SUMMARY
Suppose that we are interested in using new bio- or clinical-markers to improve prediction or diagnosis of the patient’s clinical outcome in addition to the conventional markers. The incremental value from the new markers is typically assessed by averaging across patients in the entire study population. However, when measuring the new markers is costly or invasive, an overall improvement does not justify measuring the new markers in all patients. A more practical strategy is to utilize the patient’s conventional markers to decide whether the new markers are needed for improving prediction of his/her health outcomes. In this article, we propose inference procedures for the incremental values of new markers across various subgroups of patients classified by the conventional markers. The resulting point and interval estimates can be quite useful for medical decision makers seeking to balance the predictive or diagnostic value of new markers against their associated cost and risk. Our proposals are theoretically justified and illustrated empirically with two real examples.
Keywords: Biomarker, Cardiovascular events, Diagnosis, K-fold crossvalidation, Prediction accuracy, Subgroup analysis
1. Introduction
Biological and technological advances continually generate promising new clinical- and biomarkers with the potential to improve medical care by providing more accurate, personalized predictions of health outcomes and diagnoses of clinical phenotypes. However, extensive use of new markers may provide only negligible improvements in prediction or diagnosis, while subjecting patients to additional risks and costs. It is therefore important to develop statistical methods that can quantify for individual patients the value of new markers over conventional ones, especially when measuring these markers is costly or invasive. As an example from a recent study, Wang et. al. (2006) examined extensively the incremental values of ten new biomarkers in predicting first major cardiovascular events or death in a Framingham cohort. There were 3209 participants in the study. They were followed for a median of 7.4 years, during which 207 participants died and 169 had a first major cardiovascular event. Based on various prediction precision criteria, the study investigators found that these ten contemporary biomarkers added only moderate overall predictive value to the classical risk factors including gender, age, total cholesterol, HDL cholesterol, blood pressure, smoking status, and diabetes mellitus. In contrast, other investigators studying different populations with different prediction precision measures demonstrated that certain biomarkers, for example, the high-sensitivity C-reactive protein, provide clinically useful prognostic information on top of the traditional Framingham risk score for heart diseases (Ridker et. al., 2002, 2007, Blumenthal et. al., 2007).
Despite these often controversial findings in the literature, clinical practitioners would generally not change their recommendation for the patient’s care with the extra marker information if the patient, for example, has either high or low conventional risk score. Therefore, a practically important question is how to systematically identify patients who would benefit from the additional markers instead of evaluating these markers based only on their average incremental value across the entire population (D’Agostino, 2006). In this article, we propose procedures to estimate the incremental values of new markers for diagnosis or prediction in various subgroups of patients classified by conventional markers. These, coupled with the sampling variations of the estimates, provide a useful tool for researchers and practitioners to decide when, after observing the conventional risk factors, the new markers are needed. In Section 2, we describe in detail the new procedure and provide theoretical justification. In Section 3, we illustrate our methods with two examples, one with a continuous response and the other with a binary outcome.
There are quite a few procedures in the literature for evaluating the over-all incremental value of new markers for an entire population of interest. For example, Pepe et. al. (2004) compared the ROC curves among models with and without an additional marker. Recently, Tian et. al. (2007) and Uno et. al. (2007) proposed robust inference procedures for evaluating prediction rules. Prediction or diagnostic precision measures, which may be used for comparing different prediction procedures, have also been proposed and utilized, for example, by Brier (1950), Breiman et. al. (1984), Speigelhalter (1986), Korn and Simon (1990), McLachlan (1992), Mittlböck and Schemper (1996), Ripley (1996), Zhou et. al. (2002), Pepe (2003) and Pencina et. al. (2008).
2. Estimating Subject-Specific Prediction Error Based on Risk Score Constructed from Conventional Markers
Let Y be a continuous or binary response variable, U be the set of its conventional marker values, and V be the corresponding counterpart from the new markers. Our data consist of n independent copies {(Yi, Ui, Vi), i = 1, ⋯, n} from (Y, U, V). The problem is how to use the data to identify future subjects via U, which would benefit from the new markers for better prediction of their responses Y. Suppose that there are no well-established rules for classifying subjects based on U for predicting Y. First, we may estimate a center value of Y given U nonparametrically and use this estimate to construct a predictor for Y. We then estimate the average prediction error, the “distance” between the observed response and its predicted value over all subjects which have the same marker value U. Next, we estimate the center of Y given U and V, and estimate the corresponding average prediction error conditional only on U. Inferences about the improvement from the new markers can be made via these functional estimates over U. Unfortunately, in general, we can only construct nonparametric functional estimates, which behave reasonably well, when the dimension of U is very small and the sample size n is quite large.
A practically feasible alternative to handle this problem is to consider a parametric or semi-parametric approach. To this end, let X be a p-dimensional vector, a function of U. Assume that the conditional mean of Y given U can be approximated by the following working model
| (2.1) |
where g1(·) is a smooth, strictly increasing, known function and β is an unknown vector of parameters. Note that the first component of X is one. In this article, we deal with the interesting and challenging case that β′X is a continuous variable.
To estimate the regression parameters for model (2.1) which, most likely, is an approximation to the true conditional mean of Y given U, one may use the estimator β̂ based on the simple estimating function
| (2.2) |
where {(Yi, Xi), i = 1, ⋯, n} are n-independent copies of (Y, X) (Tian et. al., 2007). Note that even when (2.1) is not the true model, β̂ converges to a constant vector β0, as n → ∞. It is not clear, however, that other standard estimators for β in (2.1) would be convergent as n gets large.
