Abstract
Background
The JAK2 V617F mutation in exon 14 is the most common mutation in chronic myeloproliferative neoplasms (MPNs); deletion of the entire exon 14 is rarely detected. In our previous study of >10,000 samples from patients with suspected MPNs tested for JAK2 mutations by reverse transcription-PCR (RT-PCR) with direct sequencing, complete deletion of exon 14 (Δexon14) constituted <1% of JAK2 mutations. This appears to be an alternative splicing mutation, not detectable with DNA-based testing.
Methodology/Principal Findings
We investigated the possibility that MPN patients may express the JAK2 Δexon14 at low levels (<15% of total transcript) not routinely detectable by RT-PCR with direct sequencing. Using a sensitive RT-PCR–based fluorescent fragment analysis method to quantify JAK2 Δexon14 mRNA expression relative to wild-type, we tested 61 patients with confirmed MPNs, 183 with suspected MPNs (93 V617F-positive, 90 V617F-negative), and 46 healthy control subjects. The Δexon14 variant was detected in 9 of the 61 (15%) confirmed MPN patients, accounting for 3.96% to 33.85% (mean = 12.04%) of total JAK2 transcript. This variant was also detected in 51 of the 183 patients with suspected MPNs (27%), including 20 of the 93 (22%) with V617F (mean [range] expression = 5.41% [2.13%–26.22%]) and 31 of the 90 (34%) without V617F (mean [range] expression = 3.88% [2.08%–12.22%]). Immunoprecipitation studies demonstrated that patients expressing Δexon14 mRNA expressed a corresponding truncated JAK2 protein. The Δexon14 variant was not detected in the 46 control subjects.
Conclusions/Significance
These data suggest that expression of the JAK2 Δexon14 splice variant, leading to a truncated JAK2 protein, is common in patients with MPNs. This alternatively spliced transcript appears to be more frequent in MPN patients without V617F mutation, in whom it might contribute to leukemogenesis. This mutation is missed if DNA rather than RNA is used for testing.
Introduction
Myeloproliferative neoplasms (MPNs) are multipotent hematopoietic stem cell disorders characterized by uncontrolled proliferation of maturing blood cells. Chronic myeloid leukemia (CML) is the most common MPN, followed by polycythemia vera (PV), essential thrombocythemia (ET), and idiopathic myelofibrosis (IMF) [1]. Whereas CML is characterized by a readily detectable translocation (Philadelphia chromosome), non-CML MPNs lack recurrent chromosomal anomalies. However, a specific molecular abnormality, the JAK2 V617F mutation, has been reported in about 95% of PV patients, 35% to 70% of ET patients, and 50% of IMF patients [2]–[4]. JAK2 exon 12 mutations, as well as other mutations in exons 13 and 14, have been reported in rare cases of non-CML MPDs negative for V617F [5]–[7].
Most testing for JAK2 mutations is performed by analyzing the genomic DNA of the JAK2 gene. We have adapted the use of mRNA as the basis for testing for JAK2 mutations and have shown that RNA allows more sensitive detection of mutations than does DNA at early stages of disease [7]–[9]. The use of RNA rather than DNA provides the additional advantage of capturing abnormalities in platelets and detecting alternatively spliced transcripts.
In a previous report of JAK2 mutation profiles in a series of >10,000 patient samples, we described detection of a novel deletion of JAK2 exon 14 (Δexon14) along with other JAK2 mutations in exons 12 through 15, using bi-directional mRNA sequencing technology [7]. The Δexon14 mutation was detected by direct sequencing in less than 1% of patients with various types of JAK2 mutations. However, direct sequencing is not amenable to detecting deletions of entire exons, owing to difficulty in interpreting results as well as the low sensitivity of this approach. Therefore, in this study we used a sensitive RT-PCR–based assay with fluorescent fragment analysis to explore the possibility that MPN patients may commonly express this JAK2 mRNA splice variant at levels that cannot be reliably detected by sequence analysis. We further sought to determine whether patients expressing this splice variant also express a corresponding truncated JAK2 protein.
