Summary
Natural killer (NK) cells are circulating lymphocytes that function in innate immunity and placental reproduction. Regulating both development and function of NK cells is an array of variable and conserved receptors that interact with major histocompatibility complex (MHC) class I molecules. Families of lectin-like and immunoglobulin-like receptors are determined by genes in the natural killer (NKC) and leukocyte receptor (LRC) complexes, respectively. As a consequence of the strong, varying pressures on the immune and reproductive systems, NK cell receptors and their MHC class I ligands evolve rapidly, are highly diverse, and exhibit dramatic species-specific differences. The variable, polymorphic family of killer cell immunoglobulin-like receptors (KIR) that regulate human NK cell development and function evolved recently, from a single-copy gene during the evolution of simian primates. Our studies of KIR and MHC class I genes in representative species show how these two unlinked but functionally intertwined genetic complexes have co-evolved. In humans, combinations of KIR and HLA class I factors are associated with infectious diseases, including HIV/AIDS, autoimmunity, reproductive success and the outcome of therapeutic transplantation. The extraordinary, and unanticipated, divergence of human NK cell receptors and MHC class I ligands from their mouse counterparts can in part explain the difficulties experienced in finding informative mouse models for human diseases. Non-human primate models have far greater potential, but to realize their promise will first require more complete definition of the genetics and function of KIR and MHC variation in non-human primate species, at a level comparable to that achieved for the human species.
Keywords: Non-human primates, NK cells, KIR, MHC, innate immunity
Natural killer (NK) cells comprise 5-25% of the lymphocytes circulating in human peripheral blood [1, 2]. NK cells are larger than circulating B and T lymphocytes, having a well-developed cytoplasm that contains cytotoxic granules. NK cells were described by morphologists as large granular lymphocytes long before their immunological properties became a subject for study in the 1970s [3-6]. At that time numerous immunologists were studying antigen specificity and major histocompatibility complex (MHC) restriction of the cytotoxic T cells that arose from long-term stimulation with antigen and cytokines in culture. The defining property that distinguished NK cells from killer T cells was their propensity to kill certain tumor cell lines, such as the K562 erythroleukemia cell line, without needing an extensive period of in vitro stimulation and proliferation. NK cells were thus seen to have a natural or inherent ability to kill damaged or dangerous cells. They are also potent secretors of cytokines, notably interferon-γ. The kinetics observed in culture reflect those of the immune response to infection: NK cells are fast-acting lymphocytes that participate in the innate immune response to primary infection, whereas the slow-acting T cells are central to the adaptive immune response.
NK cell receptors for MHC class I
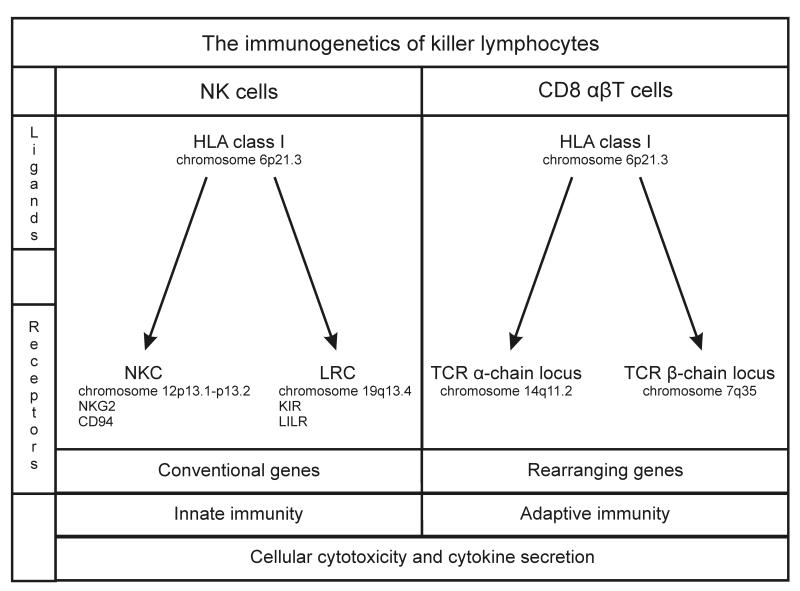
In phenotype, function and development there are many parallels between NK cells and T cells, particularly the CD8αβ T cells. A key similarity is the central role of receptors for class I major histocompatibility complex (MHC) molecules in cellular development and effector function. A key difference is that CD8αβ T cells use two somatically rearranging genes (encoding the α and β chains) to produce a vast repertoire of variable MHC class I receptors, whereas NK cells achieve a diverse but more modest repertoire through differential expression of a range of cell-surface receptors, all of which are encoded by conventional (non-rearranging) genes (Figure 1) [7]. Most of these receptors belong to one of two structural groups: either receptors resembling C-type lectins and encoded by genes in the natural killer complex (NKC) on human chromosome 12p13.1, or receptors made up of immunoglobulin-like domains and encoded by genes in the leukocyte receptor complex (LRC) on human chromosome 19q13.4 [8, 9].
Figure 1. The immunogenetics of killer lymphocytes.
Compares and contrasts the MHC class I receptors of NK cells and CD8 αβT cells.
NK cells respond to cells with perturbed expression of MHC class I
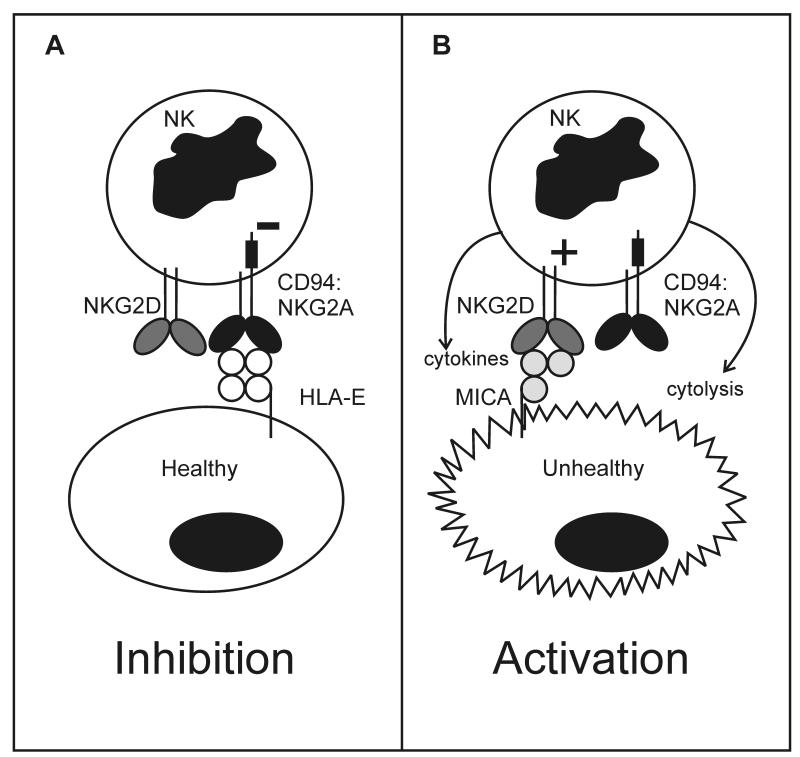
Individual NK cells express diverse combinations of activating and inhibitory receptors that recognize either MHC class I, or molecules related structurally to MHC class I. Interactions between these ligands and receptors during NK cell development create a signaling balance that prevents mature NK cells from attacking healthy cells of the body [10]. On the other hand, when MHC class I expression becomes perturbed by infection, malignant transformation or other forms of stress, the affected cells become susceptible to NK cell attack. This general property will be illustrated with a simplified example involving two NKC-encoded lectin-like receptors: NKG2D and CD94: NKG2A (Figure 2).
Figure 2. Inhibitory MHC class I receptors enable NK cells to respond to ‘missing self’ MHC class I.
Mature NK cells express inhibitory receptors that interact with MHC class I expressed by healthy cells (self MHC class I), generating signals that prevent the NK cells from responding to self. This is illustrated in the left panel for a human NK cell that expresses the inhibitory HLA-E specific receptor CD94:NKG2A and the activating receptor NKG2D, specific for MICA. Encoded by an MHC gene, the latter is structurally similar to the heavy chains of MHC class I (MIC is the acronym for MHC class I-like chain, and A refers to the first of several MIC genes and pseudogenes) but unlike them does not associate with β2-microglobulin (β2-m) [111]. The healthy cell expresses insufficient MICA to engage NKG2D and generate an activating signal, while expressing a normal amount of HLA-E that engages CD94:NKG2A and sustains NK cell inhibition. In this manner, inhibitory receptor recognition of normal self HLA class I prevents NK cells from attacking healthy cells of the body. Shown in the right panel is the situation for a cell that has become unhealthy through infection with a virus or malignant transformation. Such situations of cellular stress, induce increased cell-surface expression of MICA, while expression of HLA class I is often reduced. The former leads to engagement of NKG2D and the generation of activating signals, while the latter will reduce or eliminate engagement of CD94:NKG2A, thus attenuating inhibitory signaling. These two effects co-operate to activate the NK cell, which can then kill the unhealthy cells and secrete cytokines such as inteferon-γ that recruit and activate other immune system cells.
CD94:NKG2A is an inhibitory NK cell receptor that recognizes complexes of HLA-E, a non-classical MHC class I molecule, and peptides derived from the leader peptides of HLA-A, B and C [11]. NKG2D is an activating receptor that recognizes the class I-like molecule MICA. Poorly expressed by healthy cells, MICA is up-regulated upon viral infection or other forms of stress [12, 13]. Figure 2A shows the interaction of an NK cell that expresses CD94:NKG2A and NKG2D with a healthy cell of the body. Because the healthy cells expresses a normal complement of HLA class I and lacks expression of MICA, HLA-E will engage CD94:NKG2A and generate an inhibitory signal, whereas NKG2D will neither detect a ligand nor signal. The net result being that the NK cell does not respond to the healthy cell.
Figure 2B shows the interaction of the NK cell with an unhealthy cell. Common features of viral infection and malignant transformation are increased expression of MICA at the cell surface and decreased expression of HLA class I. As a consequence the supply of peptides derived from HLA-A, B and C leader sequences decreases, causing reduction in the amount of HLA-E reaching the cell surface. In this situation there is less engagement of CD94:NKG2A, which reduces the generation of inhibitory signals in the NK cell. Concomitantly, the NKG2D activating receptor binds to MICA and starts to produce activating signals that tip the balance towards NK cell activation, cytokine secretion and killing of the unhealthy cell. In this process of NK cell recognition of unhealthy cells, the inhibitory receptors enable NK cells to detect losses in normal MHC class I expression. For this reason, NK cells were described historically as recognizing ‘missing self’ [14]. Consistent with this mechanism, target cell lines like K562 that were instrumental to the discovery of natural killing [3-6] were ones deficient in MHC class I expression.
Conserved and variable NK cell receptors for MHC class I
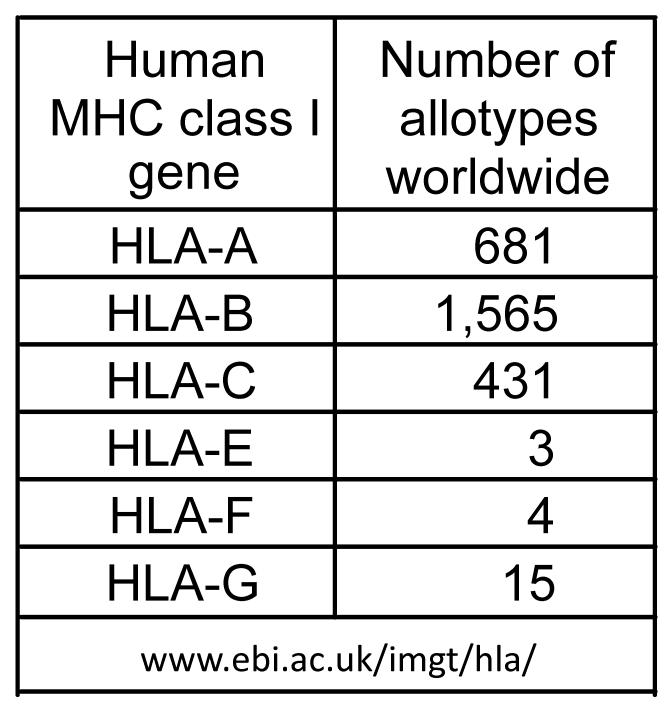
Humans have six functional genes that are either highly polymorphic (HLA-A, B and C) or highly conserved (HLA-E, F and G) (Figure 3). With the exception of HLA-F, for which a function has yet to be discovered [15, 16], all these HLA class I genes encode ligands for NK cell receptors. Like HLA class I, some of the receptors are variable and others are conserved. The receptor-ligand interaction between CD94:NKG2A and HLA-E is highly conserved. There is almost no structural variation in the CD94, NKG2A and HLA-E polypeptides [17] and the functional effects of this interaction are remarkably similar in different individuals. In primate phylogeny these receptors are relatively conserved and functional equivalents are present in mice [18].