Now, consider a future independent subject with (Y, X) = (Y0, X0). For a given β in (2.1), let Ŷ1(β′X0) be the predictor for Y0. For example, when Y0 is continuous, one may let Ŷ1(β′X0) = g1(β′X0) and when Y0 is binary, one may predict Y0 by a binary variable Ŷ1(β′X0) = I{g1(β′X0) ≥ 0.5}, where I(·) is the indicator function. Other prediction rules for the binary case will be discussed in the Example Section. To evaluate the performance of Ŷ1(β̂′X0), we first need to quantify its prediction accuracy based on a “distance” between the true Y0 and the predicted Ŷ1(β̂′X0), denoted by D{Y0, Ŷ1(β̂′X0)}. For example, a simple, physically interpretable function is the absolute prediction error with D(a, b) = |a − b|. When Y is binary, this distance function is simply the overall mis-classification rate. Choosing an appropriate distance measure is a crucial yet often difficult step in evaluating the incremental value for the cost-benefit decision. It is important to note that such quantification should have a practical/clinical interpretation. We discuss this issue in great details with real examples in Section 3 and also in the Remarks Section.
Since clinical practitioners almost always group subjects with a “risk scoring system” for medical decision making, we consider an average prediction error over a set of X’s which have “similar” g1(β̂′X) to evaluate Ŷ1(·). To be specific, let Jz = (cz, dz) be a data-independent interval centered about z, where z ranges over a set of possible values of . The average prediction error over Jz is , where the conditional expectation is taken with respect to (Y0, X0) and β̂. As n → ∞, converges to
| (2.3) |
As a process of z, this moving average process {𝒟1(z)} provides a performance profile of Ŷ1(·) over all possible values of . The proper choice of interval Jz would be made on a case-by-case basis and is illustrated with two examples in the next section.
Now, let W be a q × 1 vector, a function of U and V. Assume that a working model for the conditional mean of Y given U and V is
| (2.4) |
where g2(·) is a smooth, strictly increasing, known function and θ is an unknown vector of parameters. The first component of W is one. Again, we assume that g2(θ′W) is a continuous variable. Let θ̂ be the estimator for θ obtained from the following simple estimating function
| (2.5) |
where Wi, i = 1, ⋯, n, are n independent copies of W. Let θ0 be the limit of θ̂. Consider a future independent (Y, X, W) = (Y0, X0, W0). Let Ŷ2(θ′W0) be the predictor constructed from (2.4) with parameter value θ, the counterpart of Ŷ1(β′X0). For the aforementioned interval Jz, let the average prediction error for Ŷ2(·) over Jz be
| (2.6) |
where the expectation is taken with respect to (Y0, X0, W0). Then, as a processes in z, 𝒟1(z), 𝒟2(z) and
| (2.7) |
provide a global picture for identifying subgroups of patients who would benefit from the additional markers.
To estimate 𝒟1(z) and 𝒟2(z), one may use 𝒟̂1(z) = 𝒟̃1(z, β̂) and 𝒟̂2(z) = 𝒟̃2(z, β̂, θ̂), respectively, where
| (2.8) |
and
| (2.9) |
We then let Δ̂(z) = 𝒟̂1(z) − 𝒟̂2(z) to estimate Δ0(z). In Appendix A of the web-based supplementary material available at http://www.tibs.org/biometrics, we show that with the distance function D(a, b) = |a − b| or a function thereof, the above three estimators are uniformly consistent over an interval Ω consisting of all z’s whose intervals Jz’s are properly in the support of . Similar arguments may be used for cases with other distance functions.
To make further inferences about the added value from the new markers for predicting the response, in Appendix A of the web-based supplementary material, we show that the limiting distributions of the processes , and , are the same as those of the Gaussian processes , respectively, for z ∈ Ω, where ,
and {G1, …, Gn} are independent standard normal random variables that are independent of the data. Here, realizations from three Gaussian processes given above can be generated easily for any interval of z, where 𝒟̂1(z) and 𝒟̂2(z) are well-defined. In practice, one may not be able to construct reasonably well-behaved interval estimators for 𝒟l(z), l = 1, 2, for z is the tail parts of Ω. To this end, let Ω̂ be a set of z such that Jz ⊂ [η1, η2], where , and d1 and d2 are given positive numbers. Then, with the above large sample approximations, for z ∈ Ω̂, a (1 − α), 0 < α < 1, point-wise confidence interval for 𝒟l(z), l = 1, 2, is
| (2.10) |
Here, is the variance of the random variable and ξα is the upper 100αth percentage point of the standard normal. Furthermore, a (1 − α) simultaneous confidence band for {𝒟l(z), z ∈ Ω̂} is
| (2.11) |
where
It is important to note that in contrast to the standard subgroup analysis, our proposal takes care of the multiple comparison problems with such simultaneous confidence interval estimates via the scoring system indexed by z.
To construct interval estimators for Δ0(z), it is important to note that Δ̂(z) has a degenerate limiting distribution when for all . Therefore, to obtain reasonable interval estimators in practice, we consider the set Ω̃ ⊂ Ω̂ such that for , where d3 is a given positive number. Then, for z ∈ Ω̃, a (1 − α), 0 < α < 1, point-wise confidence interval for Δ0(z), is
| (2.12) |
Here, is the variance of the random variable 𝒲*(z). Moreover, a (1 − α) simultaneous confidence band for {Δ0(z), z ∈ Ω̃} is
| (2.13) |
where
Note that for the case with a continuous response Y, Ω̂ = Ω̃.
Now, since we use the entire data set to estimate the parameters in (2.1) and (2.4) and also to estimate the average prediction errors (2.3) and (2.6), 𝒟̂1(·) and 𝒟̂2(·) may be significantly underestimated. To reduce such potential bias, one may consider the commonly used K-fold crossvalidation scheme. Specifically, we randomly split the data into K disjoint subsets of about equal size and label them as ℐk, k = 1, ⋯, K. For each k, we use all the observations, which are not in ℐk, to estimate parameters in (2.1) and (2.4) via estimating functions (2.2) and (2.5), and then use the observations in ℐk to estimate prediction errors 𝒟1(·) and 𝒟2(·) with (2.8) and (2.9). Let the resulting estimators be denoted by 𝒟̂1k(·) and 𝒟̂2k(·), respectively. The crossvalidated estimators for 𝒟1(·), 𝒟2(·) and Δ0(·) are and Δ̃(·) = 𝒟̃1(·) − 𝒟̃2(·), respectively. Again, these estimators are uniformly consistent if K is relatively small with respect to n.
