Abstract
We previously reported a VEGF autocrine loop in head and neck squamous cell carcinoma (HNSCC) cell lines, supporting a role for VEGF in HNSCC tumorigenesis. Using a phosphotyrosine proteomics approach we screened the HNSCC cell line, SCC-9 for effectors of VEGFR2 signaling. A cluster of proteins involved in cell migration and invasion, including the p130Cas paralog, human enhancer of filamentation1 (HEF1/Cas-L/Nedd9) was identified. HEF1 silencing and overexpression studies revealed a role for VEGF in regulating cell migration, invasion, and MMP expression in a HEF1-dependent manner. Moreover, cells plated on extracellular matrix coated coverslips exhibited enhanced invadopodia formation in response to VEGF that was HEF1-dependent. Immunolocalization revealed that HEF1 colocalized to invadopodia with MT1-MMP. Analysis of HNSCC tissue microarrays for HEF1 immunoreactivity revealed a 6.5-fold increase in the odds of having a metastasis with a high HEF1 score compared to a low HEF1 score. These findings suggest that HEF1 may be prognostic for advanced stage HNSCC. They also demonstrate for the first time, that HEF1 is required for VEGF-mediated HNSCC cell migration and invasion, consistent with HEF1’s recent identification as a metastatic regulator. These results support a strategy targeting VEGF:VEGFR2 in HNSCC therapeutics.
Keywords: VEGF, Invadopodia, Cell migration, invasion, HEF1, NEDD9, CAS-L, tyrosine phosphorylation
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the 6th most prevalent cancer worldwide (Jemal et al., 2004). Within the US it is estimated that ~30,000 new cases will be diagnosed and over 7,000 people will die from HNSCC per year, with a significant number of cases presenting at an advanced stage with metastases, thus resulting in poor clinical outcomes (Ries et al., 2004). A number of growth factors have the capacity to drive HNSCC cells to a more metastatic phenotype (Cooney et al., 2005; Kyzas et al., 2005; Michi et al., 2000), including epidermal growth factor (EGF), insulin-like growth factor-1 (IGF-1) and vascular endothelial growth factor (VEGF) (Bigbee et al., 2007). We identified an IGF-1-stimulated VEGF autocrine loop in HNSCC cells (Slomiany et al., 2006) and showed that IGF-1R crosstalk to EGFR activation may contribute to enhanced tumorigenesis and invasiveness of HNSCC (Slomiany et al., 2007).
To better understand the contributions of VEGF signaling to HNSCC progression, a more detailed analysis of the molecular mechanisms regulating HNSCC cell tumorigenesis, migration and invasion is needed. VEGF action in endothelial cells leading to angiogenesis has been studied intensively (Ferrara, 2004). On the other hand, VEGF action on tumor cells has only recently been described (Bachelder et al., 2003). To that end, we applied a mass spectrometry-based screen of VEGF action as applied to the identification of EGF receptor effectors (Steen et al., 2002). The rationale for this “broad net approach” is that mass spectrometry serves as an ideal tool for discovery over conventional methods, enabling identification of site-specific posttranslational modifications and cleavages when applied to the study of signaling proteins (Kratchmarova et al., 2002).
Using three treatment groups: VEGF; VEGF plus the VEGFR2 tyrosine kinase inhibitor SU5416; untreated control, we carried out a phosphotyrosine proteomics screen of the oral squamous cell carcinoma (SCC) cell line, SCC-9. We identified a cluster of proteins involved in focal adhesions, cell migration and invasion in the VEGF treated sample including: FAK (focal adhesion kinase), paxillin, cortactin and HEF1 (human enhancer of filamentation 1 (Singh et al., 2007)). Significantly, HEF1 (Nedd9/Cas-L; neural precursor cell expressed, developmentally down-regulated 9/Crk-associated substrate-L) expression has been associated with metastases signatures in glioblastoma (Natarajan et al., 2006), breast cancer (Izumchenko et al., 2009) and melanoma (Kim et al., 2006), providing a potential link between VEGF signaling and metastatic progression. This led us to the hypothesis that HEF1 effects the invasive actions of VEGF. Based on HEF1 silencing and overexpression, we found that VEGF-induced SCC-9 cell migration and invasion were HEF1-dependent. When SCC-9 cells were seeded on extracellular matrix coated coverslips, VEGF stimulated invadopodia formation, which was blocked by HEF1 siRNA treatment. Confocal microscopy revealed a punctate distribution of actin, cortactin, and phosphotyrosine underlying the nucleus and localized to the ventral surface of VEGF treated cells. Moreover, HEF1 was present within invadopodia, colocalizing with MT1-MMP, providing the first evidence for HEF1 involvement in invadopodia formation. Analysis of a human HNSCC tissue microarray revealed elevated HEF1 staining in advanced stage HNSCC. Our findings indicate that VEGF regulates invasive signaling paradigms in HNSCC and that HEF1 is a downstream mediator of these effects. We propose a duality of action, whereby VEGF enhances tumor cell invasive signaling and provides a means for extravasation during metastasis (Ferrara, 2004).
Results
VEGF promotes phosphorylation of proteins involved in migration and invasion
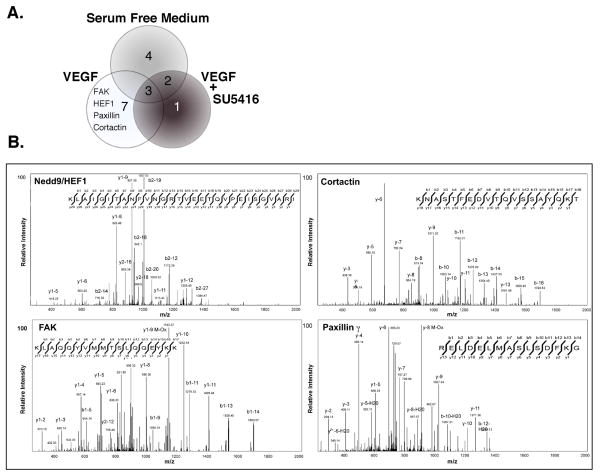
To further define VEGF signaling in head and neck cancer (Slomiany et al., 2006), we examined VEGFR2 expression in HNSCC cell lines. VEGFR2 was expressed in all cell lines tested and exhibited a time-dependent phosphorylation in response to VEGF (10 ng/ml; Supplementary Information (SI)). To define downstream effectors mediating VEGF signaling in a global fashion, we applied a phosphotyrosine proteomics screen of SCC-9 cells (serum-starved, VEGF treated, and VEGF treated plus 50 μM SU5416). Whole cell lysates from each sample were enriched for phosphotyrosine containing proteins by affinity adsorption to anti-pY (phosphotyrosine) antibodies immobilized on beads. Beads were washed and the eluted proteins digested with trypsin and analyzed by LC MS/MS. Of the more than 750 spectra obtained, 16 proteins met our significance criteria, ((Rush et al., 2005); Table 1, SI). This required discarding MS3 data due to insufficient quality, which would have identified specific sites of phosphorylation. Three proteins were common to all three treatment groups signifying they were tyrosine phosphorylated under basal conditions in SCC-9 cells. Four proteins were unique to the untreated group while two proteins were shared between the unstimulated and VEGF plus SU5416 group. Nine proteins were identified in the VEGF treated sample; 7 were specific to the VEGF stimulated sample and 6 were confirmed to contain phosphotyrosine. These included FAK (focal adhesion kinase), paxillin-γ, cortactin and HEF1 (Fig. 1b).