Figure 3. Polymorphism of human MHC class I.
For each of the six functional HLA class I genes the number of protein variants detected worldwide is listed. These data were obtained from the Immunogenetics HLA (IMGT/HLA) database (www.ebi.ac.uk/imgt/hla/) as last updated on 17th November 2009.
Contrasting with CD94:NKG2A are the killer cell immunoglobulin-like receptors (KIR) that recognize determinants of the highly polymorphic HLA-A, B, C molecules as well as HLA-G [19]. KIR are encoded by a family of LRC genes for which there is haplotypic variability in gene content as well as polymorphism of the component genes. Within the human population, the number and nature of functional interactions between KIR and HLA class I varies greatly. Even within families there is no guaranteed inheritance of both a ligand and its cognate receptor, because the KIR and HLA genes segregate on different chromosomes (Figure 1). Thus CD94:NKG2A and inhibitory KIR represent constant and variable versions of the same basic function -- though mediated by structurally different receptors -- that play complementary roles in NK cell development and function. Their distributions in phylogeny show that CD94:NKG2 is the older function and KIR the more recently evolved.
CD94:NKG2A is a heterodimeric receptor in which the NKG2A polypeptide chain provides the inhibitory signaling function. Another receptor, CD94:NKG2C has identical ligand specificity for HLA-E but has activating function provided by NKG2C [20]. Although the CD94:NKG2C does not appear to contribute to the development of missing-self recognition by NK cells, the frequency of cells expressing the receptor increases with cytomegalovirus infection [21]. This is evidence for the role that NK cells play in defense against virus infection. The first such evidence was the case of a young immunodeficient patient lacking NK cells, who suffered a succession of herpes virus infections, eventually leading to death [22].
Human KIR distinguish five epitopes of HLA class I
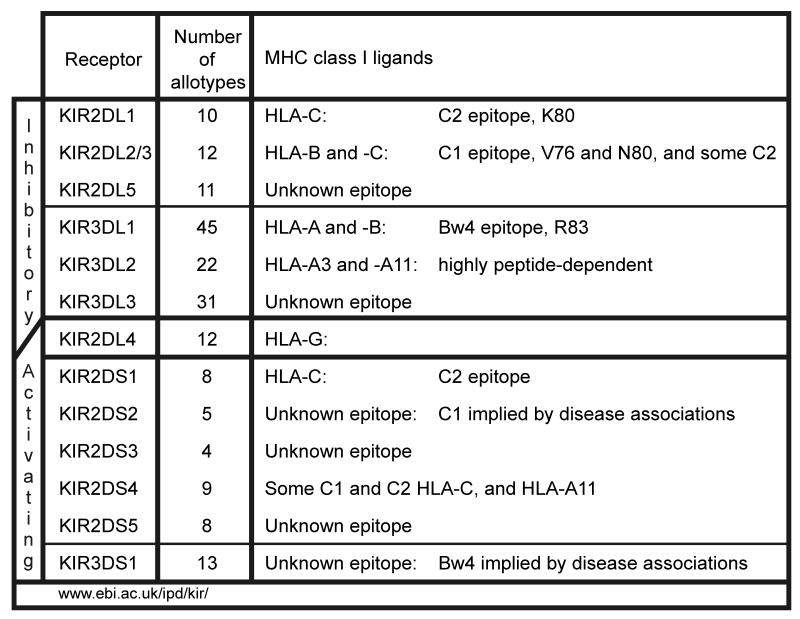
KIR have two or three extracellular Ig-like domains and are named accordingly, KIR2D or KIR3D (Figure 4). The Ig domains mutually interact to form the binding site for MHC class I [23]. KIR can either be inhibitory receptors (KIR2DL or KIR3DL) with a long cytoplasmic tail containing ITIM motifs, or activating receptors (KIR2DS or KIR3DS) with a short cytoplasmic tail and a positively charged residue in the transmembrane domain that interacts with the signaling adaptor DAP12. One exception to this rule is KIR2DL4 that has the potential for both inhibitory and activating functions [24-26]. The alleles of each KIR gene are numbered in series [27], using principles based upon those used for the HLA genes [28].
Figure 4. Function and polymorphism of human killer cell immunoglobulin-like receptors.
The receptors are grouped according to their activating and inhibitory functions, in which regard KIR2DL4 is unique having structural elements associated with both activation and inhibition. Under allotypes is given the number of protein variants for each KIR as given in the KIR database (www.ebi.ac.uk/ipd/kir/) of the Immuno Polymorphism Database (IPD), as last updated on the 20th February 2009. Under MHC class I ligands is given the reactivity of each KIR with HLA class I.
Inhibitory KIR recognize two HLA-C epitopes carried by non-overlapping subsets of HLA-C allotypes [29]. The C2 epitope defined by lysine at position 80 is recognized by KIR2DL1, whereas the C1 epitope defined by asparagine at position 80 is recognized by KIR2DL2/3 [30]. Since all HLA-C allotypes have either lysine or asparagine at position 80, they can all function as a KIR ligand. Individuals can be either C1 homozygous, C2 homozygous or C1/C2 heterozygous. This contrasts with HLA-A and B, for which only a minority of allotypes are KIR ligands. Thus HLA-C is the dominant HLA class I locus that provides KIR ligands. KIR2DS1 is an activating receptor that has similar Ig-like domains to inhibitory KIR2DL1 and also binds the C2 epitope, although with lower avidity [31]. KIR2DS2 is an activating receptor that has similar Ig-like domains to inhibitory KIR2DL2/3 but has no detectable avidity for C1 [32]. In comparison to the inhibitory KIR, the ligand specificity and functions of the activating KIR remain poorly understood.
Although all HLA-B allotypes have asparagine 80 the vast majority lack the C1 epitope and cannot interact with KIR2DL2/3. This is because they lack valine 76, a fixture of HLA-C and essential for KIR interaction [33]. Two exceptional HLA-B allotypes are HLA-B46 and HLA-B73 that both have valine 76 and are good ligands for KIR2DL2/3 [34]. HLA-B46 is the product of a gene conversion between HLA-B and HLA-C that occurred relatively recently in South-east Asia. Subsequently, HLA-B46 was driven to high frequency in South-east Asian populations under natural selection [35]. HLA-B73, in contrast, appears to be the last remnant in humans of an ancient lineage of MHC-B allotypes that remains well represented in chimpanzees [36].
The Bw4 epitope, carried by minority subsets of HLA-A and B allotypes, is recognized by the inhibitory receptor, KIR3DL1 [37, 38]. Bw4 is defined by a sequence motif at residues 77-83, in which arginine 83 is the critical residue [39, 40]. In all populations, approximately ~50% of MHC haplotypes encode a Bw4-bearing HLA-A and/or a HLA-B allotype. Thus ~75% of individuals have Bw4 and ~25% of individuals lack it. The inhibitory receptor KIR3DL2 has a relatively narrow specificity for HLA-A3 and A11 that is also dependent upon the bound peptide [41]. In most populations a minority of individuals carry this epitope. As a consequence of these distributions a substantial number of individuals have C1 as their only KIR ligand. The absence of a ligand however, does not prevent expression of the cognate KIR by NK cells [10].
KIR2DL4 recognizes HLA-G, an interaction implied to take place in endocytic vesicles [26]. The expression of HLA-G is restricted to extravillous trophoblast, the fetal tissue that interacts with maternal, uterine NK cells during placentation in reproduction [42]. Trophoblast also expresses HLA-C, but neither HLA-A nor HLA–B [43]. Interactions of trophoblast HLA-C and HLA-G with their cognate KIR on uterine NK cells are believed to control the conversion, by the trophoblast, of maternal spiral arteries into high conductance channels that provide the placenta with blood and the growing fetus with gases and nutrients [44]. Also contributing to the interactions between trophoblast and NK cells is the LILRB1 NK cell receptor, which has a broad specificity for HLA class I and particularly high avidity for HLA-G [43, 45]. This receptor is encoded in the leukocyte immunoglobulin-like receptor (LILR) gene family, which is next to the KIR locus [46]. Thus NK cells, not only function as an essential component of the immune system, they also play a crucial role in the reproductive system.
KIR2DS4, a common and widespread activating receptor, has a unique ligand-specificity for subsets of HLA-C allotypes carrying C1 and C2 epitopes and HLA-A11. The latter reactivity is the consequence of a gene conversion which gave KIR2DS4 a sequence that otherwise characterizes KIR3DL2 [47]. Ligand specificities for the activating receptors KIR2DS3 and KIR2DS5 and the inhibitory receptor KIR2DL5 remain mysterious.
Developmental interactions between MHC class I and inhibitory receptors educate NK cells to recognize missing self
For cytotoxic T cells it is well established that MHC class I molecules are essential for both their development (positive and negative selection in the thymus) and their response to infection: the recognition by αβ T cell receptors of pathogen-derived peptides that are presented by MHC class I [48]. In analogous fashion, interactions between MHC class I and cognate inhibitory NK cell receptors influence the development and effector function of NK cells. During NK cell development, the expression of CD94:NKG2A and KIR occurs at a late stage, which is also the time when the cells become competent to respond to missing self with cell killing and cytokine secretion [49]. In peripheral blood there is a subpopulation of immature NK cells that express neither CD94:NKG2A nor KIR. Whereas NK cells bearing CD94:NKG2A and/or inhibitory KIR strongly respond to MHC class I deficient cells, the receptor-null cells are non-responsive. It thus appears that during development, interactions between self MHC class I and cognate inhibitory receptors are responsible for determining the capacity of the mature NK cell’s response to cells in which the expression of self MHC class I is perturbed. This involvement of HLA class I in the development of human NK cells is described as the ‘education’ of NK cells [50].
One way to test the degree of education in an NK cell population is to examine the response to target cells lacking MHC class I, as shown in Figure 5. Comparison of NK cells from different individuals, shows that cells expressing CD94:NKG2A have a similar response to the 221 B cell line that lacks cell-surface expression of HLA class I. This uniformity is consistent with the structural conservation of HLA-E and CD94:NKG2A [17] and the capacity of leader peptides from a majority of HLA-A, B and C allotypes to bind HLA-E and form functional ligands for CD94:NKG2A. Contrasting with this constant aspect of NK cell education, is the responsiveness of NK cells expressing inhibitory KIR, as is exemplified in Figure 5 using KIR3DL1 the receptor specific for the Bw4 epitope of HLA-A and –B.
Figure 5. Developmental interaction between MHC class I and cognate inhibitory receptors ‘educate’ NK cells to respond to missing self.
Shown are the results of assays in which peripheral blood NK cells are cultured with target cells of the class I deficient 221 B cell line. Using flow cytometry selected subsets (either expressing CD94:NKG2A or KIR3DL1) of NK cells were analysed for presence of intracellular interferon-γ. Blood donors were selected based upon their HLA class I and KIR types. All donors carried the Bw4 epitope. For further details see Yawata et al [52].
