Abstract
Purpose
This study was conducted to identify novel genes with importance to the biology of adult acute myelogenous leukemia (AML).
Experimental design
We analyzed DNA from highly purified AML blasts and paired buccal cells from 95 patients for recurrent genomic microdeletions using ultra-high density Affymetrix SNP 6.0 array-based genomic profiling.
Results
Through fine mapping of microdeletions on 17q, we derived a minimal deleted region of ~0.9Mb length that harbors 11 known genes; this region includes Neurofibromin 1 (NF1). Sequence analysis of all NF1 coding exons in the 11 AML cases with NF1 copy number changes identified acquired truncating frameshift mutations in 2 patients. These NF1 mutations were already present in the hematopoetic stem cell compartment. Subsequent expression analysis of NF1 mRNA in the entire AML cohort using FACS sorted blasts as a source of RNA identified 6 patients (one with a NF1 mutation) with absent NF1 expression. The NF1 null states were associated with increased Ras-bound GTP, and shRNA-mediated NF1 suppression in primary AML blasts with wild type NF1 facilitated colony formation in methylcellulose. Primary AML blasts without functional NF1, unlike blasts with functional NF1, displayed sensitivity to rapamycin-induced apoptosis, thus identifying a dependence on mTOR signaling for survival. Finally, colony formation in methylcellulose ex vivo of NF1 null CD34+/CD38− cells sorted from AML bone marrow samples was inhibited by low dose rapamycin.
Conclusions
NF1 null states are present in 7/95=7% of adult AML and delineate a disease subset that could be preferentially targeted by Ras or mTOR-directed therapeutics.
Keywords: AML, genomic microdeletions, NF1 mutations
INTRODUCTION
Cytogenetics and mutations in selected genes are of dominant importance to the biology and clinical outcome of patients with acute myelogenous leukemia(1–7). Further, genomic changes based on karyotyping of blood or marrow specimens from patients with AML directly impact AML treatment decisions.
While the anatomy of recurrent chromosomal translocations and the functional consequences of translocation-associated fusion proteins are well studied, relatively less is known about the genes that are mutated or deregulated as a consequence of subchromosomal copy number changes. Efforts at mapping subchromosomal genomic copy number changes using array-based CGH or SNP arrays in AML have identified genomic losses and gains, and candidate genes have been proposed (8–11). Ultimately, however, the combination of high-resolution DNA copy number and LOH analysis, combined with DNA sequence and gene expression analysis and followed by functional analysis is needed to identify novel genes with pathological significance in AML.
Recent developments in genome-wide high resolution copy number analysis using SNP arrays have aided better definitions of the pathological anatomy of cancer genomes, and application of SNP array technology to hematological cancers has refined knowledge of the anatomy of clinically important chromosomal lesions(12–19). For this study, we have employed Affymetrix SNP 6.0 arrays to interrogate the genomes of a large panel of adult AML cases for recurrent microdeletions and have identified a minimal deleted region of 0.9Mb on 17q that spans NF1. Through combined sequence and expression analysis we identify NF1 null states in ~7% of adult AML and have traced NF1 mutations to the hematopoetic stem cell compartment. Importantly, in addition to Ras pathway activation, NF1 null blasts demonstrated a sensitivity to low dose rapamycin-induced apoptosis, thus identifying a dependence on mTOR signaling for survival. Furthermore, CD34+/CD38− cells isolated from NF1 null AML bone marrow were inhibited by low dose rapamycin in methylcellulose colony assays. Combined, this data provides the most complete description yet of the incidence and consequences of NF1 functional loss in AML and suggests that a subset of AML patients are good candidates for clinical applications of mTOR inhibitors.
METHODS
Patients
Between March 2005 and August 2008, 95 patients with AML that were evaluated at the University of Michigan Comprehensive Cancer Center were enrolled into this study. The study was approved by the University of Michigan Institutional Review Board (IRBMED #2004-1022) and written informed consent was obtained from all patients prior to enrollment. Of these 95 patient samples, 92 resulted in array-based data for paired samples (blasts and buccal DNA) and 3 resulted in tumor data only.
Cell Isolation
Ficoll gradient separation and cryopreservation
Peripheral blood or bone marrow mononuclear cells from AML patients were isolated by Ficoll-paque gradient centrifugation (GE Healthcare), aliquoted into fetal calf serum (FCS) with 10% DMSO, and cryopreserved in the liquid phase of a liquid nitrogen tank.
Microbead-based negative selection and subsequent flow cytometry sorting of leukemia specimens
Liquid nitrogen-cryopreserved (liquid phase) peripheral blood or bone marrow mononuclear cells derived from AML patients were washed and recovered by centrifugation and then treated with anti-human CD3 (Miltenyi Biotec #130-050-101), anti-human CD14 microbeads (if blasts were negative for CD14 expression; Miltenyi Biotec #130-050-201), anti-human CD19 (if blasts were negative for CD19 expression; Miltenyi Biotec #130-050-301) and anti-human CD235a (Miltenyi Biotec #130-050-501) per manufacturer’s recommendations. Cell suspensions were run through Miltenyi MACS LS separation columns (#130-042-401) in order to negatively enrich for AML blasts. All blast preps were analyzed by cytospins for purity. This schema always resulted in greater than 90% blast purity.
AML blast DNA used for SNP 6.0 profiling and RNA used for expression analysis was extracted from samples that were further purified as follows: post-Miltenyi column samples were washed and stained with FITC-conjugated anti-CD33, PE-conjugated anti-CD13, and APC-conjugated anti-CD45 (all antibodies: eBioscience, San Diego, CA). After final washing, propidium iodide (PI) was added to a concentration of 1 μg/ml to discriminate dead cells. Sorting of cells was done on a FACS-ARIA high-speed flow cytometer (Becton Dickinson, Mountain View, CA). Live cells (PI-negative) were gated for blasts by identifying those cells with intermediate-intensity staining for CD45 and low- to moderate-intensity side scatter(20). CD33 and CD13 were then used in order to further discriminate blasts versus erythroid lineage and mature myeloid lineage cells.
Preparation of Sample DNA
Highly pure blast cells or buccal swabs were digested overnight in 100 mM Tris pH 8.0, 50 mM EDTA, 50 mM NaCl, 0.5% SDS and 100 μg/ml of Proteinase K (Invitrogen) at 56°C. DNA was extracted using phenol-chloroform and precipitated using ammonium acetate, ethanol and glycogen.
Array Data Analysis
The DNA was prepared for hybridization to SNP 6.0 arrays according to the manufacturers’ recommendations. Affymetrix CEL files for each blast and buccal sample were analyzed using Genotyping Console software for initial quality control, followed by use of the Affymetrix “Birdseed” algorithm to generate tab-delimited SNP call files in text format. Call rates for the entire group of samples included in this report were between 93.57% and 99.45% with mean and median call rates of 98.35% and 98.58%, respectively; none of the DNA samples gave out-of-bounds results. Sample copy number heatmap displays were obtained from CEL files through use of the freely available software dChip adapted to operate in a 64-bit environment(21). To generate functional and practical displays of LOH, a two-step, internally developed, Java-based software analysis system was employed. The Pre-LOH Unification Tool (PLUT) served to align all individual patient SNP calls to their respective dbSNP rs ID numbers and genomic physical positions prior to incorporation into the LOH tool version 2, an updated version of the LOH tool able to accommodate Affy 6.0 SNP array data(22). SNP 6.0 data files from all samples described in this study have been deposited with Geo accession number GSE21107.