Table 1.
Quantification of HEF1 staining in HNSCC tumor tissue microarray.
| HEF1 Score | 0 | 1 | 2 | 3 | Raw Total |
|---|---|---|---|---|---|
| No Metastasis | 13 | 6 | 4 | 0 | 23 |
| Metastasis | 2 | 5 | 6 | 1 | 14 |
| Column Total | 15 | 11 | 10 | 1 | 37 |
| IHC HEF1 Score | |||||
| Stage | 0 | 1 | 2 | 3 | Raw Total |
| 1 | 8 | 3 | 0 | 0 | 11 |
| 2 | 1 | 0 | 1 | 0 | 2 |
| 3 | 3 | 1 | 3 | 0 | 7 |
| 4 | 8 | 7 | 4 | 4 | 23 |
| 5 | 5 | 3 | 4 | 2 | 14 |
| Column Total | 25 | 14 | 12 | 6 | 57 |
Tissue microarrays were stained for HEF1 as detailed in Methods. Additional HEF1 antibody validation using SCC-25 and SCC-9 cells can be found in Fig. 3, SI. HEF1 staining was differentially distributed across samples from subjects subdivided by the presence of metastasis. HEF1 staining was further differentially distributed across multiple stages.
Figure 1. Screen for VEGFR2 effectors using phosphotyrosine proteomics.
(A) The Venn diagram illustrates the number of proteins identified within treatment group analyzed. (B) SCC-9 cells were treated with 10 ng/ml VEGF for 10 min as indicated. Cell lysates were enriched for pY-containing proteins, subjected to proteolytic digestion and subsequent analysis by LC-MS/MS. Representative spectra were taken from the VEGF treated sample of those proteins involved in cancer cell tumorigenesis. Additional tyrosine phosphorylated proteins detected but not further characterized include valosin-containing protein, α-tubulin, HNRPF, medium-chain acyl-CoA dehydrogenase and uraDNA glycosylase (see Table 1D, SI).
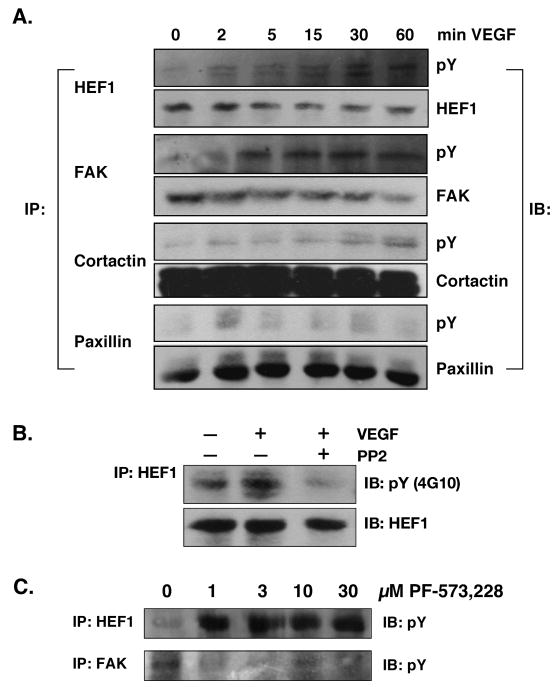
These four proteins are known to be regulated by tyrosine phosphorylation (Brabek et al., 2005); HEF1 is a FAK and Src substrate (Tachibana et al., 1997) and cortactin is a Src substrate (Tehrani et al., 2007). All four proteins were tyrosine phosphorylated in response to VEGF in SCC-9 cells (Fig. 2a) with HEF1 phosphorylation being blocked by the Src family kinase inhibitor, PP2 (Fig 2b). Interestingly, the FAK tyrosine kinase inhibitor, PF-573,228 (Slack-Davis et al., 2007) did not alter HEF1 tyrosine phosphorylation (Fig 2c), supporting a role for Src in mediating HEF1 phosphorylation (Ruest et al., 2001).
Figure 2. VEGF induces tyrosine phosphorylation of proteins identified in the phosphotyrosine proteomics screen.
(A) SCC-9 cells were serum starved overnight followed by treatment with VEGF (10 ng/ml) for the times shown. Cell lysates were immunoprecipitated (IP) with antibodies to the individual proteins and immunoblotted (IB) with anti-phosphotyrosine (anti-pY) as indicated. (B) HEF1 tyrosine phosphorylation in SCC-9 cells in response to VEGF (10 ng/ml; 15 min) was abrogated by the Src family kinase inhibitor PP2 (1 μM). (C) Effect of FAK inhibition on HEF1 tyrosine phosphorylation. HEF1 tyrosine phosphorylation was unaffected by the FAK-specific tyrosine kinase inhibitor PF-573,228. These experiments were performed at least three times with the same results.
HEF1 is required for VEGF-induced cell migration and invasion
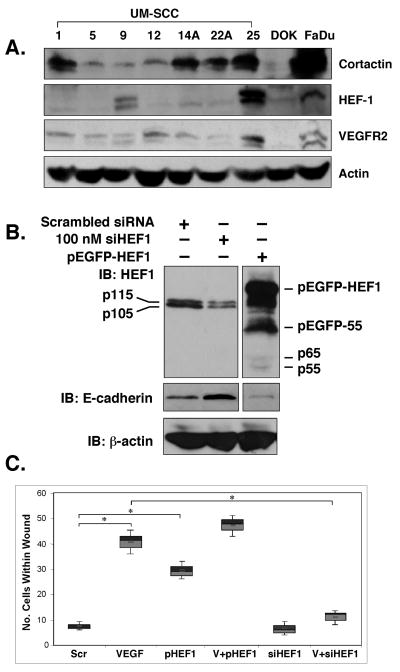
Because a role for HEF1 in VEGF signaling in HNSCC has not been described, we examined HEF1 function in HNSCC cells. SCC-9 cell HEF1 protein expression was moderate, when compared to a panel of other HNSCC cell lines, with SCC-25 cells exhibiting the highest HEF1 levels (Fig. 3a). Of note, SCC-9 cell cortactin levels were the lowest of all cell lines examined, consistent with a previous report ((Timpson et al., 2005); Fig. 3a). HEF1 silencing in SCC-9 cells significantly reduced HEF1 protein levels, while HEF1 overexpression enabled visualization of its otherwise undetectable protease-induced isoforms (Fig. 3b). Significantly, we observed that HEF1 elevation led to decreased E-cadherin expression and conversely, HEF1 silencing led to increased E-cadherin levels (Fig. 3b). Decreases in E-cadherin expression accompany loss of cell:cell contact, epithelial-mesenchymal transition (EMT) and increased cell motility (Guarino et al., 2007). Consistent with this observation, HEF1 silencing abrogated, while HEF1 overexpression enhanced SCC-9 cell migration (Fig. 3c).
Figure 3. HEF1 expression augments VEGF stimulated cell migration.
