Abstract
The development of a complete organism from a single cell involves extraordinarily complex orchestration of biological processes that vary intricately across space and time. Systems biology seeks to describe how all elements of a biological system interact in order to understand, model, and ultimately predict aspects of emergent biological processes. Embryogenesis represents an extraordinary opportunity – and challenge – for the application of systems biology. Systems approaches have already been used successfully to study various aspects of development, from complex intracellular networks to 4D models of organogenesis. Going forward, great advancements and discoveries can be expected from systems approaches applied to embryogenesis and developmental biology.
Keywords: Development, Regulatory Networks, Computational Models, Organogenesis, Complex Adaptive Systems
The Systems Biology Paradigm
Systems biology is an experimental and theoretical framework that treats biology as an informational science, and seeks to study the behavior of biological systems as a whole. In particular, the deep complexity of developmental processes motivates the use of systematic and integrative analyses to garner biological insights. In these approaches, biological information is represented as being transmitted, modulated and integrated by biological networks that are then executed by molecular 'machines' (Hood et al. 2004; Price et al. 2009). At its heart, systems biology seeks to understand the dynamic behavior of complex biological systems in sufficient detail to construct computational models that can predict how various perturbations will affect a living system. An iterative model-building process is often employed, wherein an in silico model evolves through various iterations and increases in complexity, completeness and predictive accuracy as it is informed by increasing experimental data. The constructed computational model thus serves as a large-scale hypothesis about how an integrated biological process works as a whole. That is, the model characterizes explicitly the relevant components, their relationships to one another, and the dynamics of the interacting system. For example, a systems analysis of a mouse embryo might involve as a first step the identification of all proteins and genes expressed using shot gun proteomics and transcriptomics. Then the systems biologist might try to identify how these interact using yeast two-hybrid screening or affinity purification. Based on the results, a model is built and the system is tested ‘virtually’ by imposing different constraints or perturbations (such as a knock out) – and the predictions are compared with experiments. The model is further refined based on the novel results, and new and more accurate predictions are made. This process is repeated over and over again, with understanding and predictive accuracy increasing accordingly. Ultimately what is sought is the ability to predict how perturbations to any individual component (or combination of components) will affect the system, in order to effect desired changes to the system such as to recover health from a disease state.
The need for a systems biology approach in complex biological systems such as embryogenesis can be illustrated by analogy to complex physical machines such as automobiles. The ultimate structure and function of a car is dependent upon interactions between several constituent components – from small, regulatory electronics to the engine and chassis. To fully understand the nature of a car – and to be able to fix or build one – the behavior of each part must be understood, but so too must the complex schema of how all parts interact to create the full ‘emergent behavior’ of an automobile. Biological ‘machines’ exhibit even more multi-scale complexity than do cars, and it is thus just as important to understand how the constituent parts work together in the biological setting as it is the mechanical. Systems biology strives to characterize this interactivity using rigorous and systematic approaches. This challenge is daunting, but the capabilities to succeed in this endeavor are ever increasing.
Enabling Tools and Technologies in Systems Biology
Systems biology research typically requires large amounts of high-throughput experimental measurements to be successful. Probably the most familiar and frequently used high-throughput technology today is microarrays, which provide large-scale quantitative data for both transcriptional and genomic measurement (Allison et al. 2006; Gunderson et al. 2005; Schena et al. 1995). Among the most rapidly advancing measurement technologies is nucleotide sequencing, with novel ‘Next-Generation’ sequencing methods promising improved efficiency and significantly reduced expense (Eid et al. 2009; Mardis 2008; Mir et al. 2008; Shendure and Ji 2008). Using reverse transcription techniques these sequencing technologies can also be applied for the de novo interrogation of transcription, small RNA species (Landgraf et al. 2007), alternative splicing (Pan et al. 2008), and noncoding transcripts (Core et al. 2008). Of great importance to systems biology as it applies to human development and medicine is the advent of technologies for personalized genomics – which has already enabled sequencing of several individual human genomes (Bentley et al. 2008; Kidd et al. 2008; Wang et al. 2008; Wheeler et al. 2008). Major efforts are underway in both academic and industry settings to bring the cost of a full genome sequence down to $1000 or even less (Quail et al. 2008; Rothberg and Leamon 2008). Example large projects in academia for personalized genomics include the Personal Genome Project at Harvard led by George Church (Kaiser 2008), and a multi-institution Systems Medicine project led by Leroy Hood at the Institute for Systems Biology in Seattle (Langton 2008). These programs strive to catalyze advancements in generating and analyzing personalized sequence data by validating their use in large-scale experimental efforts. Increasingly robust and inexpensive measurement technologies for nucleotide measurement – both genomics and transcriptomics – will continue to empower systems-based studies of biology highly relevant to human embryogenesis.
Data-intensive pursuits in systems biology are also aided by advancing technologies for measurement of protein concentrations, post-translational modifications, and molecular interactions. Applications of proteomics in systems biology can be broadly divided into two groups: 1) shotgun proteomics, which focuses on global discovery of proteins relevant to specific biological processes without prior knowledge of what those components may be; and 2) targeted proteomics, where the specific proteins being searched for are known and the focus is on the their quantitative measurement and dynamic characterization. Discovery of cellular or serum-borne proteins is possible through the application of diverse techniques in tandem mass spectrometry (Bantscheff et al. 2007; Gupta et al. 2007; Nesvizhskii et al. 2007), as well as two-dimensional gel electrophoresis (Froment et al. 2005; Kolkman et al. 2005). These methodologies allow large-scale examination of up to thousands of proteins, and enable the discovery of relevant species participating in biological events. Once important participating agents in a signaling or regulatory network are known, other proteomic methods enable high-resolution study of causal linkages and dynamic parameters. Techniques such as Multiple Reaction Monitoring of isotope-labeled peptides and other diverse label-based approaches enable elucidation of quantitative and dynamic relationships between signaling and regulatory proteins. Recently developed surface plasmon resonance antibody microarrays (Boozer et al. 2006; Lausted et al. 2008; Usui-Aoki et al. 2005) also enable such targeted proteomic screens for proteins that have adequate capture agents. Complementing these interrogations of protein concentrations, diverse techniques have been developed to assay protein-protein interactions and to characterize the substantial human protein ‘Interactome’ (Braun et al. 2009; Gandhi et al. 2006; Rual et al. 2005). Proteomic technologies contribute to systems-based analyses at multiple scales of biological complexity.