Cytogenetic Analysis and Fluorescence In Situ Hybridization (FISH)
Cytogenetic analysis was performed on direct and 24-h cultures of leukemic cells without mitogens from bone marrow and peripheral blood samples that were prepared and fixed using standard techniques. Cryopreserved purified blast cells used for FISH analysis were thawed and cultured at 1×106 cells/ml for 24 hr at 37°C. Following incubation, the cells were prepared as for cytogenetics samples using standard techniques. Interphase FISH analysis for the NF1 locus was performed using BAC probe RP11-241P14 labeled with the fluorophore Green (BlueGnome, UK) and a SpectrumOrange™-labeled control probe that hybridizes to the subtelomeric region at 17q25 (TelVysion17q, Abbott Molecular, IL). The BAC probe RP11-241P14 hybridizes to chromosome band 17q11.2 (from nucleotide 27,081,373 to 27,261,539); it is 180 kb in length. FISH was performed following the procedures recommended by the manufacturers. A DAPI (4′-6-diamidino-2-phenylindole) counter-stain was used to visualize interphase nuclei. The expected normal signal pattern is two orange and two green signals. Slides were examined using a Leica DMRA fluorescence microscope equipped with a filter set including TexasRed/FITC dual-band pass and DAPI filters. A total of 200 interphase cells were analyzed per patient. Negative controls were prepared from cryopreserved purified AML blast cells without evidence of deletion of NF1 by SNP 6.0 array profiling, and were run in parallel with each hybridization.
Quantitative real-time PCR analysis of genomic copy number changes at 17q
Primers and TaqMan-based probes for Q-PCR applications were purchased from Applied Biosystems. Primers/probe mixtures included Hs06413068_cn and Hs01851142_s1. PCR reactions were done using DNA extracted from sorted blasts as template. Standardization of relative copy number estimates for genomic regions of interest was done using the ΔΔCt method with the Ct values for the RAG2 locus as reference.
Exon resequencing of NF1, NPM1, Flt3, p53, N-Ras and K-ras
Primers to amplify and sequence exons 1-58 of human NF1 (labeled 1-57 plus 31a), exon 12 of human NPM1, exons 13–15 and 20 of human Flt3, exons 2 and 3 of N-ras and K-ras and exons 5–9 of human p53 and adjacent intronic sequences were designed using the primer 3 program (http://frodo.wi.mit.edu/primer3/). The sequences of the NF1 primers used are tabulated in Supplementary Table 1. High quality sequence tracings were obtained for 631 out of (11×58=638)=99% of exons covered. PCR products were generated using Repli-g (Qiagen)-amplified DNA from highly pure blast cells as templates. Amplifications were done using Taq polymerase. PCR amplicons were prepared for direct sequencing with internal nested sequencing primers using the exonuclease/shrimp alkaline phosphatase method (USB). Mutation Surveyor (SoftGenetics LLC, State College, PA) software was used to compare experimental NF1 sequences against Refseq GenBank sequences as well as by visual inspection of sequence tracings.
Measurements of normalized NF1 or IRF8 expression using Q-PCR
RNA was prepared from ~2×105–106 ultrapure blasts, purified as outlined above, using the Trizol reagent and resuspended in 50μl DEPC-treated water. Complementary DNA was made from ~20ng of RNA using the Superscript III first strand synthesis kit (Invitrogen) and oligo-dT priming. Primers and TaqMan-based probes were purchased from Applied Bio-systems (Primers-on-demand). Primer/probe mixtures included: NF1 (Hs00169714_m1), IRF8 (Hs00175238_m1) and PGK1 (Hu PGK1). Duplicate amplification reactions included primers/probes, TaqMan® 2× Universal PCR Master Mix, No AmpErase UNG and 1μl of cDNA in a 20ul reaction volume. Reactions were done on an ABI 7900HT machine. Normalization of relative copy number estimates for NF1 or IRF8 mRNA was done with the Ct values for PGK1 as reference (Ct mean gene of interest − Ct mean PGK1).
Cell fractionation of AML cases with NF1 mutations for NF1 sequence analysis
AML cells were stained with anti-CD34 and CD38 and sorted into CD34+CD38+, CD34−CD38−, CD34+CD38− and CD34−CD38+ subsets using a FACSAria (Becton Dickinson).
Ras activity measurements in AML blasts
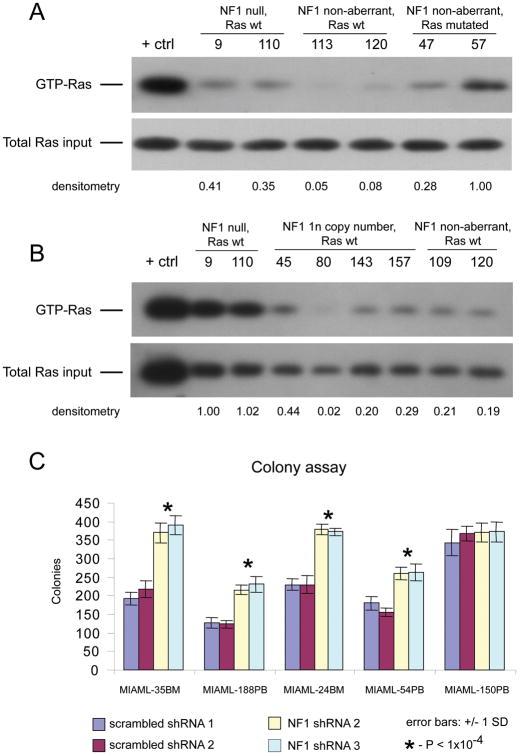
Analysis of Ras activation was performed using a Ras Activation Assay Kit from Millipore according to the manufacturer’s instructions. Briefly, two million leukemia blast cells from selected patients (see Supplementary Table 2) isolated using bead-based depletion of non-blasts, as above, were lysed in 500 μl of lysis buffer that was included in the Ras activity assay kit. The lysates were cleared through centrifugation and subsequently incubated at 4°C overnight with glutathione agarose beads bound to a GST fusion protein of the human Ras binding doman (RBD) of Raf-1. Following washings with lysis buffer, the bound GTP-Ras was liberated through boiling in SDS-PAGE loading buffer and then resolved by SDS-PAGE. A pan-anti-Ras antibody able to recognize total H-, K-, and N-Ras was used to measure Ras protein amounts. A GTPγS loaded cell supernatant was used as a positive assay control. The same amount of lysate for each sample prior to the bead pull down was analyzed in parallel by immunoblotting for total Ras. Blots were subjected to densitometric quantitation using an Alpha Imager transilluminator (Cell Biosciences, Santa Clara, CA) and the SpotDenso feature of this company’s AlphaEaseFC software. An image of each blot film was captured using identical camera exposure settings for all films. For each blot image, identically-sized density measurement rectangles were placed around each protein band of interest, and local background was subtracted for each individual rectangle. Resulting band density values were imported into Microsoft Excel. For each sample, the protein band density values for GTP-Ras and total Ras were divided by the respective values for GTP-Ras and total Ras in one selected sample (indexed as 100%) as indicated in Figure 3A–B.
Figure 3. Increased Ras activity in NF1 null blasts and increased colony formation of AML blasts with experimentally reduced NF1 protein.
A: Immunoblot results for GTP-Ras and total Ras for 6 AML cases with NF1 and Ras status as indicated (all Flt3 wild type). Results of densitometric quantification of relative band intensities compared with case #57. B: Immunoblot results for GTP-Ras and total Ras for 8 AML cases with NF1 and Ras status as indicated (all Flt3 wild type). Results of densitometric quantification of relative band intensities compared with case #9. C: Results of AML sample colony formation assays in methylcellulose for 5 AML-derived Ficoll-gradient purified cell samples with wild type Ras and wild type NF1 (all Flt3 wild type), each infected in aliquots with lentiviruses carrying either scrambled or NF1-targeted and validated shRNAs. Colony counts were done in triplicate for each shRNA for each case; error bars represent two standard deviations. P-values are for comparison of NF1 shRNA versus scrambled shRNA treated samples.