Most individuals have the KIR3DL1 gene, which is expressed by a subpopulation of their NK cells. Such expression is not dependent upon the presence of a Bw4-bearing HLA-A or HLA-B allotype that interacts with KIR3DL1 [51], as is also the case for other KIR [10]. Thus it is a common phenomenon for some NK cells to express receptors for which there is no physiological ligand. Consequently, the response to class I deficient cells can be compared for KIR3DL1-expressing NK cells that have developed in the presence and absence of a particular HLA class I ligands. Although HLA-B51, -B44, -A24 and -B13 all carry the Bw4 epitope as defined by amino-acid sequence and HLA serology, they differ in their capacity to educate KIR3DL1-expressing NK cells (Figure 5) [52]. Whereas HLA-B51 and –B44 educate KIR3DL1-expressing NK cells to a higher level than achieved for CD94:NKG2A-expressing NK cells, HLA-A24 is less effective and HLA-B13 ineffective. This variability can be attributed to polymorphisms at sites away from the Bw4 sequence motif and/or differences in bound peptide, which are demonstrated to influence the interaction of KIR and HLA class I [41, 53, 54]. Similar educating effects to those shown in Figure 5, are also observed with HLA-C and NK cells expressing the receptors for the C1 and C2 specific KIR [50, 52]. These analyses show how the polymorphism of HLA-A, B and C provides a variable education of KIR-expressing NK cells, contrasting with the constant education by HLA-E of CD94:NKG2A-expressing NK cells.
KIR3DL1 is highly polymorphic [55] (Figure 4) in ways that effect the level of cell-surface expression and the avidity of binding to Bw4 [56, 57]. As shown in Figure 5, the amino-acid substitutions that distinguish KIR3DL1 allotypes also affect the education of NK cells by Bw4. Thus NK cells expressing the 015 or 005 allotypes of KIR3DL1 are better educated than those expressing CD94:NKG2A, whereas cells expressing the 007 allotype cells are educated to similar or slightly lower level. Having no effect is 3DS1 the activating counterpart of KIR3DL1, which segregates as an allele of the same locus (KIR3DL1/S1), for which the dominant form of 3DS1 (013) is the most prevalent allele worldwide [55].
Epidemiological studies of patients infected with human immunodeficiency virus (HIV) have demonstrated how different combinations of KIR3DL1 and HLA-B alleles affect the rate at which the HIV infection progresses to acquired immunodeficiency syndrome (AIDS). From initial analysis of HLA alone, the Bw4 epitope was shown to be associated with a slower progression to AIDS than the alternative Bw6 epitope (which is not recognized by any NK cell receptor) [58]. The magnitude of the effect, however, was increased when combinations of HLA and KIR3DL1/S1 were considered [59, 60]. The greatest protection was offered by the combination of the Bw4-bearing HLA-B57 and KIR3DL1 allotypes such as 015 that are expressed at relatively high abundance at the NK cell surface. The next most protective effect was seen for the combination of the Bw4–bearing HLA-B27 and KIR3DL1 allotypes such as 005 that are expressed at relatively low abundance at the NK cell surface. These data point to the functional co-evolution between particular KIR3DL1 and HLA-B alleles.
Laboratory mice do not use KIR as NK cell receptors for MHC class I
Historically, much twentieth century immunological research was performed on animal species, such as horses and rabbit, from which substantial quantities of blood, antibodies and serum could be obtained and studied. In this regard, the mouse was frugal, but its great advantage was the ability to withstand total inbreeding by humans without losing the capacity to reproduce. The MHC was initially discovered and subsequently defined using inbred strains of mice, and later, with appreciation that T-cell responses were restricted by MHC allotypes, the laboratory mouse ascended to become the dominant animal model for immunological research. The analytical power of the mouse system has been so seductive that it is not uncommon for mouse immunologists to refer to ‘the human model’ without any trace of irony.
Emerging in the 1990s was the inconvenient truth that the inbred laboratory mouse (Mus musculus domesticus) lacked an expanded family of KIR genes that function as NK cell receptors for MHC class I. In the mouse LRC there are no KIR genes at all, but at some point in history they were moved to the X chromosome where they now comprise a pair of genes: Kirl1 and Kirl2 [61, 62]. Kirl1 is not expressed, but Kirl2 is expressed “in subregions of the mouse brain where synaptic plasticity and neurogenesis occur, including olfactory bulbs, rostral migratory stream and dentate gyrus of hippocampus” [63]. Thus for laboratory mice, the one functional KIR appears to be a component of the nervous system.
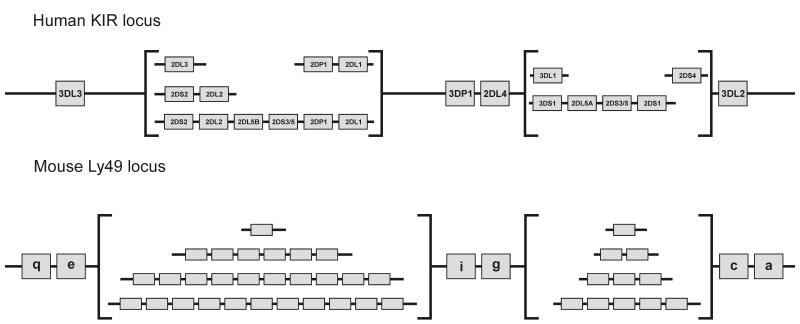
Instead of using LRC-encoded immunoglobulin-like receptors as variable NK cell receptors for polymorphic MHC class I, mice use a family of NKC-encoded lectin-like receptors called Ly49 [64]. Not only are the receptors of different structure but they bind to completely different sites on the MHC class I molecule [23, 65]. Whereas KIR interact with the top-face of the peptide-binding site, in a manner with some similarities to the αβ T cell receptor, Ly49 variants interact with sites underneath the peptide-binding site and involving contacts with the two immunoglobulin-like domains of MHC class I: β2-microglobulin and the α3-domain. Given the structural differences between KIR and Ly49, they clearly have independent origins. Thus the functional congruence of KIR and Ly49 as NK cell receptors for polymorphic MHC class I presents a text-book case of convergent evolution. As a result of the similar forces of natural selection that have operated on human KIR and mouse Ly49, the organization of the two gene families is remarkably similar (Figure 6) as is their variegated mode of expression, whereby individual NK cells express different combinations of either KIR or Ly49 in stable fashion [10, 66, 67], and the evolution of activating receptors from inhibitory receptors [68].
Figure 6. Comparison of the organization of the gene families encoding human KIR and mouse Ly49 NK cell receptors.
Human KIR and mouse Ly49 receptors serve similar functions as highly diverse NK cell receptors for MHC class I. However these functions have evolved by convergent evolution, because Ly49 are lectin-like receptors encoded in the natural killer cell complex (NKC) and KIR are immunoglobulin-like receptors encoded in the leukocyte receptor complex (LRC). Despite their independence the gene families encoding mouse Ly49 and human KIR have a similar organization with a framework defining two regions of gene-content variability. Shown in the figure are the most common gene-content motifs in the centromeric and telomeric regions of human KIR haplotypes in Caucasian populations [112, 19], and the organization of the four mouse Ly49 haplotypes sequenced [69]. In the human genome the Ly49 gene family is represented by one pseudogene [113, 114]; in the mouse genome the KIR family is represented by one pseudogene and one functional gene that is not expressed by NK cells [63]. A variety of human haplotypes with different gene-content from those shown in the figure have been described and are the results of gene duplication or deletion arising from non-homologous recombination. Such haplotypes are either rarer than those comprising the common gene-content motifs, and/or restricted to non-Caucasian populations [115-120].
Whereas the mouse NKC contains a family of functional Ly49 genes [69], the human NKC contains a single non-functional Ly49 gene [9]. Conversely, the human LRC contains a family of KIR genes, whereas the mouse LRC has none [61, 62]. Thus the ascendancy of KIR in humans is associated with the loss of Ly49 function. Conversely the ascendancy of Ly49 gene function in mice, is associated with loss of KIR function as NK cell receptors.
Species comparisons clearly demonstrate the evolutionary instability of variable NK cell receptors for MHC class I [70, 71]. The fundamental cause of this instability is almost certainly to be the diverse and variable pressures imposed upon the immune response by infectious agents. Such pressures could act directly on the NK cell response, or indirectly via the T cell response with selection of novel MHC class I variants that have altered interaction with NK cell receptors. That MHC class I genes are themselves unstable is also evident from species comparisons [72]. Only closely-related species have orthologous MHC class I genes, comparison of the human and mouse MHC having revealed little evidence for correspondence between individual genes in the two species. The last debate on this issue is the possible orthology of human HLA-E and mouse H2-Qa1, both of which bind peptides derived from the leader sequences of other MHC class I molecules from their respective species, forming ligands for CD94:NKG2 NK cell receptors [18, 73]. Although the functional similarities are striking, there is only weak phylogenetic evidence in their sequences to support an orthologous relationship. One side of the debate is that species-specific recombination between MHC class I genes (concerted evolution) has obscured the common origin, the other is that similar but independent evolutionary pressures have produced the same function in two different MHC class I genes.
Although the mouse model has been highly informative in identifying basic immune mechanisms, it has been unimpressive in producing informative and translatable models of human diseases [74]. Although the basic principal of NK cells having MHC class I receptors is the same in humans and mice, there are dramatic and qualitative differences between the human system of KIR interactions with HLA and the mouse system of Ly49 interactions with H-2. The extent of these differences, and those being uncovered in many other families of immune-system genes [74], including ligands for NKG2D [75] helps explain the inherent limitations of mouse as a model for the human. In our own work the comparison of mouse and human was instrumental in revealing the instability of NK cell receptor systems, but in order to gain better perspective on human KIR it was necessary to study other species, particularly non-human primates.
The modern family of human KIR3DL genes originated in the simian primates
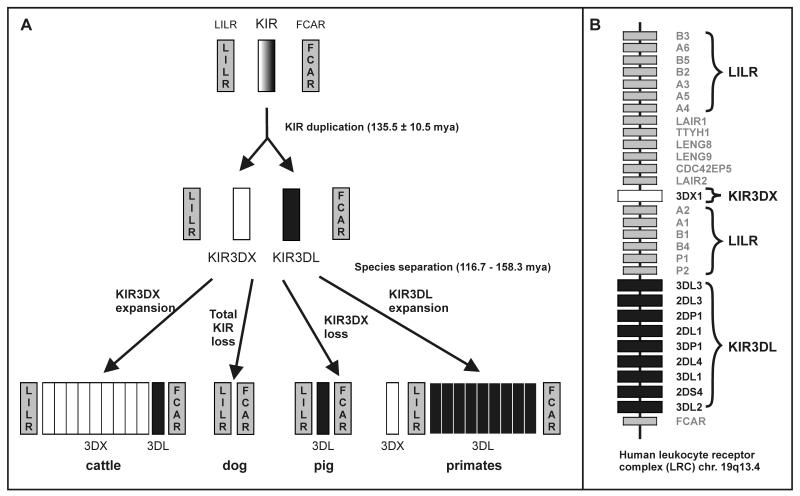
Comparison of species from several mammalian orders showed that cattle (Bos taurus) constitutes the only non-primate species to have a diverse family of KIR genes. Phylogenetic analysis of primate and cattle KIR sequences shows the first step in KIR diversification was duplication of the ancestral KIR3D gene around 136 million years ago to give daughter genes KIR3DL and KIR3DX [76] (Figure 7A). In primates the KIR3DL gene subsequently expanded to become a diverse gene family, while the KIR3DX gene became non functional, now being found in the human LRC at some distance from the functional KIR locus, within the region occupied by the LILR genes (Figure 7B). Conversely, in cattle, the KIR3DX gene expanded and diversified, while KIR3DL remained a single copy gene. While cattle and primates have retained both KIR3DL and KIR3DX genes the pig (Sus scrofa) is seen to have lost KIR3DX [77] and the dog (Canis familiaris) has lost both KIR3DX and KIR3DL [78].
Figure 7. Origin of KIR gene families in cattle and simian primates.