Multiple online databases have been created for the storage and distribution of diverse genome-scale data. A small subset of what is available for different data types includes: 1) transcriptomics from the Gene Expression Omnibus Omnibus (Edgar et al. 2002) and the Stanford Microarray Database (Sherlock et al. 2001); 2) regulatory sequences from the Eukaryotic Promoter Database (Perier et al.) and the Transcription Regulatory Element Database (Zhao et al. 2005); and 3) proteomics from the Proteomics Identification Database (Martens et al. 2005). Additionally, standardized data formats such as the Systems Biology Markup Language (Hucka et al. 2003) and CellML (Lloyd et al. 2004), as well as software packages such as Cytoscape (Shannon et al. 2003) and the Gaggle (Shannon et al. 2006) have been developed to enable rapid porting of datasets, annotations, and models between different programs and formats. Cumulatively, these technologies for both experimental measurement and data analysis embody powerful modalities for rigorous and quantitative characterization of biological systems.
Systems Studies of Embryogenesis and Development
The process of embryogenesis exhibits diverse layers of complexity, including large-scale modulation of transcription programs, and the propagation of information from the molecular level to the scale of tissues and organs. The process involves more interacting biological variables than are easily accounted for manually, from complex transcription regulatory networks to multi-dimensional morphogen gradients. Furthermore, embryogenesis is an intrinsically dynamical process in which both intracellular and tissue-level phenotypes change immensely across time and space. However, such complexity does not make the system inherently vulnerable to perturbations. In fact, biological robustness is profoundly exemplified during embryogenesis – leading to the observed fact that networks involved in development have evolved dramatic robustness to changes in kinetic parameters associated with most system components (Eldar et al. 2002; Meir et al. 2002; von Dassow et al. 2000). Improved characterization and modeling of the robustness of embryogenesis amidst genetic heterogeneity and environmental perturbations is a major ongoing challenge for developmental systems biology.
An important concept underlying much systems-based thinking within and outside of biology is the notion of ‘Complex Adaptive Systems’ (Gell-Mann 1994). These systems are referred to as ‘complex’ because they are composed of many linked components; they are modular in nature and form interconnected networks. 'Adaptive' refers to a capacity to modify behavior in response to changing environment or context. At many different scales, life – in the form of cells, tissues, organisms, and even ecosystems – exhibits the qualities of complex adaptive systems. A complex and adaptive nature is particularly characteristic of higher organisms, such as humans, in both health and disease (Coffey 1998; Schwab and Pienta 1996). Particularly, cell lineages and differentiation programs can be considered highly adaptive systems – for they achieve specific phenotypic character even in diverse and dynamic biological contexts (Theise 2006; Theise and d'Inverno 2004). Such systems are notoriously difficult to model and control accurately because their emergent behaviors are dependent on the interactions of diffuse agents, and thus the challenge of modeling such a complex and adaptive process as embryogenesis is great indeed.
Embryogenesis can be considered a ‘unidirectional’ adaptive process, in which individual system components modulate behavior in response to changing biological context, but do so in a (typically) ‘irreversible’ manner with clear directionality. Furthermore, these developmental processes exhibit discrete ‘stages.’ That is, as cells differentiate, they occupy a series of discrete phenotypic states, and are less stable outside of these defined conditions. Quantitative transcriptomic analyses have shown that differentiating cells are drawn towards ‘attractor states’ – particular configurations which exhibit high stability, and a corresponding tendency of cells to reach and remain therein (Chang et al. 2006; Huang et al. 2005; Kashiwagi et al. 2006). The phenomenon of ‘canalization’ is one feedback process for such attraction dynamics – in which differentiating cells are ‘guided’ into particular phenotypic states by internal processes, such as transcriptional regulatory circuits circuits (Lott et al. 2007) and microRNA (Hornstein and Shomron 2006). Canalization is one example of systems-level, emergent phenomena witnessed in complex adaptive processes such as embryogenesis.
Biological networks operating in the process of embryogenesis exhibit marked robustness to perturbations due to redundancy in control and feedback across a wide and diffuse network. The survival need and evolutionary selection for robustness is clear in that embryogenesis is such an immensely complex process that, remarkably, operates correctly so much of the time. Such robustness to uncertainty and noise is considered as a hallmark of living systems (Kitano 2004). A recent study found that the segment polarity network in Drosophila development was robust to large changes in the biochemical kinetic constants that govern its behavior (von Dassow et al. 2000). This intriguing study showed that picking a kinetic parameter in this network at random, even over multiple orders of magnitude, would on average work to produce proper segmentation over 90% of the time. This robustness of the overall phenotype to differences in individual kinetic properties makes the network resistant to the effects of mutations and other developmental 'noise' that alters the dynamics of catalyzed reactions occurring during embryogenesis. Similar behavior has been observed in networks governing cell growth, development, metabolism and chemotaxis (Stelling et al. 2004). Biological robustness should not be misunderstood to imply a phenotypic constancy regardless of stimuli or mutations: rather, robust networks maintain specific functionalities in the context of perturbations, while remaining sufficiently flexible to change modes of operation in a suitable manner. Certain biological systems thus exist at a 'critical' state – at the boundary of ordered and chaotic states, imbuing sufficient rigidity to resist small uncertainties, but flexible enough to adapt to major environmental changes (Nykter et al. 2008). Such design principles can potentially be exploited to study dynamic and complex biological systems such as those at work in embryogenesis.
