Abstract
Transcription factors play key roles in plant development through their interaction with cis-elements and/or other transcription factors. A HD-Zip IV family transcription factor, Gossypium barbadense Meristem Layer 1 (GbML1) has been identified and characterized here. GbML1 specifically bound to the L1 box and the promoters of GbML1 and GbRDL1. GbML1 physically interacted with a key regulator of cotton fibre development, GbMYB25. Truncated and point mutation assays indicated the START–SAD domain was required for the binding to the C terminal domain (CTD) of GbMYB25. GbML1 overexpression in Arabidopsis increased the number of trichomes on stems and leaves and increased the accumulation of anthocyanin in leaves. Taken together, the L1 box binding protein, GbML1 was identified as the first partner for GbMYB25 and the role of START domain was discovered to be a protein binding domain in plants. Our findings will help the improvement of cotton fibre production and the understanding of the key role of HD-Zip family and MYB family in plants.
Keywords: Cotton fibre, EMSA, HD-Zip IV transcription factor, L1 box, R2R3-MYB transcription factor, SAD domain, START domain, yeast two-hybrid
Introduction
Cotton plants produce natural fibres that are extensively used in the textile industry all over the world. However, the molecular mechanism of cotton fibre development remains largely unknown.
Cotton fibres are unicellular structures of epidermal origin, which is similar to Arabidopsis trichomes, and it is likely that the development of cotton fibre and Arabidopsis trichomes share a similar mechanism. For the model plant Arabidopsis, trichome development has been studied in depth and many transcription factors that constitute a signalling pathway have been reported. R2R3-MYB protein GLABRA1 (GL1), the WD40 protein TRANSPARENT TESTA GLABRA1 (TTG1), and the basic helix–loop–helix proteins GLABRA3 (GL3) or ENHANCER OF GLABRA3 (EGL3) form a transcription factor complex to determine aerial part trichome patterning in Arabidopsis (for a review, see Ishida et al., 2008). These transcription factors together regulate the trichome-specific expression of GLABRA2 (GL2), a homeobox (HOX) transcription factor that promotes leaf trichome formation. Several genes implicated in cotton fibre development were shown functionally to substitute their homologues in Arabidopsis. For example, overexpression of either GhMYB1 (L04497) or GaMYB2 could rescue the gl1 mutant in Arabidopsis (Payne et al., 1999; Wang et al., 2004). Moreover, four cotton genes homologous to Arabidopsis TTG1 were identified, and one of them could rescue the Arabidopsis ttg1 mutant (Humphries et al., 2005). Furthermore, three HOX genes (HOX1-3) homologous to GL2 in G. hirsutum were cloned and one of them (GhHOX1) could activate a fibre-specific promoter RDL1 when it was overexpressed, suggesting that HD-Zip IV family members in cotton had a role in fibre development (Wang et al., 2004). In addition, another cotton HOX gene, GaHOX1 is a functional homologue of the Arabidopsis GL2 (Guan et al., 2008). These evidences strongly support that a similar mechanism exists in regulating cotton fibres and leaf trichomes, suggesting models controlling Arabidopsis trichomes may give hints to the complicated regulatory network existing in cotton fibre development.
Cotton fibre is different from Arabidopsis trichomes in that cotton fibre is unbranched. Thus the gene controlling branching in Arabidopsis may have a new function in fibre development. For example, AtMYB106 was a negative regulator of the branching in Arabidopsis while its homologue GhMYB25 was shown to be a key regulator for cotton fibre initiation (Jakoby et al., 2008). The expression of GhMYB25 was enriched in fibre initial cells and more in fibre-bearing plants than the fibreless mutants. Overexpression of GhMYB25 in tobacco increased the branches of leaf trichomes (Wu et al., 2006). Recently, knock-down of the expression of GhMYB25 in cotton greatly reduced fibre length and initials, while overexpression led to more fibre initials (Suo et al., 2003; Machado et al., 2009). GhMYB25 was also shown to act upstream of another key regulator for fibre elongation, GhMYB109. Thus GhMYB25 plays a key role in cotton fibre development, especially at the initiation stage.
Compared with the well-established interaction map for leaf trichome development in Arabidopsis, little is known about the complicated network for regulating cotton fibre development. Given that cotton fibre arises from the outer epidermis of ovules, it is possible that L1 layer specific genes would account for the epidermal cell differentiation, and in this case, it is fibre. A previous study showed that the L1 box was needed for the expression of a fibre-specific gene, GaRDL1 (Wang et al., 2004). Moreover, a short EST designated as GhHD1 showed similarity to ATML1, which was a HD-Zip IV family gene that bound to L1 box in Arabidopsis (Lu et al., 1996; Abe et al., 2001). GhHD1 was also found to be up-regulated in fibre initial cells as GbMYB25 (Wu et al., 2006, 2007). These findings gave hints that GhHD1 might be an important regulator in the L1 layer for specifying fibre cell initials from other ovule epidermal cells. But, to our knowledge, the cloning and functional study of this gene has not been reported.
Here, the isolation and characterization of a homologue of GhHD1 in G. barbadense, designated as GbML1, are reported. The EMSA assay showed that GbML1 could bind to the L1 box and the dimmer formation contributed by the ZLZ domain was essential for DNA binding. GbML1 could bind to GbMYB25 in vitro and in vivo. Truncated and point mutation assays showed that both the START domain and the SAD domain were required to interact with the CTD of GbMYB25. The high expression of GbML1 in Arabidopsis affected the number of trichomes and the accumulation of anthocyanin.
Materials and methods
Plant material and growth conditions
Cotton plants of G. barbadense were grown in a greenhouse. Arabidopsis thaliana ecotype Columbia-0 was used in this research and grown under 16 h light (70 mmol m−2 s−1) and 8 h dark cycle at 23 °C.
Extraction of RNA and DNA
RNA and DNA were extracted from different tissues of G. barbadense plants using a modified CTAB-method as described by Wang et al. (2009). RNA and DNA from Arabidopsis plants were extracted using the Trizol reagent (Invitrogen) and the CTAB method, respectively. All extracted RNA was treated by DNase I and purified using the RNAprep Plant RNA Purification Kit (Tiangen) according to the manufacturer's instructions. The concentration of the purified RNA and DNA was quantified by a nucleic acid analyser (DU-640, Beckman). The quality and concentration of the purified RNA were further checked by RNA denaturing MOPS agarose gel.
Cloning of GbML1
The 3′ and 5′ ends of the GbML1 cDNA were obtained using the SMART™ RACE cDNA amplification kit (Clontech) according to the manufacturer's manual. PCR products were cloned into the pMD18-T vector (TakaRa, Japan) and sequenced.
The promoter region of GbML1 was cloned by genomic walking using the LA PCR™ in vitro Cloning Kit (TakaRa) according to the manufacturer's manual. Briefly, the completely digested DNA was purified and ligated to the corresponding adaptors to generate several DNA fragment libraries which were subjected to a first round of PCR amplification with Cassette Primer C1 and an outer gene-specific primer. The PCR products were diluted and subjected to a second round of PCR amplification using the Cassette Primer C2 and the inner gene-specific primer. PCR products were cloned into the pMD18-T vector (TakaRa) and sequenced.
Bioinformatics analysis
Database searches were performed using BLASTP (http://www.ncbi.nlm.nih.gov/BLAST/). Protein domains were identified using programs RPS-BLAST (NCBI) and ProfileScan (http://hits.isb.cn/cgi-bin/PFSCAN) searching the Pfam-A, prosite profiles, and smart databases (NCBI). Amino acid sequence alignments were created using ClustalW (http://www2.ebi.ac.uk/clustalw; Larkin et al., 2007). Phylogenetic analyses were carried out using the Neighbor–Joining (N–J) method with 1000 bootstrap replicates implemented in the MEGA 4 programme. The three-dimensional structures of START domains were deduced using SWISS-MODEL via the ExPASy web server using automated mode based on the structure of Molecular modeling (Peitsch, 1995; Arnold et al., 2006; Kiefer et al., 2009) visualized using Accelrys ViewerLite Version 4.2 (Accelrys).