(A) Immunoblot analysis of HEF1 expression in HNSCC cell lines. (UM-SCC lines were obtained from Dr. Tom Carey, University of Michigan. (B) HEF1 overexpression (pEGFP-HEF1) and silencing with siRNA (siHEF1) reciprocally alters E-cadherin expression. (C) Scratch/wound assays demonstrate that VEGF stimulated cell migration is HEF1-dependent. Scr Scrambled siRNA, pHEF1 HEF1 transfected only, V+HEF1 10ng/ml VEGF treatment and HEF1 transfection, V+siHEF1 10 ng/ml VEGF treated and HEF1 siRNA transfected.
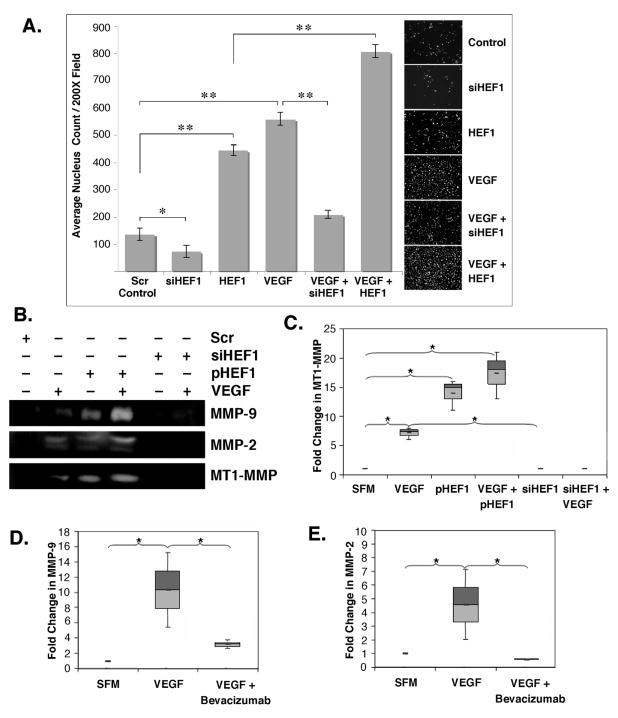
As shown in Fig. 4a, using matrigel-coated transwell filter assays, VEGF stimulated invasive activity was HEF1-dependent, supporting a role for VEGF acting via HEF1 in HNSCC metastatic signaling. Because increased invasive behavior is associated with elevated matrix metalloproteinase (MMP) activity, we tested the effect of VEGF treatment on MT1-MMP (MMP-14), MMP-2 and MMP-9 activities. VEGF treatment increased the activities of all three MMPs (Fig. 4b). MT1-MMP activity was significantly increased in cells overexpressing HEF1, and completely abrogated following HEF1 silencing (Fig. 4c). Further, VEGF stimulated MMP-9 and MMP-2 activities were inhibited with the VEGF neutralizing monoclonal antibody, bevacizumab (Fig. 4d and 4e). These findings suggest that VEGF induced MMP activity is dependent on HEF1 expression. Fashena and coworkers (Fashena et al., 2002) reported HEF1 overexpression increased MMP-1, 8, 12, 13 and 14 mRNA levels supporting a role for HEF1 in cancer progression.
Figure 4. VEGF enhances cell invasion and MMP expression via HEF1.
(A) SCC-9 cells seeded on matrigel-coated transwell filters were treated as indicated for 24 h. Filters were collected and cells quantified as detailed in Methods. VEGF stimulated invasion was increased by HEF1 transfection and attenuated by HEF1 silencing, * p<0.05; ** p<0.005. (B) MMP-2, -9, and MT1-MMP expression were increased by HEF1 transfection and VEGF addition and blocked with HEF1 siRNA. (C) HEF1 significantly increased VEGF-induced MT1-MMP expression. (D) VEGF stimulated and bevacizumab blocked MMP-9 and MMP-2 expression. All experiments were repeated a minimum of three times.
HEF1 is involved in invadopodia structure and function
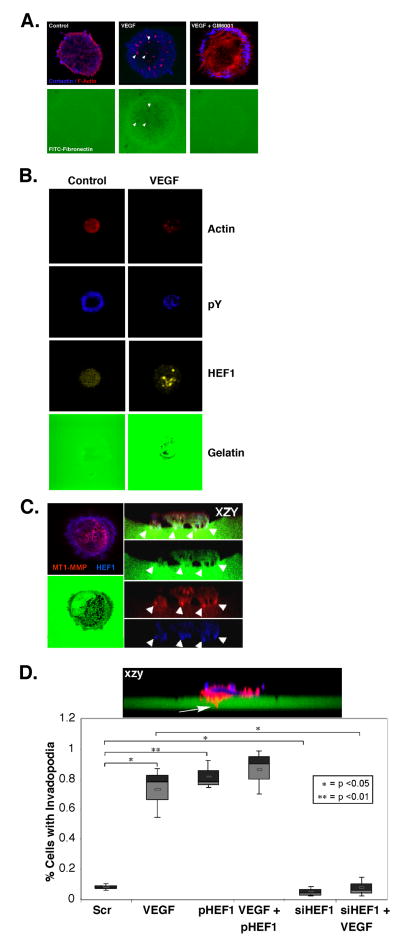
Invadopodia are cellular structures responsible for the focal delivery of MMPs to the ventral surface of cells enabling cellular invasion and metastatic progression (Gimona et al., 2008; Linder, 2007). Based on VEGF’s ability to increase SCC-9 cell motility, invasion and MMP activation, we reasoned that it may regulate these processes via invadopodia formation. Cortactin is a key component of invadopodia (Linder, 2007) and necessary for their formation (Ayala et al., 2008; Clark et al., 2007). We therefore tested whether HEF1 participates in invadopodia formation. SCC-9 cells were seeded onto FITC-fibronectin- coated coverslips and examined by confocal microscopy following stimulation with VEGF. VEGF treatment caused a redistribution of actin from a uniform cytoplasmic staining to a punctate distribution, localized below the nucleus on the ventral side of the cell, consistent with invadopodia (Fig 5a). This conclusion was supported by of foci of matrix digestion, or black holes within the FITC-labeled matrix, underlying actin punctae, that were absent in untreated cells (Fig. 5a). Pretreatment with the broad spectrum MMP inhibitor, GM6001, blocked the VEGF effects on actin distribution and matrix digestion (Ayala et al., 2008). VEGF treatment resulted in HEF1 redistribution into central zones of punctate staining, overlying areas of matrix digestion. This corresponded to the staining seen for actin and phosphotyrosine, strongly suggesting HEF1 localization within invadopodia (Fig. 5b). Over-expression of HEF1 in SCC-9 cells treated with VEGF yielded a saw tooth pattern of digestion with numerous invadopodia (Fig. 5c). MT1-MMP was present in regions of matrix digestion, colocalizing with HEF1 within invadopodia tips (Fig. 5c). These findings strongly suggest that VEGF induces the redistribution of HEF1 from generalized plasma membrane localization to the tips of invadopodia. VEGF stimulated the number of invadopodia formed and HEF1 depletion reduced invadopodia formation, in support of this process being HEF1-dependent (Fig. 5d).
Figure 5. VEGF stimulates invadopodia formation.