Methods
Patients and Samples
We tested peripheral blood samples from three groups of patients in addition to healthy normal control subjects. Group 1 comprised 61 consecutive randomly selected patients with confirmed non-CML MPN on the basis of clinical findings and complete peripheral blood and bone marrow analysis. The diagnosis of these patients was myelofibrosis in 27 (43%), polycythemia vera in 12 (19%), essential thrombocythemia in 6 (10%) and not-otherwise classified in 16 (27%). The other 2 patient groups were constructed from 183 residual de-identified samples from individuals with suspected non-CML MPNs initially submitted to Quest Diagnostics Nichols Institute for testing of JAK2 V617F as well as mutations in JAK2 exons 12 and 13: Group 2 comprised 90 samples that were negative for JAK2 mutations in V617 and exon 12 and 13, and group 3 comprised 93 samples from patients with JAK2 V617F. In addition, we tested 46 normal healthy control individuals.
Plasma was separated from peripheral blood samples and used for extraction of total RNA. The mRNA was then used for detection of the JAK2 Δexon14 variant by RT-PCR with bidirectional sequencing. All samples were also screened for the Δexon14 transcript using a sensitive assay based on RT-PCR with fluorescent fragment analysis.
Ethics statement
All work was performed according to a protocol approved by an Institutional Review Board (IRB) (Independent Review Consulting Inc. San Anselmo, California). Samples collected from group 1 and the normal control were collected with consent form.
Sequence Analysis
Total nucleic acid was extracted from patient plasma or PB/BM cells using the NucliSens (BioMerieux, Durham, NC) extraction kit. The primer pair was designed to encompass JAK2 exons 12 through 14 and part of exon 15: 5′-CTAAATGCTGTCCCCCAAAG-3′ (forward); and 5 ′-CCATGCCAACTGTTTAGCAA-3′ (reverse). The RT-PCR was performed using Superscript III one-step RT-PCR systems with Platinum Taq (Invitrogen, Carlsbad, California) under the following thermocycler conditions: initial step of 94°C for 2 minutes, followed by 40 cycles of 94°C for 15 seconds, 60°C for 30 seconds, and 68°C for 1 minute, with a final extension step of 68°C for 7 minutes. The 491-bp PCR product was then purified and sequenced in both forward and reverse directions using an ABI PRISM 3730XL Genetic Analyzer (Applied Biosystems, Foster City, CA). Sequencing data were base-called using sequencing analysis software and assembled and analyzed with SeqScape software (Applied Biosystems) using GenBank accession number NM 004972 as reference.
Detection of JAK2 Δexon14 Transcript Using Fragment Length Analysis
Total nucleic acid was extracted as described above from patient plasma or cells (peripheral blood or bone marrow). The primer pair was designed to encompass JAK2 exon 14, with the forward primer annealed in JAK2 exon 13 (5′-GAC TAC GGT CAA CTG CAT GAA A-3′) and the reverse primer annealed in exon 16 (5′-CCATGCCAACTG TTTAGCAA-3′). One of the two primers was FAM-labeled. The RT-PCR was performed using same buffer system (Invitrogen) and thermocycler conditions as the sequencing method. The JAK2 wild-type and Δexon 14 products were verified by determining the size of PCR products using the GeneScan 350ROX size standard (Applied Biosystems) and ABI PRISM 3730XL Genetic Analyzer. The wild-type product displays a 273-bp peak while the Δexon 14 splice variant displays a 185-bp peak (ie, 273–88 bp). The percentage of transcript accounted for by the JAK2 Δexon 14 splice mutant is calculated using the following formula:
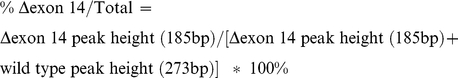 |
JAK2 Immunoprecipitation
Cells (5×105–1×106) were lysed in isotonic lysis buffer (150 mM NaCl, 20 mM Tris/HCl [pH 7.4], 0.3% Nonidet P-40, 12.5 mM β-glycerophosphate, 2 mM NaF, 200 µM Na3VO4, and 1 mM phenylmethylsulfonyl fluoride) containing 1× protease inhibitor mix (Roche Applied Science, Indianapolis, IN). Clarified lysates were subjected to immunoprecipitation using TrueBlot™ beads (Ebioscience, San Diego, CA) with an N-terminal anti-JAK2 antibody (JAK2-M126; Santa Cruz Biotechnology, Santa Cruz, CA). After incubation at 4°C for 4 to 12 hours, immune complexes were washed 4 times in lysis buffer, separated by SDS/PAGE, and analyzed by immunoblotting using C-terminal JAK2 antibody (JAK2 [D2E12]; Cell Signaling Technology, Danvers, MA) or N-terminal JAK2 antibody (JAK2 N-17; Santa Cruz Biotechnology). K562 cell line (ATCC CCL-243 Manassas VA) was used as negative control.