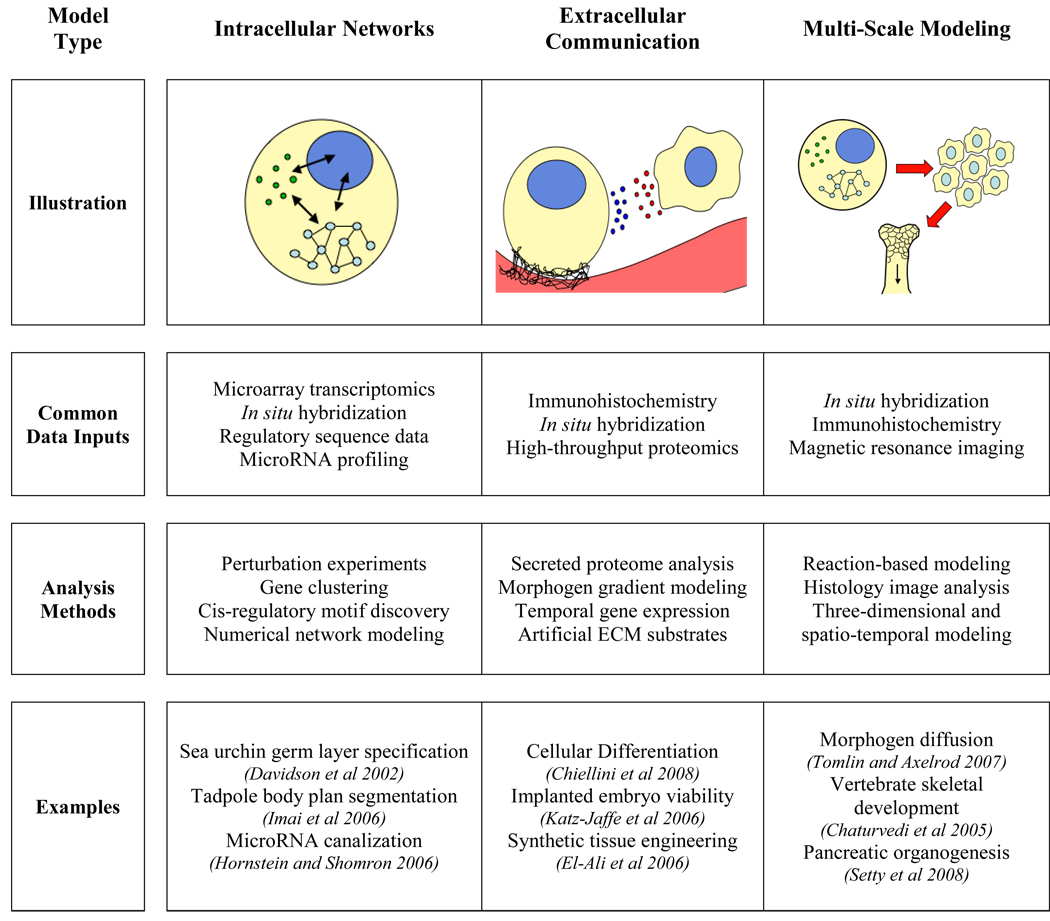
Given this multifaceted complexity, embryogenesis lends itself to characterization through systems-based approaches. Detailed experimental investigations identify a substantial ‘parts list’ in which genes, signals, and phenotypes important to embryogenesis are elucidated. Systems biology, complementary to this important process, presents a framework to collate the behaviors of these diverse biological components and to form cohesive, integrated explications of biological phenomena. Systems biology presents a powerful set of analytical tools well-suited to recording and assessing the complex phenomena of embryogenesis. Systems analysis has already been brought to bear on a number of aspects of the developmental process, including 1) intracellular networks, 2) communication signals between cells and with the environment, and 3) multi-scale integration (Box 1).
Box 1: Systems-Based Models of Embryogenesis
Common types of systems-based models of embryogenic and developmental biology, including experimental data sources, analytical frameworks, and representative examples of prior studies for each model type.
Intracellular Networks
Perhaps the most common form of systems-based study has been the characterization of intracellular networks involved in developmental processes. In silico modeling of these networks describes variously the genetic, transcriptional, and signaling events relevant to large-scale cellular and tissue phenotype. A pioneering achievement was the elucidation of a gene regulatory network that regulates the specification of endoderm and mesoderm layers in the sea urchin embryo by Eric Davidson’s group (Davidson et al. 2002). This study involved large-scale perturbation experiments, genomic and transcriptomic measurement, and computational analyses to define the transcriptional regulatory ‘circuit’ that underlies this developmental process – and was among the first systems-based characterizations of molecular networks governing embryogenesis. This network showed a basis for irreversibility in cellular development in rigorous, readily visualized form – including activating cues from maternal tissues, and internal regulatory feedback processes which progress and ‘lock-down’ the embryogenic process such that it is self-sustaining. This project also spoke to the idea of biological attractor states, as the study’s multi-component transcriptional program propels differentiating cells towards specific phenotypic trajectories. These discrete developmental states exhibit particular stability in the environmental and genetic context wherein they reside. Such preferentially stable states represent an intrinsic quality of any dynamical system with internal or external sources of feedback, and developmental systems are frequently found to have both (Alon 2007).