RT-PCR
The first-strand cDNA was synthesized from an equivalent amount of RNA using SuperScript® III First-Strand Synthesis Super Mix (Invitrogen). PCR amplification for tissue-specific expression of GbML1 and GbMYB25 was carried using gene-specific primers, GbML1-RT-F and GbML1-RT-R, GbMYB25-RT-F and GbMYB25-RT-R, respectively. Meanwhile, a housekeeping gene, Ubiquitin, was amplified and used as an internal control. In the expression analysis of transgenic plants, 1 μg of total RNA from aerial parts of 14-d-old seedlings was used as the template for the first-strand cDNA synthesis with an oligo (dT) primer. Primers and Cycles for RT-PCR are listed in Supplementary Table S1 at JXB online.
Subcellular localization analysis
The CFP and YFP coding sequences were fused in-frame to the 5′ end of GbML1 and to the 3′ end of GbMYB25 to generate CFP-GbML1 and GbMYB25-YFP fusions, respectively. These fragments were driven by the 35S promoter. The gold particles were coated by plasmid DNA. Onion epidermal cells were bombarded with the combination of these two constructs using a particle gun-mediated systerm (PDS-1000/He; Bio-Rad) and analysed by confocal microscope (Leica TCS SP5).
Transactivation assay
The coding region of GbML1 was cloned into vector pENTR-D-TOPO to generate pENTR-GbML1 construct. The GbML cDNAs were then recombined into pDEST32 bait vector by Gateway LR recombination reaction (Invitrogen) to generate 32-GbML1. The same method was used to generate 32-GbMYB25 and 32-GbMYB2 constructs. The yeast strain AH109 was co-transformed with bait and prey constructs. Yeast cells were plated onto SD/-T-L and incubated at 28 °C for 3 d. Transformed yeast cells were subsequently grown on SD/-T-L-H with different concentrations of 3-AT (0 mM, 2 mM, 5 mM, 10 mM) and SD/-T-L-H-A medium. Transactivation ability was determined by evaluating the growth of yeast cells on the selective medium.
For mapping the activation domain, different truncated parts were PCR amplified and cloned into Entry vectors. The generation of destination constructs and assessment of transcription activation activity were done as above.
Yeast two-hybrid assay
Yeast two-hybrid assay was performed using the ProQuest™ Two-Hybrid System (Invitrogen). pDEST 22 was used for GAL4 AD, and pDEST 32 was used for GAL4 BD. Coding sequence of GbML1, GbMYB25, and GbMYB2 were amplified by PCR using pfx DNA polymerase (Invitrogen) with the primers listed in Supplementary Table S1 at JXB online. All PCR products were cloned into pENTR-D-TOPO entry vectors. The destination vectors (fused to BD in pDEST 32, or fused to AD in pDEST 22) were constructed using LR reaction. Yeast AH109 cells were co-transformed with baits and preys and plated onto SD/-T-L medium. Five independent 3-d-old colonies with the same size were picked and diluted into 100 μl 1× TE, and aliquots of 10 μl yeast cells were dropped on selective medium SD/-T-L-H-A or SD/-T-L-H supplemented with different concentrations of 3-AT.
Point mutagenesis of GbML1
Point mutation of GbML1 was performed using a PCR assisted strategy. To make G291D mutation, overlapped fragments of GbML1-START–SAD were amplified using pfx DNA polymerase (Invitrogen) using two pairs of primers: GbML1-START-F and GbML1-G291D-R1, GbML1-G291D-F1 and GbML1- R. The PCR products were separated on 1.2% agarose gel and the DNA was extracted using a Gel extraction kit (Sangon). Purified PCR products were mixed, and 1 μl of the mixed DNA was used as the template in PCR amplification by pfx DNA polymerase using GbML1-START-F and GbML1-R primers for Y2H vector construction, or GbML1-START-F-EcoRI and GbML1-R-Sal for protein expression vector construction. The similar method was used to make the L94P, N107I, H429D, and S655C mutations. All constructs were checked by sequencing before use.
Protein expression and purification
GbML1 was cloned into the EcoRI and SalI sites of the pMAL-C2 vector to produce a MBP-GbML1 fusion construct (New England BioLabs). The sequenced pMAL-C2-GbML1 construct was introduced into E. coli BL21 for expression. Other truncated or mutated versions of GbML1 protein constructs were built by the same method. Fusion proteins were expressed in BL21 cells by adding 0.1 mM IPTG to culture medium for 7 h at 28 °C and purified using amylase resin (New England BioLabs).
GbMYB25 was cloned into the EcoRI and BamHI sites of the pET28-a (+) vector. The sequenced His-GbMYB25 construct was introduced into E. coli BL21 (DE3) for expression. Fusion proteins were expressed in BL21 (DE3) cells by adding 0.1 mM IPTG to culture medium for 7 h at 28 °C and purified using NI-NTA agarose (Invitrogen).
All purified recombinant protein was quantified using the Bradford assay (2-D Quant Kit, Amersham Biosciences Corp., San Francisco, CA, USA).
Electrophoretic mobility shift assay
The 3′ end biotin-labelled oligonucleotides for the L1 box and the mutated L1 box were synthesized (Sangon) and equimolar pairs were annealed using the protocol provided by Sigma. Briefly, oligo pairs were dissolved in annealing buffer (10 mM TRIS, 50 mM NaCl, 1 mM EDTA) and heated to 94 °C for 4 min and then the heat block was moved to the table and left for the tubes to cool down slowly. The oligonucleotides of the RDL1 promoter were synthesized by the Sangon Company. The size of about 100 bp promoter region of GbML1 was amplified by PCR using platium pfx taq. The RDL1 promoter oligonucleotides and PCR products of the GbML1 promoter region were labelled using the Biotin 3′ End DNA Labeling Kit (Pierce) according to the manufacturer's manual. The labelled RDL1 promoter oligonucleotides were annealed for use. EMSA was performed with the Light Shift Chemiluminescent EMSA Kit (Pierce) according to the manufacturer's instructions. The binding reactions, containing 10 mM TRIS (pH 7.5), 50 mM KCl, 1 mM DTT, 2.5% glycerol, 0.05% NP-40, 5 mM MgCl2, 0.5 mM EDTA, 5 ng μl−1 poly (dI.dC), 1 μg recombinant fusion protein, and 100 fmol biotin-labelled DNA, were kept for 30 min at room temperature before loading buffer was added. Gel electrophoresis was performed on a 10% native polyacrylamide gel. After blotting on a positively charged nylon membrane (Amersham), the DNA was linked using a transilluminator equipped with 312 nm bulbs with the membrane face down for 15 min. The biotin-labelled DNA was detected by Chemiluminescence and exposed to X-ray film (Kodak). The probes and primers used in EMSA assay are listed in the supplementary files at JXB online.
In vitro pull-down assay
The pull-down assay was performed following the protocol described by Park et al. (2009). The expression and purification procedure are as follows. A single sequenced clone was cultivated in 5 ml LB broth with appropriate antibiotics in a 15 ml tube (Corning) overnight at 37 °C. Then, an aliquot of 3 ml overnight culture was added into 250 ml LB broth with appropriate antibiotics in a 1.0 l flask and cultured at 37 °C until the OD600 was 0.6. The culture was cooled to 28 °C, 110 μl 20% IPTG (0.84 M) was added, and the culture was then shaken at 28 °C for 8 h. After induction, the culture was cooled on ice and bacteria cells were collected in a 50 ml tube (Corning). Pellets for pull-down assay were stored overnight at –80 °C. As to the input His-GbMYB25 protein, pellets were resuspended in 12 ml 1× Column binding buffer (CB, 200 mM NaCl, 20 mM TRIS-HCl, 1 mM EDTA) with 1 mM DTT, 1 mM PMSF, and 10 mM imidazole and then kept overnight in –80 °C. Pellets for pull-down were resuspended in 12 ml 1× CB with 5 mM DTT and 1 mM PMSF. All samples were sonicated 18 times with 10 s each and 10 s interval using program 7 (JYD-650, Zhisun instrument). Samples were transferred into 15 ml Corning tubes, spun at 12 000 rpm at 4 °C for 15 min, and the supernatants were transferred into new 15 ml tubes. An aliquot of 300 μl washed amylase resin was added to each sample used for pull-down assay and incubated at 4 °C for 2 h with gentle rotation. The MBP or MBP fusion protein samples were spun at 1200 rpm at 4 °C for 1 min. The pellets were washed three times with 10 ml 1× CB (rotated for 5 min each wash). After the final wash, the pellets were transferred to 1.5 ml microcentrifuge tubes with 50 μl pellets in each tube. The His-GbMYB25 prey sample was spun at 1200 rpm for 1 min. An aliquot of 1 ml His-GbMYB25 supernatant was added to each 1.5 ml tube containing MBP fusion proteins. Immobilized MBP fusion proteins and pre-cleared His-GbMYB25 lysates were incubated at 4 °C for 3 h with gentle rotation. Then the binding reaction was spun at 1200 rpm for 1 min. The pellets were washed three times with 1 ml wash buffer. After a final wash, 50 μl 2× SDS sample buffer was added to each tube, mixed, and boiled for 5 min. The supernatant was ready for Western blot. As for the His-GbMYB25 sample for input, 300 μl washed Ni-NTA agarose was added into the 15 ml tube and the His tagged protein was purified according to the manufacturer's manual.