Immunoblot Analysis
Immunoblot analysis was performed as previously described (Bruey et al, Leuk Res. 2009). Briefly, equal amounts of immunoprecipitation products were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the gels were electrophoretically transferred to nitrocellulose membranes (0.2-mm pore size; Whatman, Florham Park, NJ). The blots were blocked with 5% bovine serum albumin in Tris-buffered saline with 0.05% Tween-20 (TBS-Tween) for 2 hours. The membrane was incubated with primary antibody for 5 hours at 4°C, washed with TBS-Tween, and incubated with secondary antibody for 30 minutes at room temperature (TrueBlot™, Ebioscience). After additional washing in TBS-Tween, chemiluminescent reagent (ECL; GE Healthcare, Piscataway, NJ) was added and the image was developed on x-ray film.
Results
Detection and Prevalence of JAK2 Δexon 14 Transcript
When JAK2 RNA (not DNA) is used for direct sequencing, the Δexon 14 transcript is reliably detected if present at levels >15% to 20% of total JAK2 RNA (Figure 1 and 2). MPN patients rarely have Δexon 14 transcript levels above this threshold, and the results of direct sequence analysis can be difficult to interpret in patients with apparent low-level expression. In these cases, the Δexon14 transcript can easily be interpreted as background or poor sequencing if the background sequence is not read completely and aligned to the JAK2 sequence (Figure 2). To more accurately detect low levels of Δexon 14 transcript expression, we developed an RT-PCR–based assay with fluorescent fragment length analysis. With this method, the JAK2 Δexon14 splice variant shows a 185-bp fragment while the wild-type shows a 273-bp fragment (Figure 3). To confirm that the Δexon14 splice variant that is detected in plasma is actually present in cells, we analyzed paired cell and plasma RNA from patients previously confirmed to show expression of the Δexon14 transcript in plasma. Both plasma and cells revealed reliable results for detecting Δexon14 transcripts (Figure 4).
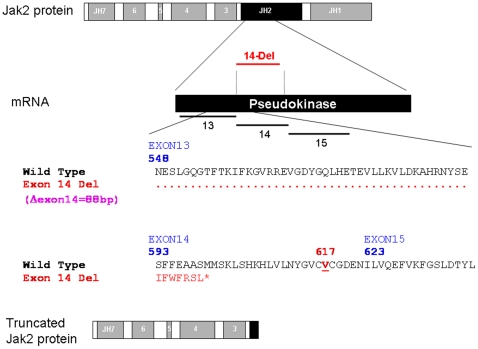
Figure 1. Schematic presentation of JAK2 Δexon 14.
Top, schematic diagram of the JAK2 protein showing JAK homology domains 1 through 7 (JH1-JH7) with the JH2 pseudokinase domain highlighted in black. The corresponding exon regions of the mRNA is shown with the exons 13, 14, and 15. Because exon 14 is consists of 88 bp, its deletion leads to frameshift and early termination of translation after coding for seven new amino acids and elimination of the V617 codon of JAK2 (lower panel). The resulting truncated JAK2 protein is shown on the bottom.