Following this seminal study, many other intracellular networks involved in development have been characterized using similar and advancing methods, with multiple reviews available on the topic (Davidson and Erwin 2006; Levine and Davidson 2005). Several projects have extended the study of germ layer specification to elucidate similar networks in other organisms, including Drosophila (Sandmann et al. 2007) and the tadpole (Imai et al. 2006). These studies have demonstrated that such networks exhibit extensive combinatoric regulation of developmental genes, and have revealed important biomolecular control points in embryogenic events. Intracellular processes in development are now commonly referred to as Genetic Regulatory Networks (GRNs), and are becoming represented by increasingly quantitative and genome-scale models (Longabaugh et al. 2005; Reeves et al. 2006). For example, recent investigations were able to define ‘clusters’ of genes related according to biological function and expression patterns through global gene expression analysis in Drosophila embryogenesis (Hornstein and Shomron 2006; Shalgi et al. 2007; Tomancak et al. 2007). Another major focus has been analysis of cis-regulatory motifs. Recent studies have combined computational and experimental approaches to identify the nature of transcriptional regulatory sites across the genome, and the connectivity between transcription factors and their cognate binding sites (Kuntz et al. 2008; Li et al. 2007; Van Loo et al. 2008). Noncoding transcripts such as microRNA also play important roles in developmental processes. Recent studies have revealed that miRNA-mediated events are a ubiquitous mechanism to augment the robustness and precision of gene regulation (Aboobaker et al. 2005; Johnston et al. 2005; Tsang et al. 2007). By examining biomolecular networks at the systems level, these methods elucidate both important individual components involved in developmental processes, and the systems-level architecture of the circuits they compose.
Cell-Cell Communication and the Extracellular Environment
The ability of cells to respond appropriately to surrounding cells and the extracellular environment serves as a major basis for developmental processes. The complex relationships observed between intracellular networks and extracellular cues warrants rigorous modeling and simulation to more fully comprehend. A major area for human systems biology and medicine going forward involves characterizing and interpreting the information content of the ‘secretome’ – the set of all biomolecules secreted by cells and tissues (Price et al. 2009). Importantly, the secretome can serve as a fully non-invasive proxy to survey cell/cell and cell/tissue interactions – including those involved in development. Learning to non-invasively monitor important aspects of embryogenesis and development via the secretome has been the subject of intensive research in recent years (Dominguez et al. 2009; Katz-Jaffe et al. 2009). These investigations used a combination of high-throughput proteomic techniques, including two-dimensional gel electrophoresis and tandem mass spectrometry, to characterize major elements of the secreted proteome. They have revealed a substantially global portrait of the secretome and elucidated biomolecules important to differentiation and development. Similar recent studies have identified secreted proteins associated with the differentiation of mesenchymal stem cells into osteoblasts and adipocytes (Chiellini et al. 2008; Schinköthe et al. 2008). The molecular processes that drive the highly multi-potent character of mesenchymal stem cells are intrinsically complex and involve many regulatory agents. In striving to consider a large repertoire of cellular factors simultaneously, systems approaches are well-suited to examine such integrated, multi-component processes.
Similar proteomic techniques have been employed to study the extracellular matrix and its application to developmental processes in cells and tissues (Xiao et al. 2009). These investigations have examined roles played by the ECM in structural support, intercellular communication, and the modulation of cell growth and behavior. Detailed understanding of the extracellular matrix is necessary for successfully culturing cells in synthetic conditions that mimic the in vivo scenario. Many model tissue systems employ this knowledge in an attempt to recapitulate natural ECM to study and rationally direct cellular differentiation on artificial substrates (El-Ali et al. 2006). In systematically characterizing the complex extracellular environments observed in developmental processes, these studies inform more precise laboratory investigations for further experimental research. Large-scale proteomics has also been applied directly to clinical medical tasks. Importantly, a recent analysis of the secretome of human embryos identified extracellular predictors of viable embryos for implantation (Katz-Jaffe et al. 2006). Extracellular phenomena represent the mechanistic bridge between intracellular networks and tissue-level events; they thus embody a crucial component of any systems-based study of embryogenesis and development.
Multiscale and Integrative Modeling
Multiscale modeling approaches strive to connect microscopic events with macroscopic phenomena, and to model the molecular processes that propagate information from the scale of genes to the scales of cells, tissues, and organs (Lewis 2008; Walker and Southgate 2009). Such models have progressed substantially from the simple mathematical models of neurons by Hodgkin and Huxley, up to modern models of morphogen diffusion in developing embryos (Tomlin and Axelrod 2007). Digital simulation of three-dimensional structures formed during embryonic development can be performed using tools such as Compucell3D (Cickovski et al. 2007). For example, this tool has been employed to model skeletal development in a vertebrate limb (Chaturvedi et al. 2005). An impressive recent project elucidated a four-dimensional in silico model of pancreatic organogenesis as it progresses in space and time (Setty et al. 2008). This model considers interactions from the level of genes to the level of organs to model and visualize pancreatic development over multiple time scales. Similar multiscale models have examined the migration of germ layers in Xenopus laevis as a function of Wnt/β-catenin signaling (Peirce et al. 2006; Robertson et al. 2007), and the effects of extracellular growth factors on epithelial development (Walker et al. 2006).