Ten microlitres of each protein sample was separated by 10% SDS-PAGE gel (Bio-Rad) and electrotransferred onto an Immobilon-P membrane (Millipore). The membrane was blocked for 1 h in PBS-T buffer with 5% skim milk, washed twice with PBS-T, and incubated for 1 h with 1:20 000 diluted anti-His (Tiangen) or anti-MBP (New England Biolabs) antibodies. Membranes were washed three times with PBS-T buffer. Then the membranes were incubated for 1 h with the 1:10 000 diluted anti-mouse HRP-conjugated (Pierce, Rockford, IL). Membranes were washed three times with PBS-T buffer. Detection was performed using the SuperSignal West Dura Extended Duration Substrate (Thermoscience, Rockford, IL), according to the manufacturer's instructions. For the strip procedure, membranes were immersed into 30 ml strip buffer (100 mM 2-mercaptoethanol, 62 mM TRIS-HCl, pH 8.0; SDS, 2%) at 55 °C with rotation. Then membranes were washed three times in PBS-T buffer and subjected to the block procedure.
Construction of transgenic plants
The full-length GbML1 coding sequence was amplified from the T-GbML1 vector with primers GbML1-OXF1 and GbML1-OXR1 by Platinum pfx DNA polymerase (Invitrogen) and subcloned into pMD18-T simple vector. The pMD18-T-GbML1 vector was digested by SacI and then semi-digested by BamHI. The full-length ORF of GbML1 was cloned into the BamHI and SacI sites of the pHB vector under the 2× 35S promoter to generate pHB-GbML1 construct for over expression study. The pHB-GbML1 construct was introduced into Arabidopsis using a flower dipping method (Zhang et al., 2006). Transgenic plants were selected on MS plates containing 20 mg l−1 hygromycin.
Results
GbML1 encodes a HD-Zip IV transcription factor binding to L1 box
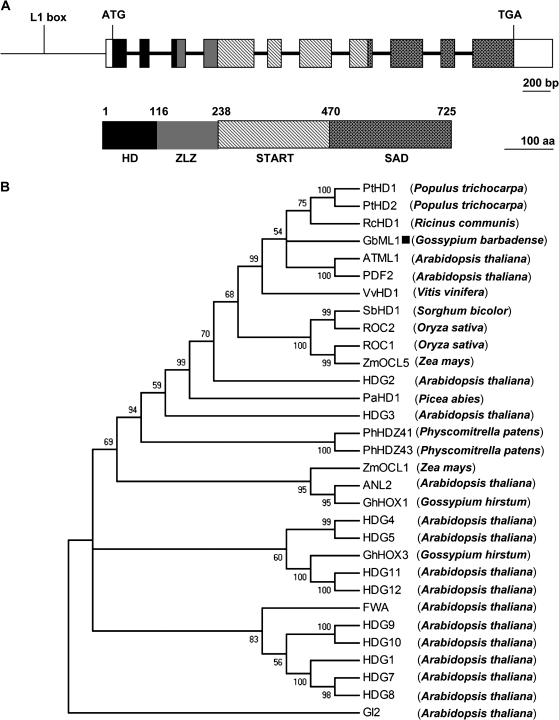
Based on the GhHD1 EST sequence (AY464063) and a fragment recovered from a screening for transcription activators in our laboratory (K Zuo et al., unpublished data), degenerated primers were designed for 5′ RACE and 3′ RACE. The full-length cDNA was obtained and designated as GbML1 which was 2803 bp long and contained an ORF encoding 725 amino acid residues (Fig. 1A). The comparison of the full-length cDNA and the genomic DNA of GbML1 revealed that the GbML1 genomic DNA contained nine introns (Fig. 1A), which was common in the HD-Zip IV family.
Fig. 1.
Molecular characterization of GbML1. (A) (Top) Genomic structure of GbML1. Exons are represented in boxes, and intron positions are indicated by bold lines. The promoter region is represented by a narrow line and the L1 box position is indicated. (Bottom) The HD domain is indicated by a black box (aa: 1–115), the ZLZ domain (aa: 116–237) is indicated by a grey box, the START domain is filled with dashed lines (238–469), the SAD domain is filled with stars (aa: 470–725), and the untranslated regions are indicated by empty boxes. The bar for the nucleotide represents 200 bp while the bar for the protein represents 100 aa. (B) Phylogenetic tree showing the relationship between GbML1 and other HD-Zip IV proteins. The tree presented here is a Neighbor–Joining tree based on amino acid sequence alignment. The numbers next to each node give bootstrap values for 1000 replicates. Alignments are derived from the following sources: Gossypium barbadense GbML1 (this study), Gossypium hirsutum GhHOX1 (AF530913) and GhHOX3 (AY626159), Arabidopsis thaliana ATML1 (U37589), PDF2 (AB056455), GL2 (Z54356), ANL2 (AF077335), FWA (AAG09302), HDG1 (AJ224338), HDG2 (AC000098), HDG3 (AC005700), HDG4 (Z97344), HDG5 (AB013394), HDG7 (AB025603) HDG8 (AC012328), HDG9 (AB005238), HDG10 (AC007894), HDG11 (AC012396), and HDG12 (AC034106), Populus trichocarpa PtHD1 (CM000340) and PtHD2 (CM000347), Vitis vinifera VvHD1 (CU459396), Ricinus communis RcHD1 (EQ973828), Zea mays ZmOCL1 (Y17898) and ZmOCL5 (AJ250987), Sorghum bicolor SbHD1 (CM000765), Oryza sativa ROC1 (AB077993) and ROC2 (AB101645), Picea abies PaHD1 (AF172931), Physcomitrella patens PhHDZ41 (BK005813) and PhHDZ43 (BK005815). The GbML1 was marked with a black square.
The deduced GbML1 protein had a calculated MW of 79.626 kDa and a pI of 5.86 (http://cn.expasy.org/cgi-bin/protparam). Domain analysis indicated that as most of the HD-Zip IV proteins, GbML1 contained four conserved domains (Fig. 1A). The N terminus (55–115) had a homeobox domain. The part immediately following (116–237) was a ZLZ motif. The C terminal part of GbML1 contained a START domain (aa: 244–467) and a SAD domain (aa: 486–717). A detailed description of the features of these domains is displayed in Supplementary Fig. S1 at JXB online.
Using genomic walking, a 1040 bp fragment upstream putative GbML1 transcription start site was obtained. In this region, there was an L1 box element (Fig. 1A).
To understand the evolutionary relationship between GbML1 and members of the HD-Zip IV family, an unrooted phylogenetic tree was generated by the Neighbor–Joining distance method, presented in Fig. 1B, using an amino acid alignment comparing the full length of proteins from different species (see Supplementary Fig. S7 at JXB online). GbML1 fell into the subclass including the dicot plants Arabidopsis thaliana (ATML1, PDF2), Populus trichocarpa (PtHD1, PtHD2), Ricinus communis (RcHD1), and Vitis vinifera (VvHD1) and the monocot plants Oryza sativa (ROC1, ROC2), Sorghum bicolor (SbHD1), and Zea mays (ZMOCL5).
