Abstract
Sucrose transporters (SUTs) are known to play critical roles in the uptake of sucrose from the apoplast in various steps of sugar translocation. Because developing pollen is symplastically isolated from anther tissues, it is hypothesized that SUTs are active in the uptake of apoplastic sucrose into pollen. To investigate this possibility, a comprehensive expression analysis was performed for members of the SUT gene family in the developing pollen of rice (Oryza sativa L.) using real-time RT-PCR combined with a laser microdissection technique. Among the five SUT genes, OsSUT1 and OsSUT3 were found to be preferentially expressed and had temporal expression patterns that were distinct from each other. Expression of OsSUT1 in pollen was confirmed by a promoter–GUS fusion assay. The physiological function of OsSUT1 in pollen was further investigated using retrotransposon insertion mutant lines. While the homozygote of disrupted OsSUT1 (SUT1–/–) could not be obtained, heterozygote plants (SUT1+/–) showed normal grain filling. Their progeny segregated into SUT1+/– and SUT1+/+ with the ratio of 1:1, suggesting that the pollen disrupted for OsSUT1 is dysfunctional. This hypothesis was reinforced in vivo by a backcross of SUT1+/– plants with wild-type plants and also by in vitro pollen germination on the artificial media. However, starch accumulation during pollen development was not affected by disruption of OsSUT1, suggesting that the sugar(s) required for starch biosynthesis is supplied by other sugar transporters.
Keywords: Gene expression, laser microdissection, pollen germination, pollen maturation, sucrose transporter (SUT), Oryza sativa L
Introduction
Sucrose is the most common sugar molecule translocated in higher plants, and the transmembrane transport of sucrose in higher plants is mediated by sucrose transporter (SUT) proteins. A SoSUT1 cDNA from spinach was the first reported SUT in higher plants (Riesmeier et al., 1992); since then, more than 60 SUT genes have been cloned. Looking back over the history of SUT research, it was fortuitous that SUTs reported in the early days were localized to vein or phloem and that the proteins were suggested to be a phloem loader, a protein that researchers had long been searching for as a central player of sucrose loading. However, early reports on the phloem loader SUTs from a number of plant species were followed by a growing body of evidence supporting the idea that SUTs have very diverse roles and that phloem loading is merely one of them. It is now more generally accepted that (plasma membrane-localized) SUTs play critical roles in the uptake of sucrose from the apoplast at various steps in sugar translocation (see Sauer, 2007, for a review).
In addition to phloem loading, SUTs have been well documented to mediate the uptake of sucrose into developing filial tissues that are symplastically isolated from maternal tissues. In developing seeds, for instance, all nutrients including sucrose are released from maternal tissues into the apoplast surrounding the filial tissues; they are then taken up by outer cell layers of the filial tissues, such as endosperm or the embryo (see Patrick and Offler, 2001, for a review). The uptake of sucrose into developing filial tissues is mediated by SUTs, and the expression and function of these SUTs have been investigated in both grain legumes and cereals. In the developing rice caryopsis, a gene for SUT, OsSUT1, is highly expressed in the aleurone (Hirose et al., 2002; Ishimaru et al., 2007), and antisense suppression of OsSUT1 results in poor grain-filling accompanied by a decreased rate of radiolabelled sucrose uptake into seed tissues (Scofield et al., 2002). Therefore, OsSUT1 is thought to have a crucial role in the uptake of sucrose by the filial tissues of developing rice caryopsis. In the pea, seed-specific overexpression of SUT was reported to enhance both the sucrose uptake and growth rates of the cotyledons of developing seeds (Rosche et al., 2002; Zhou et al., 2009).
Because pollen grains are symplastically isolated from anther tissues (e.g. filial tissues) of the seeds and accumulate large amounts of starch as in cereal endosperm, SUTs are expected to be active in pollen development. In fact, mRNA or protein of several SUTs has been detected in pollen or pollen tube (Lemoine et al., 1999; Stadler et al., 1999, Hackel et al., 2006; Lauterbach et al., 2007; Sivitz et al., 2008). In tomato, LeSUT2 has been shown to play a role in pollen tube growth using the antisense suppression technique (Hackel et al., 2006). In the disruption mutants of Arabidopsis, AtSUC1 (At1g71880), pollen having disrupted AtSUC1 is defective in vivo, as shown by segregation distortion; the mutants also have a lower rate of germination in vitro (Sivitz et al., 2008).
The rice genome encodes five SUT genes (Aoki et al., 2003); however, none of them has been shown to function in pollen grains. In this study, laser microdissection-assisted expression analysis of the rice SUT genes in developing pollen was carried out and functional characterization of one of the SUT genes (OsSUT1) was conducted using retrotransposon insertion mutant lines.
Materials and methods
Plant materials
A japonica type wild-type parental rice (Oryza sativa L.) cultivar Nipponbare and its three mutant lines containing the retrotransposon Tos17 insertion within the OsSUT1 gene were used. Insertion mutants of Tos17 were selected by BLAST searches against a dataset of Tos17 flanking sequences in the rice genome (Miyao et al., 2003). Rice plants were grown in the paddy field of the Hokuriku Research Center, Joetsu, Japan (37°06' N, 138°16' E) or in plastic pots filled with soil from the paddy field under outdoor conditions.