NF1 shRNA-mediated interference and blast colony formation assays
The lentiviral vector pLenti6/V5-TOPO, obtained from Invitrogen, was modified to express either two scrambled shRNAs (Sr1: ggcgttggctcgtttagta, and Sr2: gaattagaacgaatcaaag) or two previously validated NF1 shRNA (Sh2: ggttgcgcagttagcagtt (23) and Sh3: ataaatagcctggaaaagg (24)), according to our published procedure (25).
Ficoll-prepared cells from bone marrow or peripheral blood from five AML patients (blast percentage range 40–90%; see Supplementary Table 2) that were all wild type for NF1 were spin-inoculated with recombinant lentiviruses and incubated for 2 hours in medium containing IL-3, SCF, and GM-CSF (all cytokines: R&D Systems, Minneapolis, MN). Transduction efficiencies of leukemia specimens were measured using the background fluorescence of untransduced cells as a baseline (accepting <2% of cells above the fluorescence intensity gate) and measuring GFP intensity in transduced cells in the FITC channel. Using this criterion, transduction efficiencies for the five studied cases were at least 27.1%, 48.2%, 50.8%, 59.1% and 69%, respectively. Twenty thousand cells were subsequently plated in semisolid methylcellulose medium supplemented with multiple cytokines (Methocult H4535, StemCell Technologies, Vancouver, BC). Colony counts were obtained 7 days after seeding.
To estimate NF1 mRNA knock-down efficiency, Ficoll-prepared cells from 3 AML cases were spin-inoculated with recombinant lentiviruses and GFP positive cells purified through FACS-sorting ~36 hours post infection. RNA was prepared and NF1 and PGK1 mRNA quantified using Q-PCR as above.
Rapamycin treatment and AML blast apoptosis assays
2.5×105 bead-purified blasts were incubated in 100 μl RPMI with 10% FCS and analyzed by annexin V and PI-based flow cytometry after 24 hours. Cells were incubated in DMSO only or with 20 nM rapamycin (LC Laboratories, Woburn, MA). The fractions of cells still viable after 24h (annexin V-negative, PI-negative) were normalized to the untreated percent viable cell fraction for each patient and results plotted.
Ex vivo colony growth assays of purified CD34+/CD38− bone marrow cells in the presence of rapamycin
Ficoll-gradient prepared, cryopreserved AML marrows (see Supplementary Table 3) were thawed, washed, blocked, and stained with anti-CD34-PE and anti-CD38-APC (both: eBioscience, San Diego, CA). The CD34+/CD38− fraction of each marrow was sorted on a FACSAria flow sorter (Becton-Dickinson, San Jose, CA). Purified cells were introduced, in duplicate, into semisolid methylcellulose medium supplemented with multiple cytokines (Methocult H4535, StemCell Technologies, Vancouver, BC) at concentrations of 1000 cells/ml and 5000 cells/ml and using rapamycin (#R-5000, LC Laboratories, Woburn, MA) concentrations from 0 to 100 nM. Compact colonies were counted in duplicate at 7 days.
Ex vivo colony growth assays of AML marrow or blood cells
Ficoll-gradient prepared, cryopreserved AML marrows with >50% blasts (see Supplementary Table 4) were thawed, washed, and introduced directly, in duplicate, into methylcellulose medium (Methocult H4230, StemCell Technologies, Vancouver, BC) containing either a range of GM-CSF from 0 to 1 ng/ml only, or containing both GM-CSF and a constant 50ng/ml of SCF (#215-GM and 255-SC, respectively, both from R&D Systems, Minneapolis, MN). Final concentration of cells in methylcellulose was 20,000/ml. Colonies were counted at 7 days. Marrows were prescreened whenever possible to exclude cases with prevalent FLT3 ITD mutations or lack of IRF-8 expression to minimize confounding effects of these genes on the assay(26).
RESULTS
Patient Characteristics
Included in this analysis are data from 95 patients, of which 76 patients were previously untreated and 19 patients had relapsed AML. Sixty nine percent, 13%, and 18% of cases were primary AML, secondary AML or treatment-related AML, respectively. Data are summarized in Table 1.
Table 1.
Baseline clinical characteristics of AML patients analyzed in this study.
| Characteristic | Treatment-naive at enrollment, no. (%) | Previously treated and relapsed at enrollment, no. (%) |
|---|---|---|
| Sample size (95 total) | 76 (79) | 19 (21) |
| Age, y | ||
| Median | 64 | 65 |
| Range | 20–85 | 21–79 |
| Sex | ||
| Male | 45 (59) | 11 (58) |
| Female | 31 (41) | 8 (42) |
| FAB classification* | ||
| M0 | 10 (16) | 0 (0) |
| M1 | 13 (20) | 3 (20) |
| M2 | 12 (19) | 3 (20) |
| M4 | 24 (37) | 6 (40) |
| M5 | 5 (8) | 3 (20) |
| M6 | 0 (0) | 0 (0) |
| M7 | 0 (0) | 0 (0) |
| Immunophenotype | ||
| CD13 positive | 67 (88) | 16 (84) |
| CD33 positive | 66 (87) | 19 (100) |
| CD34 positive | 60 (79) | 10 (53) |
| CD117 positive | 70 (92) | 17 (89) |
| Cytogenetic class** | ||
| Favorable | 6 (8) | 0 (0) |
| Intermediate | 38 (51) | 16 (84) |
| Unfavorable | 31 (41) | 3 (16) |
| No. of karyotypic abnormalities | ||
| Three or more | 16 (20) | 1 (5) |
| Less than three | 59 (80) | 18 (95) |
| Pathogenesis | ||
| De novo | 51 (67) | 15 (78) |
| Prior myelodysplasia | 10 (13) | 2 (11) |
| Treatment-related | 15 (20) | 2 (11) |
The FAB subclassification was unspecified in 16 samples. The M3 subtype was not included in this study.
One sample had no cytogenetic data available
SNP 6.0 array-based data analysis
In support of the analyses outlined below, we developed and refined previously validated software tools that can be used for display of data resulting from SNP 6.0 array profiling. These are PLUT, the LOHtool version 2 and an updated version of dChipSNP(18, 21, 27). Lesion calling relied on visual inspection of copy number and LOH displays for paired tumor and buccal DNA samples within dChipSNP, a conservative and very specific method of lesion detection (see methods). To obtain additional experimental verification that this approach would reliably detect single copy number changes, we initially measured genomic copy number estimates in 10 cases with heterozygous NF1 deletions (see below) and 10 NF1 undeleted cases using a Taqman-based Q-PCR probe for NF1 (located in intron 1 at physical position 26,447,140, based on NCBI build 36) compared with a reference gene (RAG2). Data are summarized in Supplementary Table 5, demonstrating excellent agreement (~1.8-fold mean copy number differences between deleted and undeleted samples) with SNP array-based genomic copy number estimates (case #76 displayed a 17q deletion that may not have included the NF1-cn probe location and case #147 displayed a heterozygous deletion of RAG2 located on chromosome 11 at ~36.57–36.576 Mb).