In Appendix B of the web-based supplementary material, we show that for large n, the distributions of the processes and can also be approximated well by those of , respectively. Point-wise and simultaneous confidence intervals for 𝒟1(·), 𝒟2(·), and Δ0(·) can then be constructed based on the crossvalidated estimates and their large sample distributions accordingly.
3. Examples
We use two examples to illustrate the new proposals. The first example is from a clinical trial conducted by the AIDS Clinical Trials Group, ACTG 320 (Hammer et. al., 1997). The study demonstrates that for various response endpoints, on average the three-drug combination therapy consisting of indinarvir, zidovudine and lamivudine, is much better than the two drug combination without indinarvir for treating HIV infected patients. Unfortunately, even with this potent combination, some patients may not respond to treatment, but suffer from non-trivial toxicity. Therefore, for future patients’ management, it is important to have a reliable model for predicting patient’s treatment responses based on certain “baseline” markers. A general conception is to use the baseline CD4 count and HIV RNA, a measure of viral load, and the early changes of these two markers after initiation of therapy for treatment guidance (Demeter et. al., 2001). For resource-limited regions, however, the cost of obtaining RNA is relatively expensive. Therefore, a challenging question is when we need RNA in addition to CD4 for better prediction of patient’s response.
Recently Tian et. al. (2007) demonstrated that, on a population average sense, neither the baseline nor early RNA change (from baseline to week 8) would add a clinically meaningful value for predicting the long term change of CD4 (from baseline to Week 24), an important measure of the patient’s immune response. Here, we try to locate a subgroup of patients, if any, who would benefit from the expensive marker RNA. To this end, let the response Y be the change of CD4 cell counts from Week 0 to 24, let U consist of age, baseline CD4 and the early change in CD4, and let V consist of the baseline RNA and the early change in RNA. For our analysis, in Models (2.1) and (2.4), we let X = (1, U′)′, W = (1, U′, V′)′, and g1(·) and g2(·) be the identity function. Also, we let Ŷ1(β′X) = β′X, Ŷ2(θ′W) = θ′W, D(a, b) = |a − b| and interval Jz be [z − 10, z + 10] for z ∈ Ω̂ = [15, 165]. Note that the intra-patient standard deviation of the CD4 count is about 60. Therefore, a choice of Jz whose length of the similar magnitude to 60 would be appropriate from a practical point of view. Moreover, in our analysis, we let d1 = d2 = 0.01 discussed in Section 2 for choosing Ω̂. With n = 392 sets of complete observations of (Y, U, V), the regression parameter estimates for Models (2.1) and (2.4) are reported in Table 1. Note that the short term changes of CD4 and RNA are statistically highly significant.
Table 1.
Estimates of the regression parameters with their standard errors and corresponding p-values for testing zero covariate effects for the AIDS example
| Age | Baseline RNA | RNA Change | Baseline CD4 | CD4 Change | |
|---|---|---|---|---|---|
| Estimate | −0.55 | 0.08 | −12.06 | 0.03 | 0.68 |
| Std Error | 0.35 | 5.53 | 2.80 | 0.07 | 0.10 |
| P-value | 0.12 | 0.99 | 0.00 | 0.72 | 0.00 |
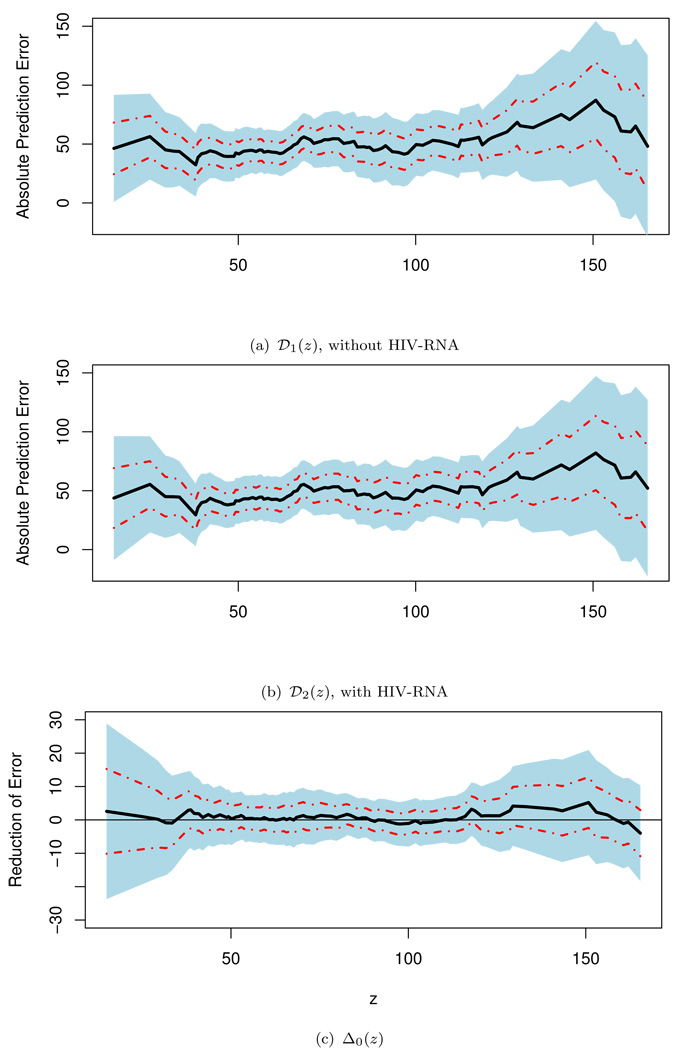
For both working models, we utilized 5-fold crossvalidation scheme discussed in Section 2 to obtain the regression parameters and then 𝒟̃1(·), 𝒟̃2(·), and Δ̃(·). In Figure 1, we present these estimated prediction errors and their differences with the corresponding 0.95 point-wise and simultaneous confidence intervals given in (2.10)–(2.13). The values of {𝒟̃1(z)} based on the model with age, baseline CD4 and early change in CD4 range from 37 to 74. The values of {𝒟̃2(z)} based on the model with additional RNA information range from 36 to 73. The estimated differences {Δ̃(z)} range from −1.7 to 6.0. These indicate that there is no clinically meaningful gain from RNA for any subgroup of patients classified by β̂′X. One may draw further statistical inference about the Δ0(·). For example, for subjects whose score g1(β̂′X) ∈ Jz = [40, 60], the estimated Δ̃(50) = 0.45 with 0.95 point-wise interval of (−3.25, 4.15) and simultaneous interval of (−7.48, 8.38). Note that the results reported here are based on Jz with interval length of 20, which is well within the intra-patient variation of CD4 measures. Various analyses have also been done with Jz’s whose lengths range from 30 to 60. All the results lead to the same conclusion. That is, statistically or clinically, we cannot identify a subgroup of patients who would benefit from the extra information of RNA for prediction of the long term CD4 change.