Genotyping mutant plants and pollen grains
The genotype of each of the three mutant lines was determined by PCR. Genomic DNA was extracted from mature leaf blades of each plant using the method of Murray and Thompson (1980), and used as template for the PCR with the primers listed in Table 1. The same PCR primers were also used for genotyping pollen samples.
Table 1.
PCR primers used in this study
| Purpose | Oligo name | Sequence |
| NC7083 genotyping | 7083-L | TATTGCTGGCATGGTGGTAA |
| 7083-R | TACAATAGCAGCATGCCACC | |
| NF8036 genotyping | 8036-L | TCCCACTGATACCAGAAGCC |
| 8036-R | TCCTTAAGTTGAAGTCCCCG | |
| NF2752 genotyping | 2752-L | CCTACTGGGATGCCTTCTGT |
| 2752-R | TGGAAGTCCTTGAGTGACCA | |
| Tos17 left border, genotyping | Tos17-L | ATTGTTAGGTTGCAAGTTAGTTAAGA |
| OsSUT1 RT-PCR | OsSUT1-L | TCATCCCTCAGGTGGTCATCG |
| OsSUT1-R | CTTGGAGATCTTGGGCAGCAG | |
| OsSUT2 RT-PCR | OsSUT2-L | GTCATACCACAGGTTATTGTGTC |
| OsSUT2-R | GAATTGCAAAGAATGGCCG | |
| OsSUT3 RT-PCR | OsSUT3-L | TCCTCTTCGACACCGACTG |
| OsSUT3-R | CAGCACGATCGAGTTAAGGAG | |
| OsSUT4 RT-PCR | OsSUT4-L | CGTTGTTCCGCAGATAGTAGTG |
| OsSUT4-R | GTGTTCTGCTCAGCCAAATCC | |
| OsSUT5 RT-PCR | OsSUT5-L | TCGGCATGGTGTCCATGAG |
| OsSUT5-R | CAATGGCAAGACCTTGGCC | |
| RUBIQ1 RT-PCR | RUBIQ1-L | GGAGCTGCTGCTGTTCTTGG |
| RUBIQ1-R | CACAATGAAACGGGACACGA |
Preparation of an OsSUT1 promoter::GUS construct and transformation into rice
A translational fusion of the OsSUT1 gene promoter with the β-glucuronidase (GUS) reporter gene in a rice transformation vector was constructed as follows. To obtain the OsSUT1 promoter::GUS construct, a DNA fragment spanning nucleotides –1675 to +19 of the translation start of OsSUT1 was amplified by PCR, and then ligated into a transformation vector, pZH2B. This vector is a modified version of pPZP202 (Hajdukiewicz et al., 1994), which carries hygromycin resistance. In this construct, the fusion protein consisted of the first seven codons of the OsSUT1 open reading frame followed by the complete GUS open reading frame. The construct was introduced into Agrobacterium tumefaciens strain EHA101 by electroporation. Transformation of this construct into rice tissue was performed by the method of Hiei et al. (1994).
GUS staining of transgenic plants
Glumaceous flowers from the OsSUT1 promoter::GUS transgenic lines were stained for GUS activity at 37 °C in the dark, overnight in X-gluc solution containing 0.5 mM K3[Fe(CN6)], 0.5 mM K4[Fe(CN6)].3H2O, 0.3% (v/v) Triton X-100, 20% (v/v) methanol, and 2 mM 5-bromo-4-chloro-3-indolyl-glucuronide cyclohexylamine salt in 100 mM sodium phosphate buffer (pH 7.0). GUS staining was examined with a stereomicroscope (SZX10, Olympus). Some of the GUS-stained samples were fixed with FAA fixative, embedded in paraffin, and examined with a microscope (BX50, Olympus).
Laser microdissection (LM) and real-time RT-PCR
Anther sampling was performed in the morning before flowering of the day commenced. Anthers were collected from glumaceous flowers and fixed in acetone at 4 °C for 3 h. The fixative was infiltrated into anther samples under vacuum for 5 min at 4 °C. The fixed tissues were then embedded in paraffin (Paraplast-XTra, Sigma), sectioned to 10 μm using a rotary microtome (RM2145, Leica) and mounted on PEN membrane frame slides (Arcturus). The sections were deparaffinized by xylene and air-died just prior to LM. Pollen grains were collected from the sections by LM (Veritas, Arcturus) and total RNA was extracted using a PicoPure RNA isolation kit (Arcturus) following the manufacturer's instructions. RNA quality was checked by microcapillary electrophoresis (BAS2000, Agilent Technologies). Approximately 100 pg reaction−1 of total RNA was used for real-time quantitative RT-PCR with a One Step SYBR PrimeScript RT-PCR Kit II (Takara Bio Inc.) and Smart Cycler II (Cepheid). The results obtained for the different RNAs were normalized to the transcript level of a rice polyubiquitin gene (RUIBQ1; Wang et al., 2000; accession number U37687), which shows highly constitutive expression in various rice tissues.