(A) SCC-9 cells seeded on FITC-gelatin-coated coverslips were treated and stained as indicated. The punctate staining pattern observed in response to VEGF stimulation was lost upon co-treatment with the MMP inhibitor, GM6001. (B) The pY staining of VEGF stimulated cells exhibited a central, punctate distribution, similar to that seen for actin and HEF1. (C) xyz and xzy projection of SCC-9 cells following VEGF stimulation and HEF1 overexpression. Note: HEF1 co-localization with MT1-MMP at invadopodia tips in this xzy visualization of ventral invadopodia projecting into the matrix. (D) SCC-9 cells seeded on FITC-labeled- FN coverslips were treated for 12 h as indicated. Invadopodia formation (inset, top) were identified and quantified.
HEF1 expression is linked to advanced stage HNSCC
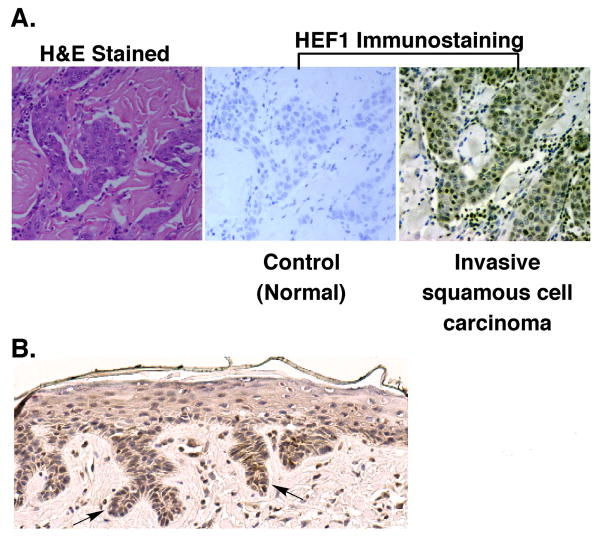
To translate our in vitro findings to human disease, we quantified HEF1 expression in HNSCC tissue specimens. HEF1 staining was greatest within invasive nests of advanced stage cancers (Fig. 6a). For analysis, samples were split into two groups based on staging data: those with known distant/lymph node metastasis and those with no or no known metastasis. These groups were similar in bivariate analysis with respect to all covariates except those containing staging information. Covariates required for our analysis include age, sex, race, gender, and grade and staging information. Subjects were included only if all the above covariates were known. Out of 200 subject biopsies, 37 met these criteria. Using a HEF1 score of 2, HEF1 staining was dichotomized into high vs. low groups to allow for a clinical decision oriented analysis. Our analyses show a 6.88-fold increase (95% CI 1.47-32.58) in the odds of having a metastasis with a high HEF1 score compared to those with a low HEF1 score. These results indicate that elevated HEF1 levels correlate with advanced stage HNSCC (Table 1).
Figure 6. HEF1 staining in human tissue.
(A) (Upper)Tissue sections were stained with H&E, secondary antibody alone (Negative Control), and primary HEF1 antibody with secondary antibody. (B) Representative staining from each level of positivity observed from the TMA.
Discussion
In this report we applied a phosphotyrosine proteomics screen to the analysis of VEGF signaling in HNSCC. Tandem MS data allowed the sequencing of a large number of peptides and the identification of 17 proteins (Table 1 Supplementary Information) among three experimental groups. HEF1, FAK, paxillin and cortactin were identified as targets of VEGF action in SCC-9 cells. These proteins have roles in cell migration and invasion (Brown and Turner, 2004; Buday and Downward, 2007; Cohen and Guan, 2005; Lua and Low, 2005; Mon et al., 2006; Sabe et al., 2006; Sloan and Anderson, 2002; van Nimwegen and van de Water, 2007), with cortactin having been reported to be dysregulated in HNSCC (Canel et al., 2006; Clark et al., 2007; Patel et al., 1996). Of note, cortactin is frequently overexpressed in breast cancer (Ormandy et al., 2003) and HNSCC (Rothschild et al., 2006) due to amplification of chromosomal region 11q13 (Patel et al., 1996; Rothschild et al., 2006). Cortactin overexpression and posttranslational modification correlate with its redistribution from the cytoplasm to focal adhesions, consistent with its role in modulating cell adhesion.
Identification of HEF1 in this screen was noteworthy, as HEF1 association with the metastatic signatures of malignant melanoma (Kim et al., 2006), and glioblastoma (Natarajan et al., 2006) and lung cancer (Ji et al., 2007) have been reported. Given that the HEF1 paralog p130Cas facilitates cancer cell migration and invasion (Nakamoto et al., 2000), we tested whether HEF1 influenced VEGF signaling in HNSCC. VEGF stimulated cell migration, invasion, MMP expression and invadopodia formation, making this the first report defining a role for VEGF in regulating HNSCC tumorigenicity. These actions were each facilitated by and dependent upon HEF1 expression. In contrast to our findings, Simpson et al., (Simpson et al., 2008) reported that HEF1 silencing increased MCF10A cell motility. Similarly, HEF1 down-regulation was reported as part of the metastatic transcriptome signature in a TGF-β model of breast cancer metastasis to lung (Minn et al., 2005). Izumchenko and coworkers (Izumchenko et al., 2009) showed that Nedd9 null mice had significantly lower tumor incidence . These differing findings in alternate models may be the result of cell type specific differences or the presence of alternate pathways regulating tumor cell progression and metastasis (Sanz-Moreno et al., 2008). Additional support for HEF1 in cell migration/invasion comes from studies of multiple cancer cell types (Singh et al., 2008), metastatic melanoma (Kim et al., 2006), breast cancer (Izumchenko et al., 2009), lung cancer (Ji et al., 2007) and head and neck cancer (Yu et al., 2008).
Cortactin (Linder, 2007) and paxillin (Badowski et al., 2008) are present in focal adhesions, podosomes and invadopodia, whereas FAK is excluded from invadopodia (Bowden et al., 2006). The dynamic affiliations of these proteins in the formation and maintenance of these organelles and whether they form a continuum is the subject of intense research (Gimona et al., 2008). HEF1 localizes to focal adhesions, the primary cilium and the centrosome but its precise role in invasion/metastasis relative to invadopodia has not been defined (OșNeill et al., 2007). The C-terminal domain of HEF1 contains a sequence that is structurally homologous to the focal adhesion targeting (FAT) domain within the C-terminus of FAK (Arold et al., 2002). This domain targets FAK to paxillin in focal adhesions. FAK contains an acceptor site (Y925) for Grb2 that is lacking in HEF1. These differences may account for the alternate functions/localizations of these proteins. Thus, if a continuum exists between these three structures, there are biochemical differences maintained that might be responsible for their differing functions. It is tempting to speculate that the presence of HEF1 in invadopodia might fulfill a unique structural role effected by selective protein interactions. It is noteworthy that Alexander et al., (Alexander et al., 2008) identified p130Cas in invadopodia. We did note that HEF1 silencing reduced VEGF-induced cell migration, invasion, MMP expression and invadopodia formation while having no effect on p130Cas levels (SI Fig. 2). Additional studies are required to define the roles of p130Cas vs. HEF1 in invadopodia structure and function.
Several lines of evidence support a role for HEF1 in invadopodia formation. 1) HEF1 depletion reduced and HEF1 overexpression increased invadopodia formation; 2) VEGF induced invadopodia formation was dependent upon HEF1 expression; 3) HEF1 depletion reduced MMP-2, MMP-9 and MT1-MMP expression; 4) confocal analysis revealed HEF1 as a resident invadopodia protein where it colocalized with MT1-MMP. Prior to this report, HEF1 had not been implicated as an invadopodia protein or a regulator of invadopodia structure/function. Our findings suggest that HEF1 associates with invadopodia and influences their formation.