Figure 1.
Point estimates for 𝒟1(·), 𝒟2(·) and Δ0(·) with corresponding 0.95 point-wise (dashed lines) and simultaneous (shaded regions) confidence intervals for the HIV example.
The data for the second example is from a population of patients screened for a clinical study, called TRACE, for treating heart failure or acute myocardial infraction (MI) (Kober et. al., 1995). There were 6676 patients screened. Each patient had six routine clinical covariates: age, creatine (CRE), occurrence of heart failure (CHF), history of diabetes (DIA), history of hypertension (HYP), and cardiogenic shock after MI (KS). Moreover, each patient had an echocardiographic assessment of left ventricular systolic function which was quantified by a measure called the wall motion index (WMI). Compared with the above six covariates, the WMI is relatively expensive to obtain. Although not every screened patient entered the clinical trial, all patients screened were followed closely for mortality.
Recently, Thune et. al. (2005) studied the prognostic importance of left ventricular systolic function in patients diagnosed with either heart failure or acute MI in addition to the patient’s medical history. It would be interesting to identify subpopulations that can benefit from the extra WMI measure for predicting clinical outcomes such as mortality. Here, we let the outcome Y be a binary variable, which is one if the patient died within five years. The five-year survival rate for this data set is approximately 42%. To evaluate the incremental value of WMI, we first fit the data using Model (2.1) with X = (1, AGE, CRE, CHF, DIA, HYP, KS), and g1(s) = exp(s)/{1 + exp(s)}. With the extra variable WMI, we fit a second logistic regression model with W = (X′, WMI)′. A total of 5921 subjects have complete predictor information. The estimates for the regression parameters with their standard errors are reported in Table 2. Note that the WMI is highly statistically significant.
Table 2.
Estimated Regression Coefficients for Model (2.1) with AGE, CRE, CHF, DIA, HYP, KS and WMI for the screened population of TRACE study
| AGE | CRE | CHF | DIA | HYP | KS | WMI | |
|---|---|---|---|---|---|---|---|
| Estimate | 0.055 | −0.010 | 0.759 | 0.718 | 0.187 | 1.153 | −1.097 |
| Std. Error | 0.004 | 0.002 | 0.067 | 0.101 | 0.073 | 0.163 | 0.083 |
| P-value | 0.000 | 0.000 | 0.000 | 0.000 | 0.010 | 0.000 | 0.000 |
Here, we consider the prediction rules
| (3.1) |
and
| (3.2) |
Note that we have also fitted the data with more complicated models, for example, by including various interaction terms. The results for the present case, however, are almost identical to that based on the above two additive models. For binary response variables, we consider the distance function D(a, b) = |a − b|, where a is the observed response and b is a binary predicted response based on the working model with the assumed event rate of g(·). This distance function is a conventionally used metric for evaluating the binary prediction rules. One may use other possible distance functions, for instance, by letting b be g(·) from (2.1) and (2.4) and D(a, b) = |a − b| or (a − b)2. Moreover, one may consider a likelihood-based criterion to evaluate Models (2.1) and (2.4). In general, the metric comparing Y and the estimated g(·) can efficiently discriminate two prediction models. On the other hand, for the present case, we are more interested in evaluating the practical performance of the specific prediction rules, not the adequacy of the model fitting (although these two are closely related). Thus, considering distance functions between a practically applicable prediction rule Ŷ and the true response Y seems more relevant. The distance function, |a − b|, consists of two discordance rates or two types of error rates. Specifically, 𝒟1(z) in (2.3) is 𝒟11(z) + 𝒟10(z), where are the discordance rates for false negative and false positive errors, respectively. Similarly, 𝒟2(z) = 𝒟20(z) + 𝒟21(z). Let Δ0(z) = 𝒟10(z) − 𝒟20(z) and Δ1(z) = 𝒟11(z) − 𝒟21(z). Oftentimes, a false negative error is more serious than a false positive one. Therefore, one may consider a weighted sum Δ0(w, z) = w0Δ0(z) + w1Δ1(z) to evaluate the prediction rules. Here, w = (w0, w1)′ and the weights w0 and w1 reflect the “cost” of making these two types of errors. It is interesting to note that the corresponding distance function for Δ0(w, z) is . For a given w, the crossvalidated point estimates Δ̃(w, z) and their interval estimates for Δ0(w, z) can be constructed as for Δ0(z) in Section 2. The large sample properties for Δ̃(w, z) are derived in Appendix A of the web-based supplementary material.