Panel A: duplication of the ancestral KIR gene gave rise to founders of the KIR3DX and KIR3DL lineages. In the evolution of cattle, KIR3DX expanded and diversified while KIR3DL remained a single-copy gene. In the evolution of simian primates, KIR3DX remained a single gene while KIR3DL became a diverse gene family [76]. In addition to mice (Mus musculus domesticus), dog (Canis familiaris) and probably cat (Felis catus) have no KIR genes in the LRC [78] and pig (Sus scrofa) [121], seals (Phoca vitulina) and sea lions (Zalophus californianus) [78] have lost KIR3DX while retaining a single KIR3DL gene. Panel B: the organization of genes in the human Leukocyte Receptor Complex (LRC). Note that 3DX1 (originally called KIR3DL0 and a non-functional gene) [77]) is situated between the two groups of leukocyte immunoglobulin-like receptors (LILR). The KIR gene content corresponds to that of the A haplotype [46].
In conclusion we see that the diversification of the KIR3DL gene to form a repertoire of NK cell receptors is a specific product of primate evolution. We have therefore compared representatives of the different groups of non-human primate species to determine when and in what manner the expansion of the KIR3DL genes occurred. In a prosimian species, the mouse lemur (Microcebus murinus), the KIR3DL is represented by a single pseudogene, and the single Ly49 gene is also a pseudogene. Instead of a diverse family of KIR or Ly49 genes the mouse lemur has an expansion of CD94 and NKG2 genes [79] and a diverse family of MHC class I genes [80]. Four MHC class I pseudogenes are present in the MHC on chromosome 6, but the six functional class I genes and three additional pseudogenes are present on chromosome 26. In neither location is there a gene corresponding to HLA-E. Thus the potential MHC class I ligands for the diverse lemur CD94:NKG2 receptors likely lie with products of the prosimian-specific MHC class I genes on chromosome 26 [79].
In contrast to prosimians, all but one species of New World monkey, Old World monkey and ape studied have a rich diversity of KIR3DL. The overall picture is thus very clear, the expanded family of KIR3DL now present in all human populations originated in the simian primates and is ~40-58 million years old [81].
CD94:NKG2 and MHC-E are common to New World monkeys and humans
Among four New World monkeys studied the owl (Aotus nancymae), spider (Ateles geoffroyi) and squirrel (Saimiri boliviensis) monkeys were seen to have a diversity of KIR3DL genes, whereas the Titi monkey (Callicebus moloch) has just one gene. Phylogenetic analysis shows that the New World monkey KIR are specific to these species and bear no particular similarity with the four sublineages of KIR3DL that are common to humans, apes and old world monkeys [82, 83]. Reflecting this distinction is absence from New World monkeys of any MHC class I loci corresponding to the HLA-A, B, and C genes that encode ligands for human KIR. HLA-E is the only functional human MHC class I gene that has a strict orthologue in New World monkeys [84], but in humans it is the source of ligands for CD94:NKG2A, not KIR. In conclusion, New World monkeys are seen to have MHC class I [72, 85] and KIR [86] that are very different from those of Old World monkeys and hominoids, and have clearly taken a distinctive evolutionary trajectory from them.
Co-evolution of MHC-A, –B and lineage II KIR in Old World monkeys
Old world monkeys such as macaques (Macaca mulatta and Macaca fasicularis) have an expansion of MHC class I genes that correspond to the human HLA-A and HLA-B genes [87, 88] and exhibit high polymorphism [89-91]. In humans the two lineage II KIR genes (KIR3DL1 and KIR3DL2) provide the HLA-A, B receptors (Figure 8). Consistent with their expansion of MHC-A and -B, macaques have an expansion and diversity of lineage II KIR genes [92-95], as also seems to be true for the West African sabaeus monkey (Chlorocebus sabaeus) [96]. Sequence motifs that correspond to the Bw4 and Bw6 epitopes distinguished by the human Bw4-specific KIR3DL1, are present in MHC-B sequences of Old World monkeys. Thus there is the potential for Bw4-specific KIR in these species. In summary, the first evidence for MHC-A and B and lineage II KIR is seen in Old World monkeys, where their co-evolution led to a unique expansion of the genes encoding both the ligands and their receptors.
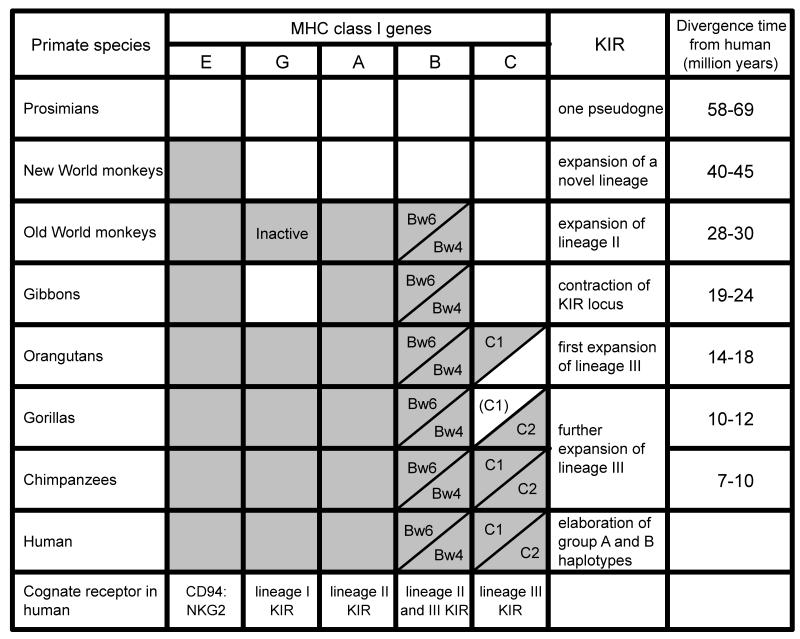
Figure 8. Co-evolution between MHC class I genes and KIR in primates.
Shows the distribution in non-human primates of genes orthologous to human HLA-A, B, C, E, and G. The species distribution of the Bw/Bw6 epitopes and the C1/C2 epitopes distinguished by KIR is also shown. Few gorilla MHC-C alleles have been defined and they all have the C2 epitope. Thus C1 may have been lost from gorilla. Under ‘KIR’ are described the changes in the KIR locus that accompany the changes in MHC class I. For each species the time since the last common ancestor with humans is given.
Co-evolution of MHC-C and lineage III KIR in great apes
In Old World monkeys, the lineage III KIR is represented by a single gene [97]. This situation changes with the orangutan, where the emergence of MHC-C is associated with the expansion of lineage III KIR and their evolution as MHC-C receptors [82]. From phylogenetic analysis and ancestral reconstruction it is likely that MHC-C evolved and differentiated from one of the multiple MHC-B observed in Old World monkeys and which carried the C1 epitope. In orangutans (Pongo pygmaeus and Pongo abelii) the MHC-C locus is not fixed, as in humans, but present on ~50% of MHC haplotypes. Consequently, 75% of orangutans have MHC-C and 25% do not. A further difference from the human situation is that all orangutan MHC-C allotypes have asparagine 80 and express the C1 epitope. Thus the C2 epitope defined by lysine 80 is absent from orangutan. Consistent with this distribution the orangutan has C1-specific KIR but no C2-specific KIR. Thus the orangutan resembles an evolutionary intermediate in which C1 epitopes and C1-specific receptors are in place, but the C2 epitope and C2-specific receptors have not yet evolved [98].
In chimpanzees (Pan trogolodytes and Pan paniscus) and gorilla (Gorilla gorilla) the MHC-C locus has become fixed, and both the C2 epitope and cognate KIR have evolved. The complexity and gene-content diversity that is apparent in the MHCs of Old World monkeys has largely gone, to be replaced by the relatively simple system of six, fixed functional MHC class I genes. Lineage III KIR is represented by nine genes in chimpanzee, none of them being fixed in the genome and which are carried in diverse combinations by KIR haplotypes. Eight of the lineage III KIR are high avidity MHC-C receptors and comprise two inhibitory and one activating C1 receptors, plus four inhibitory and one activating C2 receptors [36]. In chimpanzee some 25% of MHC-B allotypes have the C1 epitope and are functional ligands for the C1-specific KIR. The ninth lineage III KIR (activating KIR2DS4) has undergone gene conversion with a KIR3DL2-like lineage II KIR and has a hybrid specificity for HLA-A11 and some MHC-C [47]. That 8 of the 13 functional KIR genes are specific for either C1 or C2 illustrates the dominance of this form of NK cell regulation in the chimpanzee.
Erosion of the KIR locus in gibbons accompanies loss of MHC-G and lack of MHC-C
Overall the comparison of non-human primate species leads to a picture in which changes in the character of the KIR locus correlate with changes in the MHC class I genes. Furthermore there is a progression to this evolution, in which the complexity of the KIR locus increases as we have considered in order prosimians, monkeys, and great apes. Not fitting in with this progression are the gibbons, species of smaller or lesser apes that are equally related to humans and great apes [83].
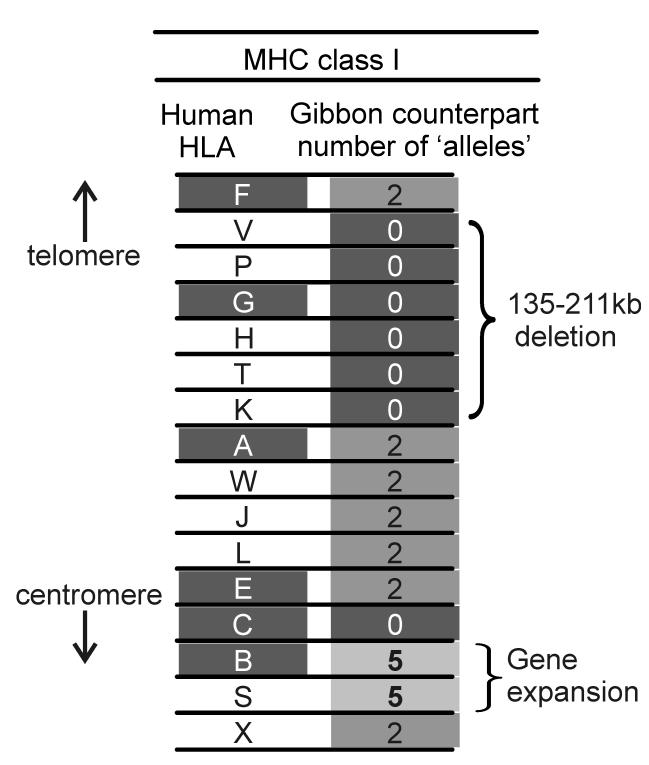
By analyzing the raw sequencing data from the shotgun sequencing project for the white-cheeked gibbon (Nomascus leucogenys), we defined the family of MHC class I genes in one gibbon individual (Figure 9). Most of these genes were like human MHC class I. Thus we found two alleles for genes orthologous to genes HLA-A, E, and F and pseudogenes HLA-W, J, L and X. Noticeably absent were counterparts to HLA-G and its flanking pseudogenes HLA-H, K, P, T and V. Thus the segment containing these MHC class I genes has been lost from the gibbon genome. Unlike Old World monkeys, where the inactivation of MHC-G is accompanied by presence of an active MHC-AG, we obtained no evidence for a gibbon equivalent of MHC-AG [99]. A further difference was found in the segment occupied by HLA-B and C in the human MHC. Here there was no gene corresponding to HLA-C, but five gibbon genes similar to HLA-B, a configuration similar to that seen in Old World monkeys. This suggests that gibbons either separated from the common ancestor of great apes and humans before the emergence of the MHC-C locus, or that MHC-C was lost by deletion in a manner similar to MHC-G [83].
Figure 9. Gibbon MHC class I genes.
Shows the set of MHC class I genes identified in the white-cheeked gibbon and their correspondence to human HLA class I genes [83]. The number of alleles for each gibbon gene is shown. The identification of five alleles for MHC-B and MHC-S is indicative of a triplication of these two genes in the gibbon MHC.