Cortactin is tyrosine phosphorylated and localizes to the base of invadopodia where it regulates Arp2/3 mediated actin polymerization and formation of invadopodia projections into the ECM at the ventral surface of cells (Linder, 2007). It has been suggested that cortactin is required for invadopodia formation and involved in MMP delivery to these sites (Ayala et al., 2008; Clark et al., 2007). Our findings suggest that HEF1 may function in combination with or independently of cortactin; this will require further studies to resolve. If one or both of these proteins can target MMPs to forming invadopodia, it may be via directing MMP containing vesicles to invadopodia. This implies a chaperone function mediated by specific protein:protein interactions with vesicle membrane proteins. MMP delivery to the ventral plasma membrane is one of the stimuli driving invadopodia formation (Weaver, 2008), with inhibition of MMP action abrogating invadopodia formation and function (Ayala et al., 2008; Clark et al., 2007).
Analysis of human HNSCC tissue microarrays revealed significantly higher HEF1 staining in advanced stage tumor tissue that we interpret as indicative of its unique role in invasive signaling. These results suggest that HEF1, as reported for melanoma, glioblastoma and lung cancer, might also be a signature protein in HNSCC metastatic progression. They further indicate that elevated HEF1 levels reflect a poor prognosis potentially serving as a biomarker. Taken together with the activities we observed for HEF1 in this report, it is likely that elevations in HEF1 enhance EMT, MMP expression, invadopodia formation and invasive spread.
In summary, using a proteomics approach to examine VEGF signaling in HNSCC, we identified HEF1 as a mediator of invasive signaling pathways. In support of this role, we demonstrated that MMP-2, MMP-9 and MT1-MMP activity were dependent upon HEF1 expression, as was the ability of VEGF to stimulate cell invasion. VEGF-induced invadopodia formation was HEF1-dependent suggesting a mechanism whereby MMPs are delivered to and released in a localized manner at invadopodia to effect cell invasion (Linder, 2007). We further showed that elevated HEF1 levels are indicative of a poor prognosis in HNSCC, providing insight into the mechanism by which HEF1 may contribute to the metastatic process. It will be important to determine the HEF1 domains and sites of tyrosine phosphorylation required for migration, invasion and invadopodia formation.
Methods and Materials
Materials and Reagents
DMEM, iodoacetamide and mitomycin C were from Sigma (St. Louis, MO). ZM323881and SU5416 were purchased from Calbiochem (La Jolla, CA). BCA reagent was obtained from Pierce (Rockford, IL). VEGF was generously provided by Genentech, Inc. (San Francisco, CA). PF-573,228 was a gift from Pfizer, Inc. (Groton, CT). 4G10 Anti-phosphotyrosine (pY) antibody and 4G10-agarose, FAK, paxillin, cortactin and HRP-conjugated antibodies and Re-Blot Plus were obtained from Millipore (Temecula, CA). HEF1 (2G9) monoclonal, HEF1 (H10) and VEGFR2/flk-1 antibodies were from Santa Cruz (Santa Cruz, CA). ECL Reagent and CH-Sepharose beads were obtained from GE Biosciences (Piscataway, NJ). HEF1 constructs were generously provided by Dr. Erica Golemis (Fox Chase Cancer Center). All other chemicals were of reagent grade or higher.
Tissue Culture
SCC-9 cells (ATCC, Manassas, VA) were maintained as described at 37° C in a humidified 5% CO2-95% air incubator (Rheinwald and Beckett, 1981). For all experiments, SCC-9 cells were seeded in 150 cm dishes, grown overnight to 80-95% confluency, and serum starved for 24 h before the treatment. Following pretreatment for 2 h with fresh serum free medium (SFM) containing inhibitors solubilized in ethanol, cells were treated with ethanol vehicle, inhibitors, and VEGF.
Immunoblot Analysis
Standard immunoblot analysis of whole cell lysates was carried out as previously described (see Supplementary Information; (Slomiany et al., 2006)).
Immunoadsorption and Trypsin Digestion of Tyrosine Phosphorylated Proteins
Cleared cell lysates were diluted 10-fold with modified RIPA buffer containing 2 mM sodium orthovanadate and 10 mM NaF to 0.1% Triton X-100 and 4G10 anti-pY antibody-agarose was added (250 μl beads per 30 mg protein). Details of the collection of the tyrosine phosphorylated proteins and treatment prior to mass spectrometric analysis can be found in Supplementary Information.
MALDI and LC/MS/MS Analysis
For MALDI analysis, samples were mixed with 10 mg/ml α-cyano-4 hydroxy cinnamic acid in 70% acetonitrile containing 0.1% TFA and analyzed using a Voyager DE STR Proteomics workstation (Applied Biosystems, Foster City CA). For electrospray analyses, samples were loaded onto a C18 nano-capillary column (3.5 μm particle size, 15 × 0.75 i.d cm) and eluted with a 245 min gradient from 5% acetonitrile/95% H20/0.2% formic acid to 95% acetonitrile/5% H20/0.2% formic acid. Samples were ionized in-line from the column with a nanospray ion source on an LTQ ion trap mass spectrometer (ThermoFinnigan, Waltham MA). Tandem mass spectra were collected in data dependent neutral loss scanning mode using a top 5 method with a dynamic exclusion repeat of 2 and a repeat duration of 0.5 min.
Wound Healing Assay
SCC-9 cells were grown to confluency and subsequently serum starved for 24 h. Following pretreatment with 10 μg/ml mitomycin C for 2 h, a scratch was introduced into the monolayer using a sterile 20-200 μl pipet tip. All treatments were added along with mitomycin C. Phase contrast images were taken at 0, 6, 12 and 24 h with a computer controlled microscope.
Invasion Assay
Serum-starved cells were plated in the top chamber of matrigel-coated transwell filters in 24 well plates at a density of 30×103 cells/well for 1 h. SFM in the upper and lower chambers was exchanged for fresh SFM containing the treatment indicated and the cells incubated for 24 h. To terminate the experiment, each filter was rinsed with PBS and stained with a 1:10,000 dilution of Hoechst dye in PBS for 10 min. The top portion of each filter was swabbed with a Q-tip and washed three times with PBS. Cells on the filter bottoms were imaged with a DAPI filter and nuclei were counted using Cell Profiler v1.0.4942 (Carpenter et al., 2006).
Gelatin Zymography
Two ml of conditioned medium (CM) was collected at various times, spin dialyzed and concentrated in Centricon microconcentrators. Alternatively, MMPs were enriched by affinity adsorption on gelatin-Sepharose (Descamps et al., 2002). Gelatin-Sepharose (Supplementary Information; 50 μl) was mixed at a 1:3 ratio with CM for 1 h, rinsed with equilibration buffer containing 0.05% Tween followed by equilibration buffer without NaCl. SDS sample buffer lacking DTT was added and the proteins were resolved on 7.5% acrylamide, SDS gels containing polymerized gelatin (2 mg/ml). MMPs were renatured using two 30 min detergent exchange washes. Gels were incubated in enzyme buffer for 6-16 h at 37°C, stained with Coomassie blue, destained and imaged using a Bio-Rad FlourS-Multimager.