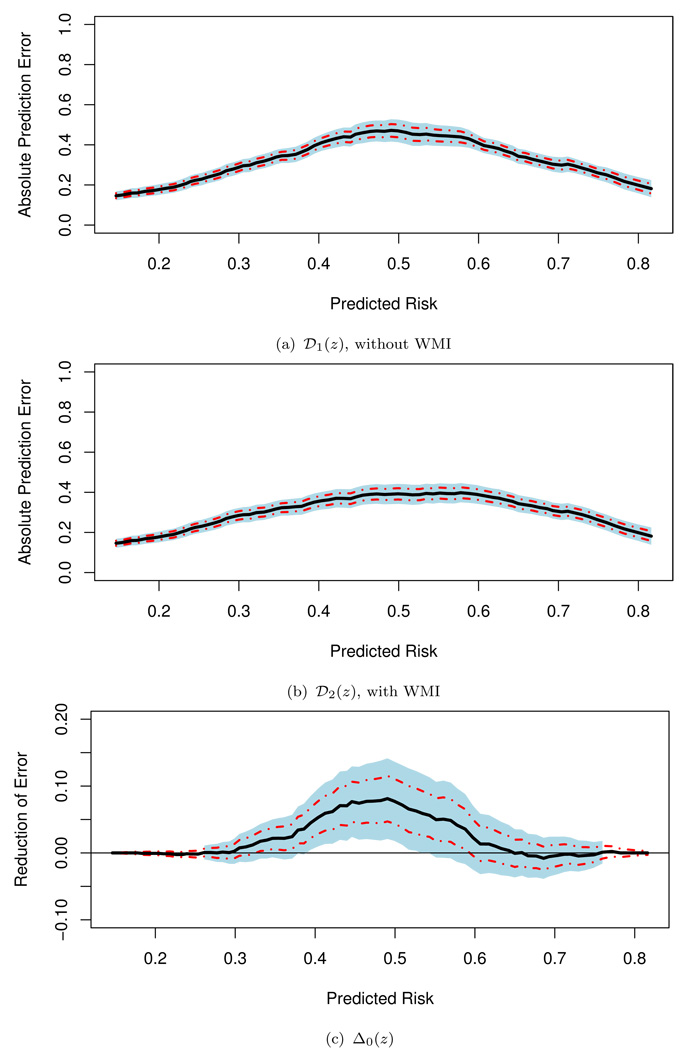
We first considered the most commonly used prediction rule with c = 0.5 and w0 = w1 = 1. The 5-fold crossvalidated estimates, obtained by letting Jz be the entire real line in (2.8) and (2.9), for the overall prediction errors E[D{Y0, Ŷ1(β̂′X0)}] and E[D{Y0, Ŷ2(β̂′W0)}] are 0.28 and 0.26, respectively, a modest overall incremental gain from the extra information of WMI for the entire population of interest. To identify which subgroup of patients who would benefit with WMI, we let Jz = [z − 0.1, z + 0.1], for z ∈ Ω̂ = [0.15, 0.82]. Here, the scale of z is the probability of developing the event within five years based on the conventional risks factors in the model. The choice of the interval length of Jz is not obvious as that for the HIV example and should be made by the entire research team (not only from the statistical point of view). For example, it may depend on the cost of obtaining the WMI measure, the distribution of the initial predicted risk score, and the clinical interpretation of the scale of such a scoring system. In our analysis, Ω̂ is chosen by letting d1 = d2 = 0.01 discussed in Section 2. To estimate 𝒟l(z), l = 1, 2, and Δ0(z), we used the 5-fold crossvalidation to obtain 𝒟̃1(·), 𝒟̃2(·) and Δ̃(·). In Figure 2, we present these point estimates and their corresponding 0.95 point-wise and simultaneous confidence intervals. For the interval estimation, we let d3 = 0.01. This results in Ω̃ = [0.26, 0.76]. Note that the point estimates Δ̃(z) for z outside Ω̂ are not reliable, and Δ̃(z) is pretty at around 0 for z ∈ Ω̂ − Ω̃, indicating that there is no evidence that WMI has a meaningful gain outside the interval Ω̃. On the other hand, with the point and interval estimates displayed in Figure 2(c), one may conclude that WMI is likely to be beneficial for patients with conventional risk scores g1(β̂′X) ranging from 0.16 to 0.74. If WMI is relatively affordable to the population of interest, then one may consider using the upper bound of the simultaneous confidence intervals to identify the subpopulation based on 0 and thus conclude that patients with g1(β̂′X) ∈ [0.16, 0.86] are likely to benefit from the WMI. On the other hand, when WMI is not quite affordable, then one may select the region conservatively and use the lower bound of the simultaneous confidence intervals based on and thus conclude that patients with g1(β̂′X) ∈ [0.29, 0.63] are likely to benefit from the WMI.
Figure 2.
Point estimates for 𝒟1(·), 𝒟2(·) and Δ0(·) with corresponding 0.95 point-wise (dashed lines) and simultaneous (shaded regions) confidence intervals for the screened population of the TRACE study (the prediction with c = 0.5).
