Abstract
To unravel the molecular mechanisms of drought responses in tomato, gene expression profiles of two drought-tolerant lines identified from a population of Solanum pennellii introgression lines, and the recurrent parent S. lycopersicum cv. M82, a drought-sensitive cultivar, were investigated under drought stress using tomato microarrays. Around 400 genes identified were responsive to drought stress only in the drought-tolerant lines. These changes in genes expression are most likely caused by the two inserted chromosome segments of S. pennellii, which possibly contain drought-tolerance quantitative trait loci (QTLs). Among these genes are a number of transcription factors and signalling proteins which could be global regulators involved in the tomato responses to drought stress. Genes involved in organism growth and development processes were also specifically regulated by drought stress, including those controlling cell wall structure, wax biosynthesis, and plant height. Moreover, key enzymes in the pathways of gluconeogenesis (fructose-bisphosphate aldolase), purine and pyrimidine nucleotide biosynthesis (adenylate kinase), tryptophan degradation (aldehyde oxidase), starch degradation (β-amylase), methionine biosynthesis (cystathionine β-lyase), and the removal of superoxide radicals (catalase) were also specifically affected by drought stress. These results indicated that tomato plants could adapt to water-deficit conditions through decreasing energy dissipation, increasing ATP energy provision, and reducing oxidative damage. The drought-responsive genes identified in this study could provide further information for understanding the mechanisms of drought tolerance in tomato.
Keywords: Drought stress, gene expression, introgression lines, microarray, tomato
Introduction
Water deficiency is one of the primary causes of the reduction in crop yield (Boyer, 1982). Increasing aridity of semi-arid regions together with limited water resources has led to an exigent necessity for improving crop drought resistances (Passioura, 2007). Elucidating the molecular mechanisms of drought tolerance is critical for increasing crop production and quality. Previous reports have indicated that plants do not passively accept environmental stresses but respond actively through perception of drought stress signals, resulting in enhanced expression of related genes to protect themselves from stress damage (Shinozaki and Yamaguchi-Shinozaki, 2007).
During the process of plant responses to drought stress, a large number of genes are induced. These genes are classified into two major groups according to their putative functional modes. The first group comprises the genes encoding structural proteins, which are downstream effectors in the stress response pathway including osmo-regulatory genes, antioxidant proteins, aquaporins, and late embryogenesis abundant (LEA) proteins (Bailly et al., 2001; Breton et al., 2003; Wang et al., 2006; Shinozaki and Yamaguchi-Shinozaki, 2007). The second group consists of the genes encoding regulatory proteins, which are early response transcriptional activators including transcription factors and protein kinases. The stress-related transcription factors mainly including bZIP, WRKY, MYB, and AP2/EREBP proteins have been proved to play important roles in the regulation of drought tolerance (Abe et al., 1997; Finkelstein and Lynch, 2000; Marè et al., 2004; Song et al., 2005). The protein kinases, including calmodulin dependent protein kinases (CDPKs), mitogen-activated protein kinases (MAPKs), receptor protein kinases (RPKs), and ribosomal protein kinases, are involved in the signal cascade amplification in response to different stress factors (Mizoguchi et al., 1995; Hong et al., 1997; Ludwig et al., 2004). The expression of drought-inducible genes can be governed by ABA-dependent or ABA-independent regulatory systems (Yamaguchi-Shinozaki and Shinozaki, 2005). There are also extensive cross-talks between responses to drought and other environmental stresses such as light and biotic stresses (Huang et al., 2008).
Changes of gene expression under drought stress cause a series of physiological and biochemical alterations. Photosynthesis, one of the primary biosynthetic pathways, is significantly affected by drought stress, which restricts the normal function of other metabolic pathways, such as nitrogen fixation (Chaves et al., 2009). The respiration pathway, which breaks complex molecules into simple compounds to provide the energy required for plant development, is accelerated under drought stress (Haupt-Herting et al., 2001). Protection systems such as the antioxidation pathway, which can reinforce plant cells to form reactive oxygen species scavengers, are also affected by drought stress (Apel and Hirt, 2004). Despite significant progress during the past decade in our understanding of pathways affected by drought stress, limited information is available regarding pathway dynamics in tomato under drought stress.
Tomato has been used as a model system and is also one of the most important economical vegetable plants in the Solanaceae family. There exist many wild relatives of cultivated tomato Solanum lycopersicum with different degrees of tolerances to abiotic and biotic stresses. S. pennellii LA716 is one of the wild crossable relatives of cultivated species and its leaves have a distinct ability to withstand desiccation in extreme arid conditions (Rick, 1973). A collection of 50 introgression lines (ILs), which covers the entire genome of their donor parent, S. pennellii, in the background of the cultivated species, S. lycopersicum M82, a drought-sensitive cultivar, were previously generated (Eshed et al, 1992; Eshed and Zamir, 1994). This together with the currently expanding tomato research platforms such as genome sequencing and microarray applications could facilitate research toward a better understanding of drought-tolerance mechanisms in tomato. Genome-wide expression profiling of plants under drought stress has been reported in many plant species, including Arabidopsis (Seki et al., 2001, 2002; Bray, 2004; Huang et al., 2008), rice (Rabbani et al., 2003; Hazen et al., 2005), barley (Talamè et al., 2007; Guo et al., 2009), maize (Hayano-Kanashiro et al., 2009), sorghum (Pratt et al., 2005), and potato (Vasquez-Robinet et al., 2008), however no such reports have been found in tomato. In this study, two highly drought-tolerant ILs were identified, and gene expression changes of these two ILs and their recurrent parent M82 were characterized under drought stress to gain a deeper understanding of the drought-tolerance mechanisms in tomato.
Materials and methods
Plant material and screening of drought-tolerant introgression lines
Seeds of 50 ILs, their donor parent, S. pennellii, and recurrent parent, S. lycopersicum cv. M82, were obtained from the Tomato Genetic Resource Center (TGRC) at the University of California, Davis. Each IL was named by the corresponding single inserted chromosome segment of S. pennellii. For example, IL1-1 (LA4027) means that one fragment of chromosome 1 of S. pennellii was introduced into the background genotype of M82. Different inserted fragments were identified using molecular makers (Eshed and Zamir 1994).
To establish the drought-screening system, around 240 uniform-sized seeds of M82 and S. pennellii were surface-sterilized with 75% (v/v) ethanol for 30 s, further sterilized using 4% (w/v) NaClO solution for 15 min, and finally rinsed with sterile distilled water. Thirty seeds were placed on each filter paper soaked with 4 ml of 15% (w/v) PEG 6000 solution or sterile distilled water in 9 cm Petri dishes. Petri dishes were placed in an illuminated growth chamber at the temperature regime of 29/25 °C (light/dark). Germination rate was scored visually as radical protrusion (2 mm) at 12 h intervals for 10 consecutive days. To investigate phenotypic changes at the seedling stage in M82 and S. pennellii under drought stress, seedlings of both genotypes were grown in black plastic pots in the greenhouse. Plants at the five-leaf stage were watered only once every 5 d. Phenotypic changes were observed and final survival rate was calculated after one month.
To screen for drought-tolerant ILs, 50 plump and uniform-sized seeds of each IL, S. pennellii, and M82 were germinated on filter papers in Petri dishes at 29 °C. The germinated seeds were then sown in germination media. At the one-true-leaf stage, 10 uniform seedlings of each genotype, in triplicate, were transplanted into 8×8 cm nutrition pots (one seedling per pot) filled with a mixture of peat, vermiculite, and perlite at a ratio of 1:1:1 (by vol.). These seedlings were randomly placed in the greenhouse with automatic control systems. Seedlings were irrigated every third day with 40 ml of modified 1/5 Johnson's solution supplemented with Fe-EDDHA (Wang et al., 2001). When these plants were at the five-true-leaf stage, all pots were immersed in water (3 cm depth) for 12 h and then kept in the greenhouse without watering at 29/23 °C (day/night) and 65% relative humidity. Seedlings were not supplied with water or nutrient solution for recovery until leaves of most lines showed permanent wilting. After 2 weeks of recovery, drought stress was imposed again and finally the lines which survived were identified. The screening experiments were performed in three different seasons (May, September, and November) during the year 2006.
To investigate drought tolerance of the two selected lines further, pot-grown seedlings of S. pennellii, M82, and the two inbred lines at the 5-leaf stage were taken out and immersed in 2%, 4%, 6% (w/v) PEG 6000 solutions, respectively, at 25 °C for 12 h. Leaf electroconductivity of each stress-treated sample was measured using the DDBJ-350 portable electric conductivity radiometer. The leaf damage rate was determined as EC1/EC2, where EC1 is the electrical conductivity of the bathing water with leaf discs after 12 h and EC2 is the electrical conductivity of the same water after boiling for 20 min.