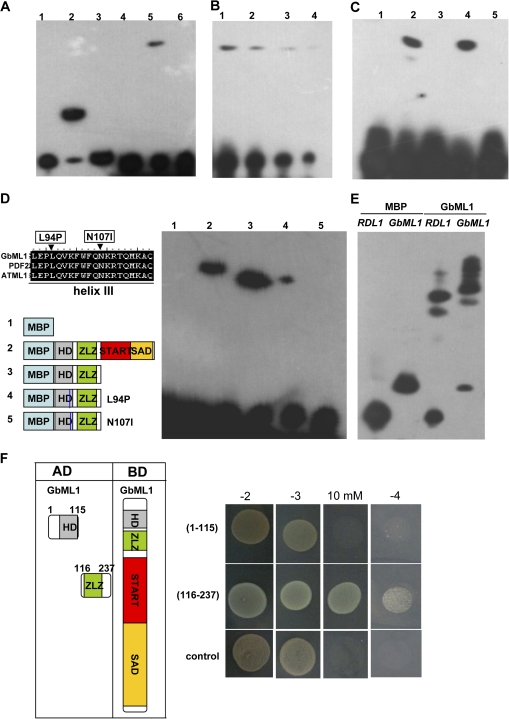
Previous studies demonstrated that recombinant ATML1 and PDF2 proteins could bind to the L1 box in vitro (Abe et al., 2001, 2003). EMSA assays were performed by using purified full-length or truncated GbML1 proteins (see Supplementary Fig. S2A at JXB online). Complex formation was observed with the GbML1 (Fig. 2A, lane 5; Fig. 2B) and HD-ZLZ proteins (Fig. 2C, lane 4) but not with the MBP (Fig. 2A, lane 4) or the HD (Fig. 2C, lane 3), START–SAD proteins (Fig. 2C, lane 5). No complex formation was observed with the mutated L1 box (Fig. 2A, lane 6). The third helix of the HD domain was shown to contact the double-stranded DNA, and was important for protein–DNA interaction. Two mutations were made in this region, L94P and N107I (Fig. 2D, left; see Supplementary Fig. S2B at JXB online). While the L94P mutation greatly reduced the interaction between the HD-ZLZ protein and the L1 box, the N107I mutation abolished the interaction (Fig. 2D, right). Moreover, GbML1 could bind to the L1 box containing parts from GaRDL1 (Li et al., 2002) and GbML1 promoters in vitro (Fig. 2E; see Supplementary Fig. S3 at JXB online). These results indicate that GbML1 can specifically bind to L1 box and may specify the gene expression in the L1 layer.
Fig. 2.
GbML1 binds to the L1 box in vitro. (A) Interaction of GbML1 protein with the L1 box. 1–3: Controls provided by the kit. 1, No protein, biotin-ENBL probe; 2, ENBL, biotin-ENBL probe; 3, ENBL, biotin-ENBL probe and 100× ENBL cold competitor. 4–6, binding assay for GbML1;. 4, MBP, biotin-L1 probe (L1: TGTAAATGCACCTGCAACACA); 5, GbML1, biotin-L1 probe; 6, GbML1, biotin ml1 probe (mL1:TGTAAGGGCACCTGCAACACA). (B) Reduced probe concentration results in weaker binding. 2, 2×dilution; 3, 4×dilution; 4, 8× dilution. (C) HD-ZLZ domains are required and sufficient for binding to the L1 box. Binding reaction includes biotin-labelled L1 box probe with different MBP fusion proteins: 1, MBP protein; 2, MBP-GbML1 protein; 3, MBP-HD protein; 4, MBP-HD-ZLZ protein; 5, MBP-START–SAD protein. (D) Point mutations in the third helix affect the binding of HD-ZLZ to L1 box. Left: Diagram of the point mutation and proteins used in EMSA. Right, EMSA assay. The number above is correlated to the number of the proteins diagramed. (E) GbML1 binds to the L1 box containing promoters from cotton. (F) ZLZ domain confers homodimer formation. left: Diagram of different constructs used for the yeast two-hybrid assay; right, yeasts harbouring BD-GbML1/AD-HD or BD-GbML1/AD-ZLZ grown on selective plates as indicated. Control medium: –2 (SD/-T-L); selective medium: –3 (SD/-T-L-H), 10 mM (–3 supplemented with 10 mM 3-AT), –4 (SD/-T-L-H-A). The control is yeast transformed with BD-GbML1/AD.
The requirement of the ZLZ motif for L1 box binding indicated that GbML1 might form dimmers. To test this possibility, the full-length GbML1 protein and the ZLZ part of the GbML1 protein was fused to the BD domain and the AD domain, respectively (Fig. 2F, left). GbML1 was shown to have weak transactivation activity in yeast when fused with the BD domain, but this activity could be inhibited by adding 5 mM 3-AT to the SD medium (see Supplementary Fig. S4 at JXB online). Yeast cells harbouring BD-GbML1 and AD-GbML1-ZLZ constructs could grow on selective medium, supporting the hypothesis that GbML1 could form a homodimer (Fig. 2F, right). By contrast, yeast cells harbouring both BD-GbML1 and AD-GbML1-HD constructs could not grow on selective medium.
Taken together, GbML1 is an HD-Zip IV transcription activator which binds to the L1 box and may be implicated into the regulation of gene expression in the L1 layer.
GbML1 binds to GbMYB25 but not to GbMYB2
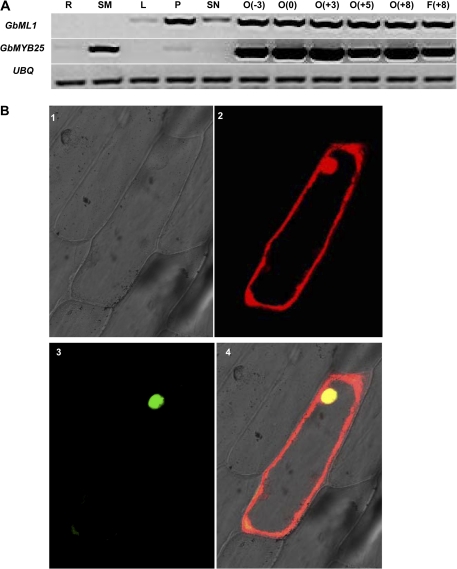
A previous study has shown that GhHD1 and GhMYB25 had enriched expression in fibre initial cells (Wu et al., 2006). To address if GbML1 and the homologue of GhMYB25 in G. barbadense (see Supplementary Fig. S5 at JXB online), GbMYB25, also had a similar expression pattern, an RT-PCR assay was performed (Fig. 3A). RT-PCR results showed that GbML1 had high expression in petal and ovules (–3 DPA, 0 DPA, +3 DPA, +5 DPA, +8 DPA) and fibres (+8DPA), weak expression in leaves and stamens, and no signal was detected in roots or stems. GbMYB25 had high expression in ovules (–3 DPA, 0 DPA, +3 DPA, +5 DPA, +8 DPA) and fibres (+8 DPA), moderate expression in stems, and weak expression in roots, petals, and stamens. No signal was detected in the leaves.
Fig. 3.
Expression patterns and subcellular localization of GbML1 and GbMYB25. (A) Expression patterns of GbML1 and GbMYB25 in different tissues. R, root; SM, stem; L, leaf; P, petal; SN, stamen; O, ovule; F, fibre. The number in the brackets indicates the number of days post-anthesis. UBQ, ubiquitin gene used as the internal control. (B) Subcellular localization of GbML1 and GbMYB25. The CFP-GbML1 and GbMYB25-YFP constructs were co-bombarded into onion epidermal cells. 1, Bright field; 2, CFP channel, 3: YFP channel; 4, merged picture.
It was further explored if GbML1 and GbMYB25 proteins had the same subcellular localization pattern. GbML1 and GbMYB25 proteins were fused in-frame to CFP and YFP, respectively. These constructs were co-bombarded into onion epidermal cells and the localization pattern was observed using confocal microscope. CFP-GbML1 protein was detected both in the cytoplasm and nucleus while GbMYB25-YFP was mainly localized into nucleus (Fig. 3B).