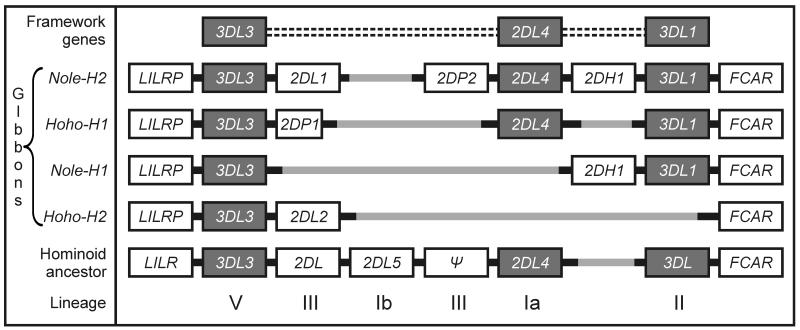
The KIR haplotypes of this same white-cheeked gibbon, as well as for a hoolock gibbon (Hoolock hoolock), were found to be short and did not preserve the three framework genes that characterize macaque, great ape and human KIR haplotypes (Figure 10). The absence of MHC-G correlates with the deletion of KIR2DL4, its cognate receptor, from two of the KIR haplotypes and the loss of inhibitory signaling motifs from the KIR2DL4 allotypes of the other two haplotypes. Absence of MHC-C correlates with the absence of a lineage III KIR from three of the four haplotypes, the only intact lineage III KIR being Nole-KIR2DL1. The small size and irregular organization of the KIR locus in gibbons is clearly due to the selection of deletion variants, because the ancestral hominoid KIR is predicted to have a full complement of framework genes as well as three additional genes. Thus during the evolution of gibbons, natural selection drove the erosion of the KIR locus. That HLA-C and HLA-G have the common property of being expressed by extravillous trophoblast and interacting with the KIR of uterine NK cells [43], raises the possibility that this selection came from pressure on the reproductive system rather than the immune system.
Figure 10. Short deleted KIR haplotypes in gibbons.
Comparison of the gene organization of four gibbon KIR haplotypes with that of the predicted ancestral haplotype for all hominoid species. The haplotypes are from two divergent gibbon species: the white-cheeked gibbon (Nomascus leucogenys leucogenys: Nole) and the eatern hoolock gibbon (Hoolock hoolock leuconedys: Hoho). Note the absence of the KIR2DL4 framework gene in two haplotypes and of the KIR3DL framework gene in one haplotypes. Gibbon evolution has been accompanied by the deletion of genes from the KIR locus [83].
The only framework gene present on all four gibbon haplotypes is KIR3DL3, for which the function remains unknown. In contrast to the degradation suffered by KIR2DL4, KIR3DL3 has the potential to be highly functional, all four variants having intact ITIMS in the cytoplasmic tail [83]. This also contrasts to human KIR3DL3, which has only one ITIM. Human KIR3DL3 is highly polymorphic and has an unusual promoter that selectively expresses KIR3DL3 in the CD56 bright NK cells that are a minority population in peripheral blood but are the majority of uterine NK cells [100, 101]. This opens up the possibility that KIR3DL3 has functions in reproduction that have been expanded and strengthened in gibbons, while the contributions of other KIR have dwindled in the absence of MHC-G and –C.
Human-specific evolution of HLA class I and KIR
Whereas chimpanzee (and orangutan) retain C1-bearing MHC-B allotypes at substantial frequency, the trend in human evolution was to lose these allotypes, thus making the C1 receptors increasingly specific for HLA-C. In the modern human population, HLA-B73 is the only remaining representative of a divergent lineage of C1-bearing HLA-B, and it is rare. However such C1-bearing HLA-B allotypes can still prove beneficial, as evidenced by HLA-B46, a product of recent recombination between HLA-B and HLA-C that has risen to high frequencies in South East Asian populations [35].
Chimpanzee and human have a comparable number of KIR genes (14 and 15, respectively) and a similar set of framework genes [97]. A major difference, however, is that all the variable chimpanzee KIR genes are present in the centromeric region, whereas human KIRs segregate between the centromeric and telomeric regions. Only three human and chimpanzee KIR are true orthologs, but of these only KIR2DL4 is in the same place in human and chimpanzee KIR haplotypes. While chimpanzee KIR2DL5 and KIR2DS4 are always in the centromeric region, human KIR2DS4 is only found in the telomeric region and human KIR2DL5 can be present in the centromeric region, the telomeric region or both. Since the time of the common ancestor there has been species specific reorganization of the KIR locus. A major effect of this reorganization is that in humans recombination at the center of the locus, between KIR2DL4 and KIR2DP1, assorts centromeric and telomeric motifs, to diversify KIR haplotypes. Similar recombination between chimpanzee KIR haplotypes does not change gene content only the linkage disequilibrium between centromeric gene-content motifs and alleles of KIR2DL4 and the telomeric framework gene, KIR3DL1/2 [70].
A major species difference is in the activating KIR that are structurally similar to inhibitory MHC-C specific KIR. Whereas chimpanzee activating and inhibitory KIR have similar specificity and strong avidity for MHC-C (unpublished data), human KIR2DS1 has low avidity for C2, and 2DS2, 2DS3 and 2DS5 have no detectable affinity for HLA class I [31]. Similarly, human KIR3DS1 that is structurally similar to KIR3DL1 does not bind with detectable avidity to the Bw4 epitope. In each of these KIR the low avidity can be attributed to one or a few amino-acid substitutions. Thus a strong trend in human evolution has been to reduce the interaction of the activating KIR with HLA class I ligands.
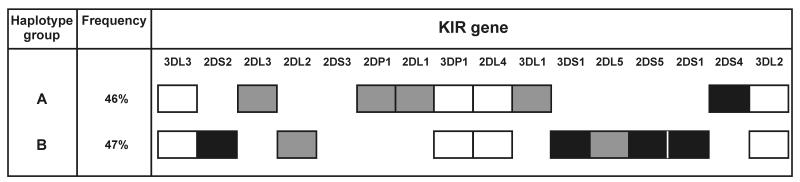
Associated with the reorganization of the human KIR locus and the attenuation of activating KIR genes has been the evolution of functionally distinctive centromeric and telomeric motifs that contain different genes and different alleles of shared genes. These motifs are the basis for the division of human KIR haplotypes into two functional groups. A and B [102], that are present in all human populations but with differing relative frequencies. Illustrating the haplotype differences is the Yucpa, a small Venezuelan Amerindian population, who have a long history of population bottleneck and selection by infectious disease [103]. Two KIR gene-content haplotypes, at frequencies of 46% and 47%, dominate this population (Figure 11). These two haplotypes share the framework genes, but differ in segregating 11 other KIR genes. The B haplotype is distinguished by having four of the five activating receptors with poor avidity for HLA class I, as well as the KIR2DL5 inhibitory receptor of unknown ligand-binding specificity. The B haplotype lacks genes for the inhibitory receptors specific for C2 and Bw4, and has a variant form (KIR2DL2) of the inhibitory C1-specific receptor. In contrast, while the A haplotype has genes for the inhibitory C1, C2 and Bw4 receptors, KIR2DS4 that has measurable avidity for some HLA-C and HLA-A11 is the only activating receptor gene. With these two highly divergent haplotypes the Yucpa have maintained all but one of the common KIR genes (KIR2DS3) found in human populations worldwide.
Figure 11. Two diverse dominant haplotypes in the Yucpa Amerindian population.
Shown is the gene content of the divergent Aand B haplotypes that are present at even frequency in the Yucpa population and account for 93% of all haplotypes. Each gene corresponds to a box. Framework genes are white, activating KIR genes are black, and the inhibitory KIR genes and the 2DP1 pseudogene are shaded grey [103].
Balancing selection is well established as a factor maintaining HLA class I polymorphism [104] and it is also strongly implicated in maintaining both the haplotype and allele diversity of the KIR system [55, 103]. One cause of this balance could be the distinctive functions that NK cells and the interactions of their KIR with HLA class I play in the immune and reproductive systems. Supporting this hypothesis, epidemiological studies show that women who are heterozygous or homozygous for B KIR haplotypes are less likely to experience pregnancy syndromes of pre-eclampsia, and spontaneous abortion than women who are homozygous for A KIR haplotypes [105-107]. Conversely, homozygosity for KIR2DL3, a component of A KIR haplotypes correlates with resolution of acute infection with hepatitis C virus [108].
The model that emerges from simple consideration of these data is that the B KIR haplotype has been selected for its beneficial effect upon the function of NK cells in human reproduction, whereas the A KIR haplotype has been selected for its beneficial effect upon the function of human NK cells in innate immunity against viral infection. Both these functions are needed if a human population is to survive [19, 103].
Towards better non-human primate models of human disease
A high proportion of the diseases that affect modern human populations involve the immune system, which is working for the rest of the body to prevent infection and cancer, or against it as in the ever increasing chronic diseases of inflammation, allergy and autoimmunity. The medical importance of variability in KIR, MHC class I, and their mutual interaction is seen from the extent of the clinical correlations and the potential impact of such knowledge on clinical practice. For example, therapeutic hematopoietic cell transplantation for acute myelogenous leukemia (AML) could be significantly improved by favoring HLA-matched donors who have one or two B KIR haplotypes over HLA-matched donors who are homozygous for A KIR haplotypes [49].
The importance of HLA variation in clinical transplantation has been known for more than 50 years, during which time there has been continuous, international effort to describe and define HLA polymorphism in the world’s human populations with increasing precision. As a result of this effort the use and success of transplantation as routine therapy has also continued to expand and improve. Technical developments emanating from the human genome project have had direct application to HLA analysis, with the results that nucleotide sequencing is now a routine method for typing and population studies. Meanwhile genome wide scans confirm and extend the dominance of HLA factors as markers of disease susceptibility. The study of human KIR variation is only 15 years old, but it has benefited from the HLA experience and the resources established by the HLA and transplantation communities. All indicators point to KIR variation having an importance approaching that of HLA if not on a par. Notably, the polymorphisms of KIR and HLA class I interact functionally, co-evolving to serve the necessary functions of defense and reproduction.
As reviewed here, the co-evolution of KIR and MHC class I has been fast, furious and specific to the simian primates. Even here we see that the New World monkeys took an evolutionary path that was dissimilar from that taken by the common ancestor of Old World monkeys, apes and humans. In similar fashion during ape evolution, the gibbons took a very different pathway than the great apes, one involving the significant loss of KIR and MHC class I components that humans have preserved. These immunological considerations point to Old World monkeys as the species most likely to provide useful and informative models of human disease. Considerations of practicality also point in the same direction.
Limiting the potential of the Old World monkey models is insufficient knowledge of the basic components of MHC and KIR variation, of the alleles, haplotypes, genotypes, and frequencies in populations, and how they all fit together functionally. The very essence of a biological ‘model’ is that it be studied to a depth in excess of that possible with the human species. That has not happened for any non-human primate. Although there is considerable information published on the MHC of Old World monkeys, the whole is not sufficiently more than the sum of the parts and the genomic sequence analysis [87, 109, 110] has yet to break open up the field, as it did for HLA. A major challenge is that Old World monkey MHC haplotypes have variable MHC class I gene content, involving numerous genes, whereas humans have fewer and fixed genes. However, this is no insurmountable problem, as shown by the work in other gene families, including human KIR. The HLA experience would also argue for international cooperation, regular workshops, an agreed nomenclature, a curated database, and quality control as essential elements in the development of a robust non-human primate model for biomedical research on human disease. Finally, it will important to compare different Old World monkey species in order to identify and choose for intensive study those that more closely reflect the human situation, and not ones like the gibbon apes who have clearly gone their own way [83].
Acknowledgments
Funding: NIH grants AI24258, AI17892 and AI22039 to P.P. We thank Yerkes Regional Primate Center at Emory University for non-human primate blood samples and acknowledge the support of the Yerkes Center Base Grant RR000165.