To determine the leaf water-holding capacity of different tomato genotypes, third leaves from the top of S. pennellii, M82, and the two selected lines at the 5-leaf stage were sampled. The detached leaves were placed in 25 °C in a dark environment and weighed every hour. Water deficit ratio of each sample was then calculated as the ratio of the leaf weight lost to the initial leaf fresh weight.
Stress treatment and total RNA isolation
Two drought-tolerant lines (IL2-5 and IL9-1) and M82 were grown as described above. Seedlings were well watered until they reached the 6-leaf stage, then drought stress was imposed by withholding water, except for control plants that were watered as usual. Triplicate samples with five seedlings of each line were collected at 09.00 h every day. The third leaf from the bottom of each plant was collected and used to determine the relative water content (RWC), while the fourth leaf from each plant was frozen in liquid nitrogen and stored at –70 °C until use. Control plants were selected with 88.13% leaf RWC (M82), 88.41% (IL2-5), and 88.61% (IL9-1), and stress treated plants with 68.86% leaf RWC (M82), 72.42% (IL2-5), and 73.93% (IL9-1), which were under moderate stress according to Hsiao (1973), for RNA extraction.
Total RNA was isolated using the TRIzol reagent (Invitrogen, USA) according to the manufacturer's instruction. The quality of RNA was checked on a denaturing formaldehyde gel and further confirmed by measuring the ratio of A260/A280.
Fluorescent probe preparation and hybridization
Gene expression profiles of two selected drought-tolerant lines and M82 under drought stress and normal irrigated conditions were determined using tomato TOM2 arrays. Hybridization was performed on each of the materials tested with three biological replicates and two technical replicates (dye-swap).
Total RNA (5 μg) along with T7-Oligo (dT)15 (Boya, China) was used for reverse transcription of double-stranded cDNA using the DNA Synthesis Kit (Promega, USA), then transcribed to cRNA in vitro using T7 RiboMAX Express Large Scale RNA Production System (Promega, USA). 2 μg of cRNA plus Random Primer 9 (New England Biolabs, USA) was used to produce cDNA by M-MLV (200 μ/μl, Invitrogen, USA). Finally, a 2 μg cDNA aliquot, along with Random Primer 9, 120 μM final concentration of each dATP, dGTP, dTTP, 60 μM final concentration dCTP and 40 μM final concentration of Cy5-dCTP, Cy3-dCTP was used to produce Cy5/Cy3-labelled cDNAs by KLENOW (Takara, Japan).
Cy5/Cy3-labelled cDNA probes were hybridized with TOM2 microarrays at 42 °C overnight. Subsequently, the arrays were washed with 0.2% SDS and 2× SSC at 42 °C for 5 min, followed by washing with 0.2% SSC for 5 min at room temperature.
Microarray scanning and data analysis
Arrays were scanned using a LuxScan 10KA confocal laser scanner (CapitalBio, China). The images were analysed with ImaGene image analysis software (BioDiscovery, CA, USA). Spots with mean signal intensities less than local background intensities plus two standard deviations of the local background in both channels were regarded as empty spots and not included in the downstream statistical analysis. A Print-tip Lowess Normalization strategy was applied to normalize the ratio values for each array using the marray package in Bioconductor (Yang et al, 2002). Significances of gene expression changes between stressed and control plants were identified using the Patterns from Gene Expression package (PaGE; Grant et al., 2005). Genes with a false discovery rate (FDR; Benjamini and Hochberg, 1995) less than 0.05 and a fold change no less than 2 were considered as differentially expressed genes. Identification of significantly affected biochemical pathways and highly enriched GO terms, as well as functional classification of differentially expressed genes, were performed using the Plant MetGenMAP system (Joung et al., 2009). The entire array dataset and the experimental descriptions are available at the Tomato Expression Database (http://ted.bti.cornell.edu; Fei et al., 2006).
RT-PCR analysis
Total leaf RNAs isolated from M82, IL2-5, and IL9-1, as well as S. pennellii, under moderate drought stress and control conditions were used for RT-PCR analysis. cDNAs were synthesized from 9 μg total RNA using the M-MLV reverse transcriptase (Promega, USA). Tomato elongation factor 1α (EF1α) was used as the internal control. Primers were designed with the Primer3 program (http://frodo.wi.mit.edu/primer3/input.htm). Sequences of all the primer pairs are listed in Supplementary Table S1 at JXB online. PCRs were performed with 25–29 cycles of 94 °C 30 s, 52 °C 30 s, and 72 °C 1 min, using 1 μl cDNA as the template. PCR products were separated by electrophoresis on a 1% agarose gel with ethidium bromide in 1× TAE buffer and visualized under UV light.
Results
Drought tolerance characteristics of recurrent and donor parents of ILs
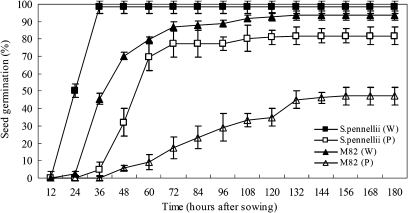
S. pennellii (LA716) has been assigned in TGRC as one of the stress-tolerant accessions. In this study, drought tolerance between S. pennellii and S. lycopersicum cv. M82 were also compared. Under normal conditions, the seeds of S. pennellii germinated faster than those of M82, however, no significant difference in the final germination rate was observed between the two genotypes. While under drought stress, the final germination rate of S. pennellii was significantly higher than that of M82 (Fig. 1). In addition, young seedlings of S. pennellii under drought stress for one month had lush green leaves except for the bottom first and second leaves; while most plants of M82 were dead and the leaves at the bottom and middle of the plants which survived turned yellow. All the above results supported that S. pennellii was more tolerant to drought stress than M82.
Fig. 1.
Seed germination patterns of S. pennellii and M82 under water irrigated and PEG treatment conditions. Vertical bars represent standard error of means. W, water irrigation; P, PEG treatment.
Identification of drought-tolerant ILs
Seedlings of M82 and a set of 50 ILs were subjected to drought stress and the survival rate of each line was investigated. Two lines, IL2-5 (LA4040) and IL9-1 (LA4078), showed significantly higher survival rates than M82 across all three seasons investigated (see Supplementary Table S2 at JXB online) while the leaf damage rates of these two lines, as well as S. pennellii, were significantly lower than that of M82 under different stress treatments (see Supplementary Table S3 at JXB online). Moreover, the water loss rates of detached leaves of the two selected lines were also significantly lower than those of M82 (see Supplementary Fig. S1 at JXB online). All the above results suggested that the drought tolerance-related QTLs were present in both lines, mainly corresponding to segments on chromosome 2 and chromosome 9 of S. pennellii, respectively. In addition, five lines with higher survival rates than M82, but lower than IL2-5 or IL9-1 were also identified (see Supplementary Table S2 at JXB online), indicating these corresponding chromosome segments may contain minor effect QTLs related to drought tolerance.
Expression profiles of drought-responsive genes in tomato
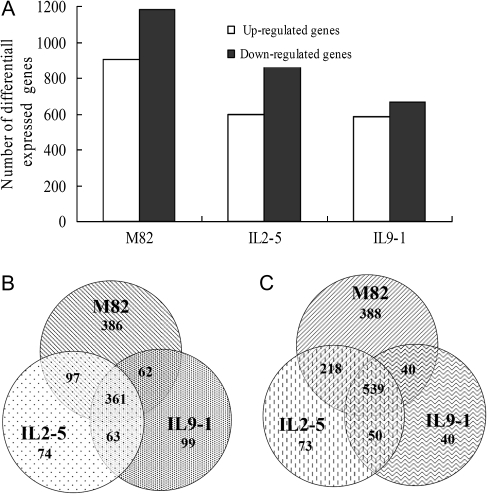
Changes of gene expression in IL2-5, IL9-1, and M82 under drought stress were investigated using tomato TOM2 microarrays. Approximately 1400, 1200, and 2000 drought-responsive genes were identified in IL2-5, IL9-1, and M82, respectively (Fig. 2A). All these drought-responsive genes were classified into different groups based on their expression patterns (Fig. 2B, C), among which 399 genes were differentially expressed only in the tolerant genotypes (see Supplementary Table S4 at JXB online). In addition, 900 genes were differentially expressed in all three genotypes tested. Drought-responsive genes were functionally classified into different categories of biological processes using a set of plant-specific GO slims (http://www.geneontology.org/GO.slims.shtml). As expected, the ‘response to stress’ and ‘response to abiotic stimulus’ were among the largest groups in all three genotypes (see Supplementary Table S5 at JXB online).