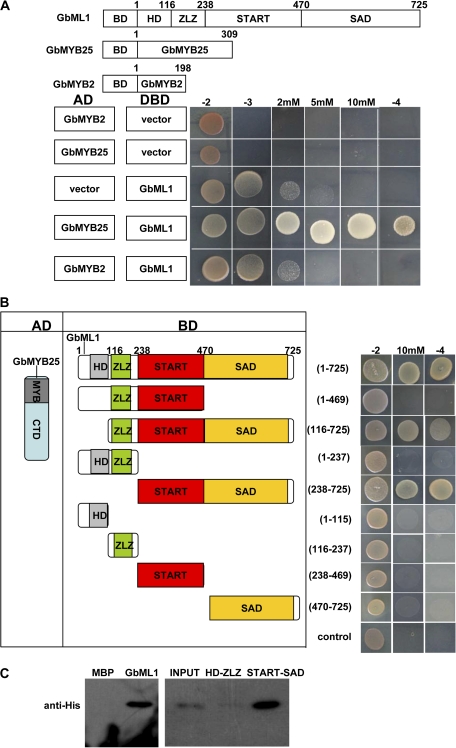
Since GbML1 and GbMYB25 had strong expression in ovules and both could localize into nucleus, it was tested whether GbML1 and GbMYB25 could interact with each other. Due to that that GbMYB25 had strong activation activity and the transactivation activity of GbML1 could be inhibited by adding 5 mM 3-AT into the medium (see Supplementary Fig. S4 at JXB online), GbML1 protein was fused to the BD domain, while GbMYB25 or GbMYB2 (a homologue for GaMYB2; see Supplementary Fig. S6 at JXB online) protein was fused to the AD domain (Fig. 4A, left).
Fig. 4.
START–SAD domain of GbML1 are required and sufficient for the interaction with GbMYB25 protein. (A) GbML1 could bind to GbMYB25 but not to GbMYB2. (Top) diagram of different constructs used in this panel. (Bottom) Y2H assay. Yeast cells harbouring the constructs shown on the left were spotted on control medium (–2) and selective medium (–3; –3 with 2 mM 3-AT, 5 mM 3-AT, or 10 mM 3-AT; –4). (B) Mapping of domains of GbML1 to bind GbMYB25. As shown on the left, different truncated versions of GbML1 protein were fused with the BD domain and co-transformed into yeast with AD-GbMYB25. The yeast cells harbouring the bait and prey were spotted on the selective medium. As shown, both the START domain and the SAD domain are required for the interaction with GbMYB25. Yeast cells habouring BD and AD-GbMYB25 constructs were used as the negative control. (C) In vitro pull-down assay of MBP, full-length or truncated GbML1 proteins with HIS-GbMYB25 fusion protein. The full-length or truncated MBP-GbML1 fusion proteins are used as baits to pull down the His-GbMYB25 fusion protein from the induced cell extracts. MBP protein is assayed as a negative control. Immunoblot detection of prey protein is with His antibody. Arrow indicates the corresponding target proteins.
Yeast cells co-transformed with BD-GbML1 and AD-GbMYB25 could grow on selective medium. Yeast cells harbouring BD and AD, and BD-GbML1 and AD, BD and AD-GbMYB25, BD-GbML1 and AD-GbMYB2 could not grow on the selective medium (Fig. 4A right). These results suggest that GbML1 could bind to GbMYB25 but not to GbMYB2.
GbML1 and GbMYB25 interactions were mapped using the yeast two-hybrid assay. The deletion of HD domain retained the interaction while the deletion of the SAD domain completely abolished the interaction (Fig. 4B). This result indicated that the SAD domain was required for the binding. The GbML1 protein was divided into two continuous but not overlapping parts, HD-ZLZ (aa 1–269) and START–SAD (aa 270–725). The HD-ZLZ part could not interact with GbMYB25 while the START–SAD part could interact with GbMYB25 as the full-length GbML1 (Fig. 4B). Subsequently, it was examined if the SAD domain alone could bind to GbMYB25. No single domain (HD, ZLZ, START, SAD) showed an interaction with GbMYB25 (Fig. 4B), suggesting that the START domain was also required for the interaction. Together, the START–SAD part was required and sufficient for the interaction between GbML1 and GbMYB25.
To confirm the interaction observed in the yeast two-hybrid assay, in vitro pull-down assays were performed. Maltose binding domain (MBP) was fused to GbML1 proteins (full-length GbML1, HD-ZLZ, START–SAD), and the MBP-GbML1 fusion proteins were expressed in and purified from Escherichia coli (E. coli) BL21. The 6× His tag was fused to the full-length GbMYB25 and the His-GbMYB25 fusion protein was expressed in E. coli BL21 (DE3). Equal lysates harbouring His-GbMYB25 were incubated with immobilized MBP or MBP-GbML1 proteins. As expected, GbMYB25 bound to GbML1 and GbML1-C but not to the control MBP protein or GbML1-NM (Fig. 4C).
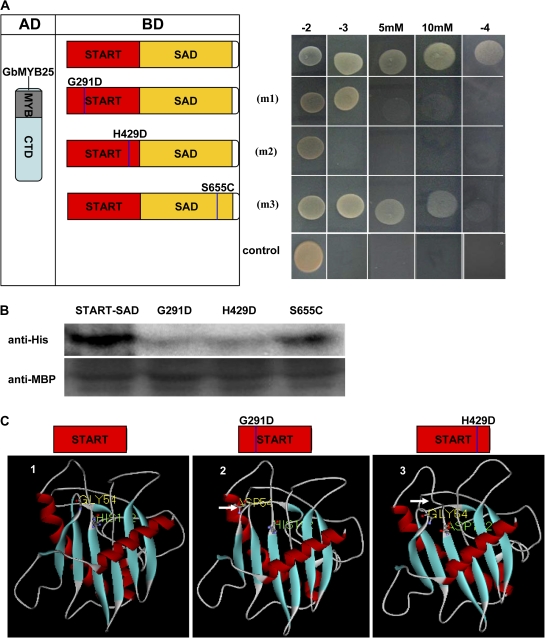
Since the START–SAD domain of GbML1 was required and was sufficient for the binding to GbMYB25, it was investigated if point mutations in some conserved regulatory amino acids might affect the interaction. Three mutations were made by the alteration of one amino acid in different positions (as indicated in Fig. 5A, left). In the yeast two-hybrid assay, the G291D mutation reduced the interaction, H429D completely abolished the interaction, and S665C slightly reduced the interaction (Fig. 5A, right). An in vitro pull-down assay was performed to assess the effect of point mutations on binding to GbMYB25. While the S665C mutation retained less amount of the GbMYB25 protein compared with the wild-type part, the G291D and H429D mutations greatly reduced the amount of bound GbMYB25 protein (Fig. 5B). This pull-down result was consistent with the results gained from the yeast two-hybrid assay, further demonstrating that the START domain was critical for the binding.
Fig. 5.
Effect of point mutations of GbML1 on protein interaction. (A) Effect of point mutations on the interaction between GbML1 and GbMYB25 in the yeast two-hybrid assay. (Left) Diagram of point mutation sites. m1 indicates a G to D mutation at the 291 site; m2 indicates an H to D mutation at the 429 site; m3 indicates an S to C mutation at the 655 amino acid site. (Right) The START–SAD domain and its mutations were fused with the BD domain and introduced into yeast with the AD-GbMYB25 construct. Yeast cells harbouring BD-START–SAD/AD were used as negative control. The growth on selective medium was shown. (B) In vitro pull-down assay to confirm the yeast two-hybrid results. MBP-GbML1-START–SAD, MBP-START–SAD-m1, MBP-START–SAD-m2, and MBP-START–SAD-m3 fusion proteins are used as baits to pull down the HIS-GbMYB25 fusion protein from the induced cell extracts. Immunoblot detection of prey protein is with the His antibody. (C) Deduced three-dimensional structure of START domains indicates the pocket structure is important for the binding. 1, Modelled structure of the native START domain from GbML1. The pocket structure is obvious on the upper side. The No. 54 amino acid (Gly) and the No. 192 amino acid (His) of the START domain are shown. 2, The G291D mutation affects the structure of the third loop. The changed conformation is shown by a white arrow. 3, The H429D mutation changes the open pocket to be closed. The white arrow indicates the changed loop.
