Abstract
The aerial organs of plants are covered by the cuticle, a polyester matrix of cutin and organic solvent-soluble waxes that is contiguous with the polysaccharide cell wall of the epidermis. The cuticle is an important surface barrier between a plant and its environment, providing protection against desiccation, disease, and pests. However, many aspects of the mechanisms of cuticle biosynthesis, assembly, and restructuring are entirely unknown. To identify candidate proteins with a role in cuticle biogenesis, a surface protein extract was obtained from tomato (Solanum lycopersicum) fruits by dipping in an organic solvent and the constituent proteins were identified by several complementary fractionation strategies and two mass spectrometry techniques. Of the ∼200 proteins that were identified, a subset is potentially involved in the transport, deposition, or modification of the cuticle, such as those with predicted lipid-associated protein domains. These include several lipid-transfer proteins, GDSL-motif lipase/hydrolase family proteins, and an MD-2-related lipid recognition domain-containing protein. The epidermal-specific transcript accumulation of several of these candidates was confirmed by laser-capture microdissection and quantitative reverse transcription-PCR (qRT-PCR), together with their expression during various stages of fruit development. This indicated a complex pattern of cuticle deposition, and models for cuticle biogenesis and restructuring are discussed.
Keywords: Cuticle, cutin, lipid, proteome, tomato fruit, wax
Introduction
The plant cuticle is a hydrophobic membrane that covers the aerial organs of land plants and provides protection against desiccation, pathogens, UV radiation, and herbivory (Riederer, 2006). It is continuous with the outer periclinal polysaccharide cell wall of the epidermis and consists of organic soluble waxes embedded in, and layered on, a non-soluble polyester matrix of ω-substituted fatty acids. The waxes include both aliphatic compounds, derived from very long chain fatty acids, and secondary metabolites, such as triterpenoids and flavonoids (Jetter et al., 2006). In the majority of species analysed to date, cutin is composed primarily of polymerized mid-chain-substituted ω-hydroxy fatty acids, although Arabidopsis thaliana is a notable exception in that α,ω-dicarboxylic fatty acids predominate in stems and leaves (Bonaventure et al., 2004; Franke et al., 2005) and ω-hydroxy fatty acids only contribute significantly to the cutin of flowers (Beisson et al., 2007; Li-Beisson et al., 2009; Panikashvili et al., 2009). In addition, the presence of glycerol in the cutin polymer is now well established (Graca et al., 2002).
Both cutin monomers and waxes are produced within the epidermal cells, and a clear picture of the molecular biology of their synthesis is emerging. This has been largely a result of the characterization of Arabidopsis mutants (Pollard et al., 2008; Samuels et al., 2008), a species whose cutin is probably rich in glycerol (Pollard et al., 2008). For example, the glycerol-3-phosphate acyltransferases GPAT4, GPAT6, and GPAT8 have been shown to be required for cutin synthesis (Li et al., 2007; Li-Beisson et al., 2009), and it was recently reported that an acyltransferase of the BAHD family, DCR, is required for cutin synthesis in Arabidopsis floral organs (Panikashvili et al., 2009). Both classes of enzymes appear to be intracellular: GPAT8 was localized to the endoplasmic reticulum (ER) (Gidda et al., 2009) and DCR was shown to be in the cytoplasm (Panikashvili et al., 2009). However, the subsequent extracellular aspects of cuticle biogenesis, including trafficking of the constituents and their assembly into a mature cuticle, as well as restructuring of cuticle architecture during growth and development, are far less well understood.
Current models hypothesize the involvement of several classes of extracellular proteins and enzymes, although few examples have yet been identified. Following biosynthesis in the ER, wax and cutin monomers, or oligomers, are exported across the plasma membrane to the apoplast, in a process dependent on ABC transporters such as the Arabidopsis proteins CER5 (Pighin et al., 2004) and WBC11 (Bird et al., 2007). Recently, a glycosylphosphatidylinositol (GPI)-anchored lipid-transfer protein, LTPG, was shown to be required for wax secretion, possibly by acting as a membrane-anchored lipid-binding protein that receives waxes as they are extruded by ABC transporters (Debono et al., 2009). Trafficking of hydrophobic lipids across the polar environment of the polysaccharide cell wall is then often attributed to soluble extracellular lipid transfer proteins (LTPs). However, their ability to bind wax or cutin monomers has not been confirmed, and no cuticle mutant has been attributed to a lesion in a gene encoding a soluble LTP (Yeats and Rose, 2008).
Polymerization of the cutin polymer during development and organ expansion may also involve extracellular proteins. The protein BODYGUARD (BDG) is secreted by epidermal cells and is required for normal cuticle development in Arabidopsis, although the bdg mutant paradoxically accumulates a larger amount of cutin (Kurdyukov et al., 2006a). While no biochemical activity for BDG has been identified, the protein is a member of the α/β-hydrolase superfamily, leading the authors to suggest that it is a putative cutin synthase. A similar function has been proposed for AgaSGNH, a GDSL-motif lipase/hydrolase family protein from Agave americana, which was reported to have protein localization and gene expression patterns that correlated with cutin biosynthesis, although it was not associated with a genetic phenotype or biochemical activity (Reina et al., 2007).
Thus, remarkably little is known as yet about key mechanisms of cuticle biogenesis, and experimental strategies to identify new proteins that associate with cutin and waxes could provide a valuable means to identify new candidates. Cuticular waxes are easily extracted free of cellular lipid contamination by brief immersion of plant organs in organic solvents such as chloroform (Jetter et al., 2006), while a small additional fraction of the recovered material is comprised of proteins (Martin and Juniper, 1970). Edman degradation peptide sequencing has previously been used to identify three proteins in plant cuticular waxes: an LTP from Brassica oleracea (Pyee et al., 1994), and an endo-β-1,3-glucanase and a chitinase (glycosyl hydrolase family 17 and 18, respectively; www.cazy.org) from the wax of Copernicia cerifera (Cruz et al., 2002). However, it was hypothesized that generating a more comprehensive inventory of proteins that are associated with the outermost surface tissues of plant organs, using a range of complementary protein fractionation strategies coupled with modern sensitive mass spectrometry-based methods, would help identify new candidate proteins with a potential role in cuticle biosynthesis. To this end, the surface proteome of developing tomato (Solanum lycopersicum) fruit was targeted as a model system. Although Arabidopsis research has greatly accelerated the discovery of new cuticle-related genes, its cuticle poses some experimental limitations since it is relatively thin, fragile, and difficult to isolate in substantial quantities. Conversely, tomato fruit cuticles are astomatous and large amounts of intact cuticular material can be isolated for chemical and biomechanical analyses. For example, the fruit accumulate of the order of 1 mg cm−2 cutin (Baker et al., 1982), compared with the stem of Arabidopsis, which has 0.5–10 μg cm−2 (Franke et al., 2005; Suh et al., 2005). Thus, the typical 6 week period of tomato fruit development represents a remarkably rapid and extensive phase of cuticle biosynthesis, in a genetically tractable species for which there are now also many genomic resources (Mueller et al., 2005; www.solgenomics.net).
The proteomic analysis of tomato fruit cuticle extracts and the identification of several secreted proteins with lipid-related domains are described. The expression patterns of the genes encoding these proteins are further analysed as to the specificity of their expression in the epidermis and during the time course of fruit development. Finally, based on these expression patterns and current models of cuticle biosynthesis, potential roles for these candidates in extracellular cutin and wax deposition and metabolism are discussed.
Materials and methods
Plant materials
Solanum lycopersicum (cv. M82) plants were grown in the field (Freeville NY, summer 2007 and 2008) and 500 immature green fruits were harvested for protein extraction. To avoid bruising and damage during handling, fruits were harvested from all stages of expansion after the fruits had lost their visible trichomes and became glossy in appearance, at ∼15–40 days post-anthesis (DPA). Prior to protein extraction, fruits were washed with deionized water and left to dry overnight. By first rinsing the fruits, it is believed that the analysis excluded phylloplane proteins that are secreted to the outer surface of the cuticle by mechanisms discussed by Shepherd and Wagner (2007). Fruits used for confocal microscopy, laser-capture microdissection, and developmental gene expression time course experiments were harvested from plants grown in the greenhouse (Ithaca, NY). To define the developmental stage of fruits during expansion, flowers were tagged at anthesis. The ripening stages were determined visually by colour change according to standard conventions (Gonzalez-Bosch et al., 1996). For RNA isolation, pericarp tissue from 3–10 fruits at each developmental stage was manually dissected, flash-frozen, ground in liquid nitrogen, and stored at –80 °C.