References
- 1.Cooper MA, Fehniger TA, Caligiuri MA. The biology of human natural killer-cell subsets. Trends Immunol. 2001;22:633–640. doi: 10.1016/s1471-4906(01)02060-9. [DOI] [PubMed] [Google Scholar]
- 2.Caligiuri MA. Human natural killer cells. Blood. 2008;112:461–469. doi: 10.1182/blood-2007-09-077438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herberman RB, Nunn ME, Lavrin DH. Natural cytotoxic reactivity of mouse lymphoid cells against syngeneic acid allogeneic tumors. I. Distribution of reactivity and specificity. Int J Cancer. 1975;16:216–229. doi: 10.1002/ijc.2910160204. [DOI] [PubMed] [Google Scholar]
- 4.Herberman RB, Nunn ME, Holden HT, Lavrin DH. Natural cytotoxic reactivity of mouse lymphoid cells against syngeneic and allogeneic tumors. II. Characterization of effector cells. Int J Cancer. 1975;16:230–239. doi: 10.1002/ijc.2910160205. [DOI] [PubMed] [Google Scholar]
- 5.Kiessling R, Klein E, Wigzell H. ‘Natural’ killer cells in the mouse. I. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Specificity and distribution according to genotype. Eur J Immunol. 1975;5:112–117. doi: 10.1002/eji.1830050208. [DOI] [PubMed] [Google Scholar]
- 6.Kiessling R, Klein E, Pross H, Wigzell H. ‘Natural‘ killer cells in the mouse. II. Cytotoxic cells with specificity for mouse Moloney leukemia cells. Characteristics of the killer cell. Eur J Immunol. 1975;5:117–121. doi: 10.1002/eji.1830050209. [DOI] [PubMed] [Google Scholar]
- 7.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 8.Trowsdale J. Genetic and functional relationships between MHC and NK receptor genes. Immunity. 2001;15:363–374. doi: 10.1016/s1074-7613(01)00197-2. [DOI] [PubMed] [Google Scholar]
- 9.Trowsdale J, Barten R, Haude A, Stewart CA, Beck S, Wilson MJ. The genomic context of natural killer receptor extended gene families. Immunol Rev. 2001;181:20–38. doi: 10.1034/j.1600-065x.2001.1810102.x. [DOI] [PubMed] [Google Scholar]
- 10.Valiante NM, Uhrberg M, Shilling HG, Lienert-Weidenbach K, Arnett KL, D’Andrea A, Phillips JH, Lanier LL, Parham P. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7:739–751. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- 11.Borrego F, Masilamani M, Kabat J, Sanni TB, Coligan JE. The cell biology of the human natural killer cell CD94/NKG2A inhibitory receptor. Mol Immunol. 2005;42:485–488. doi: 10.1016/j.molimm.2004.07.031. [DOI] [PubMed] [Google Scholar]
- 12.Burgess SJ, Maasho K, Masilamani M, Narayanan S, Borrego F, Coligan JE. The NKG2D receptor: immunobiology and clinical implications. Immunol Res. 2008;40:18–34. doi: 10.1007/s12026-007-0060-9. [DOI] [PubMed] [Google Scholar]
- 13.Lopez-Larrea C, Suarez-Alvarez B, Lopez-Soto A, Lopez-Vazquez A, Gonzalez S. The NKG2D receptor: sensing stressed cells. Trends Mol Med. 2008;14:179–189. doi: 10.1016/j.molmed.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 14.Karre K. Natural killer cell recognition of missing self. Nat Immunol. 2008;9:477–480. doi: 10.1038/ni0508-477. [DOI] [PubMed] [Google Scholar]
- 15.Boyle LH, Gillingham AK, Munro S, Trowsdale J. Selective export of HLA-F by its cytoplasmic tail. J Immunol. 2006;176:6464–6472. doi: 10.4049/jimmunol.176.11.6464. [DOI] [PubMed] [Google Scholar]
- 16.Apps R, Gardner L, Traherne J, Male V, Moffett A. Natural-killer cell ligands at the maternal-fetal interface: UL-16 binding proteins, MHC class-I chain related molecules, HLA-F and CD48. Hum Reprod. 2008;23:2535–2548. doi: 10.1093/humrep/den223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shum BP, Flodin LR, Muir DG, Rajalingam R, Khakoo SI, Cleland S, Guethlein LA, Uhrberg M, Parham P. Conservation and variation in human and common chimpanzee CD94 and NKG2 genes. J Immunol. 2002;168:240–252. doi: 10.4049/jimmunol.168.1.240. [DOI] [PubMed] [Google Scholar]
- 18.Vance RE, Jamieson AM, Raulet DH. Recognition of the class Ib molecule Qa-1(b) by putative activating receptors CD94/NKG2C and CD94/NKG2E on mouse natural killer cells. J Exp Med. 1999;190:1801–1812. doi: 10.1084/jem.190.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 20.Borrego F, Masilamani M, Marusina AI, Tang X, Coligan JE. The CD94/NKG2 family of receptors: from molecules and cells to clinical relevance. Immunol Res. 2006;35:263–278. doi: 10.1385/IR:35:3:263. [DOI] [PubMed] [Google Scholar]
- 21.Guma M, Angulo A, Vilches C, Gomez-Lozano N, Malats N, Lopez-Botet M. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood. 2004;104:3664–3671. doi: 10.1182/blood-2004-05-2058. [DOI] [PubMed] [Google Scholar]
- 22.Biron CA, Byron KS, Sullivan JL. Severe herpesvirus infections in an adolescent without natural killer cells. N Engl J Med. 1989;320:1731–1735. doi: 10.1056/NEJM198906293202605. [DOI] [PubMed] [Google Scholar]
- 23.Boyington JC, Sun PD. A structural perspective on MHC class I recognition by killer cell immunoglobulin-like receptors. Mol Immunol. 2002;38:1007–1021. doi: 10.1016/s0161-5890(02)00030-5. [DOI] [PubMed] [Google Scholar]
- 24.Yusa S, Catina TL, Campbell KS. SHP-1- and phosphotyrosine-independent inhibitory signaling by a killer cell Ig-like receptor cytoplasmic domain in human NK cells. J Immunol. 2002;168:5047–5057. doi: 10.4049/jimmunol.168.10.5047. [DOI] [PubMed] [Google Scholar]
- 25.Kikuchi-Maki A, Catina TL, Campbell KS. Cutting edge: KIR2DL4 transduces signals into human NK cells through association with the Fc receptor gamma protein. J Immunol. 2005;174:3859–3863. doi: 10.4049/jimmunol.174.7.3859. [DOI] [PubMed] [Google Scholar]
- 26.Rajagopalan S, Bryceson YT, Kuppusamy SP, Geraghty DE, van der Meer A, Joosten I, Long EO. Activation of NK cells by an endocytosed receptor for soluble HLA-G. PLoS Biol. 2006;4:e9. doi: 10.1371/journal.pbio.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marsh SG, Parham P, Dupont B, Geraghty DE, Trowsdale J, Middleton D, Vilches C, Carrington M, Witt C, Guethlein LA, Shilling H, Garcia CA, Hsu KC, Wain H. Killer-cell immunoglobulin-like receptor (KIR) nomenclature report, 2002. Hum Immunol. 2003;64:648–654. doi: 10.1016/s0198-8859(03)00067-3. [DOI] [PubMed] [Google Scholar]
- 28.Robinson J, Waller MJ, Fail SC, McWilliam H, Lopez R, Parham P, Marsh SG. The IMGT/HLA database. Nucleic Acids Res. 2009;37:D1013–1017. doi: 10.1093/nar/gkn662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colonna M, Spies T, Strominger JL, Ciccone E, Moretta A, Moretta L, Pende D, Viale O. Alloantigen recognition by two human natural killer cell clones is associated with HLA-C or a closely linked gene. Proc Natl Acad Sci U S A. 1992;89:7983–7985. doi: 10.1073/pnas.89.17.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mandelboim O, Reyburn HT, Vales-Gomez M, Pazmany L, Colonna M, Borsellino G, Strominger JL. Protection from lysis by natural killer cells of group 1 and 2 specificity is mediated by residue 80 in human histocompatibility leukocyte antigen C alleles and also occurs with empty major histocompatibility complex molecules. J Exp Med. 1996;184:913–922. doi: 10.1084/jem.184.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stewart CA, Laugier-Anfossi F, Vely F, Saulquin X, Riedmuller J, Tisserant A, Gauthier L, Romagne F, Ferracci G, Arosa FA, Moretta A, Sun PD, Ugolini S, Vivier E. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc Natl Acad Sci U S A. 2005;102:13224–13229. doi: 10.1073/pnas.0503594102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winter CC, Gumperz JE, Parham P, Long EO, Wagtmann N. Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA-C allotype recognition. J Immunol. 1998;161:571–577. [PubMed] [Google Scholar]
- 33.Mandelboim O, Reyburn HT, Sheu EG, Vales-Gomez M, Davis DM, Pazmany L, Strominger JL. The binding site of NK receptors on HLA-C molecules. Immunity. 1997;6:341–350. doi: 10.1016/s1074-7613(00)80336-2. [DOI] [PubMed] [Google Scholar]
- 34.Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J Immunol. 2008;180:3969–3979. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- 35.Barber LD, Percival L, Valiante NM, Chen L, Lee C, Gumperz JE, Phillips JH, Lanier LL, Bigge JC, Parekh RB, Parham P. The inter-locus recombinant HLA-B*4601 has high selectivity in peptide binding and functions characteristic of HLA-C. J Exp Med. 1996;184:735–740. doi: 10.1084/jem.184.2.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moesta AK, Abi-Rached L, Norman PJ, Parham P. Chimpanzees use more varied receptors and ligands than humans for inhibitory killer cell Ig-like receptor recognition of the MHC-C1 and MHC-C2 epitopes. J Immunol. 2009;182:3628–3637. doi: 10.4049/jimmunol.0803401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cella M, Longo A, Ferrara GB, Strominger JL, Colonna M. NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J Exp Med. 1994;180:1235–1242. doi: 10.1084/jem.180.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gumperz JE, Litwin V, Phillips JH, Lanier LL, Parham P. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J Exp Med. 1995;181:1133–1144. doi: 10.1084/jem.181.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanjanwala B, Draghi M, Norman PJ, Guethlein LA, Parham P. Polymorphic sites away from the Bw4 epitope that affect interaction of Bw4+ HLA-B with KIR3DL1. J Immunol. 2008;181:6293–6300. doi: 10.4049/jimmunol.181.9.6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma D, Bastard K, Guethlein LA, Norman PJ, Yawata N, Yawata M, Pando M, Thananchai H, Dong T, Rowland-Jones S, Brodsky FM, Parham P. Dimorphic motifs in Do and D1+D2 domains of KIR3DL1 combine to form receptors with high, moderate and no avidity for the complex of a peptide derived from human immunodeficiency virus and HLA-A*2402. J Immunol. 2009;183:4569–4582. doi: 10.4049/jimmunol.0901734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hansasuta P, Dong T, Thananchai H, Weekes M, Willberg C, Aldemir H, Rowland-Jones S, Braud VM. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur J Immunol. 2004;34:1673–1679. doi: 10.1002/eji.200425089. [DOI] [PubMed] [Google Scholar]
- 42.Trowsdale J, Moffett A. NK receptor interactions with MHC class I molecules in pregnancy. Semin Immunol. 2008;20:317–320. doi: 10.1016/j.smim.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 43.Apps R, Murphy SP, Fernando R, Gardner L, Ahad T, Moffett A. Human leucocyte antigen (HLA) expression of primary trophoblast cells and placental cell lines, determined using single antigen beads to characterize allotype specificities of anti-HLA antibodies. Immunology. 2009;127:26–39. doi: 10.1111/j.1365-2567.2008.03019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moffett A, Loke C. Immunology of placentation in eutherian mammals. Nat Rev Immunol. 2006;6:584–594. doi: 10.1038/nri1897. [DOI] [PubMed] [Google Scholar]