We also employed FISH with probes located within NF1 and a reference probe (see methods) on 7 cases with available material for study (cases # 7, 45, 87, 140, 147, 157 and 132 [NF1 gain]) and for each case confirmed the SNP 6.0 array findings (Supplementary Tables 6A and B). Finally, the complete list of acquired chromosomal copy number changes and acquired UPD for the 92 samples subjected to paired analysis can be found in Supplementary Table 7.
Detection of microdeletions spanning the NF1 tumor suppressor gene on 17q in AML
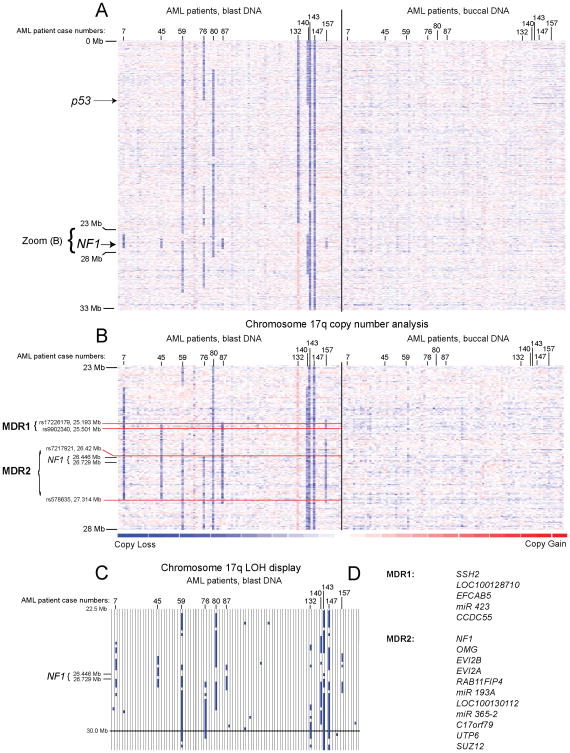
Copy number analysis for chromosome 17p and 17q detected large deletions and microdeletions in genomic regions where deletions of various lengths have previously been described(8, 9, 11), and two minimal regions of deletion (MDR) could be deduced (Figure 1A and B and Supplementary Table 8). These are 17q-MDR1 (spanning ~0.3Mb), which contained five genes as tabulated in Figure 1D, and 17q-MDR2 (spanning ~0.9Mb), which contained 11 genes as tabulated in Figure 1D. 17q-MDR2 was present in 10/95= 11% of cases and invariably included the NF1 gene. An additional case (#132) had a gain of NF1 as part of a larger gain on 17p (see Figure 1A–B). All cases of LOH at 17q-MDR2 (Figure 1C) were associated with copy loss- thus copy neutral LOH (acquired UPD) at the NF1 locus did not exist in this AML cohort. Review of cytogenetics data for the AML cases with microdeletions at 17q (#7, 45, 87, 140 and 157) demonstrated that only one case was detected using routine karyotyping.
Figure 1. Fine mapping of minimal deleted regions on chromosome 17q in AML uncovers frequent NF1 deletions (heatmap display).
Fig. 1A, copy number heatmap display for a region on chromosome 17p and 17q for DNA from AML-derived blasts and paired buccal DNA based on SNP 6.0 array profiling. The approximate location of p53 and NF1 is indicated. Fig. 1B, copy number display for a region on chromosome 17q for DNA from AML-derived blasts and paired buccal DNA from AML patients. Each column represents one patient. Minimal deleted regions and boundaries are marked. Fig. 1C, LOH display for AML blasts versus buccal DNA for the genomic region corresponding to Figure 1B. Fig. 1D, genes located within 17q-MDR 1 and 2.
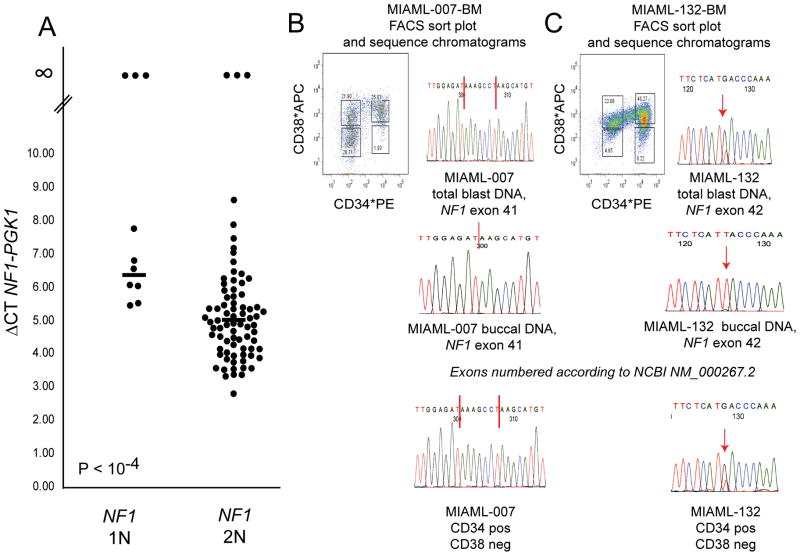
Given that NF1 is a known tumor suppressor in humans and a negative regulator of Ras signaling, and given that mutations in NF1 have been described in sporadic cases of pediatric leukemias (T-ALL and one confirmed case of AML) and frequently in cases of juvenile myelomonocytic leukemia (JMML), we proceeded to sequence all 58 coding exons of NF1 in all 11 affected AML cases using direct exon resequencing (28–31). These efforts uncovered a seven base pair insertion in NF1 in exon 41 in case #7 (heterozygous NF1 deletion) resulting in a truncated NF1 protein (NF1 delta amino acid 2058–2818) and a heterozygous frame-shift mutation in NF1 in exon 42 in case #132 (a case with 3 NF1 copies), resulting in a truncated NF1 molecule (NF1 delta amino acid 2137–2818, based on NM_000267.2) encoded by the mutated alleles (Figure 2B&C). Confirmatory sequencing of paired buccal DNA confirmed the somatic nature of these mutations (Figure 2B&C).
Figure 2. Differential expression of NF1 in AML blasts with one or two copies of NF1, detection of somatic NF1 mutations and tracking of NF1 mutations to the leukemia stem cell compartment.
Panel A: AML-derived blasts were sorted using FACS sorting, and cDNA was made from RNA using oligo-dT priming. Duplicate Q-PCR reactions for NF1 and PGK1 were performed. Displayed are delta Ct mean NF1 minus Ct mean PGK1 values grouped by NF1 status (1N versus 2N) and the mean of these values (horizontal bars). Panels B–C: DNA sequence tracings of parts of selected NF1 exons demonstrating somatically acquired NF1 frameshift mutations. B: MI-AML #7 blasts result with vertical red bars indicating the boundaries of a 7 basepair insertion and corresponding results for paired buccal DNA, C: MI-AML #132 blasts result with vertical red arrow indicating the T to G mutation and corresponding results for paired buccal DNA. Flow tracing of CD34 and CD38 sorted cell populations for AML cases #7 and 132 and associated NF1 mutation analysis results.
Given the absence of mutations in NF1 in the majority of AML cases with one retained NF1 copy, we proceeded with measurements of NF1 expression in all AML blast specimens for which sufficient intact RNA was available (Figure 2A). Interestingly, this analysis disclosed three AML cases with heterozygous NF1 deletions (including case # 7) and three AML cases without NF1 deletions with absent NF1 expression and thus complete loss of NF1 activity (in addition to cases with low relative NF1 expression), and further, reduced mean NF1 expression in the remaining AML cases with heterozygous NF1 deletion (N=7) compared with all AML cases without NF1 deletion (N=77; delta CT mean NF1-PGK1 of 6.3 versus 5.0 in heterozygously deleted NF1 versus undeleted NF1 blasts, p<0.001). A summary of clinical and genomic characteristics of the AML cases with NF1 null states and the cases with heterozygous NF1 deletion but preserved NF1 expression from the retained allele can be found in Table 2.