Fig. 2.
Number of differentially expressed genes in different tomato genotypes tested under drought stress. (A) Number of up- or down-regulated genes in M82, IL2-5, and IL9-1. (B) Venn diagram of up-regulated genes categorized in different genotypes. (C) Venn diagram of down-regulated genes classified in different genotypes.
Verification of microarray results by RT-PCR
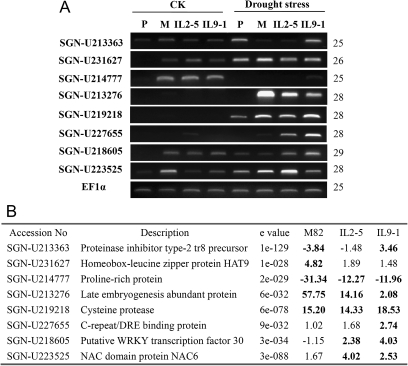
To confirm the microarray results, RT-PCR was conducted on eight randomly selected drought-responsive genes based on the microarray analysis (Fig. 3A). RT-PCR results agreed with the microarray data for 20 out of 24 (83%) data points (Fig. 3B), indicating there was a good consistency between the RT-PCR and microarray results.
Fig. 3.
Verification of microarray results by RT-PCR. (A) RT-PCR analysis of eight selected genes. eF1α was used as the internal control. Number of PCR cycles is listed on the right side. CK:, control plants watered as usual. (B) Expression ratios of the eight selected genes derived from the microarray analysis. P, S. pennellii; M, M82.
Expression profiles of the selected genes were investigated further under drought stress in S. pennellii, a drought-tolerant line and the donor parent of IL2-5 and IL9-1. As expected, the overall expression pattern of these genes under drought stress in IL2-5 and IL9-1 was more similar to that in S. pennellii than that in M82. In addition, genes (SGN-U213363, SGN-U218605, and SGN-U223525) induced only in IL2-5 and/or IL9-1 exhibited similar expression changes in S. pennellii under drought stress, indicating changes in expression of these genes in IL2-5 and IL9-1 could be largely due to the corresponding introgressed chromosome segments.
Drought-responsive transcription factors and signalling proteins in the tolerant lines
To understand the transcriptional regulation in drought stress response in the drought-tolerant genotypes, 43 genes identified in the tolerant genotypes which encoded transcription factors were characterized further (Table 1). These genes were classified into five major groups according to their putative DNA binding domains. The first group was the zinc-finger family which contained ten genes (SGN-U225317, SGN-U215566, SGN-U222278, SGN-U213245, SGN-U218605, SGN-U229870, SGN-U219949, SGN-U213244, SGN-U212725, SGN-U235916). Among them, all WRKY family genes were up-regulated under drought stress.
Table 1.
Transcription factors and signalling proteins responsive to drought only in drought-tolerant genotypes
| Accession no. | Annotation | e-value | M82 | IL2-5 | IL9-1 |
| Transcription factors | |||||
| Zinc-finger family | |||||
| SGN-U225317 | Zinc finger protein | 2e-035 | 1.62 | 2.51 | 2.09 |
| SGN-U215566 | Zinc finger protein OBP4-like | 6e-034 | 1.81 | 1.80 | 2.64 |
| SGN-U222278 | Zinc finger (C2H2 type) family protein | 1e-032 | –1.35 | –2.09 | –1.46 |
| SGN-U213245 | WIZZ | 1e-100 | 1.30 | 6.79 | 2.99 |
| SGN-U218605 | WRKY transcription factor 30 | 3e-034 | –1.15 | 2.38 | 4.03 |
| SGN-U229870 | WRKY transcription factor-c | 9e-063 | 1.44 | 2.57 | 2.71 |
| SGN-U219949 | WRKY transcription factor-c | 2e-089 | 1.51 | 2.12 | 2.34 |
| SGN-U213244 | WRKY family transcription factor | 1e-028 | 1.17 | 2.88 | 2.41 |
| SGN-U212725 | WRKY transcription factor 30 | 6e-040 | 1.84 | 1.74 | 3.31 |
| SGN-U235916 | Transcription factor WRKY5 | 1e-015 | 1.76 | 2.26 | 1.20 |
| NAC family | |||||
| SGN-U213215 | NAC domain protein NAC2 | 1e-103 | –1.18 | –2.30 | –2.70 |
| SGN-U232379 | NAC domain protein NAC2 | 1e-014 | –1.29 | –2.23 | –2.80 |
| SGN-U223525 | NAC domain protein NAC6 | 3e-088 | 1.67 | 4.02 | 2.53 |
| SGN-U233528 | Nam-like protein 1 | 1e-062 | 1.81 | 2.79 | 2.19 |
| SGN-U213216 | NAC domain protein NAC2 | 1e-051 | –1.15 | –1.92 | –2.20 |
| SGN-U223083 | NAC-domain protein | 5e-088 | –1.12 | 1.75 | 2.09 |
| SGN-U216370 | Nam-like protein 1 | 1e-114 | 1.64 | 2.35 | 1.81 |
| bHLH family | |||||
| SGN-U218964 | bHLH transcription factor GBOF-1 | 1e-039 | –1.87 | –2.10 | –2.10 |
| SGN-U215556 | bHLH family protein | 1e-057 | 1.20 | 2.08 | 1.68 |
| SGN-U215557 | bHLH family protein | 3e-057 | 1.18 | 2.27 | 1.76 |
| SGN-U219797 | bHLH family protein | 1e-044 | –1.92 | –1.89 | –2.17 |
| SGN-U235082 | bHLH transcription factor | 9e-017 | –1.59 | –1.81 | –2.10 |
| SGN-U238928 | bHLH transcription factor | 3e-029 | 1.90 | 1.59 | 2.03 |
| SGN-U217931 | Helix-loop-helix DNA-binding | 1e-033 | 1.32 | 1.67 | 2.30 |
| AP2/EREBP family | |||||
| SGN-U220658 | AP2/EREBP transcription factor | 9e-021 | 1.19 | 2.57 | 1.78 |
| SGN-U224968 | AP2/EREBP transcription factor | 1e-019 | –1.13 | 2.24 | –1.27 |
| SGN-U242104 | APETALA2 protein homolog HAP2 | 6e-059 | 1.12 | 2.26 | 1.70 |
| SGN-U226365 | AP2-like ethylene-responsive transcription factor | 2e-053 | –1.73 | –2.17 | –1.91 |
| SGN-U216297 | AP2/EREBP transcription factor | 2e-024 | 1.36 | 2.57 | 1.90 |
| SGN-U213644 | Transcription factor JERF1 | 0.0 | 1.62 | 2.34 | 2.39 |
| HSF family | |||||
| SGN-U227428 | Heat shock transcription factor HSF30 | 1e-053 | 1.87 | 2.29 | 2.66 |
| SGN-U225155 | Heat shock transcription factor HSF30 | 1e-108 | 1.92 | 2.58 | 3.02 |
| SGN-U222126 | Heat shock transcription factor | 1e-068 | 1.21 | –3.59 | 1.48 |
| SGN-U227452 | Heat shock transcription factor HSFA9 | 3e-051 | –1.15 | –2.16 | –1.82 |
| MYB family | |||||
| SGN-U218279 | Myb family transcription factor | 1e-175 | –1.17 | –2.03 | –1.72 |
| bZIP family | |||||
| SGN-U216671 | G-box binding protein | 1e-100 | –1.97 | –2.30 | –2.30 |
| Other transcription factors | |||||
| SGN-U216109 | CONSTANS interacting protein 2a (CCAAT-binding transcription factor) | 1e-119 | 1.76 | 1.96 | 2.56 |
| SGN-U217064 | CCAAT-binding transcription factor | 3e-053 | 1.33 | 2.39 | 2.64 |
| SGN-U230670 | Auxin response factor | 8e-065 | 1.94 | 1.78 | 2.04 |
| SGN-U219359 | AUX/IAA protein | 4e-031 | 1.59 | 1.79 | 2.69 |
| SGN-U220106 | SCL3 (scarecrow-like 3) | 1e-154 | –1.59 | –2.31 | –2.10 |
| SGN-U227808 | SCARECROW gene regulator | 6e-070 | 1.47 | 1.61 | 2.15 |
| SGN-U224075 | Transcription factor LIM | 3e-082 | 1.90 | 2.14 | 2.53 |
| Signalling related proteins | |||||
| SGN-U231755 | Cryptochrome 1 | 7e-068 | –1.88 | –2.08 | –1.55 |
| SGN-U226221 | Receptor-like protein kinase INRPK1c | 2e-094 | 1.37 | 2.81 | –1.64 |
| SGN-U216590 | Receptor kinase LRK10 | 1e-084 | 1.12 | 3.69 | 1.59 |
| SGN-U236017 | Receptor-like protein kinase INRPK1c | 3E-59 | 1.99 | 3.52 | 1.74 |
| SGN-U213787 | Receptor-like serine-threonine protein kinase | 1E-162 | 1.33 | 2.77 | 1.99 |
| SGN-U228947 | Somatic embryogenesis receptor kinase 1 | 1e-112 | 1.19 | 1.66 | 2.29 |
| SGN-U230845 | S-receptor kinase KIK1 precursor | 3E-58 | 1.82 | 1.68 | 2.07 |
| SGN-U220999 | receptor-like protein kinase 2 | 1E-139 | 1.37 | 2.68 | 2.8 |
| SGN-U232645 | Receptor-like serine-threonine protein kinase | 5E-78 | 1.54 | 2.5 | 2.56 |
| SGN-U213785 | Receptor-like serine-threonine protein kinase | 0 | 1.54 | 2.29 | 2.22 |
| SGN-U215877 | Mitogen-activated protein kinase 3 | 0.0 | 1.80 | 3.86 | 5.06 |
| SGN-U222625 | MAP3K gamma protein kinase | 5E-34 | 1.37 | 2.7 | 1.98 |
| SGN-U222776 | CBL-interacting protein kinase | 6e-039 | –1.63 | –2.27 | –2.23 |
| SGN-U219271 | Protein kinase family protein | 1e-102 | –1.86 | –2.21 | –1.77 |
| SGN-U225466 | calmodulin binding/cation channel/cyclic nucleotide binding | 1e-122 | 1.97 | 2.22 | –1.00 |
| SGN-U215815 | Phosphatidic acid phosphatase alpha | 1e-121 | 1.69 | 2.29 | 2.35 |
Significant difference (FDR <0.05 and fold change ≥2) in relative level is shown in bold.