Total number and maturity of the pollen grains
Glumaceous flowers one or two days prior to anthesis were collected, immediately fixed in 50% (v/v) ethanol, and stored at room temperature until use. Anthers were excised from the glumaceous flowers, stained with an I2/KI solution, and the total number of the pollen grains was counted. Only pollen grains densely stained by the I2/KI solution were counted as mature pollen.
In vitro pollen germination and collection of the germinated pollen
Pollen grains were germinated on 1% agar medium containing 20% (w/v) sucrose and 20 mg l−1 K2B4O7 following the method of Kariya (1989). Glumaceous flowers just after anthesis were sampled and gently shaken above the germination medium in Petri dishes to collect pollen grains. After incubation for 20 min at 20 °C, pollen grains were stained by an I2/KI solution and the numbers of germinated pollen grains were counted under a microscope (BX50, Olympus). In the preliminary experiment it was confirmed that the sucrose concentration of the medium and the time of incubation are optimal for Nipponbare, the cultivar used in this study, which is different from that used by Kariya (1989). Some of the germinated pollen grains were picked up with pipette tips under a stereomicroscope (SZX10, Olympus), and their DNA was extracted to determine genotype by PCR using a PicoPure DNA isolation kit (Arcturus) following the manufacturer's instructions.
Results
Expression of SUT genes in the developing pollen
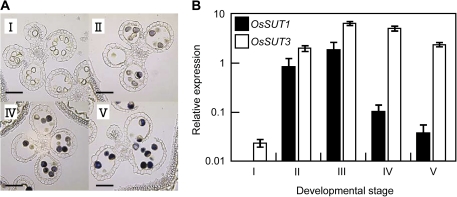
To examine which of the five members of the rice SUT gene family is expressed in the developing pollen grains, real-time RT-PCR analysis combined with laser microdissection (LM) was employed. A typical image of LM and electrophoresis of a resultant RNA sample are shown in Supplementary Fig. S1 at JXB online. LM was performed using anthers chemically fixed with pure acetone; these preparations yielded higher quality RNAs than from the more commonly used ethanol/acetic acid fixative, although preservation of specimen structural integrity was somewhat inferior (data not shown). Because anthesis of rice glumaceous flowers occurs in an orderly manner within an inflorescence, the days to the anthesis (DTA) can be estimated for each spikelet according to their position in the panicle. Thus, the developmental stages of the pollen were classified into five classes according to the estimated DTA of each glumaceous flower, as follows: stage I (5–7 DTA), stage II (4–5 DTA), stage III (2–3 DTA), stage IV (1 DTA), and stage V (0 DTA). At stage I, starch was not detectable in the pollen by I2/KI staining, and the cellular remnant of the tapetum was evident in the anther (Fig. 1A). At stage II, starch began to accumulate in the pollen (Fig. 1A). By real-time RT-PCR, only two SUT genes, OsSUT1 and OsSUT3, were detectable in the pollen; mRNA for the other three SUT genes was very low, if at all (Fig. 1B). The temporal expression patterns of the two SUT genes were different; the mRNA level of OsSUT1 was not detectable in stage I, increased suddenly in stages II and III, and then decreased through stages IV and V; whereas expression of OsSUT3 was already detectable at stage I; it then increased and maintained the highest level during stages II to V.
Fig. 1.
(A) The iodine-stained cross-sections of the anthers at different developmental stages used in the real-time RT-PCR. Bars=0.1 mm. (B) Transcript levels of OsSUT1 and OsSUT3 in the pollen at different developmental stages. The transcript levels the SUT genes determined by real-time RT-PCR were standardized against those of a constitutive ubiquitin gene RUBIQ1 and expressed on a logarithmic scale. Values are the means of three independent replications (bars indicate SE). See Materials and methods for the details of the developmental stage. (This figure is available in colour at JXB online.)
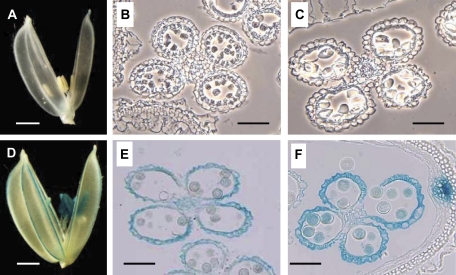
The expression of OsSUT1 was then examined by histochemical staining using transgenic plants harbouring a OsSUT1 promoter::GUS construct. No staining was detected in the glumaceous flowers at stage I or earlier (Fig. 2A, B, C); clear staining was then observed in both the anther wall and pollen at stage II and later (Fig. 2D, E, F). These results are consistent with those from the RT-PCR analysis.
Fig. 2.
Analysis of OsSUT1 expression in the glumaceous flowers of transgenic plants harbouring a OsSUT1 promoter::GUS construct. (A, D) Whole glumaceous flowers at stage I (A) and stage III (D). Bars indicate 1 mm. (B, C, E, F) Cross-sections of anthers of various developmental stages; (B) earlier than stage I (around 10 DTA), (C) stage I, (E) stage II, (F) stage IV. Bars indicate 0.1 mm. (B) and (C) are phase-contrast images to show the anatomy of the anthers better.