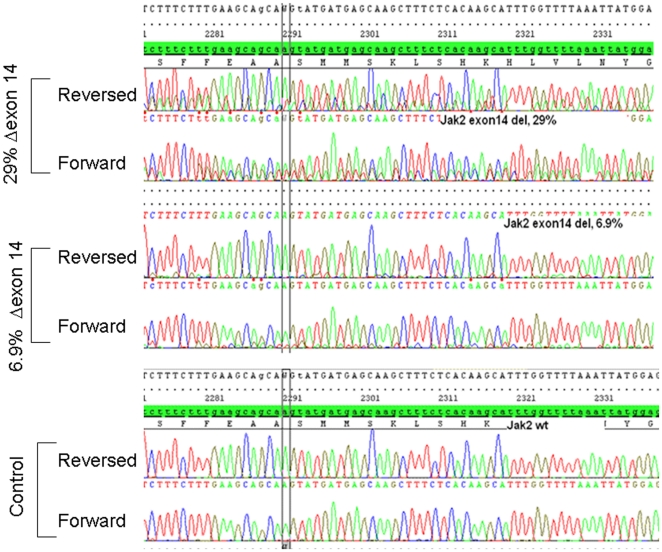
Figure 2. Detection of the JAK2 Δexon14 transcript with direct bi-directional sequencing.
Upper panels: Detection of Δexon14 is relatively easy when the transcript is present at high levels (eg, 29% of total JAK2 transcript). Detection is more difficult when the Δexon14 transcript makes up a small proportion of total JAK2 transcript (eg, 6.9%). Normal control is shown on the bottom.
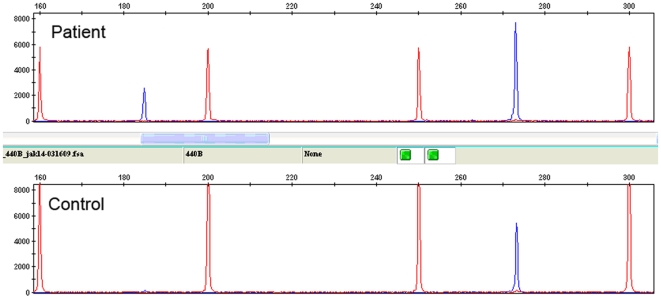
Figure 3. JAK2 Δexon14 transcript as detected using RT-PCR with fragment length analysis.
Lower panel: the expected 273-bp, full-length amplification products; upper panel: the expected 273-bp, full-length wild-type peak in addition to a peak at 185 bp corresponding to the truncated Δexon14 transcript. Size marker is shown as red peaks and amplification products are shown in blue.
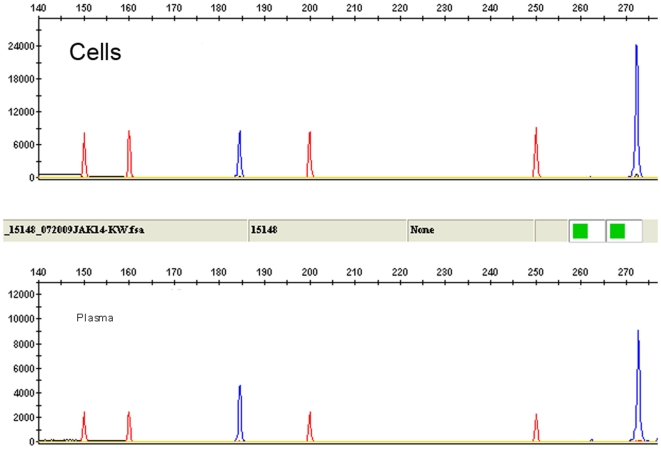
Figure 4. Source of RNA for detection of JAK2 Δexon14 transcript using RT-PCR with fragment length analysis.
Similar results are obtained whether RNA is isolated from plasma or peripheral blood cells. Size marker is shown as red peaks and amplification products are shown in blue.
Using the RT-PCR/fragment length analysis assay, we screened all groups of patients. Samples with Δexon 14 mutant to wild-type ratios greater than 15% by fragment analysis were tested and successfully confirmed by careful inspection of direct sequencing results. The Δexon14 was detected in 9 of the 61 confirmed MPN patients (15%) (Table 1), where it accounted for 3.96% to 33.85% (mean = 12.04%) of JAK2 transcript. In this group of patients, Δexon14 was detected in 33.3% of V617F-positive patients and in 57.9% of V617F-negative patients. This mutation was also detected in 51 of the 183 patients with suspected MPNs (27%) overall, including 20 of the 93 (22%) V617-positive patients (mean [range] expression = 5.41% [2.13%–26.22%]) and 31 of the 90 (34%) V617F-negative patients (mean expression = 3.88% [2.08%–12.22%]. While the difference in prevalence is not statistically significant (P = 0.07), there is a tendency of finding Δexon14 in unmutated patients. None of the 46 plasma RNA samples from the normal control group showed any expression of the Δexon14 transcript. Most patients with Δexon14-positive in each group had expression levels below 15%, therefore, it is frequently missed by direct sequencing.