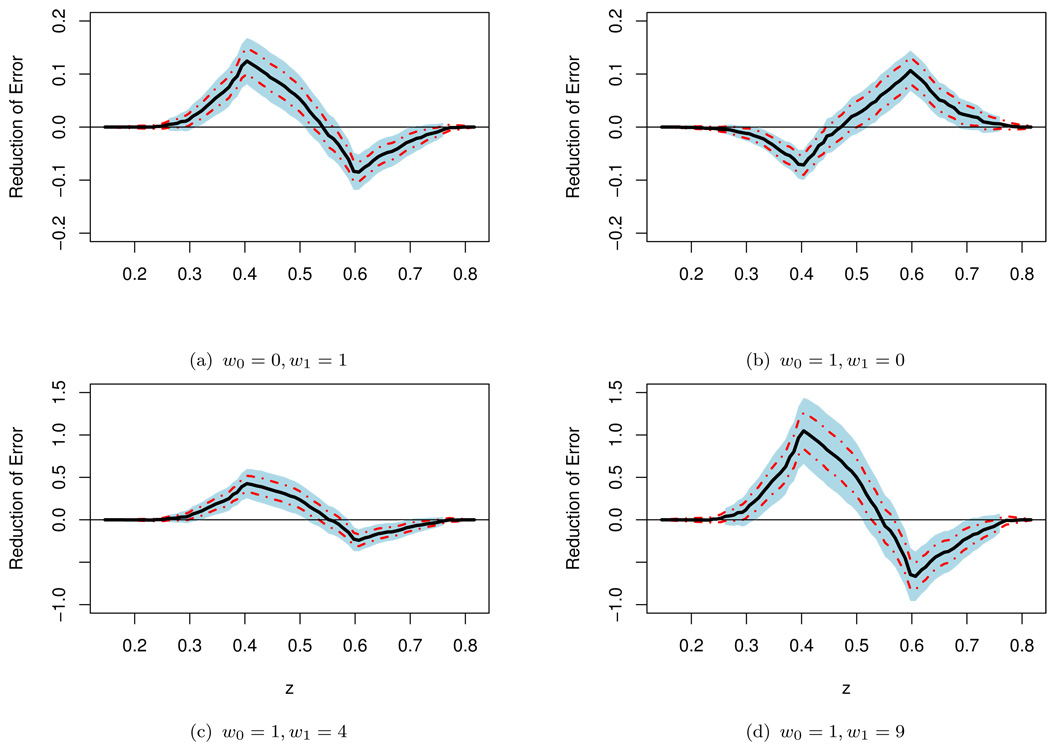
To illustrate the effect of the weighting parameter w = (w0, w1) on the incremental value of WMI, we present in Figure 3(a),(b),(c) and (d) the point and interval estimates of Δ0(w, z) for the predictors (3.1) and (3.2) with c = 0.5 and various choices of w.
Figure 3.
Point estimate Δ̃(w, ·) for Δ0(w, ·) with various weights and the corresponding 0.95 point-wise (dashed lines) and simultaneous (shaded regions) confidence intervals for the screened population of the TRACE study (the prediction with c = 0.5).
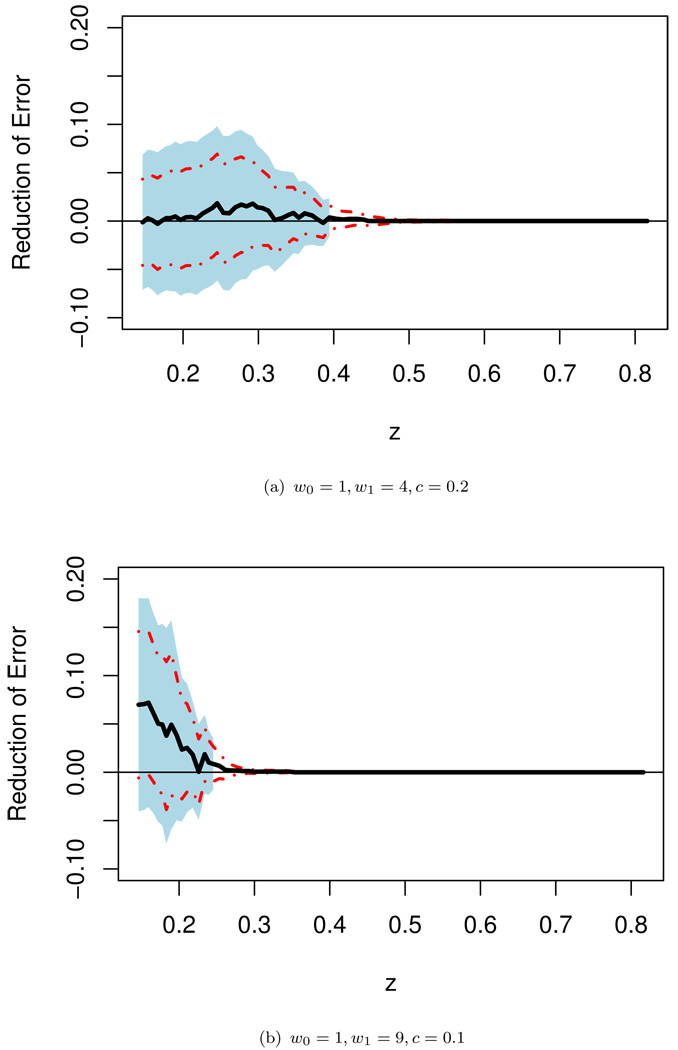
Note that when w0 ≠ w1, even if the working model is correctly specified, the prediction rule in (3.1) or (3.2) with c = 0.5 is not optimal with respect to the weighted error rate. Furthermore, with the unequal weighting criterion, for some subgroups of patients, inclusion of the extra information of WMI may significantly decrease the prediction precision. For a given w, with the weighted sum prediction precision measure, w0𝒟10(z) + w1𝒟11(z), it is straightforward to show that the optimal prediction rule based on X that minimizes the above criterion is Ŷ = I{pr(Y = 1 | X) ≥ cw}, where cw = w0/(w0 + w1). Therefore, for the present example, if g1(β̂′X) and g2(θ̂′W) are reasonably good approximations to E(Y|U) and E(Y|U, V), the predictors I(g1(β̂′X) ≥ cw) and I(g2(θ̂′W) ≥ cw) are almost optimal. In Figure 4, we present the crossvalidated point estimates along with the 0.95 interval estimates of Δ0(w, z) with w = (1, 4)′ and (1, 9)′ when “optimal” prediction rules are used for both models. It appears that there is minimal gain from WMI across all sub-populations indexed by g(β̂′X) ∈ Jz for both cases. These findings underscore the importance of selecting the cut-off value as well as the distance measure for quantifying the incremental predictive value of new biomarkers. In any event, we highly recommend to perform such sensitivity analyses to provide an over-all picture of the incremental value from the new biomarkers when there is no consensus about the weights used in the binary classification.