Disruption of OsSUT1 impairs pollen function
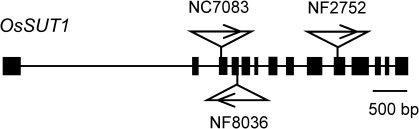
To investigate the functions of OsSUT1 and OsSUT3 in pollen, gene disruption mutants of the two genes were searched for from a population of rice mutants generated by insertion of a retrotransposon Tos17. Three mutant lines of OsSUT1 were identified, but none were identified for OsSUT3. It was therefore decided to study OsSUT1 function in pollen further using three mutant lines, NC7083, NF8036, and NF2752, in which insertions of Tos17 occurred in exons 3, 4, and 10, respectively (Fig. 3). Populations of these three mutant lines segregated into two genotypes, homozygous wild-type (SUT1+/+) and heterozygous for the Tos17 insertion (SUT1+/–); however, no homozygous plants for the insertion (SUT1–/–) were identified (data not shown). Three hypotheses were developed to explain the absence of SUT1–/– plants: (i) both male and female gametes with disrupted OsSUT1 are functional, but the SUT1–/– zygote cannot develop to maturity, (ii) a male gamete is not formed, or is dysfunctional, and (iii) a female gamete is not formed or dysfunctional. To determine which of these three hypotheses is valid, the progeny of SUT1+/– plants were grown and investigated further. The germination frequency of the seeds from SUT1+/– plants ranged between 90% and 97% in four independent observations, which was not different from that of the seeds from SUT1+/+ plants for all three mutant lines. However, homozygous plants did not develop once again, and the segregation ratio of SUT1+/+ and SUT1+/– were 1:1 in all the three mutant lines (Table 2). The SUT1+/– plants grew normally and no differences were detected between SUT1+/+ in the major agronomic traits, including days to heading, plant height, or panicle number per plant (data not shown). In addition, both the total number of spikelets and the percentage of filled grains per panicle did not differ between the two genotypes (Table 3). If the first hypothesis were true, the segregation ratio of SUT1+/+:SUT1+/– should be 1:2, with one-quarter of the caryopses (SUT1–/– zygotes) being infertile or immature. If the third hypothesis were true, the segregation ratio should be 1:1, and half of the caryopses with SUT1- female gametes would be infertile. Thus, the second hypothesis appears to be valid; in this case, the segregation ratio should be 1:1, with the number of filled grains unaffected. To confirm this, SUT1+/– plants were backcrossed to wild-type plants. A total of 74 F1 seeds were obtained using SUT1+/– zygotes of the three insertion lines as pollen parents, but none of the F1 plants inherited the OsSUT1 mutant allele, indicating that pollen function is, in fact, impaired by disruption of the OsSUT1 gene (Table 4).
Fig. 3.
A diagram showing the exon/intron structure of OsSUT1 gene and the position of the insertion of retrotransposon Tos17 in the three mutant lines used in this study. The arrowheads indicate the direction of Tos17 at the respective insertion sites.
Table 2.
Segregation of the progeny of the three SUT1+/– parent lines
| Line | Total | Genotype |
Pa | ||
| SUT1+/+ | SUT1+/– | SUT1–/– | |||
| NC7083 | 94 | 46 | 48 | 0 | 0.837 |
| NF2752 | 109 | 52 | 57 | 0 | 0.632 |
| NF8036 | 104 | 56 | 48 | 0 | 0.433 |
Probability calculated by χ2-test when 1:1 segregation of SUT1+/+ and SUT1+/– is hypothesized.
Table 3.
The number of spikelets and the filled grain percentage per panicle of SUT1+/+ or SUT1+/– plants. Values are the means of 5–7 individual plants of each genotype. ‘ns’ shows not significantly different between the two genotypes (t test).
| Line | Genotype | No. of spikelet per panicle | Filled grain percentage (%) |
| NC7083 | SUT1+/+ | 50.0±3.4 | 92.6±3.0 |
| SUT1+/– | 50.8±2.9 ns | 92.4±2.7 ns | |
| NF2752 | SUT1+/+ | 52.4±3.5 | 93.0±1.2 |
| SUT1+/– | 51.4±3.0 ns | 95.2±1.1 ns | |
| NF8036 | SUT1+/+ | 48.0±2.8 | 92.3±2.6 |
| SUT1+/– | 47.0±6.7 ns | 94.0±0.7 ns |
Table 4.
Genotypes of F1 progeny of wild-type Nipponbare backcrossed with the three SUT1+/– lines
| Female plant | Pollinator | No. of F1 | F1 genotype |
|
| SUT1+/+ | SUT1+/– | |||
| Nipponbare (SUT1+/+) | NC7083 (SUT1+/–) | 27 | 27 | 0 |
| NF2752 (SUT1+/–) | 28 | 28 | 0 | |
| NF8036 (SUT1+/–) | 19 | 19 | 0 | |
| Total | 74 | 74 | 0 | |
Pollen germination is impaired by disruption of OsSUT1, but pollen maturation is unaffected.