Microscopy
Confocal microscopy was performed as previously described (Buda et al., 2009). To illustrate the different pericarp cell types harvested by laser-capture microdissection, 10 μm paraffin sections of immature green fruits were prepared and stained with Toluidine blue O according to standard protocols (Ruzin, 1999).
Wax extraction and protein isolation
Wax extraction and purification of polar components from the wax was conducted essentially as previously described (Pyee et al., 1994). Fruits were dipped, without submerging the calyx scar, for 10 s in ∼500 ml of chloroform:methanol (2:1) that was gently stirred by a magnetic stir bar. For each set of extractions, 2–3 aliquots of 500 ml of fresh solvent were used and the extracts were pooled. The extract was then evaporated to dryness by rotary evaporation at 50 °C with reduced pressure. The residue was resuspended in 80 ml of chloroform and 40 ml of distilled water, and transferred to a separatory funnel. The upper aqueous phase was recovered and lyophilized and the residue resuspended in 500 μl of buffer [0.7 M sucrose, 0.1.M KCl, 0.5 M TRIS-HCl pH 7.5, 50 mM ethylenediaminetetra-acetic acid (EDTA), 2% β-mercaptoethanol, and 1 mM phenylmethylsulphonyl fluoride]. The protein component was then extracted into phenol and precipitated with 0.1 M ammonium acetate in methanol (Isaacson et al., 2006). Calculation of approximate protein yield by densitometry of the gel-separated samples (see below) indicated that each extraction yielded ∼8 μg of protein. Thus, assuming an average fruit surface area of 50 cm2, the yield of protein was of the order of 0.3 ng cm−2 of surface. For comparison, the wax coverage of immature green tomato fruit is of the order of 5 μg cm−2 (15 000-fold greater).
Fractionation and proteomic analysis of protein extracts
Three independent extractions were analysed using three different pre-fractionation schemes.
Isolation of individual bands from 1D polyacrylamide gels
The pelleted protein extract was resuspended in 30 μl of 1× LDS sample buffer (Invitrogen, Carlsbad, CA, USA) and separated on a 10% polyacrylamide gel (Novex 10% Bis-Tris Gel, 1.0 mm; Invitrogen) using MOPS running buffer, according to the manufacturer's instructions. The gel was fixed in 40% methanol/10% acetic acid and stained overnight with SYPRO Ruby (Invitrogen) according to the manufacturer's instructions. Gels were visualized with UV illumination and individual bands were excised (see Fig. 2A) and frozen at –80 °C.
Isolation of broad slabs from 1D polyacrylamide gels
Proteins were separated as above, except MES running buffer (Invitrogen) was used according the manufacturer's instructions. Slabs were excised (see Fig. 2B), cut into small pieces, and frozen at –80 °C.
Gel-free in-solution trypsin digest
Precipitated proteins were resuspended in 100 μl of 50 mM ammonium bicarbonate, 6 M guanidinium chloride. To this, 5 μl of dithiothreitol (DTT) stock solution (200 mM DTT in 50 mM ammonium bicarbonate) was added and the mixture boiled for 10 min. Proteins were alkylated by addition of 4 μl of 1 M iodoacetamide in 50 mM ammonium bicarbonate, followed by a 1 h room temperature incubation in the dark. To this, 40 μl of DTT stock was added and incubation was continued for an additional hour. The sample was then diluted by addition of 846 μl of 50 mM ammonium bicarbonate and digested by the addition of 5 μl of 200 ng μl−1 solution of sequencing grade trypsin (Promega, Madison, WI, USA). The reaction was incubated overnight at 37 °C and then terminated by the addition of concentrated acetic acid to lower the pH below 6.0.
Analysis of the gel-free extract was conducted by online liquid chromatography–electrospray ionization–tandem mass spectrometry (LC-ESI-MS/MS), essentially as described by Yang et al. (2007). The sample was pre-fractionated by strong cation-exchange chromatography, eluting bound peptides in five fractions with a step gradient of 25, 50, 100, 200, and 500 mM KCl. Each fraction was then analysed by LC-ESI-MS/MS as previously described. For the two gel-fractionated samples, in-gel trypsin digestion was performed as previously described (Shevchenko et al., 1996), with modifications as described by Yang et al. (2007), and tryptic peptides were recovered with C18 ZipTips (Millipore, Bedford, MA, USA), according to the manufacturer's directions. Peptides from each fraction were separated and analysed by offline LC-MALDI-TOF/TOF (liquid chromatography–matrix-assisted laser desoportion ionization time of flight tandem mass spectrometry) analysis (Yang et al., 2007).
Peak lists from the mass spectrometers were searched against the longest six-frame translation of the Sol Genomics Network (SGN) Lycopersicum Combined unigene build from May 2009 (www.solgenomics.net) using MASCOT (Perkins et al., 1999). For all experiments, the database was searched allowing for one missed cleavage, cysteine carboxyamidomethylation, and variable methionine oxidation, requiring peptide scores corresponding to ≥95% confidence. For MALDI-TOF/TOF experiments, a peptide mass tolerance of 10 ppm and fragment tolerance of 0.025 Da was used. For ESI-MS/MS experiments, these tolerances were set to 1.5 ppm and 0.6 Da, respectively. To limit the number of false-positive results, the results were filtered by requiring that each identified protein be represented by at least two unique peptides in the same or multiple analyses.
Laser-capture microdissection, RNA amplification, and cDNA synthesis
Tissue fixation and microdissection were performed based on the protocol of Nakazono et al. (2003). Pericarp tissue from 10 DPA immature green tomato fruits was manually dissected into 2 mm cubes using a razor and fixed by vacuum infiltration with 75% ethanol, 25% acetic acid. The ethanol/acetic acid was replaced with a fresh aliquot and the sample was left overnight at 4 °C. The fixative was decanted and replaced twice with a solution of 10% (w/v) sucrose in 100 mM phosphate-buffered saline (PBS). Upon penetration of the solution into the tissue, as indicated by the tissue sinking, the solution was replaced twice more with a solution of 20% (w/v) sucrose in 100 mM PBS. The tissue was then embedded in TissueTek OCT medium (Sakura Finetek USA, Torrance, CA, USA), frozen in a beaker submerged in a liquid nitrogen bath, and the resulting cryoblocks stored at –80 °C until sectioning.
A Microm HM550 cryostat (ThermoFisher Scientific, Waltham, MA, USA) was used to prepare 10 μm and 16 μm pericarp sections and the CryoJane tape-transfer system (Instrumedics, St Louis, MO, USA) was used to transfer sections to 0.5× adhesive-coated slides, where they were adhered by UV cross-linking. Slides were stored at –80 °C until later use. Immediately prior to laser-capture microdissection, slides were thawed and dehydrated as follows (all solvents at –20 °C): 1 min, 50% ethanol; 30 s, 95% ethanol; 1 min, 100% ethanol; 2 min, xylene; 2 min, fresh xylene. After air drying, cells were harvested into PALM adhesive cap tubes (Carl Zeiss, Oberkochen, Germany) using a PALM MicroBeam System (Carl Zeiss). Epidermal cells were captured from the 10 μm sections, while the larger, more vacuole-rich collenchyma cells were captured from the 16 μm sections. Total RNA was isolated from the harvested cells using an RNeasy Micro Kit (Qiagen, Valencia, CA, USA) and the mRNA amplified using the TargetAmp 2-Round aRNA Amplification Kit 2.0 (Epicentre Biotechnologies, Madison, WI, USA), according to the manufacturers’ instructions. A 1.5 μg aliquot of amplified RNA was used for cDNA synthesis using SuperScript III reverse transcriptase and random hexamer primers (Invitrogen), according to the manufacturer's instructions.
RNA isolation and cDNA synthesis for developmental time course
RNA was isolated from frozen tissue (Schneiderbauer et al., 1991) and 1.5 μg of total, DNase-treated RNA was used for cDNA synthesis using SuperScript II reverse transcriptase and oligo(dT) primers (Invitrogen), according to the manufacturer's instructions.
Quantitative PCR
Quantitative PCR experiments were performed using an iQ5 system (BioRad, Hercules, CA, USA). The cDNA samples were diluted 5-fold with water and 0.5 μl or 1 μl was used as a template for each 25 μl quantitative PCR, prepared using HotStart-IT SYBR Green qPCR Master Mix (Affymetrix, Santa Clara, CA, USA). For each gene, qPCRs were performed in technical triplicates. The sequences of oligonucleotide primers are given in Supplementary Table S1 available at JXB online. Specificity of the products was determined by gel electrophoresis, product sequencing, and high-resolution melt curve analysis. For tissue specificity, quantification was performed using REST 2008 software (Pfaffl et al., 2002) with RPL2 serving as a constitutive control, assuming PCR efficiency of 1.0 for all genes. For time course experiments, expression ratios for each gene and time point were calculated relative to RPL2 expression. For each gene, expression was linearly normalized, with a value of 0.0 assigned to the stage with lowest expression and 1.0 to the stage showing the highest expression.