Models of embryogenesis have been aided by advancements in technologies such as in situ hybridization and MRI. These tools have enabled detailed three-dimensional recording of gene expression patterns during embryogenesis and development in both humans (Matsuda 2005) and several model organisms such as Drosophila (Christiansen et al. 2006; Peng et al. 2007; Pisarev et al. 2009; Wei et al. 2006). Additionally, spatial gene expression patterns from such diverse organisms have been integrated into a large public database, enabling comparison of gene expression patterns across species (Haudry et al. 2008). These spatial models do not explicitly account for the transfer of information from the molecular to the tissue level. Nonetheless, they enable a large-scale consideration of gene expression programs – including the dynamics by which genes and gene clusters are activated, both spatially across tissues and temporally throughout developmental processes.
Future Challenges and Opportunities
A long-term objective of systems approaches to embryogenesis is the construction of in silico biological models of the developing embryo that accurately recapitulate in vivo biological events. One could potentially predict and perform in silico experiments using such computational models, thereby providing focused hypotheses for further experimental study. This exciting prospect would permit not only a rigorous description of biological activities, but also enable the prediction of responses given by cells or organisms to exogenous and therapeutic perturbations. Such technologies would empower researchers to conduct virtual trials of medical procedures or therapeutics, reducing the need for expensive and potentially high-risk experimentation.
The promises of systems biology are grand and exciting, yet there are many hurdles to be passed before such approaches come to fruition. We do not have complete knowledge of the cell and systems models of cellular processes are still at their infancy. For example, epigenetic phenomena are now understood to play crucial roles in the differentiation and maintenance of cell type, but systems-level computational models of epigenetic events in embryogenesis are only beginning to emerge (Bock and Lengauer 2008; Jones and Martienssen 2005). Similarly, recent studies have shown that different forms of noncoding transcription are ubiquitous throughout the genome (Seila et al. 2008), and it has been speculated that such activity forms a major locus for the regulation of developmental processes (Dinger et al. 2008; Mattick 2007; Taft et al. 2007). These phenomena too, due perhaps to their recent discovery, have not yet been studied extensively using systems-based approaches. Another factor that stifles the creation of most large-scale models is the lack of quantitative data. Until recently, the bulk of data in biology were qualitative and were not amenable for modeling. With the development of new high throughput methods that generate quantitative data, more systems analysis like those outlined above will emerge. Finally, it should be clear that while computational modeling will aid tremendously in generating hypotheses, predicting the effects of perturbations, and yielding insight into system behavior, it can never replace physical experiments.
In summary, systems approaches have contributed substantially to the study of early embryogenesis and development – from the level of transcriptional regulation and intracellular signaling, to cellular differentiation and tissue patterning. These innovative studies have begun to elucidate embryogenic processes that are network-based, multi-component, and distributed across several scales of biological and physical complexity. While presenting significant technological, experimental, and theoretical challenges, this pursuit promises to advance our understanding of embryogenesis and development beyond what has been attainable previously.
Acknowledgments
The authors gratefully acknowledge insightful discussions with Leroy Hood and James Eddy, and funding from the NIH Howard Temin Pathway to Independence Award in Cancer Research (NDP).
References
- Aboobaker AA, Tomancak P, Patel N, Rubin GM, Lai EC. Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proceedings of the National Academy of Sciences. 2005;102:18017–18022. doi: 10.1073/pnas.0508823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nature Reviews Genetics. 2006;7:55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- Alon U. An introduction to systems biology: design principles of biological circuits. Chapman & Hall/CRC; 2007. [Google Scholar]
- Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Analytical and Bioanalytical Chemistry. 2007;389:1017. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C, Lengauer T. Computational epigenetics. Bioinformatics. 2008;24:1. doi: 10.1093/bioinformatics/btm546. [DOI] [PubMed] [Google Scholar]
- Boozer C, Kim G, Cong S, Guan HW, Londergan T. Looking towards label-free biomolecular interaction analysis in a high-throughput format: a review of new surface plasmon resonance technologies. Current Opinion in Biotechnology. 2006;17:400–405. doi: 10.1016/j.copbio.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Braun P, Tasan M, et al. An experimentally derived confidence score for binary protein-protein interactions. Nature Methods. 2009;6:91. doi: 10.1038/nmeth.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HH, Oh PY, Ingber DE, Huang S. Multistable and multistep dynamics in neutrophil differentiation. BMC Cell Biology. 2006;7:11. doi: 10.1186/1471-2121-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi R, Huang C, et al. On multiscale approaches to three-dimensional modelling of morphogenesis. Journal of The Royal Society Interface. 2005;2:237. doi: 10.1098/rsif.2005.