Figure 4.
Point estimate Δ̃(w, ·) for Δ0(w, ·) with the “optimal” weights and the corresponding 0.95 point-wise (dashed lines) and simultaneous (shaded regions) confidence intervals for the screened population of the TRACE study.
As suggested by a reviewer of the paper, we have also applied our method to quantify the incremental value of new biomarkers based on the integrated discrimination improvement (IDI) index (Pencina et. al., 2008, Pepe et. al., 2008). The IDI index is defined as the integrated difference in Youden’s indices (Youden, 1950). It can be viewed as an improvement of the average sensitivity and specificity with the new markers. The crossvalidated point estimates and the corresponding 0.95 simultaneous interval estimates are presented in Figure 1 of the web-based supplementary material. With this utility function, it appears that the WMI would be beneficial for all patients whose conventional risk scores are between 0.10 and 0.81 for predicting subject’s five year survival. It is important to note that there are no specific prediction rules attached to this approach (similar to the area under curve as an average measure over a class of prediction rules). Therefore, at the end it is not clear which prediction rule(s) one would recommend for practical usage based on the model with the additional markers. Utilizing an over all measure of the incremental value for evaluating new markers over various subgroups of patients can be useful at the initial stage of the investigation.
4. Remarks
As for any scientific investigation, we need to define the endpoint of the study at the very first step. For the HIV example in Section 3, instead of using the change of the CD4 counts as the endpoint, one may be interested in the percent of change in CD4 counts from the baseline level. This is equivalent to considering the change in the log transformed CD4 counts as the endpoint. For the TRACE study example in Section 3, we dichotomized the patient’s survival time to make the response variable being the binary survival status at 5 years in our analysis. Naturally we may consider prediction rules for other time points. A good prediction rule for five-year survival may not be appropriate for ten-year survival. Therefore, when evaluating prediction rules for the patient’s survival time, a global distance function, for example, based on the L1 norm, between the observed and predicted survival times may not be desirable. Moreover, in practice, often the survival time cannot be observed completely and the support of its censoring variable is generally shorter than that of the survival time. As a result, it is difficult to evaluate prediction rules efficiently with, for example, the L1 distance without artificial extrapolation. On the other hand, using the approach taken by Uno et. al. (2007), one may identify patients who would benefit from the additional biomarkers via prediction of t-year survival.
After we select the endpoint of the study, we should make every effort to find the “best” models (2.1) and (2.4), which fit the data well. Model (2.1) would thus provide us a reasonable scoring system with which we can classify future patients into different subgroups via the conventional markers. Subsequently, we fit the response variable with the conventional and new marker values jointly to build model (2.4). Such an elaborate joint model may include interactions between the conventional and new markers, which would be potential contributing factors to the varying incremental values from the new markers.
The next crucial step is to choose a proper prediction precision measure to quantify the incremental value of the new markers. Different distance functions between the predicted and observed may result in quite different conclusions regarding the selection of subset of patients as illustrated by two real examples in Section 3. Since the final decision of using the new biomarkers would be based on the trade off between the risk/cost and benefit, the distance function needs to be “clinically” or heuristically interpretable. In analyzing the data from the HIV and TRACE trials, we proposed several distance functions for illustration. The choice of such functions is by no means restricted to those discussed in Section 3. For example, one may use a theoretically interesting metric such as the conventional likelihood ratio statistic or the mean square error loss to differentiate two prediction models (one with and the other without new markers). However, the magnitude of gain under such a metric is often difficult to interpret when the cost or risk is involved for decision making.
In practice the choice of the distance function even for the simple case with a binary response is rather complex. In the cardiovascular disease arena, conventionally a patient with more than 10% risk for having a serious cardiovascular event within ten years is generally regarded as having a high risk, and usually would be recommended for certain preventive treatments. However, the utility function may vary across individuals and hence different patients may have different optimal cutoff points for predicting patient-level outcomes. The weighted sum of prediction error rates presented in this article is an attempt to cope with this complicated cost-benefit issue. The complexities of choosing a distance function extend to the case with continuous responses. For example, weighting absolute prediction errors according to the observed response may lead to a more meaningful penalty in some applications compared to the un-weighted counterpart.
In this paper, we provide a useful tool for making valid inferences on the incremental value of new markers simultaneously over subsets of patients with well-defined endpoint, prediction models, utility (distance) function and the study population. We propose the use of the simultaneous confidence band for the incremental values to control the type one error and determine whether the new markers have a positive incremental value in a subset of patients. As the sample size increases, the confidence bands become tighter and one would be able to more accurately identify all subsets of interest.
Lastly, we may want to identify subsets where the incremental value is greater than a positive threshold to incorporate the cost of measuring the new markers. As such, subsets where the new markers have a positive, yet very small added predictive value would be excluded. If the new markers become less costly or invasive in the future, we may construct a new scoring system to index patients. It is likely that some old markers may not be needed on top of the new ones.
Supplementary Material
Acknowledgement
The authors are grateful to Drs. Jens Jakob Thune and James Signorovitch for helpful comments on the paper. The authors also very grateful to two reviewers and an associate editor for their insightful comments on the paper. This research was partially supported by the US NIH grants for I2B2, AIDS and General Medical Sciences.
Footnotes
Supplementary Materials
Web Appendices and figure referenced in Sections 2 and 3 are available under the Paper Information link at the Biometrics website http://www.tibs.org/biometrics.