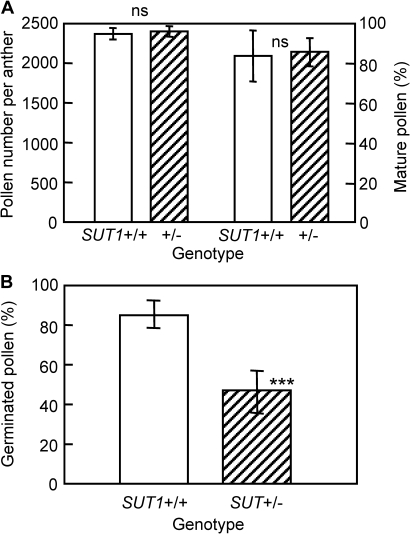
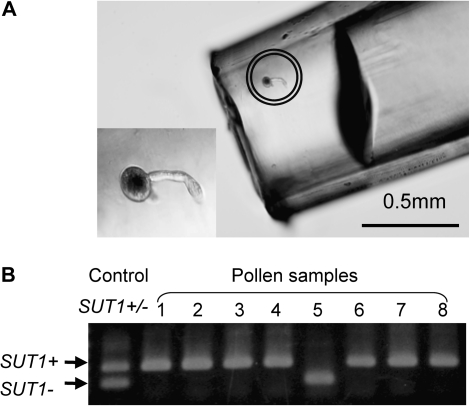
To investigate further how OsSUT1 disruption affects the development and/or function of pollen, the total number of the pollen in an anther, the rate of matured pollen, and the pollen germination rate were examined using a disruption line NF2752. The total number of pollen grains in an anther and the percentage of matured pollen, as assessed by iodine staining, were not different, irrespective of the genotype (Fig. 4A). In another observation using all three disruption lines, the rate of matured pollen did not differ irrespective of the genotypes in each disruption line (see Supplementary Table S1 at JXB online). This suggests that a male gamete with disrupted OsSUT1 is able to develop into a mature pollen grain. This also implies that disruption of OsSUT1 does not affect pollen development, but rather some other pollen function(s) during the process of fertilization, for example, pollen germination or pollen tube elongation. Pollen germination in the mutants was assessed by an in vitro pollen germination assay carried out on agar medium. The percentage of germinated pollen was found to be significantly lower in SUT1+/– plants than in wild-type SUT1+/+ plants (Fig. 4B), suggesting that disruption of OsSUT1 does in fact impair pollen germination. Germinated pollen grains of SUT1+/– plants were then individually genotyped by PCR revealing that 35 out of 40 (88%) of the germinated pollen grains had a wild-type (SUT1+) genotype (Fig. 5; Table 5).
Fig. 4.
Total number, maturity, and germination rate of the pollen of SUT1+/+ or SUT1+/– plants of the insertion mutant line NF2752. (A) The number of pollen (left) and the percentage of matured pollen (right) in an anther. Measurements of seven glumaceous flowers per plant were averaged and processed as the value of each plant, and the means of five individual plants for each genotype were plotted. Vertical bars indicate SD, and ‘ns’ shows not significantly different between the two genotypes (t test). (B) The in vitro pollen germination rate of SUT1+/+ or SUT1+/– plants. Five to seven observations per plant were averaged and processed as the value of each plant, and the means of 16 individual plants for each genotype were plotted. Vertical bars indicate SD, and the asterisks (***) show significant difference by t test (P <0.001).
Fig. 5.
Identification of the genotypes of individual pollen grain. (A) A pollen grain germinated on the agar medium was picked up with a fine pipette tip (marked in the circle). A magnified image of the pollen is lower left. (B) A typical result of genotyping by PCR, showing that seven out of eight pollen grains have wild type OsSUT1 (SUT1+) gene. A PCR product with the DNA template from the leaves of SUT1+/– plant was used as a control.
Table 5.
Genotypes of the pollens germinated on the agar media
Binominal probability of obtaining the results observed or less number of SUT1- genotype when equal ability of pollen germination for each genotype is hypothesized.
Three glumaceous flowers from each of five individual plants of NF2752 were used.
Discussion
In this study it was revealed that, in developing rice pollen, OsSUT1 and OsSUT3 are the most abundantly expressed among the five SUT genes in the rice genome. Compared with the other four SUT genes, the expression and function of OsSUT1 was the best characterized. OsSUT1 was shown to be expressed in various tissues of rice plants; for example, it has been repeatedly shown to localize to the phloem of the leaf blades, leaf sheaths, and internodes (Matsukura et al., 2000; Scofield et al., 2007a, b); it is also expressed in both filial and maternal tissues in the developing caryopsis (Furbank et al., 2001; Hirose et al., 2002). By contrast, tissue-specific expression of OsSUT3 has remained unclear, although OsSUT3 mRNA expression has been detected in various organs at low and constant levels (Aoki et al., 2003; Scofield et al., 2007b). Thus, the relatively higher expression of OsSUT3 and OsSUT1 represents a novel and distinctive feature of developing pollen. To date, there are two plant species in which multiple SUT genes have been shown to be expressed in pollen: in Arabidopsis (AtSUC1, Stadler et al., 1999; AtSUC3, Meyer et al., 2004) and in Plantago major (PmSUC1, Lauterbach et al., 2007; PmSUC3, Barth et al., 2003). In both cases, the two SUTs belong to different classes in the phylogenetic tree, which is speculated to reflect the complex roles of SUTs in pollen (Sauer, 2007). However in rice pollen, OsSUT1 and OsSUT3 belong to the same phylogenetic group (Aoki et al., 2003).