Table 2.
Clinical and genomic characteristics of AML patients with NF1 null states due to mutations or absent expression and of AML patients with heterozygous NF1 deletions and retained NF1 expression.
| AML case # | NF1 status (mutations/expression) | NF1 status | Age | Disease status | AML type | FAB type | Cytogenetics | P53 status | Flt3 status | NPM status | N-, K-Ras status |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NF1 status null | |||||||||||
| 7 | Nf1 mutated and mRNA not expressed | 1N | 53 | Relapsed | Primary | M5 | normal | wt | wt | Mutant | wt |
| 9 | mRNA not expressed | 2N | 64 | New, untreated | Primary | M4 | normal | wt | wt | wt | wt |
| 59 | mRNA not expressed | 1N | 78 | New, untreated | Primary | unknown | complex | Mutant | wt | wt | wt |
| 73 | mRNA not expressed | 2N | 75 | New, untreated | Primary | M4 | Del5q | wt | wt | wt | wt |
| 87 | mRNA not expressed | 1N | 56 | New, untreated | Primary | 0 | T(3,8,22) | wt | wt | wt | wt |
| 110 | mRNA not expressed | 2N | 27 | New, untreated | Primary | M5 | +8, inv3 | wt | wt | wt | wt |
| 132 | Nf1 mutated | 3N | 71 | New, untreated | Primary | M4 | −7, iso 17 | wt | wt | wt | wt |
| NF1 status expressed | |||||||||||
| 45 | mRNA expressed | 1N | 64 | New, untreated | Secondary | M4 | normal | wt | wt | wt | wt |
| 76 | mRNA expressed | 1N | 52 | New, untreated | Secondary | unknown | complex | wt | wt | wt | wt |
| 80 | mRNA expressed | 1N | 70 | New, untreated | Primary | M4 | complex | wt | wt | wt | wt |
| 140 | mRNA expressed | 1N | 72 | New, untreated | Primary | M4 | complex | Mutant | wt | wt | wt |
| 143 | mRNA expressed | 1N | 67 | New, untreated | Primary | M1 | complex | Mutant | wt | wt | wt |
| 147 | mRNA expressed | 1N | 55 | New, untreated | Primary | M0 | complex | Mutant | D835DY | wt | wt |
| 157 | mRNA expressed | 1N | 62 | New, untreated | Secondary | M5 | complex | Mutant | wt | wt | wt |
To address the question of the presence of NF1 mutations in leukemia progenitor cells, we repeated sorts for AML-derived bone marrow cells from the 2 cases with NF1 mutations using CD34 and CD38 as markers and analyzed the resulting CD34+/CD38− cell populations for NF1 mutations (Figure 2B&C). As can be seen in Figure 2B&C, NF1 mutations were found in the CD34+/CD38− cell population known to harbor the leukemia initiating cells.
NF1 regulates the activation state of Ras in primary AML blasts and NF1 downregulation increases blast colony formation in methylcellulose
Given that NF1 regulates the activation states of Ras, we measured GTP-bound Ras by immunoblotting in bead-purified AML blasts with various NF1 and Ras states and wild type Flt3 status (Figure 3A) and normalized data based on the signals detected in Ras mutant blasts (#57). As can be seen in Figure 3A, GTP-Ras was substantially increased in AML cases with absent NF1 expression and in cases with activating N-Ras or K-Ras mutations, respectively, if compared with blasts that were wild type for NF1 and Ras (all cases were wild type for Flt3). Thus, NF1 deficiency results in an increase in Ras activity in affected AML cases. Next we proceeded to estimate the effect of heterozygous NF1 deletions but preserved NF1 mRNA expression from the retained NF1 allele on GTP-bound Ras. As can be seen in Figure 3B, while NF1 null blasts again demonstrated an approximately 5× increase in GTP-Ras as compared with NF1 non aberrant blasts (based on newly bead-purified AML blasts from cases # 9 and 110), the effect of heterozygous NF1 deletions on the amount of GTP-Ras was not substantially different from NF1 non-aberrant blasts in three of the four cases tested, with only one case (#45) demonstrating a mild increase. Thus, within the limits of these measurements (all performed in bead-purified primary human leukemia blasts) one can conclude that complete NF1 loss is required to result in substantial Ras activation and that heterozygous NF1 states that preserve some NF1 expression are not sufficient for robust Ras activation.
Next we asked whether NF1 deficiency had an effect on blast colony formation as a readout for pro-proliferative activities of lower NF1 levels. Aliquots of blasts from five AML cases with wild type Ras and wild type Flt3 were infected with lentiviruses carrying transcriptional units for validated NF1-suppressing shRNA or scrambled shRNAs (resulting in ~ 2–3-fold reductions in NF1 mRNA in AML blasts; Supplementary Table 9). Compact colony numbers were scored after 7 days in culture. As can be seen in Figure 3B, 4 out of 5 AML cases demonstrated a substantial and significant increase in AML blast colony formation in methylcellulose when NF1 was downregulated, while the remaining AML case demonstrated already relatively elevated colony formation propensities at baseline (and was characterized by the lowest NF1 mRNA expression at baseline as compared with the other studied cases). Thus NF1 directly influences AML blast proliferation/growth (see also Figure 4B). Finally, given that baseline colony numbers were not influenced by the cell source (BM versus PB), that the colonies did not have the appearance of BFU-E, and given that CFU numbers in non-malignant blood are substantially lower than the colony numbers observed here, we conclude that the majority of colonies are AML blast-derived(32).
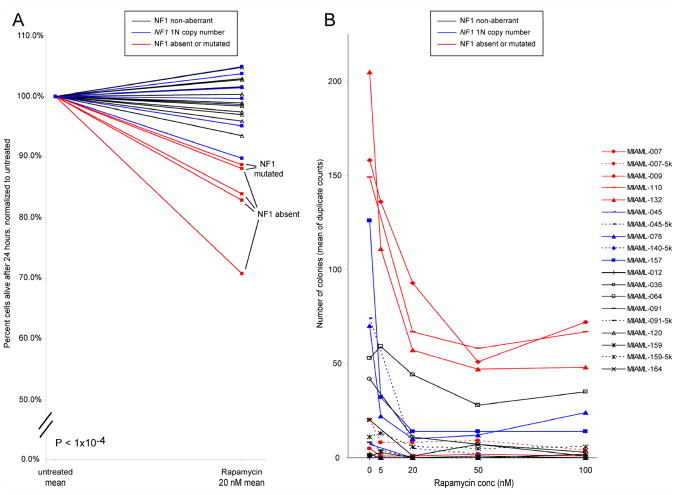
Figure 4. Sensitivity of NF1 null AML blasts to rapamycin-induced apoptosis and inhibition of colony formation of CD34+/CD38− cells by rapamycin.
A: AML blasts from AML cases with wild type NF1 (N=12; black), NF1 mono-allelic loss but preserved NF1 expression from the retained allele (N=6; blue) or NF1 null states (N=5; red) due to either NF1 mutations or absent NF1 mRNA expression were purified using negative selection and subsequently cultured for 24 hours in the presence or absence of 20nM rapamycin. Blast viability and apoptosis were measured using FACS-based annexin V-PI staining. Displayed are normalized viabilities of rapamycin-treated samples compared with paired untreated samples. B: CD34+/CD38− cells from AML cases with wild type NF1 (N=7; black), NF1 mono-allelic loss but preserved NF1 expression from the retained allele (N=4; blue) or NF1 null states (N=4; red) due to either NF1 mutations or absent NF1 mRNA expression were purified using FACS-sorting and 1000 cells plated in cytokine-supplemented methylcellulose in the absence or presence of escalating doses of rapamycin for 7 days. Colony counts represent means of duplicate measurements. Cases with sufficient cells were also plated at 5000 cells per well as indicated (−5k).