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiellini C, Cochet O, Negroni L, Samson M, Poggi M, Ailhaud G, Alessi M-C, Dani C, Amri E-Z. Characterization of human mesenchymal stem cell secretome at early steps of adipocyte and osteoblast differentiation. BMC Molecular Biology. 2008;9:26. doi: 10.1186/1471-2199-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen J, Yang Y, Venkataraman S, Richardson L, Stevenson P, Burton N, Baldock R, Davidson D. EMAGE: a spatial database of gene expression patterns during mouse embryo development. Nucleic Acids Research. 2006;34:D637. doi: 10.1093/nar/gkj006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cickovski T, Aras K, et al. From Genes to Organisms Via the Cell: A Problem-Solving Environment for Multicellular Development. Computing in Science and Engineering. 2007;9:50–60. doi: 10.1109/MCSE.2007.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffey DS. Self-organization, complexity and chaos: the new biology for medicine. Nature Medicine. 1998;4:882–885. doi: 10.1038/nm0898-882. [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA Sequencing Reveals Widespread Pausing and Divergent Initiation at Human Promoters. Science. 2008;322:1845. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH, Erwin DH. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311:796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- Davidson EH, Rast JP, et al. A Genomic Regulatory Network for Development. Science. 2002;295:1669. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- Dinger ME, Amaral PP, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Research. 2008;18:1433. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez F, Pellicer A, Simon C. The Human Embryo Proteome. Reproductive Sciences. 2009;16:188. doi: 10.1177/1933719108328612. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research. 2002;30:207. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid J, Fehr A, et al. Real-Time DNA Sequencing from Single Polymerase Molecules. Science. 2009;323:133. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- El-Ali J, Sorger P, Jensen K. Cells on chips. Nature. 2006;442:403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- Eldar A, Dorfman R, Weiss D, Ashe H, Shilo BZ, Barkai N. Robustness of the BMP morphogen gradient in Drosophila embryonic patterning. Nature. 2002;419:304–308. doi: 10.1038/nature01061. [DOI] [PubMed] [Google Scholar]
- Froment C, Uttenweiler-Joseph S, Bousquet-Dubouch MP, Matondo M, Borges JP, Esmenjaud C, Lacroix C, Monsarrat B, Burlet-Schiltz O. A quantitative proteomic approach using two-dimensional gel electrophoresis and isotope-coded affinity tag labeling for studying human 20S proteasome heterogeneity. Proteomics. 2005;5:2351–2363. doi: 10.1002/pmic.200401281. [DOI] [PubMed] [Google Scholar]
- Gandhi TKB, Zhong J, et al. Analysis of the human protein interactome and comparison with yeast, worm and fly interaction datasets. Nature Genetics. 2006;38:285–293. doi: 10.1038/ng1747. [DOI] [PubMed] [Google Scholar]
- Gell-Mann M. Complex adaptive systems. In: Cowan DP George A, Meltzer David Elliott., editors. Complexity: Metaphors, Models, and Reality. Westview Press; 1994. pp. 17–45. [Google Scholar]
- Gunderson KL, Steemers FJ, Lee G, Mendoza LG, Chee MS. A genome-wide scalable SNP genotyping assay using microarray technology. Nature Genetics. 2005;37:549–554. doi: 10.1038/ng1547. [DOI] [PubMed] [Google Scholar]
- Gupta N, Tanner S, et al. Whole proteome analysis of post-translational modifications: Applications of mass-spectrometry for proteogenomic annotation. Genome Research. 2007;17:1362. doi: 10.1101/gr.6427907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haudry Y, Berube H, et al. 4DXpress: a database for cross-species expression pattern comparisons. Nucleic Acids Research. 2008;36:D847–D853. doi: 10.1093/nar/gkm797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood L, Heath JR, Phelps ME, Lin B. Systems Biology and New Technologies Enable Predictive and Preventative Medicine. Science. 2004;306:640–643. doi: 10.1126/science.1104635. [DOI] [PubMed] [Google Scholar]
- Hornstein E, Shomron N. Canalization of development by microRNAs. Nature Genetics. 2006;38:20. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- Huang S, Eichler G, Bar-Yam Y, Ingber DE. Cell fates as high-dimensional attractor states of a complex gene regulatory network. Physical Review Letters. 2005;94:128701. doi: 10.1103/PhysRevLett.94.128701. [DOI] [PubMed] [Google Scholar]
- Hucka M, Finney A, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- Imai KS, Levine M, Satoh N, Satou Y. Regulatory blueprint for a chordate embryo. Science. 2006;312:1183–1187. doi: 10.1126/science.1123404. [DOI] [PubMed] [Google Scholar]
- Johnston RJ, Chang S, Etchberger JF, Ortiz CO, Hobert O. MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proceedings of the National Academy of Sciences. 2005;102:12449–12454. doi: 10.1073/pnas.0505530102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Martienssen R. A Blueprint for a Human Epigenome Project: The AACR Human Epigenome Workshop. AACR; 2005. pp. 11241–11246. [DOI] [PubMed] [Google Scholar]
- Kaiser J. DNA Sequencing: A Plan to Capture Human Diversity in 1000 Genomes. Science. 2008;319:395. doi: 10.1126/science.319.5862.395. [DOI] [PubMed] [Google Scholar]
- Kashiwagi A, Urabe I, Kaneko K, Yomo T. Adaptive response of a gene network to environmental changes by fitness-induced attractor selection. Plos One. 2006;1 doi: 10.1371/journal.pone.0000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz-Jaffe M, Gardner D, Schoolcraft W. Proteomic analysis of individual human embryos to identify novel biomarkers of development and viability. Fertility and Sterility. 2006;85:101–107. doi: 10.1016/j.fertnstert.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Katz-Jaffe M, McReynolds S, Gardner D, Schoolcraft W. The role of proteomics in defining the human embryonic secretome. Molecular Human Reproduction. 2009;15:271. doi: 10.1093/molehr/gap012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Cooper GM, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano H. Biological robustness. Nature Reviews Genetics. 2004;5:826–837. doi: 10.1038/nrg1471. [DOI] [PubMed] [Google Scholar]