Altogether, our analysis of the gene disruption mutant revealed OsSUT1 to be essential for pollen to fertilize the ovule normally probably through its function(s) in pollen germination and/or pollen tube growth, but not for starch accumulation during pollen maturation, although some of the data were obtained using a single disruption line. While physiological function of OsSUT1 was previously examined through the antisense suppression technique, no evidence for the function of the gene in pollen was reported (Ishimaru et al., 2001; Scofield et al., 2002). A possible explanation for this is that antisense suppression was insufficient to bring about detectable distortion of the segregation in the progeny or an increase in the number of infertile caryopsis while it was sufficient to cause poor grain-filling and retarded early growth of the seedlings. The role of SUTs in pollen germination and/or pollen tube growth has previously been reported for tomato and Arabidopsis (Hackel et al., 2006; Sivitz et al., 2008). However, OsSUT1 mutant phenotypes are distinct from those reported. In Arabidopsis, mutant plants with homozygous AtSUC1 gene disruption can be recovered, although the segregation is distorted in the progeny of the heterozygous plants (Siviz et al., 2008). In rice, however, homozygous SUT1–/– plants have never been recovered among more than 500 progeny of SUT1+/– plants examined during the course of this study. This suggests that a lack of AtSUC1 can be partially compensated for by other SUTs, including AtSUC3; while in rice, OsSUT1 is essential to pollen for normal fertilization. By contrast, a fraction of the pollen grains germinated in vitro had the SUT1– genotype, indicating that OsSUT1 is not essential for pollen germination, at least on the agar medium. Presumably, SUT1– pollen fails to fertilize due to pollen tube elongation that is slower than in SUT1+ pollen, even if it can germinate on the stigma. Indeed, an attempt was made to look at the elongation rate of the pollen tube on the agar medium, but reproducible assessment was difficult which is probably related to extraordinarily shorter longevity of rice pollen (a few minutes after dehiscence of the anther) than other plant species. However, from our analysis presented here, together with data from studies of AtSUC1 and LeSUT2, it is suggested that pollen germination represents a third mode of action for SUTs, in addition to phloem loading and seed maturation. Recently, Slewinski et al. (2009) characterized a disruption mutant of ZmSUT1, a maize SUT gene closely related to OsSUT1, and revealed that ZmSUT1 functions in phloem loading. Notably, they recovered mutation homogeneous plants of ZmSUT1 used for their study, showing that unlike OsSUT1, disruption of ZmSUT1 does not result in the complete dysfunction of pollen. However, it is not clear whether any segregation distortion was observed among the genotypes, which is potentially brought about by impaired pollen function. It is interesting that the two closely related SUT genes have different physiological functions; ZmSUT1 acts as a phloem loader but is not essential to pollen function while OsSUT1 is essential to pollen function and plays an important role in grain-filling but not in phloem loading (Ishimaru et al., 2001; Scofield et al., 2002).
It is natural to hypothesize that sucrose taken up by OsSUT1 represents an energy source and/or osmoticum for germinating pollen. However, the temporal patterns of the transcript levels of OsSUT1 seem inconsistent with the presumed roles; OsSUT1 mRNA accumulates more specifically during the starch accumulation period and decreases thereafter during pollen maturation. It has been reported that, in pollen, temporal differences can exist between the transcript levels and protein levels of a given gene. In Arabidopsis, the transcript level of AtSUC1 is highest at the tricellular stage, followed by a decrease in mature pollen (Bock et al., 2006). However, AtSUC1 protein could be detected by immunofluorescence microscopy only after the onset of germination (Stadler et al., 1999). A similar phenomenon was observed for a monosaccharide transporter AtSTP9 (Schneidereit et al., 2003). Although localization of OsSUT1 protein is yet to be examined, it can be speculated that the transcription and the translation of OsSUT1 are under similar regulation. It should be noted that these possible roles of sucrose may also be fulfilled by monosaccharides. In fact, preferential expression of monosaccharide transporters (i.e. MSTs) has been reported repeatedly; in Arabidopsis, as many as five MST genes are expressed in the pollen grain and/or pollen tube (see Büttner, 2007, for a review). Moreover, Arabidopsis pollen does germinate on agar media containing glucose but lacking sucrose (Sivitz et al., 2008). However, in their analysis of an insertion mutant of one of the MST genes (PMT1) in Petunia hybrida, Garrido et al. (2006) found no defect in pollen function. Further study is clearly needed to characterize the physiological function of MSTs in relation to the function(s) of SUTs in pollen.
It was surprising that disruption of OsSUT1 did not affect starch accumulation in pollen because OsSUT1 is highly expressed in the starch-accumulating pollen and also because pollen grains are symplastically isolated from the surrounding tissue like developing endosperm, where OsSUT1 function is crucial (Ishimaru et al., 2001; Scofield et al., 2002). Because OsSUT3 is expressed in pollen along with OsSUT1 (Fig. 1), it may be possible that OsSUT3 plays a role to supply the sucrose for starch accumulation. Here again, it is also possible that MSTs import hexoses as substrates for starch synthesis. Castro and Clément (2007) suggested that hexoses are predominantly imported and utilized for pollen maturation based on both the sugar levels and the activities of sugar metabolizing enzymes separately measured in different fractions of lily anther, i.e. anther wall, locular fluid, and pollen. In addition, expression of the genes for MSTs has been repeatedly documented as mentioned above. However, no evidence has been presented for the function of the MSTs in developing pollen. Consequently, it remains an important and open question of which protein(s) loads the sugar for starch synthesis into pollen.