Transfection of SCC-9 Cells
Cell suspensions were reconstituted in 100 μl of Lonza Nucleofection Solution V (Lonza, Gaithersburg MD). siRNA or plasmid DNA were added as indicated and nucleofection performed using program T-020 of an Lonza Nucleofector II. Cells were quickly resuspended in medium containing 20% FBS and allowed to recover. siRNA transfectants were analyzed in experiments on the third day following transfection, while plasmid transfectants were assayed the following day.
Immunohistochemistry
5 μm sections were de-paraffinized, heat treated, blocked in serum specific to the secondary antibody used and labeled with antibodies for HEF1 (Cas-L H-70, Santa Cruz Biotech, Santa Cruz CA) at a dilution of 1:50 in blocking buffer. Biotin-labeled secondary antibodies (1:200 dilution) were added for 1 h followed by washing and counterstaining the sections with H&E for 10 sec. This was followed by washing and dehydration in an ethanol series ending with xylene. Coverslips were mounted on slides and sealed for microscopy and subsequent digitization.
Invadopodia Assay
Coverslips (#1) were coated with 80 μl of warmed gelatin (comprised of fluorescently labeled:unlabeled gelatin or fibronectin at a 1:8 ratio). Coated coverslips were allowed to solidify and dry to a thin film at 23°C in the dark. Coated coverslips were cross-linked with 4% paraformaldehyde for 10 min, reduced with 1 ml of 10% Na borohydride, and washed 3 times with PBS. Coverslips were then equilibrated in SFM containing 10 μM GM6001 for at least 15 min. SCC-9 cells were trypsinized with 0.25% trypsin-EDTA, suspended and mixed with sterile 2.5% bovine serum albumin (BSA) to neutralize trypsin, and centrifuged at 500 × g for 5 min. Cells were resuspended in SFM and coverslips seeded at a low density (final GM6001 conc., 5 μM). Cells were allowed to adhere overnight and attached cells were gently washed with SFM. Cells were treated as indicated in SFM for 3-6 h. Invadopodia, defined as foci of gelatin degradation (regions of decreased gelatin fluorescence) and increased F-actin and cortactin fluorescence intensity, were quantified as: 1) % cells containing invadopodia and 2) the average number of invadopodia per cell. Three repetitions were performed to calculate the mean percentage of invadopodia/cell and a minimum of 50 cells were counted in three assays to determine the average number of invadopodia/cell.
Immunocytochemistry
Following treatment, cells were rinsed in PBS, fixed in 4% paraformaldehyde for 10 min and permeabilized with 0.1% Triton X-100 for 3 min. Washed cells were blocked with 1% BSA in PBS for at least 30 min. Cells were next incubated with 100 μl of primary antibody diluted in 1% BSA and 0.1% Triton X-100 in PBS for 1 hr, washed and incubated with secondary antibody (1:200) in the same solution for 1 h. Coverslips were then washed with PBS, inverted onto a glass slide with anti-bleaching sealant solution and sealed with fingernail polish. Labeled cells were imaged on a Leica TCS SP2 AOBS laser scanning confocal microscope with a 458, 477, 488 and 514-nm Argon laser, 543-nm He-Ne laser, and 633-nm He-Ne laser, in the xyz and xzy planes. Images were prepared for publication using Leica Imaging Software and Gnu Image Manipulation Program (GIMP) v2.6.
Tissue Microarray Analysis
HNSCC tissue microarrays (formalin fixed-paraffin embedded tissues comprised of 472 cases and 27 controls) obtained from Drs. Vyomesh Patel and J. Silvio Gutkind (NIDCR/NIH), (Molinolo et al., 2007) were stained for HEF1expression levels in order to assess HEF1 as a biomarker predictive of HNSCC metastasis (see Supplementary Information for details). HNSCC tumor array slides were analyzed by HEF1 immunohistochemistry. The number of positive cells was visually evaluated for each core and the results were expressed as a percentage of stained cells/total number of cells. According to their immunoreactivity, the tissue array cores were divided into three categories: 0, less the 10% of stained cells; 1, between 10% and 25%; 2, between 25% and 50%; 3, 75% to 100%.
Statistical Analysis
Summary statistics for all continuous variables are presented as means and standard deviations. Categorical data are summarized as frequencies and percentages. Differences in baseline characteristics between the local disease group and advanced disease group were analyzed using the Student’s T-test, Chi-Square Test or Fisherșs exact test. The local and advanced disease groups were compared based on the HEF1 score in patients that could be defined by the above criteria using a Fishers Exact Test for heterogeneity and Cochran Armitage test for trend in a 2 (2 groups) × 3 (score 1-3) table. A p-value less than 0.05 was considered statistically significant. 95% Confidence intervals are reported in parentheses. Box plot horizontal bars indicate quartiles, with the inner small bar indicating the median of the data set. Error bars in each bar graph indicate the standard deviation of the data points within that group.
Supplementary Material
Acknowledgments
We thank Amanda M. Brock for excellent technical assistance, Jennifer Bethard and Dr. Kevin Schey (MUSC Mass Spectrometry Facility), the HCC Cell and Molecular Imaging Shared Resource, the HCC Tissue Biorepository and Dr. Elizabeth Garrett-Mayer, Director (HCC Biostatistics Shared Resource) for aid and consultations. We are indebted to Drs Terry Day and M. Boyd Gillespie for human specimens and Drs. Silvio Gutkind and Alfredo Molinolo (NIDCR) for tissue microarrays. This work was supported by a MUSC Summer Health Research Fellowship, a Paul Calabresi Fellowship (PhRMA Foundation), an Abney Foundation Award, the Southeastern Pre- Doctoral Training in Clinical Research T32 (JTL), NIH grants CA78887 and CA134845 (SAR), DOD award (N6311601MD10004) to HCC and the American Health Assistance Foundation (SAR).
Footnotes
Conflict of Interest. The authors declare no conflict of interest.
Portions of this work were presented at the Annual Meeting of the AACR, 2008, San Diego, CA and the Annual Meeting of the Endocrine Society, 2008, San Francisco, CA.