To understand the mechanism of the point mutations on protein interaction, the three-dimensional structures of START domains were analysed using homology modelling based on a plant START domain structure (2r55A.pdb). A G291D mutation affected the second β sheet and the third loop which was near the pocket structure (Fig. 5C, 2). The G429H mutation affected the loop which was the component of the pocket, and formed a closed pocket structure (Fig. 5C, 3). Based on these results, GbMYB25 might bind to GbML1 in the pocket formed by the START domain, and the pocket structure was important for the interaction.
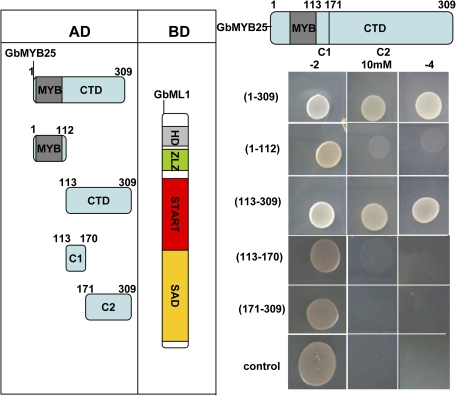
The protein–protein interaction domain of GbMYB25 (Fig. 6) was also investigated. The C terminal part (aa: 113–309) interacted with GbML1 as strongly as the complete GbMYB25. The N terminal part of GbMYB25 (aa: 1–112) containing the R2R3-MYB repeats could not interact with GbML1. Neither the C1 part (aa: 113–170) nor the C2 part (aa: 171–309) interacted with GbML1, suggesting both parts were needed for the interaction.
Fig. 6.
GbML1 binds to the C terminal domain of GbMYB25. (Left) Diagram of different preys used in mapping the interaction part of GbMYB25. CTD, C terminal domain including C1 and C2. C1 domain is a conserved region other than the MYB domain among MIXTA type proteins. (Right) Y2H assay was carried out as in Fig. 4B. The yeast two-hybrid assay indicates that the CTD of GbMYB25 binds to GbML1 while the MYB repeats do not. Both C1 and C2 domains of GbMYB25 are required for the binding to GbML1.
GbML1 ectopic overexpression in Arabidopsis causes pleiotropic developmental alterations
Arabidopsis trichomes and cotton fibres are both unicellular hairs of the epidermis, and they share a similar molecular machinery of regulation (Wang et al., 2004). Functional analysis of cotton genes in Arabidopsis has been successfully used to elucidate the mechanisms that regulate cotton fibre development. Due to the difficulties of generating transgenic cotton, functional analysis of GbML1 was carried out in Arabidopsis.
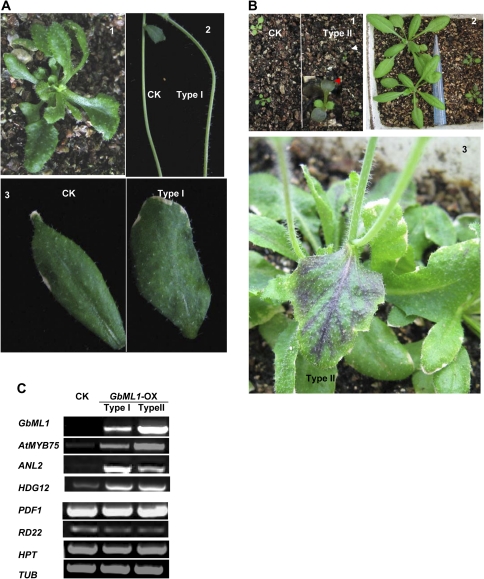
The full-length GbML1 ORF was driven under the double 35S promoter to make the overexpression construct which was later introduced into Arabidopsis. Eighteen independent GbML1 overexpression lines were obtained. In these lines, there were mainly two types of plant based on the severity of the developmental phenotypes: the first type of plant (Type I, 12/18) was developmentally similar as the plants transformed with the empty vector (CK) except that there were more trichomes on the stems and cauline leaves (Fig. 7A, 1, 2, 3). The second type of plant (Type II, 6/18) had more anthocyanin accumulation in the cotyledons (Fig. 7B, 1) and leaves (Fig. 7B, 2, 3, 4). Expression patterns of several genes related to the phenotypes were examined in Type I and Type II plants (Fig. 7C). GbML1 was highly expressed in Type II seedlings while there was only a moderate expression level of GbML1 in Type I seedlings. Genes related to anthacynin biosynthesis including AtMYB75 (Teng et al., 2005) and ANL2 (Kobo et al., 1999) were induced in the transgenic plants compared with the CK. HDG12 was also induced in GbML1 overexpressing plants. The expression level of RD22, PDF1, and the selective antibiotic gene HPT (hygromycin phosphotransferase) were similar to the CK in Type I and Type II overexpressing plants.
Fig. 7.
Phenotypes of overexpressing GbML1 in Arabidopsis plants. (A) Developmental phenotypes of Type I plants. 1, 25-d-old plant. 2, More trichomes on stems. 3, More trichomes on cauline leaves. (B) Developmental phenotypes of Type II plants. 1, Type II plants accumulate more anthocyanin on cotyledons. 2–4, Anthocyanin accumulation in rosette leaves and cauline leaves. (C) Expression of GbML1 and HPT transgenes and AtMYB75, ANL2, HDG12, PDF1, and RD22 endogenous genes are also shown. Tubulin (Tub) is used as a control for normalization.
Discussion
GbML1 binds to the L1 box
HD-Zip IV transcription factors have been discovered in many plant species (Schrick et al., 2004). In Arabidopsis, several members including GL2, ANL2, ATML1, PDF2, and HDG11 have been well characterized and their functions were mainly related to the epidermis of different organs, at least in part, via binding to the L1 box in the promoter region of downstream genes (Nakamura et al., 2006; Federico et al., 2007).
Members of the HD-Zip IV family were also been found in cotton, one of the most important crops grown all over the world for its fibre and seed oil production. Wang et al. (2004) found that, beside MYB transcription factors, there should be other factors which recognized the L1 box to regulate the fibre-specific expression pattern of RDL1. Several cotton HOX genes that fell into the group of the HD-Zip IV family were cloned, and GaHOX1 was shown to act functionally equivalent to GL2 (Guan et al., 2008). However, none of these HOX genes could bind to the L1 box in a yeast one-hybrid assay (Wang et al., 2004). In this study, a new HD-Zip IV family member was cloned and characterized from cotton (G. barbadense), GbML1. Gene structure, domain arrangement, amino acid alignment, and phylogenic analysis all suggested that GbML1 was an HD-START protein. Since GbML1 showed the highest sequence similarity to PDF2 and ATML1, the binding ability of GbML1 protein to the L1 box was also tested. The GbML1 protein bound to the L1 box but not to the mutated L1 box and this interaction required both the HD domain and the ZLZ domain. This result agreed well with previous functional studies (Sessa et al., 1993; Tron et al., 2004). Most L1-layer specific transcription factors such as ATML1 and PDF2 had L1 boxes in their promoter regions and a feedback regulation mechanism was suggested. GbML1 was a homologous gene for ATML1 and PDF2, and the promoter region of GbML1 as well as the fibre-specific gene RDL1 had L1 boxes, so the question was asked if GbML1 could also bind to these promoters to regulate their expression. A gel shift assay indicated that GbML1 bound to RDL1 and GbML1 promoter fragments containing the L1 boxes. These results showed that GbML1 was the first L1 box binding protein isolated in cotton and might have a role in epidermal cell specification.