Table 1. Prevalence and Relative Level of the ΔExon14 JAK2 Transcript in Patients with Suspected or Confirmed Myeloproliferative Neoplasms (MPNs).
| Suspected MPN (n = 183) | Confirmed MPN (n = 61) | |||||
| V617F-Negative (n = 90) | V617F-Positive (n = 93) | |||||
| n (%) | Mean (range) percentage of JAK2 transcript | n (%) | Mean (range) percentage of JAK2 transcript | n (%) | Mean (range) percentage of JAK2 transcript | |
| Δexon14 | 31 (34) | 3.88 (2.08–12.22) | 20 (22) | 5.41 (2.13–26.22) | 9 (15) | 12.04 (3.96–33.85) |
| No Δexon14 | 59 (66) | NA | 73 (78) | NA | 52 (85) | NA |
Effects of Δexon14 on JAK2 Protein
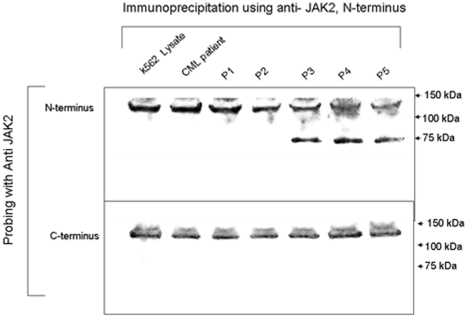
As indicated in Figure 1, deletion of exon 14 leads to a complete deletion of codon V617, a hot spot for mutation in patients with non-CML MPNs [3], [4]. More importantly, since exon 14 is composed of 88 bp, its deletion leads to a frameshift and the coding of new amino acids. However, the frame shift results in the addition of only 7 new amino acids, followed by a termination codon leading to truncation of the JAK2 protein within the pseudokinase domain. Therefore, we used immunoprecipitation and immunoblotting to confirm that the truncated JAK2 protein is expressed in cells from patients with confirmed JAK2 Δexon14 transcripts (Figure 5). Total JAK2 protein was immunoprecipitated using anti-JAK2 (N-terminal) antibody. For negative controls we used the K562 CML cell line, which does not express JAK2 Δexon14, in addition to cell samples from 1) a patient with CML; and 2) two patients with MPN who had been confirmed by RT/PCR to be negative for Δexon14 transcripts. Cells from three patients with different levels of Δexon14 transcript expression, as confirmed by RT/PCR, were used as positive samples.
Figure 5. Truncated JAK2 protein resulting from JAK2 Δexon14 mutation in patients with chronic myeloproliferative neoplasms.
Lysates were prepared from the indicated human CML K562 cell line (Lane1), a patient with chronic myelogenous leukemia (lane 2), and 5 patients with chronic MPNs (Lanes 3–7). Patient 1: non-CML CMPD, JAK2 V617F positive; Patient 2: non-CML CMPD, JAK2 V617F negative; Patient 3: non-CML CMPD, JAK2 Δexon14 positive; Patient 4: non-CML CMPD, JAK2 Δexon14 positive; Patient 5: non-CML CMPD, JAK2 Δexon14 positive. Top Panel: Probing with an anti-JAK2 N-terminal clone yielded a wild-type JAK2 band at 130 kDa in the K562 and other negative control lanes, and an additional band at 75 kDa only in patients with expression of Δexon14 transcript. Bottom Panel: An anti-JAK2 clone directed against the carboxyl-terminus of JAK2 yielded only a single band at 130 kDa.
Probing with the anti-JAK2 N-terminal clone yielded a wild-type JAK2 band at 130 kDa in the K562 and other negative control lanes, and an additional band at 75 kDa only in patients with confirmed expression of the Δexon14 transcript (Figure 5). This additional immunoprecipitation product represents the truncated JAK2 protein. The use of another anti-JAK2 clone directed against the carboxyl-terminus of JAK2, which is deleted from the truncated JAK2 protein, yielded a single band at 130 kDa, showing specificity of detection of the truncated JAK2 Δexon14 protein (Figure 5). Lysates from the K562 cell line and the CML patient showed only the wild-type band at 130 kDa. This confirms that the Δexon14 transcripts are translated.