In conclusion, by conducting the first comprehensive expression analysis of members of the rice SUT gene family, it was determined that both OsSUT1 and OsSUT3 are predominantly expressed in developing rice pollen. OsSUT1 was also suggested to be essential for normal pollen germination, but not for pollen maturation or starch accumulation; whereas the function of OsSUT3 remains undetermined.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Table S1. The percentage of matured pollen in the three gene disruption lines.
Supplementary Fig. S1. Typical images showing the procedure of LM to obtain the developing pollen and qualitative assessment of the resultant RNA.
Supplementary Material
Acknowledgments
This work was supported in part by funding from the Takano Life Science Research Foundation to TH. The authors deeply thank Naohiro Aoki (The University of Tokyo) and Graham Scofield (CSIRO Plant Industry) for valuable advice and discussion, and Kiiko Takatsuto (National Agriculture Research Centre) for expert technical help.
References
- Aoki N, Hirose T, Scofield GN, Whitfeld PR, Furbank RT. The sucrose transporter gene family in rice. Plant and Cell Physiology. 2003;44:223–232. doi: 10.1093/pcp/pcg030. [DOI] [PubMed] [Google Scholar]
- Barth I, Meyer S, Sauer N. PmSUC3: characterization of a SUT2/SUC3-type sucrose transporter from Plantago major. The Plant Cell. 2003;15:1375–1385. doi: 10.1105/tpc.010967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock KW, Honys D, Ward JM, Padmanaban S, Nawrocki EP, Hirschi KD, Twell D, Sze H. Integrating membrane transport with male gametophyte development and function through transcriptomics. Plant Physiology. 2006;140:1151–1168. doi: 10.1104/pp.105.074708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner M. The monosaccharide transporter (-like) gene family in Arabidopsis. FEBS Letters. 2007;581:2318–2324. doi: 10.1016/j.febslet.2007.03.016. [DOI] [PubMed] [Google Scholar]
- Castro A, Clément C. Sucrose and starch catabolism in the anther of Lilium during its development: a comparative study among the anther wall, locular fluid and microspore/pollen fractions. Planta. 2007;225:1573–1582. doi: 10.1007/s00425-006-0443-5. [DOI] [PubMed] [Google Scholar]
- Furbank RT, Scofield GN, Hirose T, Wang XD, Patrick JW, Offler CE. Cellular localisation and function of a sucrose transporter OsSUT1 developing rice grains. Australian Journal of Plant Physiology. 2001;28:1187–1196. [Google Scholar]
- Garrido D, Busscher J, van Tunen AJ. Promoter activity of a putative pollen monosaccharide transporter in Petunia hybrida and characterisation of a transposon insertion mutant. Protoplasma. 2006;228:3–11. doi: 10.1007/s00709-006-0171-5. [DOI] [PubMed] [Google Scholar]
- Hackel A, Schauer N, Carrari F, Fernie AR, Grimm B, Kuhn C. Sucrose transporter LeSUT1 and LeSUT2 inhibition affects tomato fruit development in different ways. The Plant Journal. 2006;45:180–192. doi: 10.1111/j.1365-313X.2005.02572.x. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. The small, versatile pPZP family of agrobacterium binary vectors for plant transformation. Plant Molecular Biology. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence-analysis of the boundaries of the T-DNA. The Plant Journal. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- Hirose T, Takano M, Terao T. Cell wall invertase in developing rice caryopsis: Molecular cloning of OsCIN1 and analysis of its expression in relation to its role in grain filling. Plant and Cell Physiology. 2002;43:452–459. doi: 10.1093/pcp/pcf055. [DOI] [PubMed] [Google Scholar]
- Ishimaru T, Nakazono M, Masumura T, Abiko M, San-oh Y, Nishizawa N, Kondo M. A method for obtaining high integrity RNA from developing aleurone cells and starchy endosperm in rice (Oryza sativa L.) by laser microdissection. Plant Science. 2007;173:321–326. [Google Scholar]
- Ishimaru K, Hirose T, Aoki N, et al. Antisense expression of a rice sucrose transporter OsSUT1 in rice (Oryza sativa L.) Plant and Cell Physiology. 2001;42:1181–1185. doi: 10.1093/pcp/pce148. [DOI] [PubMed] [Google Scholar]
- Kariya K. Sterility caused by cooling treatment at the flowering stage in rice plants. 3. Establishment of a method of in vitro pollen germination. Japanese Journal of Crop Science. 1989;58:96–102. [Google Scholar]
- Lauterbach C, Niedermeier M, Besenbeck R, Stadler R, Sauer N. Immunolocalization of the PmSUC1 sucrose transporter in Plantago major flowers and reporter-gene analyses of the PmSUC1 promoter suggest a role in sucrose release from the inner integument. Plant Biology. 2007;9:357–365. doi: 10.1055/s-2006-924659. [DOI] [PubMed] [Google Scholar]