Colony formation of AML blasts in methylcellulose supplemented with escalating doses of GM-CSF with or without SCF
Hypersensitivity of myeloid cells derived from patients with JMMoL has been described and murine cells with engineered complete NF1 inactivation are hypersensitive to GM-CSF(30, 33, 34). Therefore, we measured colony formation of AML blasts from NF1 null cases (N=4), heterozygous (1N) NF1 cases with preserved NF1 expression from the retained allele (N=3) and NF1 non-aberrant cases (N=8) in methylcellulose only supplemented with either escalating doses of GM-CSF or escalating doses of GM-CSF and a fixed dose of SCF. As can be seen in Supplementary Figures 1 and 2, most of the cases with the ability to form colonies under these conditions displayed some sensitivity to GM-CSF, although one tested NF1 null case and one 1N NF1 case with preserved NF1 mRNA expression from the retained allele appeared particularly sensitive.
NF1 null AML-derived blasts are sensitive to rapamycin-induced apoptosis
An additional important function of NF1 as a negative regulator of mTOR signaling has been described(24). Based on this observation, we reasoned that AML blasts that may have arisen in the setting of NF1 deficiency would be more dependent on mTOR signaling for survival and growth than NF1 wild type blasts. We therefore treated purified primary AML blasts with rapamycin and measured apoptosis induction after overnight cultures. Data are summarized in Figure 4A. Importantly, AML blasts without functional NF1 (N=5) were substantially and significantly (p<0.001) more sensitive to mTOR inhibition than NF1 wild type blasts (N=12) or blasts with one preserved NF1 copy (N=6) and retained NF1 expression.
Ex vivo colony formation of NF1 null AML-derived CD34+/CD38- cells is inhibited by rapamycin
Next we asked whether the cellular compartment that harbors the leukemia initiating cells in AML (CD34+/CD38− cells) would be sensitive to rapamycin treatment. CD34+/CD38− cells from AML bone marrow samples that were either NF1 null or had lost one NF1 gene copy but displayed NF1 expression from the retained allele or were 2N at the NF1 locus and expressed NF1 (non-aberrant cases) were sorted using FACS and cells subsequently plated in supplemented methylcellulose medium in the presence of varying concentrations of rapamycin (0–100nM). Colony formation was assessed after 7 days in culture. As can be seen in Figure 4B, colony formation in the absence of rapamycin treatment (untreated samples) was robust in 3 out of 4 cases each that were either NF1 null (red lines) or 1N NF1 with retained NF1 expression (blue lines) while only 2 out of 7 NF1 non-aberrant cases displayed appreciable colony formation, suggesting that lower or absent NF1 expression facilitates ex vivo growth of affected cells (and complementing the experimental observations displayed in Figure 3C). Furthermore, cells from all informative NF1 null cases or 1N NF1 cases with retained NF1 expression and, to a lesser degree, cells from the 2 informative NF1 non-aberrant cases displayed sensitivity to low dose rapamycin ex vivo. Thus in summary, a fraction of blast cells and leukemia- initiating cells from AML patients with NF1 null states are growth-inhibited or killed by low dose rapamycin.
Discussion
In this study, we have employed ultra-high density SNP arrays to interrogate the genomes of highly purified leukemic blasts and paired buccal DNA from 95 AML patients for chromosomal copy number changes. Through comparison of blast-derived and paired buccal DNA in this analysis, we were able to unequivocally identify small somatically acquired losses at unprecedented resolution. Focusing on small deletions (sub-megabase to a few megabases) that occurred recurrently or that occurred at positions that are spanned by recurrent larger deletions, we have identified a minimal deleted region of ~0.9Mb on chromosome 17q that spans NF1, a disease-causing gene in neurofibromatosis. NF1 copy number alterations were found in 11/95=12% of patients in this cohort (10 cases with heterozygous NF1 deletions and one case with a large chromosomal gain that harbored mutated NF1 alleles) and in 6/86 patients recently described by Walter et al. 2009. Sequence-based analysis identified 2 AML cases with mutated acquired NF1 alleles that predicted for truncated NF1 proteins. As these NF1 mutations were also identified in CD34+/CD38− cell populations, we can presume that NF1 mutations contribute to the biological phenotype of leukemia-initiating cells, possibly through control of proliferation. In this context it appears important that AML blasts with shRNA-mediated NF1 knock-down demonstrated improved growth and colony formation ex vivo and that most NF1 null AML-derived CD34+/CD38− cells grew robustly in cytokine-supplemented methylcellulose.
While sequencing of all coding exons of NF1 in 9 additional AML cases with heterozygous NF1 deletions did not reveal inactivating gene mutations, expression analysis of NF1 in a large collection of AML cases demonstrated lower expression in cases with heterozygous NF1 deletions as opposed to wild type NF1 cases. The accuracy of this expression analysis is further strengthened by the fact that we used FACS-sorted AML blasts as a source of RNA, effectively eliminating contributions of non-blast cells to the NF1 mRNA quantitations. Lower NF1 mRNA expression may contribute to AML pathogenesis, given prior evidence for NF1 haploinsufficiency in some biological systems(35–37). Our study of AML cases with heterozygous NF1 deletions and preserved NF1 expression from the retained allele uncovered some evidence for haploinsufficiency only in the setting of ex vivo blast colony formation. The activity of Ras was not substantially influenced in blasts with heterozygous NF1 deletions that preserved NF1 expression from the retained allele. Importantly, six AML cases (three cases with heterozygous NF1 deletions and three cases without NF1 deletions) did not express NF1 mRNA, thus identifying a subset of adult AML with NF1 null states. Where measured, NF1 null states were associated with substantially (~5-fold) increased Ras activity. Identification of AML cases with Ras activation due to complete NF1 inactivation may be of clinical importance, given i) findings of increased sensitivity of AML blasts from such patients to Cytarabine (1-β-D-arabinofuranosylcytosine)(38, 39) and ii) the potential for identifying additional subsets of AML for Ras-directed therapeutics.
In this study, we have identified for the first time a sensitivity of NF1 null AML blasts to rapamycin-induced apoptosis. Blasts with preserved NF1 expression independent of NF1 copy number status did not undergo rapamycin-induced apoptosis in agreement with prior observations (40). Subsequent analysis of the ex vivo colony formation potential of CD34+/CD38− cells isolated from AML cases with various NF1 states uncovered an inhibitory effect of low dose rapamycin on NF1 null blasts and on blasts with heterozygous NF1 deletion and preserved expression from the retained allele, although some of the informative NF1 non-aberrant cases were also inhibited(41). Together, these findings of sensitivity of NF1 null AML blasts to rapamycin extends observations of regulation of mTOR by NF1 to leukemia and opens possibilities for patient selection for clinical mTOR inhibitor applications based on NF1 status (42–44).