Discussion
Our findings confirm that many individuals with non-CML MPNs, especially those lacking V617F, express low levels of the JAK2 Δexon14. The paucity of reports of the Δexon14 variant in MPN patients most likely derives from the fact that JAK2 mutation assays typically rely on DNA rather than RNA. Furthermore, this abnormality cannot be detected with methods that rely on use of specific probes. While bi-directional sequencing of mRNA transcript can detect most mutations, including splice variants, this approach lacks sensitivity (Figure 2). Therefore, a fluorescent fragment analysis method such as the one describe here must be used to detect low levels of this variant. The fact that the JAK2 Δexon14 transcript is a fraction of the total JAK2 transcript is, most likely, due to either alternative splicing or a mutation in the DNA in small fraction of the clone. Further studies are needed to fully determine the mechanism of this phenomenon. We did not sequence the DNA in cases with 100% JAK2 Δexon14 transcript and the possibility of a mutation in the DNA in these patients is likely and should be ruled out.
The fact that the JAK2 Δexon14 is detected only in patients with MPNs, and more likely in patients negative for V617F (57.9% vs. 33.3%)(P = 0.07), suggests that it may play a significant role in the pathophysiology of MPNs. Current knowledge of JAK2 functional domains and known mutations may shed light on the potential biological effects of this unique splice variant. All currently identified JAK2 mutations, including point mutations and indels in exons 12 through 15, reside in the pseudokinase domain (JH2)-coding region of JAK2 and do not affect the reading frame [5]–[7]. The JH2 domain is usually in close proximity to the kinase domain (JH1), inhibiting its activation if not bound to an active receptor [10]–[13]. Mutations in the JH2 domain may thus lead to constitutive activation of the JAK2 protein; it is believed that mutations in the JH2 domain cause constitutive activation in the downstream JAK2-STAT signaling pathways as long as the JAK2 is bound to the receptor [14]. Deletion within the JH2 domain and complete deletion of the kinase domain (JH1) is unexpected and raises questions on its mechanism in activating the JAK2-STAT pathway. The Δexon14 mutation preserves the JAK2 FERM domain (JH4-7), which is responsible for association with growth factor (eg, erythropoietin and thrombopoietin) receptors [15]. It is thus possible that the truncated JAK2 dimerizes with wild-type JAK2 to influence its structure, activating its kinase domain and the JAK2-STAT pathway. It is also possible that the Δexon14 mutation causes activation of STAT5 by altering receptor binding of the FERM domain. Clearly, further studies are needed to understand the mechanism by which the truncated JAK2 is involved in any activation of the JAK2-STAT pathway. Ex-vivo or in-vivo experiments showing the effects of expressing JAK2 Δexon14 transcripts on tumorogenesis should be performed.
In conclusion, the JAK2 Δexon14 splice variant is a relatively common anomaly in patients with MPNs, possibly more so in those lacking the V617F mutation. Further functional analyses of the Δexon14 JAK2 protein are needed to better understand how this abnormality may contribute to the pathophysiology of disease and its potential role as a marker for diagnosis, prognosis, or prediction of response to therapy.
Acknowledgments
The authors thank Jeff Radcliff (Quest Diagnostics, San Juan Capistrano, CA) for editorial suggestions.
Footnotes
Competing Interests: Work was performed at Quest Diagnostics (WM, XZ, XW, ZZ, CHY, JMB, and MA are all employed by Quest Diagnostics Incorporated). Quest Diagnostics had no impact or influence on the performed research and does not alter the authors' adherence to PLoS ONE policy on sharing data and material. A patent has been filed on finding by Quest Diagnostics and testing for JAK2 various mutations is offered at Quest Diagnostics. This does not alter the compliance of the authors' with PLoS ONE policies on sharing data and materials.
Funding: There are no current external funding sources for this study. Work was performed at Quest Diagnostics (WM, XZ, XW, ZZ, CHY, JMB, and MA are all employed by Quest Diagnostics Incorporated). Quest Diagnostics had no impact or influence on the performed research.