The second group represented by the NAC family contained five NAC genes (SGN-U213215, SGN-U232379, SGN-U223525, SGN-U213216, SGN-U223083) and two NAM-like genes (SGN-U233528, SGN-U216370). The two NAM-like genes and two NAC genes were induced by drought treatment.
The third group included transcription factors from the basic helix-loop-helix (bHLH) family. Seven bHLH genes were identified as drought-responsive genes in this study. Of these, four genes were up-regulated (SGN-U215556, SGN-U215557, SGN-U238928, SGN-U217931).
The fourth group contained the AP2/EREBP family genes. Six genes (SGN-U220658, SGN-U224968, SGN-U216297, SGN-U242104 SGN-U226365, SGN-U213644) belonging to this family were identified. All of them except one (SGN-U226365) were up-regulated.
The fifth group consisted of heat shock transcription factors (HSF). Two up-regulated (SGN-U227428, SGN-U225155) and two down-regulated (SGN-U222126, SGN-U227452) HSFs were identified in this study.
The remaining transcription factors belonged to the MYB family (SGN-U218279), basic leucine zipper (bZIP) family (SGN-U216671) and other types, including CAAAT-binding proteins (SGN-U216109, SGN-U217064), auxin-related proteins (SGN-U230670, SGN-U219359), GRAS proteins (SGN-U220106, SGN-U227808), and a LIM domain protein (SGN-U224075).
To investigate the signal transduction process in tomato under water-stressed conditions, differentially expressed signalling-related genes were subjected to analysis. Sixteen genes involved in signalling pathways were identified in the drought-tolerant genotypes (Table 1). Among them, nine encoding RPKs (SGN-U226221, SGN-U216590, SGN-U228947, SGN-U236017, SGN-U213787, SGN-U230845, SGN-U220999, SGN-U232645, SGN-U213785) were up-regulated. The remaining differentially expressed genes encoded a MAPK (SGN-U215877), a CBL-interacting protein kinase (SGN-U222776), a calmodulin binding protein (SGN-U225466), a protein kinase family protein (SGN-U219271), and a phosphatase protein (SGN-U215815), most of which were also up-regulated.
Drought-responsive transcription and signalling regulators in both tolerant and sensitive genotypes
In the present study, a total of 82 drought-responsive transcription factors were identified in all three genotypes (see Supplementary Table S6 at JXB online). These proteins mainly fell into eight families. Among them, the expression levels of four zinc-finger genes, two MYB genes, one homeobox gene, one HSF gene, and other types of transcription factors (SGN-U232883, SGN-U222176, SGN-U219331, SGN-U223492, BT012912, SGN-U226862) were changed by more than 10-fold.
About 50 signalling-related genes differentially expressed in all three genotypes were also found. Among them, the expression levels of two hormone-related genes, one light-related gene, two phosphatase genes and one serine/threonine protein kinase gene were changed by more than 10-fold under drought stress (see Supplementary Table S6 at JXB online).
Organism growth and development related gene expression characteristics in the drought-tolerant genotypes
Among genes that were responsive to drought stress only in the tolerant genotypes, eight were involved in cell growth and differentiation, and anatomical structure morphogenesis. These genes encoded one epidermal cell wax-related gene (SGN-U213739), two cell wall structure-related genes (SGN-U213444, SGN-U214839), one brassinosteroids synthesis-related gene (SGN-U221333), one lipid molecular-related gene (SGN-U215749), one cell integrity of microspores-related gene (SGN-U222621), one programmed cell death-related gene (SGN-U216827), and one cell elongation related gene (SGN-U213594), and most of them were induced by drought stress (Table 2).
Table 2.
Organism growth and development-related drought-responsive genes only in drought-tolerant genotypes
| Accession no. | Annotation | e-value | M82 | IL2-5 | IL9-1 |
| SGN-U222621 | Nodulin MtN3 family protein | 5e-066 | –1.71 | –2.20 | –1.73 |
| SGN-U216827 | Phytophthora-inhibited protease 1 | 0.0 | 1.36 | 2.35 | 1.10 |
| SGN-U213594 | DWARF1/DIMINUTO | 0.0 | 1.24 | 1.25 | 2.18 |
| SGN-U214839 | Endo-1,4-beta-glucanase | 0.0 | 1.39 | 2.16 | 2.78 |
| SGN-U215749 | 1-phosphatidylinositol-4-phosphate 5-kinase | 0.0 | –1.63 | –1.88 | –3.10 |
| SGN-U213739 | CER1 (ECERIFERUM 1) | 0.0 | 1.29 | 1.69 | 2.18 |
| SGN-U213444 | Xyloglucan endotransglucosylase-hydrolase XTH5 | 0.0 | –1.29 | –2.23 | –2.76 |
| SGN-U221333 | Steroid 5 alpha reductase DET2 | 1e-154 | 1.38 | 3.60 | 5.56 |
Significant difference (FDR <0.05 and fold change ≥2) in relative level is shown in bold.
Biochemical pathways affected by drought stress in tomato
In order to assess the functional roles of drought-responsive genes involved in biochemical pathways, and to study the biochemical adaptations to drought stress in tomato, special and common biochemical metabolisms affected by drought stress in the tolerant and sensitive genotypes were identified.
Six metabolic pathways were specifically modulated by drought-responsive genes in the tolerant genotypes but not in the sensitive genotype (Table 3). Among these drought-responsive genes, one encoding fructose-bisphosphate aldolase (SGN-U232066) in the gluconeogenesis pathway was down-regulated in IL2-5 (see Supplementary Fig. S2 at JXB online). Genes encoding an aldehyde oxidase (SGN-U213960) involved in the tryptophan degradation pathway and an adenylate kinase (SGN-U232826) functioning in the salvage pathways of purine and pyrimidine nucleotides were up-regulated in IL2-5 (see Supplementary Fig. S2 at JXB online). Several genes encoding key enzymes in the pathways responsible for the removal of superoxide radicals, starch degradation, and methionine biosynthesis were significantly differentially expressed in IL9-1 (see Supplementary Fig. S2 at JXB online). They included up-regulated catalases (SGN-U212687, SGN-U224934) and cystathionine beta-lyase (SGN-U219944), and down-regulated beta-amylase (SGN-U220865, SGN-U221601).
Table 3.
