Abstract
Bacteria evolved restriction endonucleases to prevent interspecies DNA transmission and bacteriophage infection. Here, we show that human cells possess an analogous mechanism. APOBEC3A is induced by interferon following DNA detection, and it deaminates foreign double-stranded DNA cytidines to uridines. These atypical DNA nucleosides are converted by the uracil DNA glycosylase UNG2 to abasic lesions, which lead to foreign DNA degradation. This mechanism is evident in cell lines and primary monocytes, where up to 97% of cytidines in foreign DNA are deaminated. In contrast, cellular genomic DNA appears unaffected. Several other APOBEC3s also restrict foreign gene transfer. Related proteins exist in all vertebrates, indicating that foreign DNA restriction may be a conserved innate immune defense mechanism. The efficiency and fidelity of genetic engineering, gene therapy, and DNA vaccination are likely to be influenced by this anti-DNA defense system.
Intracellular foreign DNA threatens the wellbeing and proper function of cells. If the DNA is pathogenic in origin, it could produce cytotoxic gene products or replicate to propagate an infection. Even non-pathogenic DNA, for instance DNA from dying cells internalized by phagocytosis, poses a threat to the cellular genome through insertional mutagenesis, which may disrupt gene expression.
Foreign DNA is sensed by Toll-like receptor (TLR)-dependent and TLR-independent mechanisms1,2. TLR9 senses single-stranded DNA in endosomal compartments and signals to induce the production of type 1 interferons (IFNs) and pro-inflammatory cytokines and chemokines. Several double-stranded DNA sensor(s) have been identified, DAI, AIM2 and, recently, RNA polymerase III, but only the latter appears required for IFN production3-8. Regardless, once IFN is produced, it stimulates the transcription of many genes whose products orchestrate a wide variety of innate immune responses9. For instance, Tetherin/BST2 blocks viral budding, PKR inhibits translation, RNaseL degrades intracellular RNA, and ADAR1 deaminates double-stranded RNA adenosines to inosines10-15. In contrast, comparatively little is known about cellular proteins that mediate the clearance of foreign intracellular DNA.
The enzymatic conversion of DNA cytidines to uridines currently has two established roles in immunity (Supplementary Table 1). Activation-induced deaminase (AID) deaminates antibody gene DNA and triggers antibody gene diversification by somatic hypermutation and class switch recombination16,17. The APOBEC3s (A3s): A3A, A3B, A3C, A3DE, A3F, A3G and A3H, are DNA cytidine deaminases that have been shown to inhibit the replication of a diverse set of retroviruses and retrotransposons18,19. A3F and A3G, the best-studied members of this family, strongly inhibit the replication of Vif-defective HIV-1 by deaminating viral cDNA during retrovirus reverse transcription. In contrast, A3A does not restrict HIV-1, but it has been shown to limit the retrotransposition of L1 and Alu elements and the replication of parvoviruses20-26 (Supplementary Fig. 1 and Supplementary Tables 1 and 2). Other DNA viruses such as hepatitis B virus (HBV) and human papillomavirus (HPV) may also be targets of A3 proteins including A3A27,28. In all of these instances, the A3 proteins are thought to engage normal viral replication intermediates, which are intricate protein and nucleic acid complexes. Such intermediates are distinct from naked, foreign DNA, which enters cells from multiple exogenous sources.
The fact that human cells have innate mechanisms to sense double-stranded DNA and the fact that APOBEC3s are potent DNA deaminases with established roles in immunity led us to hypothesize that one or more of these proteins acts downstream of DNA-sensing molecules to mediate the clearance of foreign DNA. Here, we show that A3A and several other human A3 proteins are potent foreign DNA restriction factors. We find that A3A is induced by DNA detection and IFN in phagocytes and that it triggers the degradation of foreign DNA by a cytidine deamination and uracil excision mechanism. These data reveal that foreign DNA restriction is a distinct and important physiological function of the APOBEC3 proteins.
RESULTS
APOBEC3A is expressed in phagocytes and induced by IFN
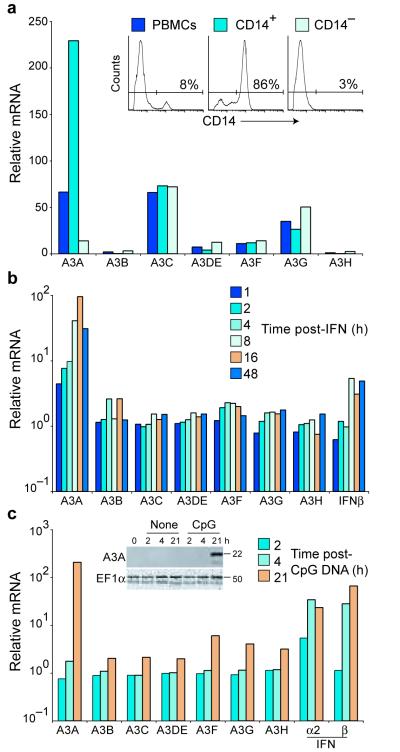
We reasoned that if A3s were indeed DNA restriction factors, then at least one of these proteins ought to be expressed in cells frequently exposed to foreign DNA and induced by DNA detection. We focused on A3A because of reports suggesting that it is expressed predominantly in monocytes, macrophages, and neutrophils25,29. These phagocytic cells ingest large quantities of foreign materials, including nucleic acids. To confirm and extend these studies, we obtained fresh human peripheral blood mononuclear cells (PBMCs), purified CD14-positive monocytes and macrophages, and used a panel of specific PCR assays to quantify A3 expression. Several APOBEC3s were expressed in PBMCs, with A3A, A3C and A3G consistently showing the highest basal mRNA levels (Fig. 1a). The relative A3A expression level increased 3-fold in the CD14-enriched cell population and decreased proportionately in the CD14-depleted fraction, whereas A3C and A3G mRNA levels were similar in the different cell populations. These data indicated that A3A is expressed predominantly in CD14-positive phagocytic cells.