References
- 1.Nelson ME, Steensma DP. JAK2 V617F in myeloid disorders: what do we know now, and where are we headed? Leuk Lymphoma. 2006;47:177–194. doi: 10.1080/10428190500301348. [DOI] [PubMed] [Google Scholar]
- 2.Morgan KJ, Gilliland DG. A role for JAK2 mutations in myeloproliferative diseases. Ann Rev Med. 2008;59:213–222. doi: 10.1146/annurev.med.59.061506.154159. [DOI] [PubMed] [Google Scholar]
- 3.Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- 4.Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- 5.Scott LM, Tong W, Levine RL, Scott MA, Beer PA, et al. JAK2 exon 12 mutations in polycythemia vera and idiopathic erythrocytosis. N Engl J Med. 2007;356:459–468. doi: 10.1056/NEJMoa065202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pietra D, Li S, Brisci A, Passamonti F, Rumi E, et al. Somatic mutations of JAK2 exon 12 in patients with JAK2 (V617F)-negative myeloproliferative disorders. Blood. 2008;111:1686–1689. doi: 10.1182/blood-2007-07-101576. [DOI] [PubMed] [Google Scholar]
- 7.Ma W, Kantarjian H, Zhang X, Yeh C-H, Zhang ZJ, et al. Mutation profile of JAK2 transcripts in patients with chronic myeloproliferative neoplasias. J Mol Diagn. 2009;11:49–53. doi: 10.2353/jmoldx.2009.080114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma W, Kantarjian H, Verstovsek S, Jilani I, Gorre M, et al. Hemizygous/homozygous and heterozygous JAK2 mutation detected in plasma of patients with myeloproliferative diseases: correlation with clinical behavior. Br J Haematol. 2006;134:341–343. doi: 10.1111/j.1365-2141.2006.06174.x. [DOI] [PubMed] [Google Scholar]
- 9.Ma W, Tseng R, Gorre M, Jilani I, Keating M, et al. Plasma RNA as an alternative to cells for monitoring molecular response in patients with chronic myeloid leukemia. Haematologica. 2007;92:170–175. doi: 10.3324/haematol.10360. [DOI] [PubMed] [Google Scholar]
- 10.Giordanetto F, Kroemer RT. Prediction of the structure of human Janus kinase 2 (JAK2) comprising JAK homology domains 1 through 7. Protein Eng. 2002;15:727–737. doi: 10.1093/protein/15.9.727. [DOI] [PubMed] [Google Scholar]
- 11.Dusa A, Staerk J, Elliot J, Pecquet C, Poirel HA, et al. Substitution of pseudokinase domain residue Val-617 by large non-polar amino acids causes activation of JAK2. J Biol Chem. 2008;283:12941–12948. doi: 10.1074/jbc.M709302200. [DOI] [PubMed] [Google Scholar]
- 12.Zhao L, Ma Y, Seemann J, Huang LJ. A regulating role of JAK2 FERM domain in hyperactivation of JAK2(V617F). Biochem J Nov. 2009;24 doi: 10.1042/BJ20090615. doi: 10.1042/BJ20090615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee TS, Ma W, Zhang X, Kantarjian H, Albitar M. Structural effects of clinically observed mutations in JAK2 exons 13-15: comparison with V617F and exon 12 mutations. BMC Struct Biol Sep 10; 2009;9:58. doi: 10.1186/1472-6807-9-58. doi: 10.1186/1472-6807-9-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saharinen P, Silvennoinen O. The pseudokinase domain is required for suppression of basal activity of JAK2 and JAK3 tyrosine kinases and for cytokine-inducible activation of signal transduction. J Biol Chem. 2002;277:47954–47963. doi: 10.1074/jbc.M205156200. [DOI] [PubMed] [Google Scholar]
- 15.Funakoshi-Tago M, Pelletier S, Moritake H, Parganas E, Ihle JN. JAK2 FERM domain interaction with the erythropoietin receptor regulates JAK2 kinase activity. Mol Cell Biol. 2008;28:1792–1801. doi: 10.1128/MCB.01447-07. [DOI] [PMC free article] [PubMed] [Google Scholar]