- Lemoine R, Burkle L, Barker L, Sakr S, Kuhn C, Regnacq M, Gaillard C, Delrot S, Frommer W. Identification of a pollen-specific sucrose transporter-like protein NtSUT3 from tobacco. FEBS Letters. 1999;454:325–330. doi: 10.1016/s0014-5793(99)00843-1. [DOI] [PubMed] [Google Scholar]
- Matsukura C, Saitoh T, Hirose T, Ohsugi R, Perata P, Yamaguchi J. Sugar uptake and transport in rice embryo. Expression of companion cell-specific sucrose transporter (OsSUT1) induced by sugar and light. Plant Physiology. 2000;124:85–93. doi: 10.1104/pp.124.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S, Lauterbach C, Niedermeier M, Barth I, Sjolund RD, Sauer N. Wounding enhances expression of AtSUC3, a sucrose transporter from Arabidopsis sieve elements and sink tissues. Plant Physiology. 2004;134:684–693. doi: 10.1104/pp.103.033399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao A, Tanaka K, Murata K, Sawaki H, Takeda S, Abe K, Shinozuka Y, Onosato K, Hirochika H. Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. The Plant Cell. 2003;15:1771–1780. doi: 10.1105/tpc.012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MG, Thompson WF. Rapid isolation of high molecular-weight plant DNA. Nucleic Acids Research. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick J, Offler C. Compartmentation of transport and transfer events in developing seeds. Journal of Experimental Botany. 2001;52:551–564. [PubMed] [Google Scholar]
- Riesmeier J, Willmitzer L, Frommer W. Isolation and characterization of a sucrose carrier cDNA from spinach by functional expression in yeast. EMBO Journal. 1992;11:4705–4713. doi: 10.1002/j.1460-2075.1992.tb05575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosche E, Blackmore D, Tegeder M, Richardson T, Schroeder H, Higgins TJV, Frommer WB, Offler CE, Patrick JW. Seed-specific overexpression of a potato sucrose transporter increases sucrose uptake and growth rates of developing pea cotyledons. The Plant Journal. 2002;30:165–175. doi: 10.1046/j.1365-313x.2002.01282.x. [DOI] [PubMed] [Google Scholar]
- Sauer N. Molecular physiology of higher plant sucrose transporters. FEBS Letters. 2007;581:2309–2317. doi: 10.1016/j.febslet.2007.03.048. [DOI] [PubMed] [Google Scholar]
- Schneidereit A, Scholz-Starke J, Buttner M. Functional characterization and expression analyses of the glucose-specific AtSTP9 monosaccharide transporter in pollen of Arabidopsis. Plant Physiology. 2003;133:182–190. doi: 10.1104/pp.103.026674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield GN, Aoki N, Hirose T, Takano M, Jenkins CLD, Furbank RT. The role of the sucrose transporter, OsSUT1, in germination and early seedling growth and development of rice plants. Journal of Experimental Botany. 2007a;58:483–495. doi: 10.1093/jxb/erl217. [DOI] [PubMed] [Google Scholar]
- Scofield GN, Hirose T, Aoki N, Furbank RT. Involvement of the sucrose transporter, OsSUT1, in the long-distance pathway for assimilate transport in rice. Journal of Experimental Botany. 2007b;58:3155–3169. doi: 10.1093/jxb/erm153. [DOI] [PubMed] [Google Scholar]
- Scofield GN, Hirose T, Gaudron JA, Upadhyaya NM, Ohsugi R, Furbank RT. Antisense suppression of the rice sucrose transporter gene, OsSUT1, leads to impaired grain filling and germination but does not affect photosynthesis. Functional Plant Biology. 2002;29:815–826. doi: 10.1071/PP01204. [DOI] [PubMed] [Google Scholar]
- Sivitz AB, Reinders A, Ward JM. Arabidopsis sucrose transporter AtSUC1 is important for pollen germination and sucrose-induced anthocyanin accumulation. Plant Physiology. 2008;147:92–100. doi: 10.1104/pp.108.118992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slewinski TL, Meeley R, Braun DM. Sucrose transporter1 functions in phloem loading in maize leaves. Journal of Experimental Botany. 2009;60:881–892. doi: 10.1093/jxb/ern335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler R, Truernit E, Gahrtz M, Sauer N. The AtSUC1 sucrose carrier may represent the osmotic driving force for anther dehiscence and pollen tube growth in Arabidopsis. The Plant Journal. 1999;19:269–278. doi: 10.1046/j.1365-313x.1999.00527.x. [DOI] [PubMed] [Google Scholar]
- Wang JL, Jiang JD, Oard JH. Structure, expression and promoter activity of two polyubiquitin genes from rice (Oryza sativa L.) Plant Science. 2000;156:201–211. doi: 10.1016/s0168-9452(00)00255-7. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Chan K, Wang T, Hedley C, Offler C, Patrick J. Intracellular sucrose communicates metabolic demand to sucrose transporters in developing pea cotyledons. Journal of Experimental Botany. 2009;60:71–85. doi: 10.1093/jxb/ern254. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.