Bioinformatics and software
The area-proportional Venn diagram was constructed using BioVenn (Hulsen et al., 2008; http://www.cmbi.ru.nl/cdd/biovenn/). Normalized gene expression profile data were converted into a heat map using Cluster 3.0 (bonsai.ims.u-tokyo.ac.jp/∼mdehoon/software/cluster/software.htm) and Java TreeView (Saldanha, 2004). Alignment of protein sequences was performed with Clustal W (Thompson et al., 1994), and a Neighbor–Joining tree was constructed using MEGA4 (Tamura et al., 2007). The alignment parameters and the settings for the phylogenetic reconstruction were the defaults of the MEGA4 package.
Results
Protein isolation and identification of candidate genes
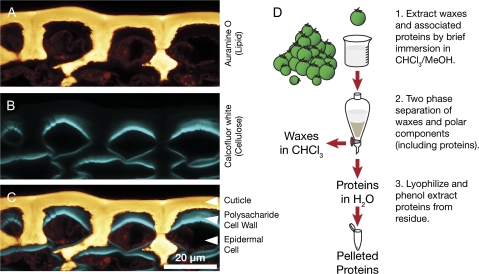
As illustrated in Fig. 1A–C, the fluorescently stained cuticle (Fig. 1A) covers the surface of the tomato fruit but is separated from the epidermal cells by a subcuticular polysaccharide cell wall (Fig. 1B). Previous studies have indicated that wax, rather than cutin, is the major barrier to the diffusion of polar molecules, including water (Leide et al., 2007; Isaacson et al., 2009), and presumably proteins, across the cuticle. It was reasoned that, despite the relatively low abundance of wax in the tomato fruit cuticle, which is of the order of 5 μg cm−2 compared with 1 mg cm−2 for the cutin polymer (Baker et al., 1982), a brief immersion of the fruits in an organic solvent would allow the isolation of proteins directly associated with cuticular wax, as well as those localized within the subcuticular epidermal cell wall and possibly epidermal intracellular proteins, depending on the degree to which the cells were compromised. A standard protocol was therefore used to remove waxes by immersion of intact plant organs in an organic solvent to obtain extracts for profiling of the fruit surface proteome, as was previously attempted on a smaller scale with Brassica oleracea leaves (Pyee et al., 1994).
Fig. 1.
Epidermis structure and experimental design. Confocal microscopy of cryosectioned tomato breaker stage fruit epidermis, co-stained with the fluorescent lipid stain Auramine O (A) and the cellulose stain Calcofluor white M2R (B). The merged image (C) illustrates the cuticle and epidermal cell wall in the context of the epidermal cell layer. (D) Schematic representation of the extraction protocol used to isolate proteins from the cuticle and epidermal cell wall.
In order to target proteins that might be associated with cuticle biosynthesis more specifically, young, rapidly expanding tomato fruits were used in this study, since this represents the phase of most rapid cuticle deposition (Baker et al., 1982; Mintz-Oron et al., 2008). After extraction of cuticular waxes and other co-extracted components, the wax was separated from the more polar proteins by partitioning of polar constituents into an aqueous phase and wax into chloroform. The aqueous phase was then lyophilized and proteins were further purified from the residue by phenol extraction and precipitation (Fig. 1D).
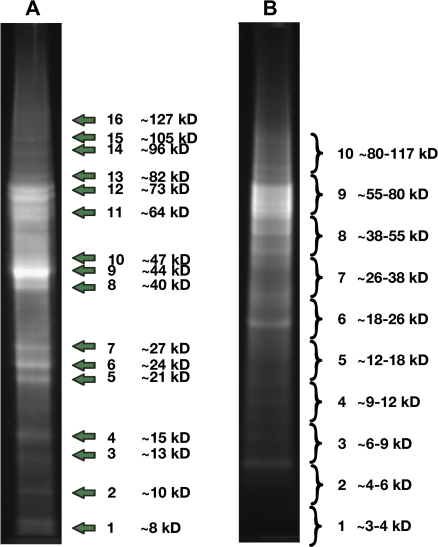
In the initial analysis, the protein extract was separated by denaturing polyacrylamide gel electrophoresis (PAGE) and the 16 most distinct bands (Fig. 2A) were excised and subjected to in-gel tryptic digestion, followed by offline LC-MALDI-TOF/TOF analysis. This use of reverse phase liquid chromatography to separate tryptic peptides and robotic mixing of chromatographic fractions with a MALDI matrix (Bodnar et al., 2003) combines the capacity for analysing complex mixtures offered by online LC-ESI-MS/MS analysis with the increased precision and reduced sensitivity to ion suppression that is offered by MALDI-TOF/TOF analysis (Yang et al., 2007). Using this approach, a total of 44 different proteins were identified from the 16 bands following MASCOT searching of the mass spectra against a database of translated tomato unigene sequences (Supplementary Table S2 at JXB online). Since an initial analysis using the spectra obtained from each band separately revealed some redundancy in the proteins identified in each band, as well as the presence of many proteins in each band (data not shown), the spectra from all bands were combined for this search.
Fig. 2.
Denaturing polyacrylamide gel electrophoresis of protein extracts. Proteins were separated and distinct bands (A), or broad slabs covering the indicated ranges (B), were isolated. (This figure is available in colour at JXB online.)
Using a second experimental strategy and a new protein isolate, proteins were pre-fractionated by PAGE, but, rather than cutting distinct bands, 10 contiguous gel slabs were excised and subjected to in-gel trypsin digestions (Fig. 2B). It was noted that the banding pattern did not closely resemble that seen in the first analysis (Fig. 2A). This probably reflects the fact that a different buffer system was used (MES), which favours the resolution of smaller proteins at the expense of larger proteins, or that the proteins may be subjected to varying degrees of post-extraction proteolysis. When spectra from these 10 slabs were combined and a MASCOT search of the tomato predicted protein database was performed, a total of 25 proteins were identified (Supplementary Table S3 at JXB online).
A third protein extract was prepared as before, but, rather than fractionating the sample by denaturing PAGE, the entire protein extract was subjected to in-solution tryptic digestion. The resulting solution of tryptic peptides was then pre-fractionated by step elution of a strong cation exchange solid phase extraction cartridge and each fraction was subjected to LC-ESI-MS/MS. A database search using MASCOT with the spectra from this analysis identified 192 unique proteins (Supplementary Table S4 at JXB online). In addition to identifying more proteins, in the cases where a protein was identified by both a gel-based and gel-free approach, the latter strategy generally resulted in greater percentage protein coverage and total ion scores.
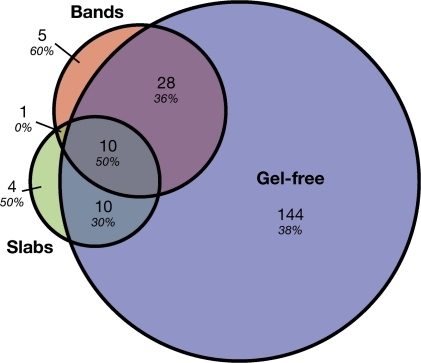
In summary, proteins corresponding to 202 distinct tomato unigenes were identified. The three analyses showed a substantial amount of overlap, as shown by the relatively high degree of redundancy between the sets of proteins identified in each analysis (Fig. 3). Notably, only 5% of the proteins were identified only by the gel-based analysis and not by the gel-free approach. Given that so little is known about extracellular cuticle assembly and restructuring, the subset of proteins that are potentially secreted to the cell wall were of particular interest. Of the 202 proteins identified, 78 (39%) had secretory signal peptides (SPs) as predicted by SignalP 3.0 (Bendtsen et al., 2004), and these were sorted into 40 putative functional families based on BLAST annotations (Table 1). Several of the putative secreted protein families had lipid-related domains, or similarity to proteins that have previously been implicated in cuticle biology. For example, five LTPs, and an MD-2-related lipid recognition domain-containing (ML) protein that is predicted to bind lipids, were identified. Also of interest were two GDSL-motif lipase/hydrolase family proteins. In addition to the proteins with putative roles in lipid metabolism, many defence-related proteins were also identified, including several PR-1 proteins, protease inhibitors, chitinases, and endo-β-1,3-glucanases. A large number of proteins belonging to the category of cell wall-modifying and structural proteins, such as expansin, xyloglucan endotransglucosylase-hydrolase, and extensin, were also identified.