Differentially expressed genes involved in biochemical pathways affected by drought stress in the tolerant genotypes
| Gene accession no | Gene description | e-value | M82 | IL2-5 | IL9-1 |
| Gluconeogenesis | |||||
| SGN-U232066 | Fructose-bisphosphate aldolase | 2e-073 | –1.48 | –2.03 | –1.30 |
| Salvage pathways of purine and pyrimidine nucleotides | |||||
| SGN-U232826 | Adenylate kinase | 3e-053 | 1.63 | 2.02 | 1.23 |
| Tryptophan degradation | |||||
| SGN-U213960 | Aldehyde oxidase | 1e-179 | 1.77 | 2.13 | 1.61 |
| Starch degradation | |||||
| SGN-U220865 | Beta-amylase 1 | 1e-149 | 1.21 | –1.12 | –2.38 |
| SGN-U221601 | Beta-amylase 1 | 1e-165 | 1.02 | –1.56 | –2.28 |
| Methionine biosynthesis | |||||
| SGN-U219944 | Cystathionine beta-lyase | 1e-110 | 1.11 | 1.66 | 3.29 |
| Removal of superoxide radicals | |||||
| SGN-U212687 | Catalase isozyme 1 | 0.0 | 1.49 | 1.41 | 2.09 |
| SGN-U224934 | Catalase 3 | 5e-093 | 1.98 | 1.64 | 2.29 |
Significant difference (FDR <0.05 and fold change ≥2) in relative level of gene expression is shown in bold.
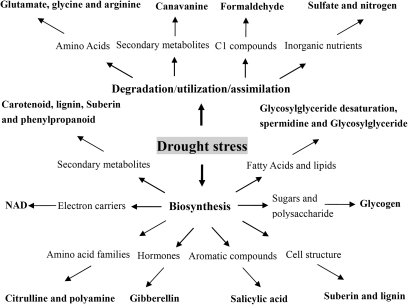
A total of 19 biochemical pathways were affected by drought stress in all three genotypes (see Supplementary Table S7 at JXB online). These pathways were responsible for the biosynthesis or degradation of diverse metabolites including secondary metabolites, electron carriers, amino acids, hormones, aromatic compounds, cell structure components, sugars and polysaccharides, fatty acids and lipids, C1 compounds, and inorganic nutrients (Fig. 4), indicating the complexity of metabolic changes involved in plant responses to drought stress.
Fig. 4.
Biochemical pathways significantly affected by drought stress in all three tomato genotypes.
Four genes encoding enzymes of the carotenoid biosynthesis pathway (phytoene synthase, lycopene β-cyclase, β-carotene hydroxylase, zeaxanthin epoxidase) were significantly down-regulated by drought stress. Almost all genes in nitrogen, sulphate, and formaldehyde degradation pathways, lignin, suberin, spermidine, polyamine, phenylpropanoid, and salicylic acid biosynthesis pathways, and the glycosylglyceride desaturation pathway were also significantly down-regulated in all three genotypes under drought stress, while the majority of genes in the pathways of glutamate, arginine, and canavanine degradation, and citrulline biosynthesis were up-regulated.
Discussion
In this study, two drought-tolerant ILs and their recurrent parent, a drought-sensitive tomato genotype, were used to characterize the differences of transcriptome dynamics under drought stress. Expression of drought-responsive genes in the drought-tolerant genotypes might be affected by inserted chromosome segments of S. pennellii and these genes might contribute to the increased drought tolerance, while a large number of genes differentially expressed in all three genotypes might play basal roles in the response to drought stress in tomato.
Transcription regulation and signal transduction under drought stress in tomato
Transcription factors and signalling regulators are considered to be the most important category of genes involved in regulating the expression of downstream drought-responsive genes (Bray, 2004; Trewavas and Malho, 1997). Comparison of gene expression profiling between the tolerant ILs and M82 under drought stress is essential for the elucidation of drought response networks in tomato.
A number of differentially expressed genes encoding transcriptional factors were identified in the tolerant genotypes. Among them, WRKY family proteins have been shown to be highly induced by various environmental stresses and involved in the regulation of diverse biological processes in plants (Zou et al., 2007; Wei et al., 2008). AP2/EREBP family transcription factors and NAC genes play significant roles in plant tolerance to various environmental stresses (Hu et al., 2006; Trujillo et al., 2008; Zhang et al., 2009; Zheng et al., 2009). In addition, bHLH genes are involved in the ABA response regulation under stress conditions (Abe et al., 2003; Li et al., 2007). HSF family genes are known as regulators to prevent the accumulation of damaged proteins and to maintain cellular homeostasis (Storozhenko et al., 1998; Wang et al., 2004). MYB family transcription factors, auxin-related transcription factors, and GRAS regulators are also involved in stress responses (Leymarie et al., 1996; Cominelli et al., 2005; Fode et al., 2008). All these groups of transcription factors identified in the tolerant genotypes showed complicated transcriptional regulatory networks of drought responses in tomato.
In the present study, a set of drought-regulated signalling-related proteins were also identified in the tolerant lines. Among them, RPK family proteins are critical components in the mediation of plant responses to dehydration (Hong et al., 1997). CBL-interacting protein kinases negatively regulate ABA-induced stomatal movement in Arabidopsis (Cheong et al., 2007). MAPKs are activated in response to drought and other environmental stresses as a link between upstream receptors and downstream targets (Jonak et al., 1996; Shou et al., 2004). The relationship between several signal-related genes expressed in the tolerant genotypes (Table 1) and drought tolerance has not previously been documented. The identification of these genes in the tolerant genotypes revealed that various signal molecules act upon improving drought tolerance of tomato.
In this study, a large number of transcription factors and signalling related proteins were also identified to be responsive to drought stress in both tolerant and sensitive genotypes. Of these, homeodomain-leucine zipper proteins can lead to desiccation tolerance by regulating drought response pathways (Deng et al., 2002). Gigantea proteins regulate cold and oxidative stress responses (Cao et al., 2005b, 2006). Diverse protein kinases play key roles in signal transduction, most of which are serine/threonine kinases (Ludwig et al., 2004). The hormone, light, and pathogenesis signalling-related genes were also found to be differentially expressed in all three genotypes, indicating that multiple signal regulations exist in tomato under drought stress.
Gene expression characteristics involved in organism growth and development processes of tomato under drought stress
Plants usually form a tolerant morphological structure to reduce the impact of adversity. Our microarray analysis indicated that the drought-tolerant lines may adapt to water deficit partially through organ morphogenesis-related regulation, such as the formation of lipid molecules (Ishihara et al., 1998), biosynthesis of epicuticular wax (Aarts et al., 1995), and regulation of cell wall structure (Cho et al., 2006) and plant height (Cao et al., 2005a). Most of the drought-responsive genes involved in organism growth and development were not previously reported to be associated with drought responses. The discovery of these genes indicated that regulation of plant growth and development are important in the acclimation to water deficit in tomato.
Specifically affected biochemical pathways in the drought-tolerant genotypes
Understanding the biochemical mechanisms in metabolic adaptations to water deficit can be used as a mean for engineering plants with improved drought tolerance. In the present study, one gene was found encoding fructose-bisphosphate aldolase in the gluconeogenesis pathway that was down-regulated under drought stress in the drought-tolerant line. It has been reported that, in Arabidopsis, the expression of fructose-bisphosphate aldolase is also down-regulated by drought stress (Kilian et al., 2007). Gluconeogenesis consumes plenty of energy thus down-regulation of the fructose-bisphosphate aldolase could inhibit gluconeogenesis for keeping energy in drought-stressed plants.
It has been reported that the development of defence responses during the adaptation to drought is related to an increased IAA content (Pustovoitova et al., 2004). In this study, one gene encoding indole-3-acetaldehyde oxidase, a key enzyme in the tryptophan degradation pathway, was found to be up-regulated in the tolerant lines, which possibly promoted the IAA biosynthesis in tomato plants to adapt to water deficit.
The maintenance of mitochondrial ATP synthesis during water stress is essential for preserving plastid function (Atkin and Macherel, 2009). Our results showed that up-regulation of an adenylate kinase participating in ATP biosynthesis in the tolerant genotype could provide more ATP for maintaining cellular activities under drought stress.
The endogenous H2O2 concentration in grains was always consistent with the activity of β-amylase, an enzyme in the starch degradation pathway (Wei et al., 2009). The down-regulation of the β-amylase gene in IL9-1 may be related to the balance of the internal H2O2 concentration, since the expression of the catalase gene used for eliminating H2O2 has been found to be highly regulated in this line.