NF1 mutations have long been implicated in the pathogenesis of JMML but evidence for a role of NF1 in adult AML has been sporadic. Using a very large collection of >600 cases of hematological malignancies and focusing on a subset of 34 cases with MDS/AML and AML1/RUNX1 mutations, Niimi et al. identified one case of high grade MDS and one case of secondary AML following MDS with NF1 sequence changes predicting for truncated NF1 alleles, although evidence that these changes were acquired in tumor cells was not provided(45). Recently, Balgobind et al. examined 103 pediatric T-ALL cases and 71 pediatric AML cases with MLL-rearrangements and identified 2 AML cases with 17q microdeletions and NF1 mutations, of which one was confirmed to be acquired in tumor cells(28). An expression analysis of NF1 in this AML cohort was not reported. In contrast with these two reports, the seven AML cases in this cohort that were NF1 null did not have MLL rearrangements and all were primary AML.
In summary, this study advances our knowledge of the incidence of NF1 aberrations and NF1 null states in AML and provides experimental evidence that NF1 null states are functionally relevant in AML. Finally, we demonstrate that AML cases with absent NF1 function are sensitive to mTOR inhibition, thus providing a rationale for the study of mTOR inhibitors in NF1 null AML subsets.
TRANSLATIONAL RELEVANCE.
Acute myelogenous leukemia (AML) has an incidence of ~12,000 new cases in the US per year with ~9,000 untimely deaths. Cytogenetics and mutations in selected genes are of importance to the biology, therapy selection and clinical outcome of patients with AML. Ras pathway activation due to either Ras mutations or upstream gene mutations is common in AML, but knowledge about the role of the tumor suppressor NF1, a negative Ras regulator, in AML is still evolving. In this study, we provide a detailed analysis of NF1 aberrations in a large cohort of AML and have identified NF1 null AML blasts and NF1 null CD34+/CD38− cells to be sensitive to mTOR inhibition, thus providing a refined molecular rationale for clinical applications of mTOR inhibitors in AML.
Supplementary Material
Footnotes
Individual contributions
Peter Ouillette, Yin Wang, Yan Liu, Yang Liu, Whitney Wright and Sami Malek performed the laboratory research.
Kerby Shedden, Cheng Li and Sajid Shakhan assisted with statistical methods.
Brian Parkin, Amanda Dressel, Lisa Kujawski, Paula Bockenstedt, Ammar Al-Zoubi, Moshe Talpaz, Harry Erba and Sami Malek enrolled patients and contributed and analyzed the clinical data.
Diane Roulston and Anjali Purkayastha assisted with cytogenetics/FISH data interpretation.
Judith Karp assisted with questions related to AML biology and classification.
Sami Malek conceived the study.
Peter Ouillette, Brian Parkin and Sami Malek wrote the paper.
Conflict of interest disclosure
None to declare
References
- 1.Schlenk RF, Dohner K, Krauter J, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. The New England journal of medicine. 2008;358:1909–18. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 2.Pedersen-Bjergaard J, Andersen MK, Andersen MT, Christiansen DH. Genetics of therapy-related myelodysplasia and acute myeloid leukemia. Leukemia. 2008;22:240–8. doi: 10.1038/sj.leu.2405078. [DOI] [PubMed] [Google Scholar]
- 3.Byrd JC, Mrozek K, Dodge RK, et al. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100:4325–36. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- 4.Grimwade D, Walker H, Oliver F, et al. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children’s Leukaemia Working Parties. Blood. 1998;92:2322–33. [PubMed] [Google Scholar]
- 5.Rowley JD. Molecular genetics in acute leukemia. Leukemia. 2000;14:513–7. doi: 10.1038/sj.leu.2401600. [DOI] [PubMed] [Google Scholar]
- 6.Renneville A, Roumier C, Biggio V, et al. Cooperating gene mutations in acute myeloid leukemia: a review of the literature. Leukemia. 2008;22:915–31. doi: 10.1038/leu.2008.19. [DOI] [PubMed] [Google Scholar]
- 7.Fitzgibbon J, Smith LL, Raghavan M, et al. Association between acquired uniparental disomy and homozygous gene mutation in acute myeloid leukemias. Cancer research. 2005;65:9152–4. doi: 10.1158/0008-5472.CAN-05-2017. [DOI] [PubMed] [Google Scholar]
- 8.Suela J, Alvarez S, Cifuentes F, et al. DNA profiling analysis of 100 consecutive de novo acute myeloid leukemia cases reveals patterns of genomic instability that affect all cytogenetic risk groups. Leukemia. 2007;21:1224–31. doi: 10.1038/sj.leu.2404653. [DOI] [PubMed] [Google Scholar]
- 9.Rucker FG, Bullinger L, Schwaenen C, et al. Disclosure of candidate genes in acute myeloid leukemia with complex karyotypes using microarray-based molecular characterization. J Clin Oncol. 2006;24:3887–94. doi: 10.1200/JCO.2005.04.5450. [DOI] [PubMed] [Google Scholar]
- 10.Paulsson K, Heidenblad M, Strombeck B, et al. High-resolution genome-wide array-based comparative genome hybridization reveals cryptic chromosome changes in AML and MDS cases with trisomy 8 as the sole cytogenetic aberration. Leukemia. 2006;20:840–6. doi: 10.1038/sj.leu.2404145. [DOI] [PubMed] [Google Scholar]
- 11.Walter MJ, Payton JE, Ries RE, et al. Acquired copy number alterations in adult acute myeloid leukemia genomes. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12950–5. doi: 10.1073/pnas.0903091106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raghavan M, Smith LL, Lillington DM, et al. Segmental uniparental disomy is a commonly acquired genetic event in relapsed acute myeloid leukemia. Blood. 2008;112:814–21. doi: 10.1182/blood-2008-01-132431. [DOI] [PubMed] [Google Scholar]
- 13.Paulsson K, Cazier JB, Macdougall F, et al. Microdeletions are a general feature of adult and adolescent acute lymphoblastic leukemia: Unexpected similarities with pediatric disease. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:6708–13. doi: 10.1073/pnas.0800408105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawamata N, Ogawa S, Zimmermann M, et al. Cloning of genes involved in chromosomal translocations by high-resolution single nucleotide polymorphism genomic microarray. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:11921–6. doi: 10.1073/pnas.0711039105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gondek LP, Tiu R, O’Keefe CL, Sekeres MA, Theil KS, Maciejewski JP. Chromosomal lesions and uniparental disomy detected by SNP arrays in MDS, MDS/MPD, and MDS-derived AML. Blood. 2008;111:1534–42. doi: 10.1182/blood-2007-05-092304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mullighan CG, Goorha S, Radtke I, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–64. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 17.Saddler C, Ouillette P, Kujawski L, et al. Comprehensive biomarker and genomic analysis identifies p53 status as the major determinant of response to MDM2 inhibitors in chronic lymphocytic leukemia. Blood. 2008;111:1584–93. doi: 10.1182/blood-2007-09-112698. [DOI] [PubMed] [Google Scholar]
- 18.Ouillette P, Erba H, Kujawski L, Kaminski M, Shedden K, Malek SN. Integrated genomic profiling of chronic lymphocytic leukemia identifies subtypes of deletion 13q14. Cancer research. 2008;68:1012–21. doi: 10.1158/0008-5472.CAN-07-3105. [DOI] [PubMed] [Google Scholar]