- 45.Clements CS, Kjer-Nielsen L, McCluskey J, Rossjohn J. Structural studies on HLA-G: implications for ligand and receptor binding. Hum Immunol. 2007;68:220–226. doi: 10.1016/j.humimm.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 46.Wilson MJ, Torkar M, Haude A, Milne S, Jones T, Sheer D, Beck S, Trowsdale J. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci U S A. 2000;97:4778–4783. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Graef T, Moesta AK, Norman PJ, Abi-Rached L, Vago L, Aguilar AM Older, Gleimer M, Hammond JA, Guethlein LA, Bushnell DA, Robinson PJ, Parham P. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C. J Exp Med. 2009;206:2557–2572. doi: 10.1084/jem.20091010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu Rev Immunol. 2003;21:139–176. doi: 10.1146/annurev.immunol.21.120601.141107. [DOI] [PubMed] [Google Scholar]
- 49.Cooley S, Xiao F, Pitt M, Gleason M, McCullar V, Bergemann TL, McQueen KL, Guethlein LA, Parham P, Miller JS. A subpopulation of human peripheral blood NK cells that lacks inhibitory receptors for self-MHC is developmentally immature. Blood. 2007;110:578–586. doi: 10.1182/blood-2006-07-036228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anfossi N, Andre P, Guia S, Falk CS, Roetynck S, Stewart CA, Breso V, Frassati C, Reviron D, Middleton D, Romagne F, Ugolini S, Vivier E. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25:331–342. doi: 10.1016/j.immuni.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 51.Gumperz JE, Valiante NM, Parham P, Lanier LL, Tyan D. Heterogeneous phenotypes of expression of the NKB1 natural killer cell class I receptor among individuals of different human histocompatibility leukocyte antigens types appear genetically regulated, but not linked to major histocompatibililty complex haplotype. J Exp Med. 1996;183:1817–1827. doi: 10.1084/jem.183.4.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yawata M, Yawata N, Draghi M, Partheniou F, Little AM, Parham P. MHC class I-specific inhibitory receptors and their ligands structure diverse human NK-cell repertoires toward a balance of missing self-response. Blood. 2008;112:2369–2380. doi: 10.1182/blood-2008-03-143727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malnati MS, Peruzzi M, Parker KC, Biddison WE, Ciccone E, Moretta A, Long EO. Peptide specificity in the recognition of MHC class I by natural killer cell clones. Science. 1995;267:1016–1018. doi: 10.1126/science.7863326. [DOI] [PubMed] [Google Scholar]
- 54.Thananchai H, Gillespie G, Martin MP, Bashirova A, Yawata N, Yawata M, Easterbrook P, McVicar DW, Maenaka K, Parham P, Carrington M, Dong T, Rowland-Jones S. Cutting Edge: Allele-specific and peptide-dependent interactions between KIR3DL1 and HLA-A and HLA-B. J Immunol. 2007;178:33–37. doi: 10.4049/jimmunol.178.1.33. [DOI] [PubMed] [Google Scholar]
- 55.Norman PJ, Abi-Rached L, Gendzekhadze K, Korbel D, Gleimer M, Rowley D, Bruno D, Carrington CV, Chandanayingyong D, Chang YH, Crespi C, Saruhan-Direskeneli G, Fraser PA, Hameed K, Kamkamidze G, Koram KA, Layrisse Z, Matamoros N, Mila J, Park MH, Pitchappan RM, Ramdath DD, Shiau MY, Stephens HA, Struik S, Verity DH, Vaughan RW, Tyan D, Davis RW, Riley EM, Ronaghi M, Parham P. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat Genet. 2007;39:1092–1099. doi: 10.1038/ng2111. [DOI] [PubMed] [Google Scholar]
- 56.Gardiner CM, Guethlein LA, Shilling HG, Pando M, Carr WH, Rajalingam R, Vilches C, Parham P. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J Immunol. 2001;166:2992–3001. doi: 10.4049/jimmunol.166.5.2992. [DOI] [PubMed] [Google Scholar]
- 57.Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med. 2006;203:633–645. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Flores-Villanueva PO, Yunis EJ, Delgado JC, Vittinghoff E, Buchbinder S, Leung JY, Uglialoro AM, Clavijo OP, Rosenberg ES, Kalams SA, Braun JD, Boswell SL, Walker BD, Goldfeld AE. Control of HIV-1 viremia and protection from AIDS are associated with HLA-Bw4 homozygosity. Proc Natl Acad Sci U S A. 2001;98:5140–5145. doi: 10.1073/pnas.071548198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin MP, Gao X, Lee JH, Nelson GW, Detels R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, Trowsdale J, Wilson M, O’Brien SJ, Carrington M. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 60.Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, Colombo S, Brown EE, Shupert WL, Phair J, Goedert JJ, Buchbinder S, Kirk GD, Telenti A, Connors M, O’Brien SJ, Walker BD, Parham P, Deeks SG, McVicar DW, Carrington M. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hoelsbrekken SE, Nylenna O, Saether PC, Slettedal IO, Ryan JC, Fossum S, Dissen E. Cutting Edge: Molecular Cloning of a Killer Cell Ig-Like Receptor in the Mouse and Rat. J Immunol. 2003;170:2259–2263. doi: 10.4049/jimmunol.170.5.2259. [DOI] [PubMed] [Google Scholar]
- 62.Welch AY, Kasahara M, Spain LM. Identification of the mouse killer immunoglobulin-like receptor-like (Kirl) gene family mapping to chromosome X. Immunogenetics. 2003;54:782–790. doi: 10.1007/s00251-002-0529-6. [DOI] [PubMed] [Google Scholar]
- 63.Bryceson YT, Foster JA, Kuppusamy SP, Herkenham M, Long EO. Expression of a killer cell receptor-like gene in plastic regions of the central nervous system. J Neuroimmunol. 2005;161:177–182. doi: 10.1016/j.jneuroim.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 64.Lanier LL. NK cell receptors. Annu Rev Immunol. 1998;16:359–393. doi: 10.1146/annurev.immunol.16.1.359. [DOI] [PubMed] [Google Scholar]
- 65.Natarajan K, Dimasi N, Wang J, Mariuzza RA, Margulies DH. Structure and function of natural killer cell receptors: multiple molecular solutions to self, nonself discrimination. Annu Rev Immunol. 2002;20:853–885. doi: 10.1146/annurev.immunol.20.100301.064812. [DOI] [PubMed] [Google Scholar]
- 66.Saleh A, Davies GE, Pascal V, Wright PW, Hodge DL, Cho EH, Lockett SJ, Abshari M, Anderson SK. Identification of probabilistic transcriptional switches in the Ly49 gene cluster: a eukaryotic mechanism for selective gene activation. Immunity. 2004;21:55–66. doi: 10.1016/j.immuni.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 67.Davies GE, Locke SM, Wright PW, Li H, Hanson RJ, Miller JS, Anderson SK. Identification of bidirectional promoters in the human KIR genes. Genes Immun. 2007;8:245–253. doi: 10.1038/sj.gene.6364381. [DOI] [PubMed] [Google Scholar]
- 68.Abi-Rached L, Parham P. Natural selection drives recurrent formation of activating killer cell immunoglobulin-like receptor and Ly49 from inhibitory homologues. J Exp Med. 2005;201:1319–1332. doi: 10.1084/jem.20042558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carlyle JR, Mesci A, Fine JH, Chen P, Belanger S, Tai LH, Makrigiannis AP. Evolution of the Ly49 and Nkrp1 recognition systems. Semin Immunol. 2008;20:321–330. doi: 10.1016/j.smim.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 70.Khakoo SI, Rajalingam R, Shum BP, Weidenbach K, Flodin L, Muir DG, Canavez F, Cooper SL, Valiante NM, Lanier LL, Parham P. Rapid evolution of NK cell receptor systems demonstrated by comparison of chimpanzees and humans. Immunity. 2000;12:687–698. doi: 10.1016/s1074-7613(00)80219-8. [DOI] [PubMed] [Google Scholar]
- 71.Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol. 2002;20:217–251. doi: 10.1146/annurev.immunol.20.092501.134942. [DOI] [PubMed] [Google Scholar]
- 72.Adams EJ, Parham P. Species-specific evolution of MHC class I genes in the higher primates. Immunol Rev. 2001;183:41–64. doi: 10.1034/j.1600-065x.2001.1830104.x. [DOI] [PubMed] [Google Scholar]
- 73.Joly E, Rouillon V. The orthology of HLA-E and H2-Qa1 is hidden by their concerted evolution with other MHC class I molecules. Biol Direct. 2006;1:2. doi: 10.1186/1745-6150-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 75.Bahram S, Inoko H, Shiina T, Radosavljevic M. MIC and other NKG2D ligands: from none to too many. Curr Opin Immunol. 2005;17:505–509. doi: 10.1016/j.coi.2005.07.016. [DOI] [PubMed] [Google Scholar]
- 76.Guethlein LA, Abi-Rached L, Hammond JA, Parham P. The expanded cattle KIR genes are orthologous to the conserved single-copy KIR3DX1 gene of primates. Immunogenetics. 2007;59:517–522. doi: 10.1007/s00251-007-0214-x. [DOI] [PubMed] [Google Scholar]
- 77.Sambrook JG, Bashirova A, Andersen H, Piatak M, Vernikos GS, Coggill P, Lifson JD, Carrington M, Beck S. Identification of the ancestral killer immunoglobulin-like receptor gene in primates. BMC Genomics. 2006;7:209. doi: 10.1186/1471-2164-7-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hammond JA, Guethlein LA, Abi-Rached L, Moesta AK, Parham P. Evolution and survival of marine carnivores did not require a diversity of killer cell Ig-like receptors or Ly49 NK cell receptors. J Immunol. 2009;182:3618–3627. doi: 10.4049/jimmunol.0803026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Averdam A, Petersen B, Rosner C, Neff J, Roos C, Eberle M, Aujard F, Munch C, Schempp W, Carrington M, Shiina T, Inoko H, Knaust F, Coggill P, Sehra H, Beck S, Abi-Rached L, Reinhardt R, Walter L. A novel system of polymorphic and diverse NK cell receptors in primates. PLoS Genet. 2009;5:e1000688. doi: 10.1371/journal.pgen.1000688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Flugge P, Zimmermann E, Hughes AL, Gunther E, Walter L. Characterization and phylogenetic relationship of prosimian MHC class I genes. J Mol Evol. 2002;55:768–775. doi: 10.1007/s00239-002-2372-7. [DOI] [PubMed] [Google Scholar]
- 81.Chatterjee HJ, Ho SY, Barnes I, Groves C. Estimating the phylogeny and divergence times of primates using a supermatrix approach. BMC Evol Biol. 2009;9:259. doi: 10.1186/1471-2148-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guethlein LA, Aguilar AM Older, Abi-Rached L, Parham P. Evolution of killer cell Ig-like receptor (KIR) genes: definition of an orangutan KIR haplotype reveals expansion of lineage III KIR associated with the emergence of MHC-C. J Immunol. 2007;179:491–504. doi: 10.4049/jimmunol.179.1.491. [DOI] [PubMed] [Google Scholar]
- 83.Abi-Rached L, Kuhl H, Roos C, ten Hallers B, Zhu B, Carbone L, de Jong PJ, Mootnick AR, Knaust F, Reinhardt R, Parham P, Walter L. A small, variable, and irregular killer cell Ig-like receptor locus accompanies the absence of MHC-C and MHC-G in gibbons. J Immunol. 2010;184:1379–1391. doi: 10.4049/jimmunol.0903016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Knapp LA, Cadavid LF, Watkins DI. The MHC-E locus is the most well conserved of all known primate class I histocompatibility genes. J Immunol. 1998;160:189–196. [PubMed] [Google Scholar]