Fig. 3.
Venn diagram of proteins found in the three proteomic analyses and signal peptide prediction. The total number in each unique or overlapping set is shown, with the percentage of each set with a predicted signal peptide (SignalP 3.0) indicated in italics.
Table 1.
Proteins identified by MASCOT with predicted signal peptides
| Annotation/gene familya | SGN unigene | Identified in analysisb | Best hit |
|
| Total ion score | Percentage coverage | |||
| Lipid and putative cuticle related | ||||
| GDSL-motif lipase/hydrolase family protein | SGN-U583101 | A | 37 | 7.5 |
| SGN-U579520 | ABC | 173 | 22 | |
| Inducible plastid lipid-associated protein | SGN-U577010 | C | 76 | 15.9 |
| Lipid transfer protein (LTP) | SGN-U577838 | C | 43 | 11.7 |
| SGN-U579033 | C | 149 | 46.7 | |
| SGN-U579687 | C | 252 | 55.7 | |
| SGN-U580659 | C | 69 | 43.5 | |
| SGN-U581465c | C | 171 | 33.1 | |
| MD-2-related lipid recognition domain-containing (ML) protein | SGN-U577903 | ABC | 93 | 8.6 |
| Defence related | ||||
| Allergen V5/Tpx-1-related family protein | SGN-U578890 | C | 105 | 13.8 |
| Bet v I allergen family protein | SGN-U577856 | AC | 67 | 13.6 |
| Chitinase (GH family 18 and 19)d | SGN-U580366 | BC | 245 | 23.3 |
| SGN-U579068 | C | 244 | 27.3 | |
| SGN-U579551 | C | 219 | 19.8 | |
| SGN-U579696 | C | 72 | 8.7 | |
| SGN-U581507 | C | 91 | 14.4 | |
| Chitin-binding lectin | SGN-U562887 | C | 46 | 7.1 |
| Defensin | SGN-U577872 | BC | 341 | 47.4 |
| SGN-U591780 | C | 62 | 17.5 | |
| Endo β-1,3 glucanase (GH family 17) | SGN-U590837 | C | 102 | 7.8 |
| Hevein-like protein | SGN-U567805 | C | 103 | 12.3 |
| SGN-U579235 | C | 863 | 68.2 | |
| Osmotin-like protein | SGN-U574403 | AC | 473 | 40.7 |
| SGN-U579414 | C | 505 | 31 | |
| SGN-U581103 | C | 574 | 30.6 | |
| Peroxidase | SGN-U581155 | AC | 588 | 21.9 |
| SGN-U583085 | BC | 550 | 23.5 | |
| SGN-U564185 | C | 49 | 7.4 | |
| SGN-U566251 | C | 200 | 25.1 | |
| SGN-U571844 | C | 102 | 8.5 | |
| SGN-U575184 | C | 243 | 36.8 | |
| SGN-U578562 | C | 149 | 6.8 | |
| SGN-U580369 | C | 199 | 17.3 | |
| SGN-U580709 | C | 211 | 34.3 | |
| SGN-U583086 | C | 461 | 23.8 | |
| Peroxiredoxin | SGN-U579538 | C | 74 | 7.4 |
| Polygalacturonase inhibitor protein | SGN-U579059 | AC | 44 | 11.6 |
| PR-1 | SGN-U578279 | C | 180 | 48.7 |
| SGN-U579345 | C | 93 | 12.2 | |
| SGN-U579426 | C | 276 | 34.8 | |
| SGN-U579545 | C | 771 | 52.8 | |
| SGN-U579883 | C | 160 | 25 | |
| Protease | SGN-U578421 | AC | 71 | 6.2 |
| SGN-U582837 | AC | 134 | 8.3 | |
| SGN-U578351 | C | 76 | 4.7 | |
| SGN-U578475 | C | 102 | 7.4 | |
| SGN-U579972 | C | 159 | 6.1 | |
| Protease inhibitor protein | SGN-U573941 | ABC | 194 | 18.3 |
| SGN-U574346 | AC | 83 | 7.3 | |
| SGN-U577283 | C | 194 | 14.8 | |
| SGN-U578389 | C | 163 | 20 | |
| SGN-U578863 | C | 62 | 9.6 | |
| SGN-U585465 | C | 134 | 15.5 | |
| Snakin-like protein | SGN-U578258 | C | 168 | 9 |
| Carbohydrate cell wall metabolism related | ||||
| α-Galactosidase (GH family 27) | SGN-U571081 | C | 102 | 5.9 |
| β-Glucosidase (GH family 1) | SGN-U580766 | A | 49 | 3.9 |
| Expansin | SGN-U577727 | C | 124 | 11.6 |
| Ole e 1 allergen/extensin like | SGN-U563658 | C | 85 | 13.8 |
| Other | ||||
| ADP/ATP translocator like | SGN-U577960 | C | 91 | 5.1 |
| Ascorbate peroxidase | SGN-U578449 | C | 98 | 14 |
| Enolase | SGN-U579393 | C | 581 | 28.9 |
| Formate dehydrogenase | SGN-U579280 | C | 65 | 7.3 |
| Fructokinase | SGN-U586194 | AC | 250 | 9.2 |
| Fructose-bisphosphate aldolase | SGN-U578572 | AC | 209 | 17.6 |
| Glyceraldehyde 3-phosphate dehydrogenase | SGN-U580213 | ABC | 438 | 38.2 |
| Glycine-rich RNA-binding protein | SGN-U578513 | B | 37 | 19.4 |
| Histone H2B | SGN-U579310 | C | 57 | 19.4 |
| Leucine-rich repeat transmembrane protein kinase | SGN-U579197 | C | 122 | 25.2 |
| Malate dehydrogenase | SGN-U565569 | C | 399 | 25.8 |
| Protein disulphide isomerase-like (PDIL) protein | SGN-U575297 | C | 61 | 6.5 |
| SGN-U577569 | C | 108 | 8.8 | |
| Ribulose bisphosphate carboxylase large chain | SGN-U565452 | ABC | 50 | 4.8 |
| SOUL haem-binding protein | SGN-U584870 | A | 48 | 9.2 |
| Strictosidine synthase family protein | SGN-U583542 | AC | 175 | 12.4 |
| Transketolase | SGN-U577918 | C | 109 | 4.2 |
| Unknown | SGN-U593950 | B | 23 | 11.3 |
| SGN-U565851 | C | 41 | 10.7 | |
| SGN-U566943 | C | 52 | 18.2 | |
Gene family groupings and annotation based on BLAST search of the NCBI non-redundant database.
Analyses [(A) is gel band-based analysis, (B) is gel slab-based analysis, (C) is gel-free analysis] from which members of the protein family were identified. The analysis that yielded the highest protein total ion score is shown in bold.
The longest-six frame translation of SGN-U581465, corresponding to CAJ19705, has an incorrect start codon that was manually adjusted before SignalP analysis
Glycosyl hydrolase (GH) families, www.cazy.org. The SGN annotation refers to the unigene identifier in the Sol Genomics Network database (www.solgenomics.net).
Several compelling candidates with homology to previously reported cuticle-related proteins were found in the set of proteins that were identified by only a single peptide and thus did not meet the stringent filtering criteria (Supplementary Tables S1, S3 at JXB online). While it was decided not to include these in the list of proteins that were confidently identified, their homology to previously identified cuticle-related proteins warranted further investigation. Thus, a glucose–methanol–choline (GMC) oxidoreductase family protein (SGN-U570812) with high similarity to the Arabidopsis protein HOTHEAD (HTH) (57% amino acid identity), which is involved in cuticle biosynthesis (Krolikowski et al., 2003; Kurdyukov et al., 2006b), and three additional GDSL-motif lipase/hydrolase family proteins (SGN-U577181, SGN-U583107, and SGN-U585129) were included in the expression and phylogenetic studies below.