- 19.Dunbar AJ, Gondek LP, O’Keefe CL, et al. 250K single nucleotide polymorphism array karyotyping identifies acquired uniparental disomy and homozygous mutations, including novel missense substitutions of c-Cbl, in myeloid malignancies. Cancer research. 2008;68:10349–57. doi: 10.1158/0008-5472.CAN-08-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borowitz MJ, Guenther KL, Shults KE, Stelzer GT. Immunophenotyping of acute leukemia by flow cytometric analysis. Use of CD45 and right-angle light scatter to gate on leukemic blasts in three-color analysis. American journal of clinical pathology. 1993;100:534–40. doi: 10.1093/ajcp/100.5.534. [DOI] [PubMed] [Google Scholar]
- 21.Lin M, Wei LJ, Sellers WR, Lieberfarb M, Wong WH, Li C. dChipSNP: significance curve and clustering of SNP-array-based loss-of-heterozygosity data. Bioinformatics. 2004;20:1233–40. doi: 10.1093/bioinformatics/bth069. [DOI] [PubMed] [Google Scholar]
- 22.Ross CW, Ouillette PD, Saddler CM, Shedden KA, Malek SN. Comprehensive analysis of copy number and allele status identifies multiple chromosome defects underlying follicular lymphoma pathogenesis. Clin Cancer Res. 2007;13:4777–85. doi: 10.1158/1078-0432.CCR-07-0456. [DOI] [PubMed] [Google Scholar]
- 23.Ozawa T, Araki N, Yunoue S, et al. The neurofibromatosis type 1 gene product neurofibromin enhances cell motility by regulating actin filament dynamics via the Rho-ROCK-LIMK2-cofilin pathway. The Journal of biological chemistry. 2005;280:39524–33. doi: 10.1074/jbc.M503707200. [DOI] [PubMed] [Google Scholar]
- 24.Johannessen CM, Reczek EE, James MF, Brems H, Legius E, Cichowski K. The NF1 tumor suppressor critically regulates TSC2 and mTOR. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:8573–8. doi: 10.1073/pnas.0503224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Liu Y, Wu C, et al. Epm2a suppresses tumor growth in an immunocompromised host by inhibiting Wnt signaling. Cancer cell. 2006;10:179–90. doi: 10.1016/j.ccr.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 26.Zhu C, Saberwal G, Lu Y, Platanias LC, Eklund EA. The interferon consensus sequence-binding protein activates transcription of the gene encoding neurofibromin 1. The Journal of biological chemistry. 2004;279:50874–85. doi: 10.1074/jbc.M405736200. [DOI] [PubMed] [Google Scholar]
- 27.Kujawski L, Ouillette P, Erba H, et al. Genomic complexity identifies patients with aggressive chronic lymphocytic leukemia. Blood. 2008;112:1993–2003. doi: 10.1182/blood-2007-07-099432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balgobind BV, Van Vlierberghe P, van den Ouweland AM, et al. Leukemia-associated NF1 inactivation in patients with pediatric T-ALL and AML lacking evidence for neurofibromatosis. Blood. 2008;111:4322–8. doi: 10.1182/blood-2007-06-095075. [DOI] [PubMed] [Google Scholar]
- 29.Side L, Taylor B, Cayouette M, et al. Homozygous inactivation of the NF1 gene in bone marrow cells from children with neurofibromatosis type 1 and malignant myeloid disorders. The New England journal of medicine. 1997;336:1713–20. doi: 10.1056/NEJM199706123362404. [DOI] [PubMed] [Google Scholar]
- 30.Bollag G, Clapp DW, Shih S, et al. Loss of NF1 results in activation of the Ras signaling pathway and leads to aberrant growth in haematopoietic cells. Nature genetics. 1996;12:144–8. doi: 10.1038/ng0296-144. [DOI] [PubMed] [Google Scholar]
- 31.Shannon KM, O’Connell P, Martin GA, et al. Loss of the normal NF1 allele from the bone marrow of children with type 1 neurofibromatosis and malignant myeloid disorders. The New England journal of medicine. 1994;330:597–601. doi: 10.1056/NEJM199403033300903. [DOI] [PubMed] [Google Scholar]
- 32.Moresi R, Tesei S, Costarelli L, et al. Age- and gender-related alterations of the number and clonogenic capacity of circulating CD34+ progenitor cells. Biogerontology. 2005;6:185–92. doi: 10.1007/s10522-005-7954-5. [DOI] [PubMed] [Google Scholar]
- 33.Emanuel PD, Bates LJ, Castleberry RP, Gualtieri RJ, Zuckerman KS. Selective hypersensitivity to granulocyte-macrophage colony-stimulating factor by juvenile chronic myeloid leukemia hematopoietic progenitors. Blood. 1991;77:925–9. [PubMed] [Google Scholar]
- 34.Largaespada DA, Brannan CI, Jenkins NA, Copeland NG. Nf1 deficiency causes Ras-mediated granulocyte/macrophage colony stimulating factor hypersensitivity and chronic myeloid leukaemia. Nature genetics. 1996;12:137–43. doi: 10.1038/ng0296-137. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Taylor BR, Shannon K, Clapp DW. Quantitative effects of Nf1 inactivation on in vivo hematopoiesis. The Journal of clinical investigation. 2001;108:709–15. doi: 10.1172/JCI12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ingram DA, Yang FC, Travers JB, et al. Genetic and biochemical evidence that haploinsufficiency of the Nf1 tumor suppressor gene modulates melanocyte and mast cell fates in vivo. The Journal of experimental medicine. 2000;191:181–8. doi: 10.1084/jem.191.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chao RC, Pyzel U, Fridlyand J, et al. Therapy-induced malignant neoplasms in Nf1 mutant mice. Cancer cell. 2005;8:337–48. doi: 10.1016/j.ccr.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 38.Neubauer A, Maharry K, Mrozek K, et al. Patients with acute myeloid leukemia and RAS mutations benefit most from postremission high-dose cytarabine: a Cancer and Leukemia Group B study. J Clin Oncol. 2008;26:4603–9. doi: 10.1200/JCO.2007.14.0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Illmer T, Thiede C, Fredersdorf A, et al. Activation of the RAS pathway is predictive for a chemosensitive phenotype of acute myelogenous leukemia blasts. Clin Cancer Res. 2005;11:3217–24. doi: 10.1158/1078-0432.CCR-04-2232. [DOI] [PubMed] [Google Scholar]
- 40.Xu Q, Simpson SE, Scialla TJ, Bagg A, Carroll M. Survival of acute myeloid leukemia cells requires PI3 kinase activation. Blood. 2003;102:972–80. doi: 10.1182/blood-2002-11-3429. [DOI] [PubMed] [Google Scholar]
- 41.Recher C, Beyne-Rauzy O, Demur C, et al. Antileukemic activity of rapamycin in acute myeloid leukemia. Blood. 2005;105:2527–34. doi: 10.1182/blood-2004-06-2494. [DOI] [PubMed] [Google Scholar]
- 42.McGillicuddy LT, Fromm JA, Hollstein PE, et al. Proteasomal and genetic inactivation of the NF1 tumor suppressor in gliomagenesis. Cancer cell. 2009;16:44–54. doi: 10.1016/j.ccr.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perl AE, Kasner MT, Tsai DE, et al. A phase I study of the mammalian target of rapamycin inhibitor sirolimus and MEC chemotherapy in relapsed and refractory acute myelogenous leukemia. Clin Cancer Res. 2009;15:6732–9. doi: 10.1158/1078-0432.CCR-09-0842. [DOI] [PubMed] [Google Scholar]
- 44.Yee KW, Zeng Z, Konopleva M, et al. Phase I/II study of the mammalian target of rapamycin inhibitor everolimus (RAD001) in patients with relapsed or refractory hematologic malignancies. Clin Cancer Res. 2006;12:5165–73. doi: 10.1158/1078-0432.CCR-06-0764. [DOI] [PubMed] [Google Scholar]
- 45.Niimi H, Harada H, Harada Y, et al. Hyperactivation of the RAS signaling pathway in myelodysplastic syndrome with AML1/RUNX1 point mutations. Leukemia. 2006;20:635–44. doi: 10.1038/sj.leu.2404136. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.