, Pei X, Hwang EL, Firestein S. Absence of adenylyl cyclase 3 perturbs peripheral olfactory projections in mice. J Neurosci. 2007;27:6675–6683. doi: 10.1523/JNEUROSCI.0699-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]