Gene expression analysis of identified proteins
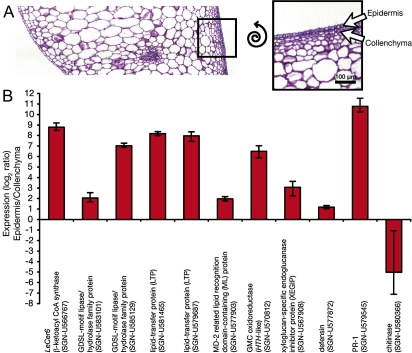
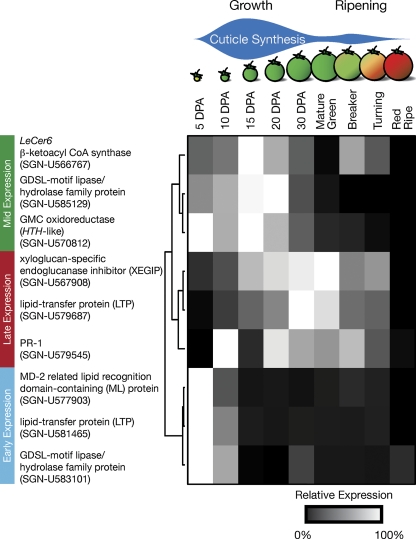
Since the initial aim of this study was to identify surface proteins with a possible role in cuticle formation, the further characterization of candidates with a previously reported cuticle association, or those with lipid-related domains, was of particular interest. Previous studies have shown that many genes encoding cuticle biosynthetic enzymes are specifically expressed in the epidermis (Suh et al., 2005; Mintz-Oron et al., 2008) and so the cell type-specific expression of several candidate genes was investigated. Epidermis and collenchyma cells from immature green fruits were harvested using laser-capture microdissection (Fig. 4A), RNA was isolated, amplified, and qRT-PCR was performed. The expression of the epidermis-specific (Mintz-Oron et al., 2008) cuticle biosynthesis gene LeCer6 was used as a positive control, and the expression of transcripts encoding four defence-related proteins, a class that represented a substantial portion of the identified proteins, was also monitored. One of these, xyloglucan-specific endoglucanase inhibitor protein (XEGIP), was only identified by a single peptide (Supplementary Fig. S2 at JXB online), but its well characterized expression and biological activity warranted its inclusion as a positive control for defence-related transcripts (Qin et al., 2003). Of the 10 genes selected for further characterization, five showed much greater expression in the epidermis relative to the collenchyma (90- to 1700-fold), three showed more modest epidermal enrichment (4- to 8-fold), and two showed low expression ratios (2- and 0.03-fold), suggesting that their transcripts were not epidermis specific (Fig. 4B). The positive control LeCer6 and the six cuticle-related candidate genes all showed epidermal enrichment of >4-fold, while the four defence-related transcripts showed mixed epidermal specificity: the XEGIP and PR-1 transcripts were more highly expressed in the epidermis while the defensin and chitinase both showed low expression ratios, indicating weak epidermal specificity and collenchyma-specific expression, respectively.
Fig. 4.
Tissue-specific expression of selected genes by qRT-PCR of RNA from microdissected cells. Epidermal cells and collenchyma cells were harvested from immature green tomato fruits by laser-capture microdissection as illustrated (A), and extracted, amplified RNA was used for qRT-PCR expression analysis of selected genes (B). The error bars are the standard error as determined by REST 2008 using three technical replicates.
Since deposition of wax and cutin follows a specific temporal pattern during fruit development, typified by maximal accumulation during fruit growth followed by a second phase of cuticle deposition during ripening (Baker et al., 1982; Bauer et al., 2004), the expression of the eight epidermis up-regulated genes during fruit growth and ripening was further characterized using qRT-PCR (Fig. 5). LeCer6 expression was again used as a positive control, as it encodes a part of the fatty acid elongation complex required for aliphatic wax biosynthesis (Vogg et al., 2004) and so its expression would be expected to correlate with wax deposition. The expression pattern of LeCer6 was most similar to that of the GDSL-motif lipase/hydrolase family gene SGN-U585129 and GMC oxidoreductase, as all three were maximally expressed during the most rapid phase of fruit expansion, peaking at 15–20 DPA (Fig. 5). The two defence-related transcripts, XEGIP and the PR-1 SGN-U579545, as well as the LTP SGN-U579687, showed related expression patterns with broad peaks of expression spanning the late phases of fruit growth and early ripening. It was noted that the expression pattern of XEGIP corresponded well with a previously reported northern blot analysis of its expression (Qin et al., 2003). Finally, the genes encoding the ML protein, the LTP SGN-U581465, and the GDSL-motif lipase/hydrolase family protein SGN-U583101 all showed similar expression patterns, with high levels of transcript in very young fruit and a substantial reduction by 15 DPA.
Fig. 5.
Time course expression of selected genes during fruit growth and ripening. Gene expression was determined by qRT-PCR relative to the constitutive control RPL2 and normalized as described in the Materials and methods. The two phases of cuticle deposition are indicated above the fruit development stages considered.
Phylogenetic analysis of the GDSL-motif lipase/hydrolase family proteins
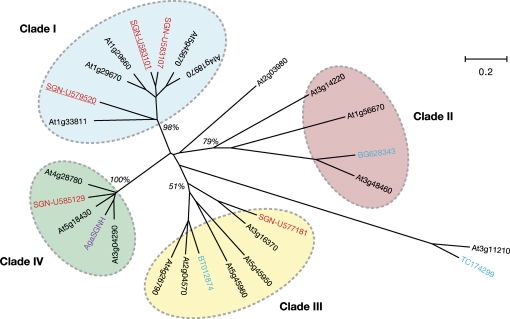
Two of the confidently identified proteins and three proteins identified by a single peptide belong to the GDSL-motif lipase/hydrolase family, which is widely distributed in both eukaryotes and prokaryotes (Akoh et al., 2004). Plant GDSL-motif lipase/hydrolases comprise large gene families; for example, there are 113 predicted members in Arabidopsis, although few have a known function. Several lines of circumstantial evidence suggest a role for these enzymes in cutin metabolism. First, biochemically characterized isozymes have been shown to have acyl hydrolase activity, and the presence of an SP suggests that many are secreted (Akoh et al., 2004). Secondly, microarray analysis of Arabidopsis stem peels revealed that a subset of 18 members of the gene family is preferentially expressed in this cuticle-synthesizing tissue (Suh et al., 2005). Furthermore, one of these, At2g04570, is highly induced by expression of the cuticle-associated transcription factor WIN1/SHN1 (Kannangara et al., 2007). Microarrays with RNA from isolated tomato peel also identified three tomato GDSL-motif lipase/hydrolase family proteins that are preferentially expressed in the epidermis (Mintz-Oron et al., 2008). Finally, the transcripts corresponding to a GDSL-motif lipase/hydrolase family protein, AgaSGNH, from Agave americana were shown to be highly abundant in the epidermis during leaf elongation, when cutin is being rapidly synthesized (Reina et al., 2007).
Phylogenetic analysis of the five GDSL-motif lipase/hydrolase family proteins described in this present study, as well as the 18 epidermis-specific Arabidopsis sequences, AgaSGNH, and the three tomato sequences previously identified by Mintz-Oron et al. (2008) indicated that the candidate cuticle-related GDSL-motif lipase/hydrolase family proteins can be grouped into four clades (Fig. 6). The sequences identified in this study align within Clades I, III, and IV. Co-expression analysis of the Arabidopsis members of Clades I and III using CressExpress (Srinivasasainagendra et al., 2008; www.cressexpress.org) showed high levels of co-expression with nine cutin biosynthesis-related genes (ATT1, LACS2, LCR, GPAT4, GPAT8, GPAT6, CYP86A4, CYP86A7, and CYP77A6), which were used as bait (Supplementary Table S5 at JXB online). While co-expression is less pronounced in Clade IV, its smaller size and higher degree of conservation make it an attractive source of candidate cuticle-related GDSL-motif lipase/hydrolase family proteins, particularly in light of the expression patterns of AgaSGNH and SGN-U585129 that coincide with cutin deposition.
Fig. 6.
Phylogenetic analysis of GDSL-motif lipase/hydrolase family proteins. The Arabidopsis genes (AGI numbers, www.arabidopsis.org, in black) are those showing >2-fold enrichment in epidermal peels relative to whole stem tissue (Suh et al., 2005). The blue TIGR plant transcript assembly numbers (plantta.jcvi.org) and NCBI EST accessions were identified as tomato transcripts enriched in the peel relative to tomato flesh (Mintz-Oron et al., 2008). AgaSGNH (purple) was identified as an epidermal-specific transcript in Agave americana (Reina et al., 2007). SGN unigenes (www.solgenomics.net) described in the current study are shown in red and are underlined if they were identified with confidence (spectra matching two or more peptides, see text). Bootstrap support of the four arbitrarily numbered clades is indicated in italics (500 replications). The bootstrap support of Clade III increases to 92% if the outlying At3g11210 and TC174299 sequences are discarded. Branch lengths are proportional to distance, as indicated by the scale legend.