Cystathionine beta-lyase is a key enzyme to catalyse cystathionine to form homocysteine in methionine biosynthesis and is involved in the regulation of plant growth and development (Maimann et al., 2000). Methionine and ATP are used for the synthesis of S-adenosylmethionine that is an essential substance for living cells as a methyl group donor and a precursor of ethylene, polyamines, and nicotianamine (Tabor and Tabor, 1984; Moffatt and Weretilnyk, 2001). Up-regulation of cystathionine beta-lyase identified in this study could possibly improve drought tolerance through affecting the S-adenosylmethionine metabolism.
It is well known that catalase catalyses the decomposition of hydrogen peroxide to water and oxygen to improve the stress tolerance of plants (Mohamed et al., 2003; Nair et al., 2008). The catalase was up-regulated in IL9-1, suggesting the enhanced drought tolerance of IL 9-1 may be partially due to the higher expression of catalase.
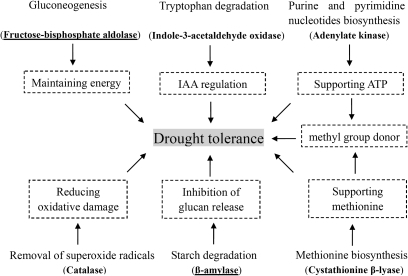
In summary, the biochemical pathways affected by the change in gene expression mentioned above might co-operate to improve drought tolerance of tomato (Fig. 5). Of these, the decreased energy dissipation through the inhibition of gluconeogenesis, ATP energy provision through the promotion of purine and pyrimidine nucleotide biosynthesis, and the reduction of oxidative damage through the removal of superoxide radicals may be closely related to the enhanced drought tolerance. The IAA regulation from the tryptophan degradation pathway and methyl group donor provision caused by the methionine biosynthesis pathway might indirectly help tomato plants to enhance drought tolerance. The altered starch degradation by down-regulated β-amylase was firstly found to be associated with drought tolerance.
Fig. 5.
Biochemical pathways leading to drought tolerance. Enzymes encoded by drought-responsive genes were shown in bold (down-regulated genes were bold and underlined while up-regulated genes were only in bold). Possible functions for increased drought tolerance by the regulation of specifically changed biochemical pathways were shown in boxes with broken lines.
Biochemical pathways affected by drought stress in all three tomato genotypes
In this study, various regulations of plant biochemical responses were found to be affected by drought stress in all three genotypes. Among them, carotenoids can reduce and eliminate the reactive oxygen damage, serve as the precursors of ABA synthesis, and also participate in photosynthesis as the pigments of chlorophyll (Goodwin and Britton, 1988; Milborrow, 2001; Treutter, 2006). Lignin is beneficial to water transportation and resistance of invasion caused by stress (Maximova et al., 2001). Suberin, a highly hydrophobic substance, can prevent water from penetrating the tissue (Graca and Santos, 2007). Biosyntheses of amino acids, hormones, and lipids, have also been shown to be involved in the drought response in plants (Akashi et al., 2001; Singh et al., 2002; Capell et al., 2004; Xiao et al., 2006). The remaining changed degradation pathways identified in this study, such as sulphate, formaldehyde, glutamate, and glycine, have not been documented previously to be related to the drought response. All the above altered metabolic pathways in the three genotypes revealed that tomato plants respond to water deficit through interactions of complex metabolic networks.
Characteristic gene expression profile between drought and salt stress in tomato
In the present study, it was found that one WRKY (SGN-U218605) and one AP2/EREBP (SGN-U213644) drought-responsive genes were also responsive to salt stress in tomato (Ouyang et al., 2007), indicating that there are common features in the upstream gene regulation in response to drought and salt stresses. In addition, several enzymes, such as adenylate kinase and catalase, were specially induced by drought while repressed under salt stress in tomato (Zhou et al., 2007). The nitrogen reduction and fixation pathway was significantly affected under drought stress by the dramatic down-regulation of its rate-limiting enzyme, nitrate reductase, which was also significantly down-regulated by salt stress in tomato (Ouyang et al., 2007). It is interesting that differentially expressed genes in the tolerant lines tend to have different expression patterns in comparison to those under salt stress, while drought-responsive genes identified in all three genotypes have somewhat similar expression patterns, such as genes involved in the metabolism of secondary metabolites, amino acids, and polyamines. These results indicate that there are some connections as well as differences between drought and salt responses.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Table S1. Primer information of genes for RT-PCR analysis.
Supplementary Table S2. Seedling survival rates of two selected ILs, M82, and S. pennellii under drought stress.
Supplementary Table S3. Leaf damage rates of the two selected ILs, M82, and S. pennellii under different drought stress conditions.
Supplementary Table S4. Genes responsive to drought only in the tolerant genotypes.
Supplementary Table S5. Functional classification of drought-responsive genes.
Supplementary Table S6. Transcription factors and signalling genes responsive to drought stress in all three genotypes.
Supplementary Table S7. Biochemical pathways and their corresponding genes affected by drought stress in all three genotypes.
Supplementary Fig. S1. Water deficit ratio of detached leaves of the two selected ILs, M82, and S. pennellii at 25°C.
Supplementary Fig. S2. Biochemical pathways affected by drought stress only in the tolerant genotypes.
Supplementary Material
Acknowledgments
We would like to thank Dr Amber Hotto for the critical review of this manuscript and Ms Julia Sharwood for proofreading. We are grateful to the Tomato Genetic Resource Center (TGRC) for supplying tomato seeds of 50 introgression lines, S. pennellii, and M82. This work was supported by grants from the Ministry of Science and Technology of China (973 Project, 2009CB119000) to HL, the National Science Foundation of China (NSFC Grants no. 30871712 and no. 30921002) to ZY, and the National Science Foundation (DBI-0501778) to ZF.
References
- Aarts MGM, Keijzer CJ, Stiekema WJ, Pereira A. Molecular characterization of the CER1 gene of Arabidopsis involved in epicuticular wax biosynthesis and pollen fertility. The Plant Cell. 1995;7:2115–2127. doi: 10.1105/tpc.7.12.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signalling. The Plant Cell. 2003;15:63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. The Plant Cell. 1997;9:1859–1868. doi: 10.1105/tpc.9.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi K, Miyakel C, Yokota A. Citrulline, a novel compatible solute in drought-tolerant wild watermelon leaves, is an efficient hydroxyl radical scavenger. FEBS Letters. 2001;508:438–442. doi: 10.1016/s0014-5793(01)03123-4. [DOI] [PubMed] [Google Scholar]
- Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annual Review of Plant Biology. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- Atkin OK, Macherel D. The crucial role of plant mitochondria in orchestrating drought tolerance. Annals of Botany. 2009;103:581–597. doi: 10.1093/aob/mcn094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly C, Audigier C, Ladonne F, Wagner MH, Coste F, Corbineau F, Côme D. Changes in oligosaccharide content and antioxidant enzyme activities in developing bean seeds as related to acquisition of drying tolerance and seed quality. Journal of Experimental Botany. 2001;52:701–708. doi: 10.1093/jexbot/52.357.701. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. 1995;57:289–300. [Google Scholar]
- Boyer JS. Plant productivity and environment. Science. 1982;218:443–448. doi: 10.1126/science.218.4571.443. [DOI] [PubMed] [Google Scholar]
- Bray EA. Genes commonly regulated by water-deficit stress in Arabidopsis thaliana. Journal of Experimental Botany. 2004;55:2331–2341. doi: 10.1093/jxb/erh270. [DOI] [PubMed] [Google Scholar]
- Breton G, Danyluk J, Charron JBF, Sarhan F. Expression profiling and bioinformatic analyses of a novel stress-regulated multispanning transmembrane protein family from cereals and Arabidopsis. Plant Physiology. 2003;132:64–74. doi: 10.1104/pp.102.015255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao SQ, Jiang ST, Zhang RX. The role of GIGANTEA gene in mediating the oxidative stress response and in Arabidopsis. Plant Growth Regulation. 2006;48:261–270. [Google Scholar]