GbML1 binds to the C part of GbMYB25 via the START–SAD domain
GhMYB25 was a key regulator of cotton fibre development. The expression of GhMYB25 was enriched in the fibre initial cells relative to the non-fibre ovular epidermal cells, by laser-capture microdissection microarrays (Wu et al., 2006). In addition, functional study in cotton directly demonstrated the key roles of MYB transcription factors in fibre initiation and elongation. Antisense-mediated suppression of GhMYB109 led to the great reduction in fibre length (Pu et al., 2008). RNAi repression of GhMYB25 expression in cotton resulted in short fibres and dramatic reductions in trichomes on other parts of the plants. Ectopic overexpression of GhMYB25 increased cotton fibre initiation and leaf trichome number (Machado et al., 2009).
MYB transcription factors appeared to act together with other transcription factors to control epidermal cell specification. For example, the expression of RDL1 was controlled by MYB transcription factors and transcription factors that bound to the L1 box, since, when mutated, either the MYB recognition motif or the L1 box greatly reduced the promoter activity. Similar expression patterns of GbML1 and GbMYB25 raised the question whether they could interact with each other to control similar developmental events. The yeast two-hybrid assay and the pull-down assay showed that GbML1 physically interacted with GbMYB25. Given that GbML1 had weak transcription activation activity while GbMYB25 was a strong activator, the binding of these two factors could form a strong transcription activator with L1 box binding selectivity to activate gene expression in the L1 layer. To our surprise, it was the START domain together with the SAD domain that interacted with the C-terminal part of the R2R3 transcription factors. Point mutation in the fourth loop of the START domain abolished the binding and other point mutations could partially affect the binding, indicating that the binding to CTD-MYB would largely be possible due to the 3D structure of the START–SAD domain. In animals, the START domain was found to bind to different chemicals such as cholesterol (Tsujishita and Hurley, 2000), phosphatidylcholine (Roderick et al., 2002), carotenoid (Tabunoki et al., 2002), and ceramide (Hanada et al., 2003). Only one study in animals demonstrated that the START domain could also act in protein interactions (Kanno et al., 2007). In plants, a new type of START domain was demonstrated to act as ABA receptors (Ma et al., 2009; Park et al., 2009). However, the function of the classic START domain was still elusive. In this study, it was discovered, at least for GbML1, that the START domain could serve as a protein binding domain. Given that every member in the HD-Zip III family and the HD-Zip IV family had a classic START domain, our results would give a hint to the functional analysis of members in these two families. The protein binding activity of the START domain would not exclude possible chemical binding activity. Indeed, as to the newly discovered ABA receptors, the chemicals ABA and parabactine bound to the START pocket and changed the conformation of the protein structure to facilitate the binding to PP2C proteins to transduce the signals. So it would be interesting to see if some chemicals could bind to GbML1, and block the binding of GbML1 and GbMYB25, to terminate the activation processes in tissues other than ovules.
GbML1 as a target for genetic improvement of cotton fibre
A previous study showed that overexpression of GaMYB2 could rescue the Arabidopsis gl1 phenotype. However, only a single seed trichome was observed from a low proportion of these transgenic plants which indicated that other members are required for sufficient specifying seed coat epidermal cells into trichomes (Lee et al., 2007). Interestingly, both the L1 box and the MYBCORE cis elements were required for the expression of a cotton fibre-specific gene GaRDL1. Previous study demonstrated that GaMYB2 can bind to the MYBCORE element and activate the expression of RDL1. Our data demonstrated that GbML1 could bind the L1 box and the RDL1 promoter in vitro. Though GbML1 was a weak transactivator, it could bind to GbMYB25, which had strong transactivation activity. Thus the GbML1 and GbMYB25 together formed a strong activator that specifically binds to the L1 box. The overexpression of GbML1 did not result in the seed trichome phenotype which might be due to the lack of the binding partner GbMYB25 in Arabidopsis ovules.
Overexpression of GbML1 in Arabidopsis mainly caused an alteration at the epidermis, including increased leaf trichomes, stem trichomes, and anthocyanin. Arabidopsis trichomes and cotton fibres were thought to have a common regulatory mechanism. Anthocyanin accumulation and trichomes shared some common regulators, such as GL3, EGL3, TTG1, and TTG2. Thus the phenotype of ectopic expression of GbML1 in Arabidopsis at least supported the fact that GbML1 was an important regulator of the specification of epidermal cells. Given that GbML1 was mainly expressed in ovules and cotton fibres arising from the ovule epidermis, GbML1's role in cotton might control fibre cell differentiation. Further transgenic analysis in cotton would be needed for this hypothesis.
The regulation of cotton fibre development is complicated and thus overexpression a single gene may not be sufficient enough to improve the quality and quantity of fibres. Combined manipulation of multiple regulators such as GbML1, GbMYB25, and GaMYB2 genes in cotton ovules may be a more efficient way for cotton fibre improvement.
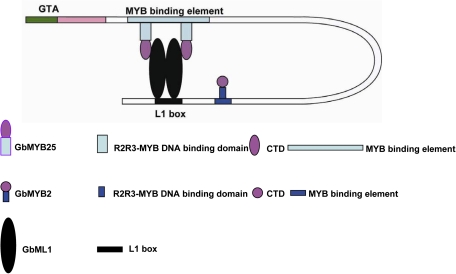
A suggested model for GbML1 and R2R3-MYB transcription factors in the regulation of gene expression
In summary, our results suggest that GbML1 and GbMYB25 can act together to activate L1-layer specific genes. At first, the ZLZ domains of GbML1 transcription factors form either homo- and/or hetero-dimers. The dimerized GbML1 transcription factors recognize and bind to the L1 box. Subsequently, the R2R3-MYB transcription factor GbMYB25 binds to the START–SAD domain of the GbML1 via its C terminal domains, which is also an activation domain. R2R3-MYB transcription factors such as GaMYB2 may also bind to the MYB binding element through its MYB recognition motif (Fig. 8). These factors together provide a regulatory complex to control fibre cell initiation and development.
Fig. 8.
A suggested model for regulating gene expression by interaction of HD-Zip IV and R2R3 MYB transcription factors in controlling cotton fibre development.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Alignment of different domains of GbML1.
Supplementary Fig. S2. Purified proteins used in EMSA.
Supplementary Fig. S3. The property of the sequence in the GbML1 promoter region.
Supplementary Fig. S4. Yeast transactivation assay of GbML1, GbMYB25, and GbMYB2.
Supplementary Fig. S5. Alignment of GbMYB25 with GhMYB25.
Supplementary Fig. S6. Alignment of GbMYB2, GaMYB2. and GhMYB2.
Supplementary Fig. S7. Alignment used to generate the phylogeny tree.
Supplementary Table S1. Primers and probes used in this research.
Supplementary Material
Acknowledgments
We thank Dr. Hongquan Yang for providing the pHB, PA7-CFP, and PA7-YFP vectors. We thank Dr Yanchun Zhang and Jingyi Zhang for the help with the confocal microscopy, and Fangyuan Zhang for drawing the model. This research was supported by the China ‘973’ Program (grant numbers 2004CB117303-3 and 2007CB108805), China “863” Program (grant number 2010AA100503), China Transgenic Research Program (grant number 2008ZX08002-001) and the Shanghai Leading Academic Discipline Project (project number B209).