References
- Alexander NR, Branch KM, Parekh A, Clark ES, Iwueke IC, Guelcher SA, et al. Extracellular matrix rigidity promotes invadopodia activity. Curr Biol. 2008;18:1295–9. doi: 10.1016/j.cub.2008.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arold ST, Hoellerer MK, Noble MEM. The Structural Basis of Localization and Signaling by the Focal Adhesion Targeting Domain. Structure. 2002;10:319–327. doi: 10.1016/s0969-2126(02)00717-7. [DOI] [PubMed] [Google Scholar]
- Ayala I, Baldassarre M, Giacchetti G, Caldieri G, Tete S, Luini A, et al. Multiple regulatory inputs converge on cortactin to control invadopodia biogenesis and extracellular matrix degradation. J Cell Sci. 2008;121:369–78. doi: 10.1242/jcs.008037. [DOI] [PubMed] [Google Scholar]
- Bachelder RE, Lipscomb EA, Lin X, Wendt MA, Chadborn NH, Eickholt BJ, et al. Competing autocrine pathways involving alternative neuropilin-1 ligands regulate chemotaxis of carcinoma cells. Cancer Res. 2003;63:5230–3. [PubMed] [Google Scholar]
- Badowski C, Pawlak G, Grichine A, Chabadel A, Oddou C, Jurdic P, et al. Paxillin Phosphorylation Controls Invadopodia/Podosomes Spatiotemporal Organization. Mol Biol Cell. 2008;19:633–645. doi: 10.1091/mbc.E06-01-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigbee WL, Grandis JR, Siegfried JM. Multiple Cytokine and Growth Factor Serum Biomarkers Predict Therapeutic Response and Survival in Advanced-Stage Head and Neck Cancer Patients. Clin Cancer Res. 2007;13:3107–3108. doi: 10.1158/1078-0432.CCR-07-0746. [DOI] [PubMed] [Google Scholar]
- Bowden ET, Onikoyi E, Slack R, Myoui A, Yoneda T, Yamada KM, et al. Co-localization of cortactin and phosphotyrosine identifies active invadopodia in human breast cancer cells. Exp Cell Res. 2006;312:1240–53. doi: 10.1016/j.yexcr.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Brabek J, Constancio SS, Siesser PF, Shin N-Y, Pozzi A, Hanks SK. Crk-Associated Substrate Tyrosine Phosphorylation Sites Are Critical for Invasion and Metastasis of Src-Transformed Cells. Mol Cancer Res. 2005;3:307–315. doi: 10.1158/1541-7786.MCR-05-0015. [DOI] [PubMed] [Google Scholar]
- Brown MC, Turner CE. Paxillin: adapting to change. Physiol Rev. 2004;84:1315–39. doi: 10.1152/physrev.00002.2004. [DOI] [PubMed] [Google Scholar]
- Buday L, Downward J. Roles of cortactin in tumor pathogenesis. Biochim Biophys Acta. 2007;1775:263–73. doi: 10.1016/j.bbcan.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Canel M, Secades P, Rodrigo JP, Cabanillas R, Herrero A, Suarez C, et al. Overexpression of focal adhesion kinase in head and neck squamous cell carcinoma is independent of fak gene copy number. Clin Cancer Res. 2006;12:3272–9. doi: 10.1158/1078-0432.CCR-05-1583. [DOI] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark ES, Whigham AS, Yarbrough WG, Weaver AM. Cortactin is an essential regulator of matrix metalloproteinase secretion and extracellular matrix degradation in invadopodia. Cancer Res. 2007;67:4227–35. doi: 10.1158/0008-5472.CAN-06-3928. [DOI] [PubMed] [Google Scholar]
- Cohen LA, Guan JL. Mechanisms of focal adhesion kinase regulation. Curr Cancer Drug Targets. 2005;5:629–43. doi: 10.2174/156800905774932798. [DOI] [PubMed] [Google Scholar]
- Cooney MM, Tserng KY, Makar V, McPeak RJ, Ingalls ST, Dowlati A, et al. A phase IB clinical and pharmacokinetic study of the angiogenesis inhibitor SU5416 and paclitaxel in recurrent or metastatic carcinoma of the head and neck. Cancer Chemother Pharmacol. 2005;55:295–300. doi: 10.1007/s00280-004-0871-5. [DOI] [PubMed] [Google Scholar]
- Descamps FJ, Martens E, Opdenakker G. Analysis of gelatinases in complex biological fluids and tissue extracts. Lab Invest. 2002;82:1607–8. doi: 10.1097/01.lab.0000038556.54069.73. [DOI] [PubMed] [Google Scholar]
- Fashena SJ, Einarson MB, O’Neill GM, Patriotis C, Golemis EA. Dissection of HEF1-dependent functions in motility and transcriptional regulation. J Cell Sci. 2002;115:99–111. doi: 10.1242/jcs.115.1.99. [DOI] [PubMed] [Google Scholar]
- Ferrara N. Vascular Endothelial Growth Factor: Basic Science and Clinical Progress. Endocr Rev. 2004;25:581–611. doi: 10.1210/er.2003-0027. [DOI] [PubMed] [Google Scholar]
- Gimona M, Buccione R, Courtneidge SA, Linder S. Assembly and biological role of podosomes and invadopodia. Current Opinion in Cell Biology. 2008;20:235–241. doi: 10.1016/j.ceb.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Guarino M, Rubino B, Ballabio G. The role of epithelial-mesenchymal transition in cancer pathology. Pathology. 2007;39:305–18. doi: 10.1080/00313020701329914. [DOI] [PubMed] [Google Scholar]
- Izumchenko E, Singh MK, Plotnikova OV, Tikhmyanova N, Little JL, Serebriiskii IG, et al. NEDD9 Promotes Oncogenic Signaling in Mammary Tumor Development. Cancer Res. 2009;69:7198–7206. doi: 10.1158/0008-5472.CAN-09-0795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Tiwari RC, Murray T, Ghafoor A, Samuels A, Ward E, et al. Cancer statistics, 2004. CA Cancer J Clin. 2004;54:8–29. doi: 10.3322/canjclin.54.1.8. [DOI] [PubMed] [Google Scholar]
- Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–810. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- Kim M, Gans JD, Nogueira C, Wang A, Paik JH, Feng B, et al. Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell. 2006;125:1269–81. doi: 10.1016/j.cell.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Kratchmarova I, Kalume DE, Blagoev B, Scherer PE, Podtelejnikov AV, Molina H, et al. A proteomic approach for identification of secreted proteins during the differentiation of 3T3-L1 preadipocytes to adipocytes. Molecular & Cellular Proteomics. 2002;1:213–22. doi: 10.1074/mcp.m200006-mcp200. [DOI] [PubMed] [Google Scholar]
- Kyzas PA, Stefanou D, Batistatou A, Agnantis NJ. Potential autocrine function of vascular endothelial growth factor in head and neck cancer via vascular endothelial growth factor receptor-2. Mod Pathol. 2005;18:485–94. doi: 10.1038/modpathol.3800295. [DOI] [PubMed] [Google Scholar]
- Linder S. The matrix corroded: podosomes and invadopodia in extracellular matrix degradation. Trends in Cell Biology. 2007;17:107–117. doi: 10.1016/j.tcb.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Lua BL, Low BC. Cortactin phosphorylation as a switch for actin cytoskeletal network and cell dynamics control. FEBS Lett. 2005;579:577–85. doi: 10.1016/j.febslet.2004.12.055. [DOI] [PubMed] [Google Scholar]