Discussion
In this study, the use of modern mass spectrometry-based proteomic techniques and a diverse set of protein fractionation strategies resulted in a large set of proteins putatively associated with the cuticle of the developing tomato fruit. During the first step of surface protein extraction, care was taken to minimize the time the fruits were submerged in the solvent, in order to reduce cell lysis and increase the proportion of secreted proteins. Bioinformatic analysis suggested that 39% of the cognate genes are predicted to encode N-terminal SPs that would direct their secretion, and this represents a substantial enrichment. For comparison, when the Arabidopsis predicted proteome (TAIR release 8, www.arabidopsis.org) is subjected to the same analysis, only 19% of proteins are predicted to have an SP (data not shown). Moreover, it is likely that the N-termini containing the SPs of some of the identified proteins are absent from the sequence databases, since a full genome sequence is not yet available for tomato. However, the presence of known intracellular proteins can be taken to indicate the lysis of some epidermal cells. In addition, it is emphasized that a subset of extracellular proteins will not be extracted or successfully fractionated with the protocols used here due to the recalcitrant nature of the cell wall proteome (reviewed in Lee et al., 2004; Isaacson and Rose, 2006). Moreover, computational tools for predicting SPs are imperfect, and so the presence, or absence, of a predicted SP is not a de facto indication of protein extracellular or intracellular localization, respectively. Nonetheless, the enrichment observed suggests that the protein extracts will provide a valuable starting point for researchers interested in cuticle assembly and restructuring.
The three fractionation strategies employed were generally complementary and helped to confirm findings in other analyses, as indicated by the significant overlap between the sets of proteins found in each analysis (Fig. 3). However, the gel-free approach has clear advantages in terms of the number of proteins that were identified and the higher identification confidence scores, as indicated by MASCOT total ion score. Conversely, the gel slab-based analysis (Fig. 2B) yielded the fewest identified proteins (Fig. 3). Since the initial goal was to identify candidate proteins that might be involved in cuticle metabolism, several proteins attracted attention because they had lipid-associated domains, or shared sequence similarity with proteins that are known, or proposed to have, roles in cuticle biogenesis.
Putative lipid-binding proteins
Of the five LTPs that were identified, four belong to family 1 of LTPs, and one to family 2 (SGN-U577838) (Yeats and Rose, 2008). The cDNA le16, corresponding to SGN-U579033, was previously identified as being up-regulated by drought and abscisic acid (ABA) (Plant et al., 1991), and the same gene, as well as the gene corresponding to SGN-U581465, was later shown to encode the tomato Lyc e 3 allergen (Le et al., 2006). In a microarray analysis of tomato peel transcripts, SGN-U579687 was seen to be more highly expressed in the exocarp than in the inner pericarp (Mintz-Oron et al., 2008), a result that supports the finding that this transcript is more highly expressed in the epidermis than in the underlying collenchyma.
Aside from LTPs, SGN-U577903, which encodes an ML protein, and has predicted extracellular localization and lipid-binding activity, is also a candidate for contributing to cuticle biogenesis. The ML domain is shared by proteins from diverse eukaryotic species and takes its name from MD-2, a soluble extracellular protein in humans that binds lipopolysaccharide (LPS) in the first step of a signalling cascade that triggers the innate immune response (Jerala, 2007). Other members of this family include the human cholesterol-binding-protein NPC2 (Friedland et al., 2003) and the dust mite allergen Der f 2, which was recently also shown to bind LPS (Ichikawa et al., 2009). Structurally, the domain is composed of two β-sheets that enclose a deep lipid-binding pocket (Ohto et al., 2007), although no ligand is known or function proposed for the protein family in plants. The transcript abundance of the ML protein was ∼4-fold greater in the epidermis than in the underlying collenchyma cells (Fig. 4B), and its expression, like that of the LTP SGN-U581465, was highest at the earliest stage of fruit development before rapidly declining (Fig. 5). This precedes the extensive cutin and wax deposition that occurs during the phase of greatest fruit expansion, from 10 DPA to 30 DPA. However, this does not necessarily lead to rejection of the LTP or ML proteins as candidates for wax or cutin transporters: both proteins have extremely stable folds that may result in the protein remaining functional far longer than steady-state mRNA levels are maintained. While no structural or biochemical characterization of any plant-derived ML proteins has been reported, it is suggested that the large lipid-binding cavity of this domain may accommodate the pentacyclic triterpenoids that are abundant in tomato cuticular wax. In contrast, the family 1 LTPs that have been previously proposed as lipid-binding proteins that transport wax across the cell wall are unable to bind planar sterols that are structurally analogous to triterpenoids (Cheng et al., 2004).
Putative HTH orthologue
The GMC oxidoreductase gene that was tentatively identified and shown to have epidermis-specific expression, SGN-U570812, has 57% amino acid identity with the Arabidopsis gene HTH. The hth mutant has a fused floral organ phenotype that is attributed to a defective cuticle (Krolikowski et al., 2003), and a biochemical activity for HTH has been proposed based on hth cutin polymer composition (Kurdyukov et al., 2006b). Mutant plants accumulate increased amounts of ω-hydroxy fatty acids and lower levels of α,ω-dicarboxylic fatty acids that predominate in Arabidopsis cutin. Thus, the authors propose that HTH oxidizes ω-hydroxy fatty acids to ω-oxo fatty acids prior to formation of α,ω-dicarboxylic fatty acids.
In tomato, α,ω-dicarboxylic fatty acids comprise only ∼1% by weight of cutin monomer composition (Leide et al., 2007; Isaacson et al., 2009). Nevertheless, this monomer may play an important structural role in determining the degree of cross-linking between either cutin chains and the polysaccharide cell wall, or other cutin chains. Expression analysis of the HTH-like SGN-U570812 indicated that it is highly expressed (∼90-fold) in the epidermis relative to the collenchyma (Fig. 4) and that its expression during fruit development coincides with the rapid expansion and cuticle deposition that occurs 10–20 DPA (Fig. 5).
GDSL-motif lipase/hydrolase family proteins
The genes encoding two confidently identified and three tentatively identified GDSL-motif lipase/hydrolase family proteins were of particular interest, and qRT-PCR characterization of the expression patterns of the five genes was attempted. Despite repeated attempts, this was only successful for SGN-U583101 and SGN-U585129. Both were shown to be more highly expressed in the epidermis than in the underlying collenchyma, although the ratio for SGN-U585129 was much greater (Fig. 4B). The time course of their expression during fruit development was also distinct as SGN-U583101 was highly expressed only very early in fruit development, while SGN-U585129 was expressed throughout fruit expansion (Fig. 5).
GDSL-motif lipase/hydrolase family proteins have previously been proposed as cutin synthases (Reina et al., 2007), or enzymes involved in modification or recycling of the cutin polymer (Pollard et al., 2008). Thus, in the absence of genetic or biochemical evidence for their activity, three specific biochemical activities that may be required for cutin metabolism can be imagined. As cutin synthases, they may incorporate either cutin monomers or oligomers. Secondly, controlled hydrolysis of cutin during organ expansion may be required, so they may act as cutin hydrolases. A third hypothetical enzyme activity is that of a cutin transacylase, wherein the cutin polymer could be loosened by simultaneous cleavage and religation (transacylation) of ester bonds, allowing for organ expansion during growth. This activity, combined with synthesis of cutin oligomers by intracellular enzymes such as GPATs and BAHD family acyltransferases, could be sufficient for cutin polymer synthesis, allowing the oligomers to be ‘stitched’ into the growing cutin polymer matrix only by exchange of existing ester bonds.
In conclusion, using a proteomic approach, a diverse collection of proteins with putative roles in lipid metabolism was identified. Several of these have gene expression patterns that correlate with cuticle biosynthesis; that is they are specifically expressed in the epidermis and their expression coincides with or precedes the deposition of wax and cutin. The results further suggest that there are discrete phases of cuticular lipid metabolism and/or trafficking, which are associated with different gene classes and, even more interestingly, distinct members of the same gene family (e.g. GDSL-motif lipase/hydrolase family proteins and LTPs). However these remain candidates for cuticle biogenesis, and reverse-genetic experiments are currently underway for functional confirmation. Analysis of the phenotypes of these plants coupled with in vitro demonstration of their proposed biochemical activities will advance the goal of better understanding of cuticle biosynthesis.
Supplementary data
Supplementary data are available at JXB online.