- Cao SQ, Xu QT, Cao YJ, Qian K, An K, Zhu Y, Hu BZ, Zhao HF, Kuai BK. Loss-of-function mutations in DET2 gene lead to an enhanced resistance to oxidative stress in Arabidopsis. Physiologia Plantarum. 2005a;123:57–66. [Google Scholar]
- Cao SQ, Ye M, Jiang ST. Involvement of GIGANTEA gene in the regulation of the cold stress response in Arabidopsis. Plant Cell Reports. 2005b;24:683–690. doi: 10.1007/s00299-005-0061-x. [DOI] [PubMed] [Google Scholar]
- Capell T, Bassie L, Christou P. Modulation of the polyamine biosynthetic pathway in transgenic rice confers tolerance to drought stress. Proceedings of National Academy of Sciences, USA. 2004;101:9909–9914. doi: 10.1073/pnas.0306974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves MM, Flexas J, Pinheiro C. Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Annals of Botany. 2009;103:551–560. doi: 10.1093/aob/mcn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong YH, Pandey GK, Grant JJ, Batistic O, Li L, Kim BG, Lee SC, Kudla J, Luan S. Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. The Plant Journal. 2007;52:223–239. doi: 10.1111/j.1365-313X.2007.03236.x. [DOI] [PubMed] [Google Scholar]
- Cho SK, Kim JE, Park JA, Eom TJ, Kim WT. Constitutive expression of abiotic stress-inducible hot pepper CaXTH3, which encodes a xyloglucan endotransglucosylase/hydrolase homolog, improves drought and salt tolerance in transgenic Arabidopsis plants. FEBS Letters. 2006;580:3136–3144. doi: 10.1016/j.febslet.2006.04.062. [DOI] [PubMed] [Google Scholar]
- Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, Leonhardt N, Dellaporta SL, Tonelli C. A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Current Biology. 2005;15:1196–2000. doi: 10.1016/j.cub.2005.05.048. [DOI] [PubMed] [Google Scholar]
- Deng X, Phillips J, Meijer AH, Salamini F, Bartels D. Characterization of five novel dehydration-responsive homeodomain leucine zipper genes from the resurrection plant Craterostigma plantagineum. Plant Molecular Biology. 2002;49:601–610. doi: 10.1023/a:1015501205303. [DOI] [PubMed] [Google Scholar]
- Eshed Y, Abu-Abied M, Saranga Y, Zamir D. Lycopersicon esculentum lines containing small overlapping introgressions from L. pennellii. Theoretical and Applied Genetics. 1992;83:1027–1034. doi: 10.1007/BF00232968. [DOI] [PubMed] [Google Scholar]
- Eshed Y, Zamir D. A genomic library of Lycopersicon pennellii in L. esculentum: a tool for fine mapping of genes. Euphytica. 1994;79:175–179. [Google Scholar]
- Fei ZJ, Tang XM, Alba R, Giovannoni J. Tomato Expression Database (TED): a suite of data presentation and analysis tools. Nucleic Acids Research. 2006;34:D766–D770. doi: 10.1093/nar/gkj110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. The Plant Cell. 2000;12:599–609. doi: 10.1105/tpc.12.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fode B, Siemsen T, Thurow C, Weigel R, Gatz C. The Arabidopsis GRAS protein SCL14 interacts with class II TGA transcription factors and is essential for the activation of stress-inducible promoters. The Plant Cell. 2008;20:3122–3135. doi: 10.1105/tpc.108.058974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin TW, Britton G. Distribution and analysis of carotenoids. In: Goodwin TW, editor. Plant pigments. FL: Academic Press; 1988. pp. 61–132. [Google Scholar]
- Graca J, Santos S. Suberin: a biopolyester of plants’ skin. Macromolecular Bioscience. 2007;7:128–135. doi: 10.1002/mabi.200600218. [DOI] [PubMed] [Google Scholar]
- Grant GR, Liu JM, Stoeckert CJ., Jr A practical false discovery rate approach to identifying patterns of differential expression in microarray data. Bioinformatics. 2005;21:2684–2690. doi: 10.1093/bioinformatics/bti407. [DOI] [PubMed] [Google Scholar]
- Guo PG, Baum M, Grando S, Ceccarelli S, Bai GH, Li RH, von Korff M, Varshney RK, Graner A, Valkoun J. Differentially expressed genes between drought-tolerant and drought-sensitive barley genotypes in response to drought stress during the reproductive stage. Journal of Experimental Botany. 2009;60:3531–3544. doi: 10.1093/jxb/erp194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt-Herting S, Klug K, Fock HP. A new approach to measure gross CO2 fluxes in leaves: gross CO2 assimilation, photorespiration, and mitochondrial respiration in the light in tomato under drought stress. Plant Physiology. 2001;126:388–396. doi: 10.1104/pp.126.1.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayano-Kanashiro C, Calderón-Vázquez C, Ibarra-Laclette E, Herrera-Estrella L, Simpson J. Analysis of gene expression and physiological responses in three Mexican maize landraces under drought stress and recovery irrigation. PLoS ONE. 2009;4 doi: 10.1371/journal.pone.0007531. e7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen SP, Pathan MS, Sanchez A, Baxter I, Dunn M, Estes B, Chang HS, Zhu T, Kreps JA, Nguyen HT. Expression profiling of rice segregating for drought tolerance QTLs using a rice genome array. Functional and Integrative Genomics. 2005;5:104–116. doi: 10.1007/s10142-004-0126-x. [DOI] [PubMed] [Google Scholar]
- Hong SW, Jon JH, Kwak JM, Nam HG. Identification of a receptor-like protein kinase gene rapidly induced by abscisic acid, dehydration, high salt, and cold treatments in Arabidopsis thaliana. Plant Physiology. 1997;113:1203–1212. doi: 10.1104/pp.113.4.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao TC. Plant response to water stress. Annual Review of Plant Physiology. 1973;24:519–570. [Google Scholar]
- Huang DQ, Wu WR, Abrams SR, Cutler AJ. The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. Journal of Experimental Botany. 2008;59:2991–3007. doi: 10.1093/jxb/ern155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu HH, Dai MQ, Yao JL, Xiao BZ, Li XH, Zhang QF, Xiong LZ. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proceedings of the National Academy of Sciences, USA. 2006;103:12987–12992. doi: 10.1073/pnas.0604882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara H, Shibasaki Y, Kizuki N, Wada T, Yazaki Y, Asano T, Oka Y. Type I phosphatidylinositol-4-phosphate 5-kinases. Journal of Biological Chemistry. 1998;273:8741–8748. doi: 10.1074/jbc.273.15.8741. [DOI] [PubMed] [Google Scholar]
- Jonak C, Kiegerl S, Ligterink W, Barker PJ, Huskisson NS, Hirt H. Stress signalling in plants: a mitogen-activated protein kinase pathway is activated by cold and drought. Proceedings of National Academy of Sciences, USA. 1996;93:11274–11279. doi: 10.1073/pnas.93.20.11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung JG, Corbett AM, Fellman SM, Tieman DM, Klee HJ, Giovannoni JJ, Fei ZJ. Plant MetGenMAP: an integrative analysis system for plant systems biology. Plant Physiology. 2009;151:1758–1768. doi: 10.1104/pp.109.145169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D'Angelo C, Bornberg-Bauer E, Kudla J, Harter K. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. The Plant Journal. 2007;50:347–363. doi: 10.1111/j.1365-313X.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- Leymarie J, Damerval C, Marcotte L, Combes V, Vartanian N. Two-dimensional protein patterns of Arabidopsis wild-type and auxin-insensitive mutants, axr1, axr2, reveal interactions between drought and hormonal responses. Plant and Cell Physiology. 1996;37:966–975. doi: 10.1093/oxfordjournals.pcp.a029046. [DOI] [PubMed] [Google Scholar]
- Li HM, Sun JQ, Xu YX, Jiang HL, Wu XY, Li CY. The bHLH-type transcription factor AtAIB positively regulates ABA response in Arabidopsis. Plant Molecular Biology. 2007;65:655–665. doi: 10.1007/s11103-007-9230-3. [DOI] [PubMed] [Google Scholar]
- Ludwig AA, Romeis T, Jones JDG. CDPK-mediated signalling pathways: specificity and cross-talk. Journal of Experimental Botany. 2004;55:181–188. doi: 10.1093/jxb/erh008. [DOI] [PubMed] [Google Scholar]
- Maimann S, Wagner C, Kreft O, Zeh M, Willmitzer L, Höfgen R, Hesse H. Transgenic potato plants reveal the indispensable role of cystathionine β-lyase in plant growth and development. The Plant Journal. 2000;23:747–758. doi: 10.1046/j.1365-313x.2000.00842.x. [DOI] [PubMed] [Google Scholar]