- Michi Y, Morita I, Amagasa T, Murota S. Human oral squamous cell carcinoma cell lines promote angiogenesis via expression of vascular endothelial growth factor and upregulation of KDR/flk-1 expression in endothelial cells. Oral Oncol. 2000;36:81–8. doi: 10.1016/s1368-8375(99)00059-7. [DOI] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinolo AA, Hewitt SM, Amornphimoltham P, Keelawat S, Rangdaeng S, Meneses Garcia A, et al. Dissecting the Akt/mammalian target of rapamycin signaling network: emerging results from the head and neck cancer tissue array initiative. Clin Cancer Res. 2007;13:4964–73. doi: 10.1158/1078-0432.CCR-07-1041. [DOI] [PubMed] [Google Scholar]
- Mon NN, Ito S, Senga T, Hamaguchi M. FAK signaling in neoplastic disorders: a linkage between inflammation and cancer. Ann N Y Acad Sci. 2006;1086:199–212. doi: 10.1196/annals.1377.019. [DOI] [PubMed] [Google Scholar]
- Nakamoto T, Yamagata T, Sakai R, Ogawa S, Honda H, Ueno H, et al. CIZ, a zinc finger protein that interacts with p130(cas) and activates the expression of matrix metalloproteinases. Mol Cell Biol. 2000;20:1649–58. doi: 10.1128/mcb.20.5.1649-1658.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan M, Stewart JE, Golemis EA, Pugacheva EN, Alexandropoulos K, Cox BD, et al. HEF1 is a necessary and specific downstream effector of FAK that promotes the migration of glioblastoma cells. Oncogene. 2006;25:1721–32. doi: 10.1038/sj.onc.1209199. [DOI] [PubMed] [Google Scholar]
- O’Neill GM, Seo S, Serebriiskii IG, Lessin SR, Golemis EA. A New Central Scaffold for Metastasis: Parsing HEF1/Cas-L/NEDD9. Cancer Res. 2007;67:8975–8979. doi: 10.1158/0008-5472.CAN-07-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormandy CJ, Musgrove EA, Hui R, Daly RJ, Sutherland RL. Cyclin D1, EMS1 and 11q13 amplification in breast cancer. Breast Cancer Res Treat. 2003;78:323–35. doi: 10.1023/a:1023033708204. [DOI] [PubMed] [Google Scholar]
- Patel AM, Incognito LS, Schechter GL, Wasilenko WJ, Somers KD. Amplification and expression of EMS-1 (cortactin) in head and neck squamous cell carcinoma cell lines. Oncogene. 1996;12:31–5. [PubMed] [Google Scholar]
- Rheinwald JG, Beckett MA. Tumorigenic keratinocyte lines requiring anchorage and fibroblast support cultures from human squamous cell carcinomas. Cancer Res. 1981;41:1657–63. [PubMed] [Google Scholar]
- Ries L, Eisner M, Kosary C, Hankey B, Miller B, Clegg L, et al. National Cancer Institute; Bethesda, MD: 2004. [Google Scholar]
- Rothschild BL, Shim AH, Ammer AG, Kelley LC, Irby KB, Head JA, et al. Cortactin overexpression regulates actin-related protein 2/3 complex activity, motility, and invasion in carcinomas with chromosome 11q13 amplification. Cancer Res. 2006;66:8017–25. doi: 10.1158/0008-5472.CAN-05-4490. [DOI] [PubMed] [Google Scholar]
- Ruest PJ, Shin N-Y, Polte TR, Zhang X, Hanks SK. Mechanisms of CAS substrate domain tyrosine phosphorylation by FAK and Src. Mol Cell Biol. 2001;21:7641–7652. doi: 10.1128/MCB.21.22.7641-7652.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- Sabe H, Onodera Y, Mazaki Y, Hashimoto S. ArfGAP family proteins in cell adhesion, migration and tumor invasion. Curr Opin Cell Biol. 2006;18:558–64. doi: 10.1016/j.ceb.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Sanz-Moreno V, Gadea G, Ahn J, Paterson H, Marra P, Pinner S, et al. Rac Activation and Inactivation Control Plasticity of Tumor Cell Movement. Cell. 2008;135:510–523. doi: 10.1016/j.cell.2008.09.043. [DOI] [PubMed] [Google Scholar]
- Simpson KJ, Selfors LM, Bui J, Reynolds A, Leake D, Khvorova A, et al. Identification of genes that regulate epithelial cell migration using an siRNA screening approach. Nat Cell Biol. 2008;10:1027–1038. doi: 10.1038/ncb1762. [DOI] [PubMed] [Google Scholar]
- Singh MK, Cowell L, S S, O’Neill GM, Golemis EA. Molecular basis for HEF1/NEDD9/Cas-L action as a multifunctional co-ordinator of invasion, apoptosis and cell cycle. Cell Biochem Biophys. 2007;48:54–72. doi: 10.1007/s12013-007-0036-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh MK, Dadke D, Nicolas E, Serebriiskii IG, Apostolou S, Canutescu A, et al. A Novel Cas Family Member, HEPL, Regulates FAK and Cell Spreading. Mol Biol Cell. 2008;19:1627–1636. doi: 10.1091/mbc.E07-09-0953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack-Davis JK, Martin KH, Tilghman RW, Iwanicki M, Ung EJ, Autry C, et al. Cellular Characterization of a Novel Focal Adhesion Kinase Inhibitor. J Biol Chem. 2007;282:14845–14852. doi: 10.1074/jbc.M606695200. [DOI] [PubMed] [Google Scholar]
- Sloan EK, Anderson RL. Genes involved in breast cancer metastasis to bone. Cell Mol Life Sci. 2002;59:1491–502. doi: 10.1007/s00018-002-8524-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomiany MG, Black LA, Kibbey MM, Day TA, Rosenzweig SA. IGF-1 induced vascular endothelial growth factor secretion in head and neck squamous cell carcinoma. Biochem Biophys Res Commun. 2006;342:851–8. doi: 10.1016/j.bbrc.2006.02.043. [DOI] [PubMed] [Google Scholar]
- Slomiany MG, Black LA, Kibbey MM, Tingler MA, Day TA, Rosenzweig SA. Insulin-like growth factor-1 receptor and ligand targeting in head and neck squamous cell carcinoma. Cancer Lett. 2007;248:269–79. doi: 10.1016/j.canlet.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Steen H, Kuster B, Fernandez M, Pandey A, Mann M. Tyrosine phosphorylation mapping of the epidermal growth factor receptor signaling pathway. J Biol Chem. 2002;277:1031–9. doi: 10.1074/jbc.M109992200. [DOI] [PubMed] [Google Scholar]
- Tachibana K, Urano T, Fujita H, Ohashi Y, Kamiguchi K, Iwata S, et al. Tyrosine phosphorylation of Crk-associated substrates by focal adhesion kinase. A putative mechanism for the integrin-mediated tyrosine phosphorylation of Crk-associated substrates. J Biol Chem. 1997;272:29083–29090. doi: 10.1074/jbc.272.46.29083. [DOI] [PubMed] [Google Scholar]
- Tehrani S, Tomasevic N, Weed S, Sakowicz R, Cooper JA. Src phosphorylation of cortactin enhances actin assembly. PNAS. 2007;104:11933–11938. doi: 10.1073/pnas.0701077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpson P, Lynch DK, Schramek D, Walker F, Daly RJ. Cortactin Overexpression Inhibits Ligand-Induced Down-regulation of the Epidermal Growth Factor Receptor. Cancer Res. 2005;65:3273–3280. doi: 10.1158/0008-5472.CAN-04-2118. [DOI] [PubMed] [Google Scholar]
- van Nimwegen MJ, van de Water B. Focal adhesion kinase: a potential target in cancer therapy. Biochem Pharmacol. 2007;73:597–609. doi: 10.1016/j.bcp.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Weaver AM. Cortactin in tumor invasiveness. Cancer Letters. 2008;265:15912–20. doi: 10.1016/j.canlet.2008.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y-H, Kuo H-K, Chang K-W. The Evolving Transcriptome of Head and Neck Squamous Cell Carcinoma: A Systematic Review. PLoS ONE. 2008;3:e3215. doi: 10.1371/journal.pone.0003215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.