Table S1. PCR primers use for gene expression analysis.
Table S2. Complete MASCOT results for gel band analysis.
Table S3. Complete MASCOT results for gel slab analysis.
Table S4. Complete MASCOT results for gel-free analysis.
Table S5. Co-expression results of Arabidopsis GDSL-motif lipase/hydrolase family proteins and known cutin-biosynthesis genes.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge Dr Mike Scanlon for generously providing access to the PALM MicroBeam System. This work was supported by NSF Plant Genome Research Program grant (DBI-0606595) and THY was supported in part by an NIH chemistry/biology interface training grant (grant no. T32 GM008500). LC-ESI-MS/MS analysis was performed by Dr Sheng Zhang and the Cornell Proteomics and Mass Spectrometry Core Facility.
Glossary
Abbreviations
- BDG
BODYGUARD
- DPA
days post-anthesis
- DTT
dithiothreitol
- EDTA
ethylenediaminetetra-acetic acid
- ER
endoplasmic reticulum
- GMC
glucose–methanol–choline
- GPAT
glycerol-3-phosphate acyltransferase
- HTH
HOTHEAD
- LC-ESI-MS/MS
liquid chromatography–electrospray ionization–tandem mass spectrometry
- LC-MALDI-TOF/TOF
liquid chromatography–matrix-assisted laser desoportion ionization time of flight tandem mass spectrometry
- LPS
lipopolysaccharide
- LTP
lipid-transfer protein
- PBS
phosphate-buffered saline
- ML
MD-2-related lipid recognition domain-containing protein
- SP
signal peptide
- XEGIP
xyloglucan-specific endoglucanase inhibitor protein
References
- Akoh CC, Lee GC, Liaw YC, Huang TH, Shaw JF. GDSL family of serine esterases/lipases. Progress in Lipid Research. 2004;43:534–552. doi: 10.1016/j.plipres.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Baker EA, Bukovac MJ, Hunt GM. Composition of tomato fruit cuticle as related to fruit growth and development. In: Cutler DF, Alvin KL, Price CE, editors. The plant cuticle. London: Academic Press; 1982. pp. 33–44. [Google Scholar]
- Bauer S, Schulte E, Thier H. Composition of the surface wax from tomatoes: II. Quantification of the components at the ripe red stage and during ripening. European Food Research and Technology. 2004;219:487–491. [Google Scholar]
- Beisson F, Li Y, Bonaventure G, Pollard M, Ohlrogge JB. The acyltransferase GPAT5 is required for the synthesis of suberin in seed coat and root of Arabidopsis. The Plant Cell. 2007;19:351–368. doi: 10.1105/tpc.106.048033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. Journal of Molecular Biology. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Bird D, Beisson F, Brigham A, Shin J, Greer S, Jetter R, Kunst L, Wu X, Yephremov A, Samuels L. Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. The Plant Journal. 2007;52:485–498. doi: 10.1111/j.1365-313X.2007.03252.x. [DOI] [PubMed] [Google Scholar]
- Bodnar WM, Blackburn RK, Krise JM, Moseley MA. Exploiting the complementary nature of LC/MALDI/MS/MS and LC/ESI/MS/MS for increased proteome coverage. Journal of the American Society of Mass Spectrometry. 2003;14:971–979. doi: 10.1016/S1044-0305(03)00209-5. [DOI] [PubMed] [Google Scholar]
- Bonaventure G, Beisson F, Ohlrogge J, Pollard M. Analysis of the aliphatic monomer composition of polyesters associated with Arabidopsis epidermis: occurrence of octadeca-cis-6, cis-9-diene-1,18-dioate as the major component. The Plant Journal. 2004;40:920–930. doi: 10.1111/j.1365-313X.2004.02258.x. [DOI] [PubMed] [Google Scholar]
- Buda GJ, Isaacson T, Matas AJ, Paolillo DJ, Rose JKC. Three-dimensional imaging of plant cuticle architecture using confocal scanning laser microscopy. The Plant Journal. 2009;60:378–385. doi: 10.1111/j.1365-313X.2009.03960.x. [DOI] [PubMed] [Google Scholar]
- Cheng CS, Samuel D, Liu YJ, Shyu JC, Lai SM, Lin KF, Lyu PC. Binding mechanism of nonspecific lipid transfer proteins and their role in plant defense. Biochemistry. 2004;43:13628–13636. doi: 10.1021/bi048873j. [DOI] [PubMed] [Google Scholar]
- Cruz MAL, Gomes VM, Fernandes KVS, Machado OLT, Xavier J. Identification and partial characterization of a chitinase and a beta-1,3-glucanase from Copernicia cerifera wax. Plant Physiology and Biochemistry. 2002;40:11–16. [Google Scholar]
- Debono A, Yeats TH, Rose JKC, Bird D, Jetter R, Kunst L, Samuels L. Arabidopsis LTPG is a glycosylphosphatidylinositol-anchored lipid transfer protein required for export of lipids to the plant surface. The Plant Cell. 2009;21:1230–1238. doi: 10.1105/tpc.108.064451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke R, Briesen I, Wojciechowski T, Faust A, Yephremov A, Nawrath C, Schreiber L. Apoplastic polyesters in Arabidopsis surface tissues—a typical suberin and a particular cutin. Phytochemistry. 2005;66:2643–2658. doi: 10.1016/j.phytochem.2005.09.027. [DOI] [PubMed] [Google Scholar]
- Friedland N, Liou HL, Lobel P, Stock AM. Structure of a cholesterol-binding protein deficient in Niemann–Pick type C2 disease. Proceedings of the National Academy of Sciences, USA. 2003;100:2512–2517. doi: 10.1073/pnas.0437840100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidda SK, Shockey JM, Rothstein SJ, Dyer JM, Mullen RT. Arabidopsis thaliana GPAT8 and GPAT9 are localized to the ER and possess distinct ER retrieval signals: functional divergence of the dilysine ER retrieval motif in plant cells. Plant Physiology and Biochemistry. 2009;47:867–879. doi: 10.1016/j.plaphy.2009.05.008. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Bosch C, Brummell DA, Bennett AB. Differential expression of two endo-1,4-beta-glucanase genes in pericarp and locules of wild-type and mutant tomato fruit. Plant Physiology. 1996;111:1313–1319. doi: 10.1104/pp.111.4.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graca J, Schreiber L, Rodrigues J, Pereira H. Glycerol and glyceryl esters of omega-hydroxyacids in cutins. Phytochemistry. 2002;61:205–215. doi: 10.1016/s0031-9422(02)00212-1. [DOI] [PubMed] [Google Scholar]
- Hulsen T, de Vlieg J, Alkema W. BioVenn—a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa S, Takai T, Yashiki T, Takahashi S, Okumura K, Ogawa H, Kohda D, Hatanaka H. Lipopolysaccharide binding of the mite allergen Der f 2. Genes to Cells. 2009;14:1055–1065. doi: 10.1111/j.1365-2443.2009.01334.x. [DOI] [PubMed] [Google Scholar]
- Isaacson T, Damasceno CM, Saravanan RS, He Y, Catalá C, Saladié M, Rose JKC. Sample extraction techniques for enhanced proteomic analysis of plant tissues. Nature Protocols. 2006;1:769–774. doi: 10.1038/nprot.2006.102. [DOI] [PubMed] [Google Scholar]
- Isaacson T, Kosma DK, Matas AJ, et al. Cutin deficiency in the tomato fruit cuticle consistently affects resistance to microbial infection and biomechanical properties, but not transpirational water loss. The Plant Journal. 2009;60:363–377. doi: 10.1111/j.1365-313X.2009.03969.x. [DOI] [PubMed] [Google Scholar]
- Isaacson T, Rose JKC. The plant cell wall proteome, or secretome. In: Finnie C, editor. Plant proteomics. Oxford: Blackwell; 2006. pp. 175–209. [Google Scholar]
- Jerala R. Structural biology of the LPS recognition. International Journal of Medical Microbiology. 2007;297:353–363. doi: 10.1016/j.ijmm.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Jetter R, Kunst L, Samuels AL. Composition of plant cuticular waxes. In: Riederer M, Müller C, editors. Biology of the plant cuticle. Oxford: Blackwell; 2006. pp. 145–181. [Google Scholar]
- Kannangara R, Branigan C, Liu Y, Penfield T, Rao V, Mouille G, Hofte H, Pauly M, Riechmann JL, Broun P. The transcription factor WIN1/SHN1 regulates cutin biosynthesis in Arabidopsis thaliana. The Plant Cell. 2007;19:1278–1294. doi: 10.1105/tpc.106.047076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krolikowski KA, Victor JL, Wagler TN, Lolle SJ, Pruitt RE. Isolation and characterization of the Arabidopsis organ fusion gene HOTHEAD. The Plant Journal. 2003;35:501–511. doi: 10.1046/j.1365-313x.2003.01824.x. [DOI] [PubMed] [Google Scholar]