- Marè C, Mazzucotelli E, Crosatti C, Francia E, Stanca AM, Cattivelli L. Hv-WRKY38: a new transcription factor involved in cold- and drought-response in barley. Plant Molecular Biology. 2004;55:399–416. doi: 10.1007/s11103-004-0906-7. [DOI] [PubMed] [Google Scholar]
- Maximova N, Österberg M, Koljonen K, Stenius P. Lignin adsorption on cellulose fibre surfaces: effect on surface chemistry, surface morphology and paper strength. Cellulose. 2001;8:113–125. [Google Scholar]
- Milborrow BV. The pathway of biosynthesis of abscisic acid in vascular plant: a review of the present state of knowledge of ABA biosynthesis. Journal of Experimental Botany. 2001;52:1145–1164. [PubMed] [Google Scholar]
- Mizoguchi T, Hayashida N, Yamaguchi-Shinozaki K, Kamada H, Shinozaki K. Two genes that encode ribosomal-protein S6 kinase homologs are induced by cold or salinity stress in Arabidopsis thaliana. FEBS Letters. 1995;358:199–204. doi: 10.1016/0014-5793(94)01423-x. [DOI] [PubMed] [Google Scholar]
- Moffatt BA, Weretilnyk EA. Sustaining S-adenosyl-l-methionine-dependent methyltransferase activity in plant cells. Physiologia Plantarum. 2001;113:435–442. [Google Scholar]
- Mohamed EA, Iwaki T, Munir I, Tamoi M, Shigeoka S, Wadano A. Overexpression of bacterial catalase in tomato leaf chloroplasts enhances photo-oxidative stress tolerance. Plant, Cell and Environment. 2003;26:2037–2046. [Google Scholar]
- Nair AS, Abraham TK, Jaya DS. Studies on the changes in lipid peroxidation and antioxidants in drought stress induced cowpea (Vigna unguiculata L.) varieties. Journal of Environmental Biology. 2008;29:689–691. [PubMed] [Google Scholar]
- Ouyang B, Yang T, Li HX, Zhang L, Zhang YY, Zhang JH, Fei ZJ, Ye ZB. Identification of early salt stress response genes in tomato root by suppression subtractive hybridization and microarray analysis. Journal of Experimental Botany. 2007;58:507–520. doi: 10.1093/jxb/erl258. [DOI] [PubMed] [Google Scholar]
- Passioura J. The drought environment: physical, biological and agricultural perspectives. Journal of Experimental Botany. 2007;58:113–117. doi: 10.1093/jxb/erl212. [DOI] [PubMed] [Google Scholar]
- Pratt LH, Liang C, Shah M, et al. Sorghum expressed sequence tags identify signature genes for drought, pathogenesis, and skotomorphogenesis from a milestone set of 16,801 unique transcripts. Plant Physiology. 2005;139:869–884. doi: 10.1104/pp.105.066134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pustovoitova TN, Zhdanova NE, Zholkevich VN. Changes in the levels of IAA and ABA in cucumber leaves under progressive soil drought. Russian Journal of Plant Physiology. 2004;51:513–517. [Google Scholar]
- Rabbani MA, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y, Yoshiwara K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiology. 2003;133:1755–1767. doi: 10.1104/pp.103.025742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rick CM. Potential genetic resources in tomato species: clues from observation in native habitats. In: Srb AM, editor. Handbook of genetics. Vol. 2. FL: Plenum Press; 1973. pp. 255–269. [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K. Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. The Plant Cell. 2001;13:61–72. doi: 10.1105/tpc.13.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. The Plant Journal. 2002;31:279–292. doi: 10.1046/j.1365-313x.2002.01359.x. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. Journal of Experimental Botany. 2007;58:221–227. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- Shou HX, Bordallo P, Wang K. Expression of the Nicotiana protein kinase (NPK1) enhanced drought tolerance in transgenic maize. Journal of Experimental Botany. 2004;55:1013–1019. doi: 10.1093/jxb/erh129. [DOI] [PubMed] [Google Scholar]
- Singh SC, Sinha RP, Häder DP. Role of lipids and fatty acids in stress tolerance in cyanobacteria. Acta Protozool. 2002;41:297–308. [Google Scholar]
- Song CP, Agarwal M, Ohta M, Guo Y, Halfter U, Wang PC, Zhu JK. Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. The Plant Cell. 2005;17:2384–2396. doi: 10.1105/tpc.105.033043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storozhenko S, Pauw PD, Montagu MV, Inzé D, Kushnir S. The heat-shock element is a functional component of the Arabidopsis APXl gene promoter. Plant Physiology. 1998;118:1005–1014. doi: 10.1104/pp.118.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor CW, Tabor H. Methionine adenosyltransferase (S-adenosylmethionine synthetase) and S-adenosylmethionine decarboxylase. Advances in Enzymology and Related Areas of Molecular Biology. 1984;56:251–282. doi: 10.1002/9780470123027.ch4. [DOI] [PubMed] [Google Scholar]
- Talamè V, Ozturk NZ, Bohnert HJ, Tuberosa R. Barley transcript profiles under dehydration shock and drought stress treatments: a comparative analysis. Journal of Experimental Botany. 2007;58:229–240. doi: 10.1093/jxb/erl163. [DOI] [PubMed] [Google Scholar]
- Treutter D. Significance of flavonoids in plant resistance: a review. Environmental Chemistry Letters. 2006;4:147–157. [Google Scholar]
- Trewavas AJ, Malhó R. Signal perception and transduction: the origin of the phenotype. The Plant Cell. 1997;9:1181–1195. doi: 10.1105/tpc.9.7.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo LE, Sotolongo M, Menéndez C, Ochogavıía ME, Coll Y, Hernández I, Borrás-Hidalgo O, Thomma BPHJ, Vera P, HerncLndez L. SodERF3, a novel sugarcane ethylene responsive factor (ERF), enhances salt and drought tolerance when overexpressed in tobacco plants. Plant and Cell Physiology. 2008;49:512–525. doi: 10.1093/pcp/pcn025. [DOI] [PubMed] [Google Scholar]
- Vasquez-Robinet C, Mane SP, Ulanov AV, et al. Physiological and molecular adaptations to drought in Andean potato genotypes. Journal of Experimental Botany. 2008;59:2109–2123. doi: 10.1093/jxb/ern073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WX, Vinocur B, Shoseyov O, Altman A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends in Plant Science. 2004;9:244–252. doi: 10.1016/j.tplants.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Wang YC, Jiang J, Zhao Xi, Liu GF, Yang CP, Zhan LP. A novel LEA gene from Tamarix androssowii confers drought tolerance in transgenic tobacco. Plant Science. 2006;171:655–662. [Google Scholar]
- Wang YH, Garvin DF, Kochian LV. Nitrate-induced genes in tomato roots. Array analysis reveals novel genes that may play a role in nitrogen nutrition. Plant Physiology. 2001;127:345–359. doi: 10.1104/pp.127.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei K, Jin X, Chen X, Wu FB, Zhou WH, Qiu BY, Qiu L, Wang XD, Li CD, Zhang GP. The effect of H2O2 and abscisic acid (ABA) interaction on beta-amylase activity under osmotic stress during grain development in barley. Plant Physiology Biochemistry. 2009;47:778–784. doi: 10.1016/j.plaphy.2009.05.005. [DOI] [PubMed] [Google Scholar]
- Wei W, Zhang YX, Han L, Guan ZQ, Chai TY. A novel WRKY transcriptional factor from Thlaspi caerulescens negatively regulates the osmotic stress tolerance of transgenic tobacco. Plant Cell Reports. 2008;27:795–803. doi: 10.1007/s00299-007-0499-0. [DOI] [PubMed] [Google Scholar]
- Xiao JH, Li HX, Zhang JH, Chen RG, Zhang YY, Ouyang B, Wang TT, Ye ZB. Dissection of GA 20-oxidase members affecting tomato morphology by RNAi-mediated silencing. Plant Growth Regulation. 2006;50:179–189. [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends in Plant Science. 2005;10:88–94. doi: 10.1016/j.tplants.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Research. 2002;30 doi: 10.1093/nar/30.4.e15. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang GY, Chen M, Li LC, Xu ZS, Chen XP, Guo JM, Ma YZ. Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. Journal of Experimental Botany. 2009;60:3781–3796. doi: 10.1093/jxb/erp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XN, Chen B, Lu GJ, Han B. Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochemical and Biophysical Research Communications. 2009;379:985–989. doi: 10.1016/j.bbrc.2008.12.163. [DOI] [PubMed] [Google Scholar]
- Zhou SP, Wei S, Boone B, Levy S. Microarray analysis of genes affected by salt stress in tomato. African Journal of Environmental Science and Technology. 2007;1:014–026. [Google Scholar]
- Zou XL, Shen QXJ, Neuman D. An ABA inducible WRKY gene integrates responses of creosote bush (Larrea tridentata) to elevated CO2 and abiotic stresses. Plant Science. 2007;172:997–1004. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.