- 85.Cardenas PP, Suarez CF, Martinez P, Patarroyo ME, Patarroyo MA. MHC class I genes in the owl monkey: mosaic organisation, convergence and loci diversity. Immunogenetics. 2005;56:818–832. doi: 10.1007/s00251-004-0751-5. [DOI] [PubMed] [Google Scholar]
- 86.Cadavid LF, Lun CM. Lineage-specific diversification of killer cell Ig-like receptors in the owl monkey, a New World primate. Immunogenetics. 2009;61:27–41. doi: 10.1007/s00251-008-0342-y. [DOI] [PubMed] [Google Scholar]
- 87.Daza-Vamenta R, Glusman G, Rowen L, Guthrie B, Geraghty DE. Genetic divergence of the rhesus macaque major histocompatibility complex. Genome Res. 2004;14:1501–1515. doi: 10.1101/gr.2134504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fukami-Kobayashi K, Shiina T, Anzai T, Sano K, Yamazaki M, Inoko H, Tateno Y. Genomic evolution of MHC class I region in primates. Proc Natl Acad Sci U S A. 2005;102:9230–9234. doi: 10.1073/pnas.0500770102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kita YF, Hosomichi K, Kohara S, Itoh Y, Ogasawara K, Tsuchiya H, Torii R, Inoko H, Blancher A, Kulski JK, Shiina T. MHC class I A loci polymorphism and diversity in three Southeast Asian populations of cynomolgus macaque. Immunogenetics. 2009;61:635–648. doi: 10.1007/s00251-009-0390-y. [DOI] [PubMed] [Google Scholar]
- 90.Otting N, Doxiadis GG, Bontrop RE. Definition of Mafa-A and -B haplotypes in pedigreed cynomolgus macaques (Macaca fascicularis) Immunogenetics. 2009;61:745–753. doi: 10.1007/s00251-009-0412-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wiseman RW, Karl JA, Bimber BN, O’Leary CE, Lank SM, Tuscher JJ, Detmer AM, Bouffard P, Levenkova N, Turcotte CL, Szekeres E, Jr., Wright C, Harkins T, O’Connor DH. Major histocompatibility complex genotyping with massively parallel pyrosequencing. Nat Med. 2009;15:1322–1326. doi: 10.1038/nm.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Grendell RL, Hughes AL, Golos TG. Cloning of rhesus monkey killer-cell Ig-like receptors (KIRs) from early pregnancy decidua. Tissue Antigens. 2001;58:329–334. doi: 10.1034/j.1399-0039.2001.580507.x. [DOI] [PubMed] [Google Scholar]
- 93.Hershberger KL, Shyam R, Miura A, Letvin NL. Diversity of the killer cell Ig-like receptors of rhesus monkeys. J Immunol. 2001;166:4380–4390. doi: 10.4049/jimmunol.166.7.4380. [DOI] [PubMed] [Google Scholar]
- 94.Bimber BN, Moreland AJ, Wiseman RW, Hughes AL, O’Connor DH. Complete characterization of killer Ig-like receptor (KIR) haplotypes in Mauritian cynomolgus macaques: novel insights into nonhuman primate KIR gene content and organization. J Immunol. 2008;181:6301–6308. doi: 10.4049/jimmunol.181.9.6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bostik P, Kobkitjaroen J, Tang W, Villinger F, Pereira LE, Little DM, Stephenson ST, Bouzyk M, Ansari AA. Decreased NK cell frequency and function is associated with increased risk of KIR3DL allele polymorphism in simian immunodeficiency virus-infected rhesus macaques with high viral loads. J Immunol. 2009;182:3638–3649. doi: 10.4049/jimmunol.0803580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hershberger KL, Kurian J, Korber BT, Letvin NL. Killer cell immunoglobulin-like receptors (KIR) of the African-origin sabaeus monkey: evidence for recombination events in the evolution of KIR. Eur J Immunol. 2005;35:922–935. doi: 10.1002/eji.200425408. [DOI] [PubMed] [Google Scholar]
- 97.Sambrook JG, Bashirova A, Palmer S, Sims S, Trowsdale J, Abi-Rached L, Parham P, Carrington M, Beck S. Single haplotype analysis demonstrates rapid evolution of the killer immunoglobulin-like receptor (KIR) loci in primates. Genome Res. 2005;15:25–35. doi: 10.1101/gr.2381205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Guethlein LA, Flodin LR, Adams EJ, Parham P. NK Cell Receptors of the Orangutan (Pongo pygmaeus): A Pivotal Species for Tracking the Coevolution of Killer Cell Ig-Like Receptors with MHC-C. J Immunol. 2002;169:220–229. doi: 10.4049/jimmunol.169.1.220. [DOI] [PubMed] [Google Scholar]
- 99.Golos TG, Bondarenko GI, Dambaeva SV, Breburda EE, Durning M. On the role of placental Major Histocompatibility Complex and decidual leukocytes in implantation and pregnancy success using non-human primate models. Int J Dev Biol. 2010;54:431–443. doi: 10.1387/ijdb.082797tg. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jones DC, Hiby SE, Moffett A, Trowsdale J, Young NT. Nature of allelic sequence polymorphism at the KIR3DL3 locus. Immunogenetics. 2006;58:614–627. doi: 10.1007/s00251-006-0130-5. [DOI] [PubMed] [Google Scholar]
- 101.Trundley AE, Hiby SE, Chang C, Sharkey AM, Santourlidis S, Uhrberg M, Trowsdale J, Moffett A. Molecular characterization of KIR3DL3. Immunogenetics. 2006;57:904–916. doi: 10.1007/s00251-005-0060-7. [DOI] [PubMed] [Google Scholar]
- 102.Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, Tyan D, Lanier LL, Parham P. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–763. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 103.Gendzekhadze K, Norman PJ, Abi-Rached L, Graef T, Moesta AK, Layrisse Z, Parham P. Co-evolution of KIR2DL3 with HLA-C in a human population retaining minimal essential diversity of KIR and HLA class I ligands. Proc Natl Acad Sci U S A. 2009;106:18692–18697. doi: 10.1073/pnas.0906051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Solberg OD, Mack SJ, Lancaster AK, Single RM, Tsai Y, Sanchez-Mazas A, Thomson G. Balancing selection and heterogeneity across the classical human leukocyte antigen loci: a meta-analytic review of 497 population studies. Hum Immunol. 2008;69:443–464. doi: 10.1016/j.humimm.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hiby SE, Walker JJ, O’Shaughnessy KM, Redman CW, Carrington M, Trowsdale J, Moffett A. Combinations of maternal KIR and fetal HLA-C genes influence the risk of preeclampsia and reproductive success. J Exp Med. 2004;200:957–965. doi: 10.1084/jem.20041214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hiby SE, Regan L, Lo W, Farrell L, Carrington M, Moffett A. Association of maternal killer-cell immunoglobulin-like receptors and parental HLA-C genotypes with recurrent miscarriage. Hum Reprod. 2008;23:972–976. doi: 10.1093/humrep/den011. [DOI] [PubMed] [Google Scholar]
- 107.Moffett A, Hiby S. Influence of activating and inhibitory killer immunoglobulin-like receptors on predisposition to recurrent miscarriages. Hum Reprod. 2009;24:2048–2049. doi: 10.1093/humrep/dep216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, Cheng J, Goedert JJ, Vlahov D, Hilgartner M, Cox S, Little AM, Alexander GJ, Cramp ME, O’Brien SJ, Rosenberg WM, Thomas DL, Carrington M. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 109.Kulski JK, Anzai T, Shiina T, Inoko H. Rhesus macaque class I duplicon structures, organization, and evolution within the alpha block of the major histocompatibility complex. Mol Biol Evol. 2004;21:2079–2091. doi: 10.1093/molbev/msh216. [DOI] [PubMed] [Google Scholar]
- 110.Watanabe A, Shiina T, Shimizu S, Hosomichi K, Yanagiya K, Kita YF, Kimura T, Soeda E, Torii R, Ogasawara K, Kulski JK, Inoko H. A BAC-based contig map of the cynomolgus macaque (Macaca fascicularis) major histocompatibility complex genomic region. Genomics. 2007;89:402–412. doi: 10.1016/j.ygeno.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 111.Shiina T, Tamiya G, Oka A, Takishima N, Yamagata T, Kikkawa E, Iwata K, Tomizawa M, Okuaki N, Kuwano Y, Watanabe K, Fukuzumi Y, Itakura S, Sugawara C, Ono A, Yamazaki M, Tashiro H, Ando A, Ikemura T, Soeda E, Kimura M, Bahram S, Inoko H. Molecular dynamics of MHC genesis unraveled by sequence analysis of the 1,796,938-bp HLA class I region. Proc Natl Acad Sci U S A. 1999;96:13282–13287. doi: 10.1073/pnas.96.23.13282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Uhrberg M, Parham P, Wernet P. Definition of gene content for nine common group B haplotypes of the Caucasoid population: KIR haplotypes contain between seven and eleven KIR genes. Immunogenetics. 2002;54:221–229. doi: 10.1007/s00251-002-0463-7. [DOI] [PubMed] [Google Scholar]
- 113.Westgaard IH, Berg SF, Orstavik S, Fossum S, Dissen E. Identification of a human member of the Ly-49 multigene family. Eur J Immunol. 1998;28:1839–1846. doi: 10.1002/(SICI)1521-4141(199806)28:06<1839::AID-IMMU1839>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 114.Barten R, Trowsdale J. The human Ly-49L gene. Immunogenetics. 1999;49:731–734. doi: 10.1007/s002510050675. [DOI] [PubMed] [Google Scholar]
- 115.Hsu KC, Chida S, Geraghty DE, Dupont B. The killer cell immunoglobulin-like receptor (KIR) genomic region: gene-order, haplotypes and allelic polymorphism. Immunol Rev. 2002;190:40–52. doi: 10.1034/j.1600-065x.2002.19004.x. [DOI] [PubMed] [Google Scholar]
- 116.Gomez-Lozano N, de Pablo R, Puente S, Vilches C. Recognition of HLA-G by the NK cell receptor KIR2DL4 is not essential for human reproduction. Eur J Immunol. 2003;33:639–644. doi: 10.1002/eji.200323741. [DOI] [PubMed] [Google Scholar]
- 117.Martin MP, Bashirova A, Traherne J, Trowsdale J, Carrington M. Cutting edge: expansion of the KIR locus by unequal crossing over. J Immunol. 2003;171:2192–2195. doi: 10.4049/jimmunol.171.5.2192. [DOI] [PubMed] [Google Scholar]
- 118.Gomez-Lozano N, Estefania E, Williams F, Halfpenny I, Middleton D, Solis R, Vilches C. The silent KIR3DP1 gene (CD158c) is transcribed and might encode a secreted receptor in a minority of humans, in whom the KIR3DP1, KIR2DL4 and KIR3DL1/KIR3DS1 genes are duplicated. Eur J Immunol. 2005;35:16–24. doi: 10.1002/eji.200425493. [DOI] [PubMed] [Google Scholar]
- 119.Norman PJ, Abi-Rached L, Gendzekhadze K, Hammond JA, Moesta AK, Sharma D, Graef T, McQueen KL, Guethlein LA, Carrington CV, Chandanayingyong D, Chang YH, Crespi C, Saruhan-Direskeneli G, Hameed K, Kamkamidze G, Koram KA, Layrisse Z, Matamoros N, Mila J, Park MH, Pitchappan RM, Ramdath DD, Shiau MY, Stephens HA, Struik S, Tyan D, Verity DH, Vaughan RW, Davis RW, Fraser PA, Riley EM, Ronaghi M, Parham P. Meiotic recombination generates rich diversity in NK cell receptor genes, alleles, and haplotypes. Genome Res. 2009;19:757–769. doi: 10.1101/gr.085738.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Traherne JA, Martin M, Ward R, Ohashi M, Pellett F, Gladman D, Middleton D, Carrington M, Trowsdale J. Mechanisms of copy number variation and hybrid gene formation in the KIR immune gene complex. Hum Mol Genet. 2009 doi: 10.1093/hmg/ddp538. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sambrook J, Sehra H, Coggill P, Humphray S, Palmer S, Sims S, Takamatsu H-H, Wileman T, Archibald A, Beck S. Identification of a single killer immunoglobulin-like receptor (KIR) gene in the porcine leukocyte receptor complex on chromosome 6q. Immunogenetics. 2006;58:481–486. doi: 10.1007/s00251-006-0110-9. [DOI] [PubMed] [Google Scholar]