- Kolkman A, Dirksen EHC, Slijper M, Heck AJR. Double Standards in Quantitative Proteomics Direct Comparative Assessment of Difference in Gel Electrophoresis and Metabolic Stable Isotope Labeling*. Molecular & Cellular Proteomics. 2005;4:255–266. doi: 10.1074/mcp.M400121-MCP200. [DOI] [PubMed] [Google Scholar]
- Kuntz SG, Schwarz EM, DeModena JA, De Buysscher T, Trout D, Shizuya H, Sternberg PW, Wold BJ. Multigenome DNA sequence conservation identifies Hox cis-regulatory elements. Genome Research. 2008;18:1955. doi: 10.1101/gr.085472.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, et al. A Mammalian microRNA Expression Atlas Based on Small RNA Library Sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langton T. Leading U.S. Bioscience Pioneers Enter International Collaboration with Government of Luxembourg to Accelerate Biomedical Research. Institute for Systems Biology Newsletter. 2008 [Google Scholar]
- Lausted C, Hu Z, Hood L. Quantitative Serum Proteomics from Surface Plasmon Resonance Imaging. Molecular & Cellular Proteomics. 2008;7:2464. doi: 10.1074/mcp.M800121-MCP200. [DOI] [PubMed] [Google Scholar]
- Levine M, Davidson EH. Gene regulatory networks for development. Proceedings of the National Academy of Sciences. 2005;102:4936–4942. doi: 10.1073/pnas.0408031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. From Signals to Patterns: Space, Time, and Mathematics in Developmental Biology. Science. 2008;322:399. doi: 10.1126/science.1166154. [DOI] [PubMed] [Google Scholar]
- Li L, Zhu Q, He X, Sinha S, Halfon MS. Large-scale analysis of transcriptional cis-regulatory modules reveals both common features and distinct subclasses. Genome Biology. 2007;8:R101. doi: 10.1186/gb-2007-8-6-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd CM, Halstead MDB, Nielsen PF. CellML: its future, present and past. Progress in Biophysics and Molecular Biology. 2004;85:433–450. doi: 10.1016/j.pbiomolbio.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Longabaugh WJR, Davidson EH, Bolouri H. Computational representation of developmental genetic regulatory networks. Developmental biology. 2005;283:1–16. doi: 10.1016/j.ydbio.2005.04.023. [DOI] [PubMed] [Google Scholar]
- Lott SE, Kreitman M, Palsson A, Alekseeva E, Ludwig MZ. Canalization of segmentation and its evolution in Drosophila. Proceedings of the National Academy of Sciences. 2007;104:10926. doi: 10.1073/pnas.0701359104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER. The impact of next-generation sequencing technology on genetics. Trends in Genetics. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Martens L, Hermjakob H, Jones P, Adamski M, Taylor C. PRIDE: The proteomics identifications database. Proteomics. 2005;5:3537–3545. doi: 10.1002/pmic.200401303. [DOI] [PubMed] [Google Scholar]
- Matsuda T. Visible Embryo: A 3D Image Database of Human Embryo Collection at Kyoto University. IEICE technical report. 2005;105:3–4. [Google Scholar]
- Mattick JS. A new paradigm for developmental biology. Journal of Experimental Biology. 2007;210:1526. doi: 10.1242/jeb.005017. [DOI] [PubMed] [Google Scholar]
- Meir E, Von Dassow G, Munro E, Odell GM. Robustness, flexibility, and the role of lateral inhibition in the neurogenic network. Current Biology. 2002;12:778–786. doi: 10.1016/s0960-9822(02)00839-4. [DOI] [PubMed] [Google Scholar]
- Mir KU, Qi H, Salata O, Scozzafava G. Sequencing by Cyclic Ligation and Cleavage (CycLiC) directly on a microarray captured template. Nucleic Acids Research. 2008 doi: 10.1093/nar/gkn906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Vitek O, Aebersold R. Analysis and validation of proteomic data generated by tandem mass spectrometry. Nature Methods. 2007;4:787–797. doi: 10.1038/nmeth1088. [DOI] [PubMed] [Google Scholar]
- Nykter M, Price ND, Aldana M, Ramsey SA, Kauffman SA, Hood LE, Yli-Harja O, Shmulevich I. Gene expression dynamics in the macrophage exhibit criticality. Proceedings of the National Academy of Sciences. 2008;105:1897. doi: 10.1073/pnas.0711525105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nature Genetics. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Peirce S, Skalak T, Papin J. Multiscale biosystems integration: Coupling intracellular network analysis with tissue-patterning simulations. Systems Biology. 2006;50 [Google Scholar]
- Peng H, Long F, Zhou J, Leung G, Eisen M, Myers E. Automatic image analysis for gene expression patterns of fly embryos. BMC Cell Biology. 2007;8:S7. doi: 10.1186/1471-2121-8-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perier RC, Junier T, Bucher P. The Eukaryotic Promoter Database EPD. Nucleic Acids Research. 1998;26:353–357. doi: 10.1093/nar/26.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev A, Poustelnikova E, Samsonova M, Reinitz J. FlyEx, the quantitative atlas on segmentation gene expression at cellular resolution. Nucl. Acids Res. 2009;37:D560–D566. doi: 10.1093/nar/gkn717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price ND, Edelman LB, Lee I, Yoo H, Hwang D, Carlson G, Galas DJ, Heath JR, Hood L. Systems biology and systems medicine. In: Ginsburg G, Willard H, editors. Genomic and Personalized Medicine: From Principles to Practice. Amsterdam: Elsevier; 2009. [Google Scholar]
- Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, Swerdlow H, Turner DJ. A large genome center's improvements to the Illumina sequencing system. Nature Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves GT, Muratov CB, Schüpbach T, Shvartsman SY. Quantitative models of developmental pattern formation. Developmental Cell. 2006;11:289–300. doi: 10.1016/j.devcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Robertson S, Smith C, et al. Multiscale computational analysis of Xenopus laevis morphogenesis reveals key insights of systems-level behavior. BMC Systems Biology. 2007;1:46. doi: 10.1186/1752-0509-1-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothberg JM, Leamon JH. The development and impact of 454 sequencing. Nature Biotechnology. 2008;26:1117–1124. doi: 10.1038/nbt1485. [DOI] [PubMed] [Google Scholar]
- Rual J, Venkatesan K, et al. Towards a proteome-scale map of the human protein–protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Sandmann T, Girardot C, Brehme M, Tongprasit W, Stolc V, Furlong EEM. A core transcriptional network for early mesoderm development in Drosophila melanogaster. Genes & Development. 2007;21:436. doi: 10.1101/gad.1509007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative Monitoring of Gene Expression Patterns with a Complementary DNA Microarray. Science. 1995;270:467. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Schinköthe T, Bloch W, Schmidt A. In vitro secreting profile of human mesenchymal stem cells. Stem Cells and Development. 2008;17:199. doi: 10.1089/scd.2007.0175. [DOI] [PubMed] [Google Scholar]
- Schwab ED, Pienta KJ. Cancer as a complex adaptive system. Medical Hypotheses. 1996;47:235–241. doi: 10.1016/s0306-9877(96)90086-9. [DOI] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent Transcription from Active Promoters. Science. 2008;322:1849. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty Y, Cohen IR, Dor Y, Harel D. Four-dimensional realistic modeling of pancreatic organogenesis. Proceedings of the National Academy of Sciences. 2008;105:20374. doi: 10.1073/pnas.0808725105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalgi R, Lieber D, Oren M, Pilpel Y. Global and local architecture of the mammalian microRNA-transcription factor regulatory network. PLoS Computational Biology. 2007;3:e131. doi: 10.1371/journal.pcbi.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon PT, Reiss DJ, Bonneau R, Baliga NS. The Gaggle: An open-source software system for integrating bioinformatics software and data sources. BMC Bioinformatics. 2006;7:176. doi: 10.1186/1471-2105-7-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J, Ji H. Next-generation DNA sequencing. Nature Biotechnology. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- Sherlock G, Hernandez-Boussard T, et al. The Stanford Microarray Database. Nucleic Acids Research. 2001;29:152–155. doi: 10.1093/nar/29.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelling J, Sauer U, Szallasi Z, Doyle FJ, Doyle J. Robustness of cellular functions. Cell. 2004;118:675–685. doi: 10.1016/j.cell.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Taft RJ, Pheasant M, Mattick JS. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays. 2007;29 doi: 10.1002/bies.20544. [DOI] [PubMed] [Google Scholar]
- Theise ND. Implications of 'postmodern biology' for pathology: the Cell Doctrine. Laboratory Investigation. 2006;86:335. doi: 10.1038/labinvest.3700401. [DOI] [PubMed] [Google Scholar]
- Theise ND, d'Inverno M. Understanding cell lineages as complex adaptive systems. Blood Cells, Molecules and Diseases. 2004;32:17–20. doi: 10.1016/j.bcmd.2003.09.010. [DOI] [PubMed] [Google Scholar]
- Tomancak P, Berman B, Beaton A, Weiszmann R, Kwan E, Hartenstein V, Celniker S, Rubin G. Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biology. 2007;8:R145. doi: 10.1186/gb-2007-8-7-r145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlin CJ, Axelrod JD. Biology by numbers: mathematical modelling in developmental biology. Nature Reviews Genetics. 2007;8:331–340. doi: 10.1038/nrg2098. [DOI] [PubMed] [Google Scholar]
- Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Molecular Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui-Aoki K, Shimada K, Nagano M, Kawai M, Koga H. A novel approach to protein expression profiling using antibody microarrays combined with surface plasmon resonance technology. Proteomics. 2005;5:2396–2401. doi: 10.1002/pmic.200401171. [DOI] [PubMed] [Google Scholar]
- Van Loo P, Aerts S, Thienpont B, De Moor B, Moreau Y, Marynen P. ModuleMiner-improved computational detection of cis-regulatory modules: are there different modes of gene regulation in embryonic development and adult tissues? Genome Biology. 2008;9:R66. doi: 10.1186/gb-2008-9-4-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Dassow G, Meir E, Munro EM, Odell GM. The segment polarity network is a robust developmental module. Nature. 2000;406:188–192. doi: 10.1038/35018085. [DOI] [PubMed] [Google Scholar]
- Walker D, Wood S, Southgate J, Holcombe M, Smallwood R. An integrated agent-mathematical model of the effect of intercellular signalling via the epidermal growth factor receptor on cell proliferation. Journal of Theoretical Biology. 2006;242:774–789. doi: 10.1016/j.jtbi.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Walker DC, Southgate J. The virtual cell--a candidate co-ordinator for'middle-out'modelling of biological systems. Briefings in Bioinformatics. 2009 doi: 10.1093/bib/bbp010. [DOI] [PubMed] [Google Scholar]
- Wang J, Wang W, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Angerer R, Angerer L. A database of mRNA expression patterns for the sea urchin embryo. Developmental Biology. 2006;300:476–484. doi: 10.1016/j.ydbio.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DA, Srinivasan M, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- Xiao Z, Blonder J, Zhou M, Veenstra T. Proteomic analysis of extracellular matrix and vesicles. Journal of Proteomics. 2009;72:34–45. doi: 10.1016/j.jprot.2008.11.011. [DOI] [PubMed] [Google Scholar]
- Zhao F, Xuan Z, Liu L, Zhang MQ. TRED: a Transcriptional Regulatory Element Database and a platform for in silico gene regulation studies. Nucleic Acids Research. 2005;33:D103. doi: 10.1093/nar/gki004. [DOI] [PMC free article] [PubMed] [Google Scholar]