- Kurdyukov S, Faust A, Nawrath C, et al. The epidermis-specific extracellular BODYGUARD controls cuticle development and morphogenesis in Arabidopsis. The Plant Cell. 2006a;18:321–339. doi: 10.1105/tpc.105.036079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdyukov S, Faust A, Trenkamp S, Bar S, Franke R, Efremova N, Tietjen K, Schreiber L, Saedler H, Yephremov A. Genetic and biochemical evidence for involvement of HOTHEAD in the biosynthesis of long-chain α-,ω-dicarboxylic fatty acids and formation of extracellular matrix. Planta. 2006b;224:315–329. doi: 10.1007/s00425-005-0215-7. [DOI] [PubMed] [Google Scholar]
- Le LQ, Lorenz Y, Scheurer S, Fötisch K, Enrique E, Bartra J, Biemelt S, Vieths S, Sonnewald U. Design of tomato fruits with reduced allergenicity by dsRNAi-mediated inhibition of ns-LTP (Lyc e 3) expression. Plant Biotechnology Journal. 2006;4:231–242. doi: 10.1111/j.1467-7652.2005.00175.x. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Saravanan RS, Damasceno CMB, Yamane H, Kim BD, Rose JKC. Digging deeper into the plant cell wall proteome. Plant Physiology and Biochemistry. 2004;42:979–988. doi: 10.1016/j.plaphy.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Leide J, Hildebrandt U, Reussing K, Riederer M, Vogg G. The developmental pattern of tomato fruit wax accumulation and its impact on cuticular transpiration barrier properties: effects of a deficiency in a beta-ketoacyl-coenzyme a synthase (LeCER6) Plant Physiology. 2007;144:1667–1679. doi: 10.1104/pp.107.099481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Beisson F, Koo AJ, Molina I, Pollard M, Ohlrogge J. Identification of acyltransferases required for cutin biosynthesis and production of cutin with suberin-like monomers. Proceedings of the National Academy of Sciences, USA. 2007;104:18339–18344. doi: 10.1073/pnas.0706984104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Pollard M, Sauveplane V, Pinot F, Ohlrogge J, Beisson F. Nanoridges that characterize the surface morphology of flowers require the synthesis of cutin polyester. Proceedings of the National Academy of Science, USA. 2009;106:22008–22013. doi: 10.1073/pnas.0909090106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JT, Juniper BE. The cuticles of plants. London: Edward Arnold; 1970. 147. [Google Scholar]
- Mintz-Oron S, Mandel T, Rogachev I, et al. Gene expression and metabolism in tomato fruit surface tissues. Plant Physiology. 2008;147:823–851. doi: 10.1104/pp.108.116004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller LA, Solow TH, Taylor N, et al. The SOL Genomics Network: a comparative resource for Solanaceae biology and beyond. Plant Physiology. 2005;138:1310–1317. doi: 10.1104/pp.105.060707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazono M, Qiu F, Borsuk LA, Schnable PS. Laser-capture microdissection, a tool for the global analysis of gene expression in specific plant cell types: identification of genes expressed differentially in epidermal cells or vascular tissues of maize. The Plant Cell. 2003;15:583–596. doi: 10.1105/tpc.008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto U, Fukase K, Miyake K, Satow Y. Crystal structures of human MD-2 and its complex with antiendotoxic lipid IVa. Science. 2007;316:1632–1634. doi: 10.1126/science.1139111. [DOI] [PubMed] [Google Scholar]
- Panikashvili D, Shi JX, Schreiber L, Aharoni A. The Arabidopsis DCR encoding a soluble BAHD acyltransferase is required for cutin polyester formation and seed hydration properties. Plant Physiology. 2009;151:1773–1789. doi: 10.1104/pp.109.143388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Research. 2002;30 doi: 10.1093/nar/30.9.e36. e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pighin JA, Zheng H, Balakshin LJ, Goodman IP, Western TL, Jetter R, Kunst L, Samuels AL. Plant cuticular lipid export requires an ABC transporter. Science. 2004;306:702–704. doi: 10.1126/science.1102331. [DOI] [PubMed] [Google Scholar]
- Plant AL, Cohen A, Moses MS, Bray EA. Nucleotide sequence and spatial expression pattern of a drought- and abscisic acid-induced gene of tomato. Plant Physiology. 1991;97:900–906. doi: 10.1104/pp.97.3.900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard M, Beisson F, Li Y, Ohlrogge JB. Building lipid barriers: biosynthesis of cutin and suberin. Trends in Plant Science. 2008;13:236–246. doi: 10.1016/j.tplants.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Pyee J, Yu H, Kolattukudy PE. Identification of a lipid transfer protein as the major protein in the surface wax of broccoli (Brassica oleracea) leaves. Archives of Biochemistry and Biophysics. 1994;311:460–468. doi: 10.1006/abbi.1994.1263. [DOI] [PubMed] [Google Scholar]
- Qin Q, Bergmann CW, Rose JKC, Saladié M, Kolli VS, Albersheim P, Darvill AG, York WS. Characterization of a tomato protein that inhibits a xyloglucan-specific endoglucanase. The Plant Journal. 2003;34:327–338. doi: 10.1046/j.1365-313x.2003.01726.x. [DOI] [PubMed] [Google Scholar]
- Riederer M. Introduction: biology of the plant cuticle. In: Riederer M, Müller C, editors. Biology of the plant cuticle. Oxford: Blackwell; 2006. pp. 1–10. [Google Scholar]
- Reina JJ, Guerrero C, Heredia A. Isolation, characterization, and localization of AgaSGNH cDNA: a new SGNH-motif plant hydrolase specific to Agave americana L. leaf epidermis. Journal of Experimental Botany. 2007;58:2717–2731. doi: 10.1093/jxb/erm136. [DOI] [PubMed] [Google Scholar]
- Ruzin SE. Plant microtechnique and microscopy. Oxford: Oxford University Press; 1999. [Google Scholar]
- Saldanha AJ. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Samuels L, Kunst L, Jetter R. Sealing plant surfaces: cuticular wax formation by epidermal cells. Annual Review of Plant Biology. 2008;59:683–707. doi: 10.1146/annurev.arplant.59.103006.093219. [DOI] [PubMed] [Google Scholar]
- Schneiderbauer A, Sandermann HJ, Ernst D. Isolation of functional RNA from plant tissues rich in phenolic compounds. Analytical Biochemistry. 1991;197:91–95. doi: 10.1016/0003-2697(91)90360-6. [DOI] [PubMed] [Google Scholar]
- Shepherd RW, Wagner GJ. Phylloplane proteins: emerging defenses at the aerial frontline? Trends in Plant Science. 2007;12:51–56. doi: 10.1016/j.tplants.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Analytical Chemistry. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Srinivasasainagendra V, Page GP, Mehta T, Coulibaly I, Loraine AE. CressExpress: a tool for large-scale mining of expression data from Arabidopsis. Plant Physiology. 2008;147:1004–1016. doi: 10.1104/pp.107.115535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh MC, Samuels AL, Jetter R, Kunst L, Pollard M, Ohlrogge J, Beisson F. Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiology. 2005;139:1649–1665. doi: 10.1104/pp.105.070805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogg G, Fischer S, Leide J, Emmanuel E, Jetter R, Levy AA, Riederer M. Tomato fruit cuticular waxes and their effects on transpiration barrier properties: functional characterization of a mutant deficient in a very-long-chain fatty acid β-ketoacyl-CoA synthase. Journal of Experimental Botany. 2004;55:1401–1410. doi: 10.1093/jxb/erh149. [DOI] [PubMed] [Google Scholar]
- Yang Y, Zhang S, Howe K, Wilson DB, Moser F, Irwin D, Thannhauser TW. A comparison of nLC-ESI-MS/MS and nLC-MALDI-MS/MS for GeLC-based protein identification and iTRAQ-based shotgun quantitative proteomics. Journal of Biomolecular Techniques. 2007;18:226–237. [PMC free article] [PubMed] [Google Scholar]
- Yeats TH, Rose JKC. The biochemistry and biology of extracellular plant lipid-transfer proteins (LTPs) Protein Science. 2008;17:191–198. doi: 10.1110/ps.073300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.