References
- Blumenthal R, Michos E, Nasir K. Further improvements in CHD risk prediction for women. JAMA. 2007;297:641–643. doi: 10.1001/jama.297.6.641. [DOI] [PubMed] [Google Scholar]
- Breiman L, Friedman J, Stone C, Olshen R. Classification and regression trees. Chapman & Hall/CRC; 1984. [Google Scholar]
- Brier G. Verification of forecasts expressed in terms of probability. Monthly Weather Review. 1950;78:1–3. [Google Scholar]
- D’Agostino RB. Risk prediction and finding new independent prognostic factors. Journal of Hypertension. 2006;24:643–645. doi: 10.1097/01.hjh.0000217845.57466.cc. [DOI] [PubMed] [Google Scholar]
- Demeter L, Hughes M, Coombs R, Jackson J, Grimes J, Bosch R, Fiscus S, Spector S, Squires K, Fischl M, Hammer S. Predictors of virologic and clinical outcomes in HIV-1-infected patients receiving concurrent treatment with indinavir, zidovudine, and lamivudine. AIDS Clinical Trials Group Protocol 320. Annals of Internal Medicine. 2001;135:954–964. doi: 10.7326/0003-4819-135-11-200112040-00007. [DOI] [PubMed] [Google Scholar]
- Hammer S, Squires K, Hughes M, Grimes J, Demeter L, Currier J, Eron J, Feinberg J, Balfour H, Deyton L, Chodakewitz J, Fischl M. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. New England Journal of Medicine. 1997;337:725–733. doi: 10.1056/NEJM199709113371101. [DOI] [PubMed] [Google Scholar]
- Kober L, Torp-Pedersen C, Carlsen J, Bagger H, Eliasen P, Lyngborg K, Videbak J, Cole D, Auclert L, Pauly N, Aliot E, Persson S, Camm A. A clinical trial of the angiotensin-converting-enzyme inhibitor trandolapril in patients with left ventricular dysfunction after myocardial infarction. New England Journal of Medicine. 1995;333:1670–1676. doi: 10.1056/NEJM199512213332503. [DOI] [PubMed] [Google Scholar]
- Korn E, Simon R. Measures of explained variation for survival data. Statistics in Medicine. 1990;9:487–503. doi: 10.1002/sim.4780090503. [DOI] [PubMed] [Google Scholar]
- McLachlan J. Discriminant analysis and statistical pattern recognition. John Wiley & Sons; 1992. [Google Scholar]
- Mittlböck M, Schemper M. Explained Variation for Logistic Regression. Statistics in Medicine. 1996;15:1987–1997. doi: 10.1002/(SICI)1097-0258(19961015)15:19<1987::AID-SIM318>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Pencina J, D’Agostino R, Sr, D’Agostino R, Jr, Vasan R. Evaluating the added predictive ability of a new marker: From area under the ROC curve to reclassification and beyond. Statistics in Medicine. 2008;27:157–172. doi: 10.1002/sim.2929. [DOI] [PubMed] [Google Scholar]
- Pepe MS. The statistical evaluation of medical tests for classification and prediction. Oxford University Press; 2003. [Google Scholar]
- Pepe MS, Janes H, Longton G, Leisenring W, Newcomb P. Limitations of the odds ratio in gauging the performance of a diagnostic, prognostic, or screening marker. American Journal of Epidemiology. 2004;159:882–890. doi: 10.1093/aje/kwh101. [DOI] [PubMed] [Google Scholar]
- Pepe MS, Feng Z, Gu JW. Comments on Evaluating the added predictive ability of a new marker: From area under the ROC curve to reclassification and beyond by M. J. Pencina et al. Statistics in Medicine. 2008;27:173–181. doi: 10.1002/sim.2991. [DOI] [PubMed] [Google Scholar]
- Ridker P, Rifai N, Rose L, Buring J, Cook N. Comparison of C-reactive protein and low-density lipoprotein cholesterol levels in the prediction of first cardiovascular events. New England Journal of Medicine. 2002;347:1557–1565. doi: 10.1056/NEJMoa021993. [DOI] [PubMed] [Google Scholar]
- Ridker P, Buring J, Rifai N, Cook N. Development and validation of improved algorithms for the assessment of global cardiovascular risk in women: the Reynolds risk score. JAMA. 2007;297:611–619. doi: 10.1001/jama.297.6.611. [DOI] [PubMed] [Google Scholar]
- Ripley B. Pattern recognition and neural networks. Cambridge University Press; 1996. [Google Scholar]
- Speigelhalter D. Probabilistic predictin in patient management and clinical trials. Statistics in Medicine. 1986;5:421–433. doi: 10.1002/sim.4780050506. [DOI] [PubMed] [Google Scholar]
- Thune J, Carlsen C, Buch P, Seibak M, Burchardtb H, Torp-Pedersen C, Kober L. Different prognostic impact of systolic function in patients with heart failure and/or acute myocardial infarction. European Journal of Heart Failure. 2005;7:852–858. doi: 10.1016/j.ejheart.2005.01.019. [DOI] [PubMed] [Google Scholar]
- Tian L, Cai T, Goetghebeur E, Wei LJ. Model evaluation based on the distribution of estimated absolute prediction error. Biometrika. 2007;94:297–311. [Google Scholar]
- Uno H, Cai T, Tian L, Wei LJ. Evaluating prediction rules for t-Year survivors with censored regression models. Journal of American Statistical Association. 2007;102:527–537. [Google Scholar]
- Wang T, Gona P, Larson M, Tofler G, Levy D, Newton-Cheh C, Jacques P, Rifai N, Selhub J, Robins S, Benjamin E, D’Agostino R, Vasan R. Multiple biomarkers for the prediction of first major cardiovascular events and death. New England Journal of Medicine. 2006;355:2631–2639. doi: 10.1056/NEJMoa055373. [DOI] [PubMed] [Google Scholar]
- Youden W. An index for rating diagnostic tests. Cancer. 1950;3:32–35. doi: 10.1002/1097-0142(1950)3:1<32::aid-cncr2820030106>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Zhou XH, Obuchowski NA, McClish DK. Statistical methods in diagnostic medicine. John Wiley & Sons; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.