Glossary
Abbreviations
- bp
base pair
- DPA
days post anthesis
- EMSA
electrophoretic mobility shift assay
- HD
homeodomain
- MW
molecular weight
- pI
isoelectric point
- RACE
Rapid Amplification of cDNA Ends
- SAD domain
START associated domain
- START domain
StAR-related lipid Transfer domain
- Wt
wild type
- ZLZ
zipper-loop-zipper motif
References
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. Role of arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. The Plant Cell. 1997;9:1859–1868. doi: 10.1105/tpc.9.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe M, Takahashi T, Komeda Y. Identification of a cis-regulatory element for L1 layer-specific gene expression, which is targeted by an L1-specific homeodomain protein. The Plant Journal. 2001;26:487–494. doi: 10.1046/j.1365-313x.2001.01047.x. [DOI] [PubMed] [Google Scholar]
- Abe M, Katsumata H, Komeda Y, Takahashi T. Regulation of shoot epidermal cell differentiation by a pair of homeodomain proteins in Arabidopsis. Development. 2003;30:635–643. doi: 10.1242/dev.00292. [DOI] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL Workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Federico DA, Pablo AM, Carlos AD, Raquel LC. The true story of the HD-Zip family. Trends in Plant Science. 2007;12:419–426. doi: 10.1016/j.tplants.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Guan XY, Li QJ, Shan CM, Wang S, Mao YB, Wang LJ, Chen XY. The HD-Zip IV gene GaHOX1 from cotton is a functional homologue of the Arabidopsis GLABRA2. Physiologia Plantarum. 2008;134:174–182. doi: 10.1111/j.1399-3054.2008.01115.x. [DOI] [PubMed] [Google Scholar]
- Hanada K, Kumagai K, Yasuda S, Miura Y, Kawano M, Fukasawa M, Nishijima M. Molecular machinery for non-vesicular trafficking of ceramide. Nature. 2003;426:803–809. doi: 10.1038/nature02188. [DOI] [PubMed] [Google Scholar]
- Humphries JA, Walker AR, Timmis JN, Orford SJ. Two WD-repeat genes from cotton are functional homologues of the Arabidopsis thaliana TRANSPARENT TESTA GLABRA1 (TTG1) gene. Plant Molecular Biology. 2005;57:67–81. doi: 10.1007/s11103-004-6768-1. [DOI] [PubMed] [Google Scholar]
- Ishida T, Kurata T, Okada K, Wada T. A genetic regulatory network in the development of trichomes and root hairs. Annual Review of Plant Biology. 2008;59:365–386. doi: 10.1146/annurev.arplant.59.032607.092949. [DOI] [PubMed] [Google Scholar]
- Jakoby MJ, Falkenhan D, Mader MT, Brininstool G, Wischnitzki E, Platz N, Hudson A, Hülskamp M, Larkin J, Schnittger A. Transcriptional profiling of mature Arabidopsis trichomes reveals that NOECK encodes the MIXTA-like transcriptional regulator MYB106. Plant Physiology. 2008;148:1583–1602. doi: 10.1104/pp.108.126979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno K, Wu MK, Agate DS, Fanelli BJ, Wagle N, Scapa EF, Ukomadu C, Cohen DE. Interacting proteins dictate function of the minimal START domain phosphatidylcholine transfer protein/StarD2. Journal of Biological Chemistry. 2007;282:30728–30736. doi: 10.1074/jbc.M703745200. [DOI] [PubMed] [Google Scholar]
- Kiefer F, Arnold K, Künzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Research. 2009;37:387–392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo H, Peeters AJ, Aarts MG, Pereira A, Koornneef M. ANTHOCYANINLESS2, a homeobox gene affecting anthocyanin distribution and root development in Arabidopsis. The Plant Cell. 1999;11:1217–1226. doi: 10.1105/tpc.11.7.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lee JJ, Woodward AW, Chen ZJ. Gene expression changes and early events in cotton fibre development. Annals of Botany. 2007;100:1391–1401. doi: 10.1093/aob/mcm232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CH, Zhu YQ, Meng YL, Wang JW, Xu KX, Zhang TZ, Chen XY. Isolation of genes preferentially expressed in cotton fibres by cDNA filter arrays and RT-PCR. Plant Science. 2002;163:1113–1120. [Google Scholar]
- Lu P, Porat R, Nadeau JA, O'Neill SD. Identification of a meristem L1 layer-specific gene in Arabidopsis that is expressed during embryonic pattern formation and defines a new class of homeobox genes. The Plant Cell. 1996;8:2155–2168. doi: 10.1105/tpc.8.12.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- Machado AC, Wu YR, Yang YM, Llewellyn DJ, Dennis ES. The MYB transcription factor GhMYB25 regulates early fibre and trichome development. The Plant Journal. 2009;59:52–62. doi: 10.1111/j.1365-313X.2009.03847.x. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Katsumata H, Abe M, Yabe N, Komeda Y, Yamamoto KT, Takahashi T. Characterization of the class IV homeodomain-leucine zipper (HD-Zip IV) gene family in Arabidopsis. Plant Physiology. 2006;141:1363–1375. doi: 10.1104/pp.106.077388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne T, Clement J, Arnold D, Lloyd A. Heterologous myb genes distinct from GL1 enhance trichome production when overexpressed in Nicotiana tabacum. Development. 1999;126:671–682. doi: 10.1242/dev.126.4.671. [DOI] [PubMed] [Google Scholar]
- Peitsch MC. Protein modeling by e-mail. Bio/Technology. 1995;13:658–660. [Google Scholar]
- Pu L, Li Q, Fan X, Yang W, Xue Y. The R2R3 MYB transcription factor GhMYB109 is required for cotton fibre development. Genetics. 2008;180:811–820. doi: 10.1534/genetics.108.093070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roderick SL, Chan WW, Agate DS, Olsen LR, Vetting MW, Rajashankar KR, Cohen DE. Structure of the human phosphatidylcholinetransfer protein in complex with its ligand. Nature Structural Biology. 2002;9:507–511. doi: 10.1038/nsb812. [DOI] [PubMed] [Google Scholar]
- Schrick K, Nguyen D, Karlowski WM, Mayer KF. START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome Biology. 2004;5:R41. doi: 10.1186/gb-2004-5-6-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa G, Morelli G, Ruberti I. The Athb-1 and -2 HD-Zip domains homodimerize forming complexes of different DNA binding specificities. The EMBO Journal. 1993;12:3507–3517. doi: 10.1002/j.1460-2075.1993.tb06025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo J, Liang X, Pu L, Zhang Y, Xue Y. Identification of GhMYB109 encoding a R2R3 MYB transcription factor that expressed specifically in fibre initials and elongating fibres of cotton (Gossypium hirsutum L.) Biochimica et Biophysica Acta. 2003;1630:25–34. doi: 10.1016/j.bbaexp.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Tabunoki H, Sugiyama H, Tanaka Y, Fujii H, Banno Y, Jouni ZE, Kobayashi M, Sato R, Maekawa H, Tsuchida K. Isolation, characterization, and cDNA sequence of a carotenoid binding protein from the silk gland of Bombyx mori larvae. Journal of Biological Chemistry. 2002;277:32133–32140. doi: 10.1074/jbc.M204507200. [DOI] [PubMed] [Google Scholar]
- Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S. Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiology. 2005;139:1840–1852. doi: 10.1104/pp.105.066688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tron AE, Welchen E, Gonzalez DH. Engineering the loop region of a homeodomain leucine zipper protein promotes efficient binding to a monomeric DNA binding site. Biochemistry. 2004;43:15845–15851. doi: 10.1021/bi048254a. [DOI] [PubMed] [Google Scholar]
- Tsujishita Y, Hurley JH. Structure and lipid transport mechanism of a StAR-related domain. Nature Structural Biology. 2000;7:408–414. doi: 10.1038/75192. [DOI] [PubMed] [Google Scholar]
- Wang S, Wang JW, Yu N, Li CH, Luo B, Gou JY, Wang LJ, Chen XY. Control of plant trichome development by a cotton fibre MYB gene. The Plant Cell. 2004;16:2323–2334. doi: 10.1105/tpc.104.024844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Qiu C, Zhang F, Guo B, Miao Z, Sun X, Tang K. Molecular cloning, expression profiling and functional analyses of a cDNA encoding isopentenyl diphosphate isomerase from Gossypium barbadense. Bioscience Reports. 2009;29:111–119. doi: 10.1042/BSR20070052. [DOI] [PubMed] [Google Scholar]
- Wu Y, Llewellyn DJ, White R, Ruggiero K, Al-Ghazi Y, Dennis ES. Laser capture microdissection and cDNA microarrays used to generate gene expression profiles of the rapidly expanding fibre initial cells on the surface of cotton ovules. Planta. 2007;226:1475–1490. doi: 10.1007/s00425-007-0580-5. [DOI] [PubMed] [Google Scholar]
- Wu Y, Machado AC, White RG, Llewellyn DJ, Dennis ES. Expression profiling identifies genes expressed early during lint fibre initiation in cotton. Plant and Cell Physiology. 2006;47:107–127. doi: 10.1093/pcp/pci228. [DOI] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin SS, Niu QW, Chua NH. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nature Protocols. 2006;1:641–646. doi: 10.1038/nprot.2006.97. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.