Figure 1.
APOBEC3A is expressed in monocytes and macrophages and induced by interferon and CpG DNA. (a) Relative basal A3 mRNA levels in PBMCs or the specified cell subpopulations, with a value of 1 assigned to the level of A3H mRNA in unsorted PBMCs. Inset: flow cytometry histograms showing the efficiency of CD14+ cell enrichment. (b and c) A3 and IFNβ mRNA levels in PBMCs treated with recombinant IFN or CpG DNA oligonucleotide for the indicated time. mRNA levels are relative to those measured in untreated cells. Inset in c: immunoblot of A3A in PBMCs treated with a CpG DNA oligonucleotide. The same membrane probed with anti-eEF1α is shown as a loading control.
Several A3s are IFN-inducible29-34. To systematically quantify the relative magnitudes and kinetics of this IFN response, fresh PBMCs were treated with IFN and RNA was isolated at multiple time points for quantification by real-time PCR. A3A was strongly IFN-responsive with mRNA levels consistently peaking 100-fold above those in untreated cells (Fig. 1b). Other A3s showed modest 2- to 3-fold increases. We also found that incubating PBMCs with the TLR9 ligand CpG single-stranded DNA caused a 200-fold increase in A3A, and a 2- to 4-fold increase in the expression of other A3s (Fig. 1c). This dramatic level of induction places A3A among the highest IFN-responsive genes (Supplementary Fig. 2). A3A induction was also exceptionally clear by immunoblotting using an A3A-specific polyclonal antibody and by fluorescence-based DNA cytidine deaminase activity assays (Fig. 1c inset and Supplementary Fig. 3).
APOBEC3A reduces gene transfer efficiency
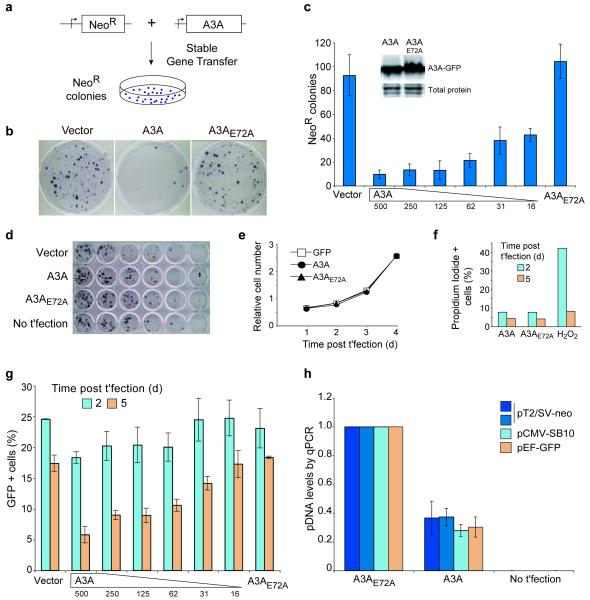
The finding that A3A is expressed in cells that ingest pathogens and extracellular debris, and the fact that its expression is induced by DNA detection suggested that it might mediate the clearance of foreign DNA. To directly test this hypothesis, we quantified the impact of A3A expression on several measures of foreign DNA integrity and stability. First, we measured the impact of A3A on stable gene transfer efficiency (Figs. 2a-c). HEK-293 cells, which lack detectable A3A mRNA (data not shown), were co-transfected with a neomycin-resistance (NeoR) reporter plasmid and an expression construct for A3A or A3A-E72A (a catalytically inactive mutant; Supplementary Fig. 4), or a GFP control. Two days were allowed for integration of the reporter plasmid into the chromosomal DNA and for expression of the NeoR gene product. The cells were then plated into neomycin (G418)-containing medium, and drug resistant colonies were allowed to grow. The resulting NeoR colonies were stained and counted.
Figure 2.
Foreign DNA restriction by APOBEC3A. (a) Schematic of plasmid-based stable gene transfer experiments. (b) Representative plates of NeoR colonies obtained in a stable gene transfer experiment in HEK-293 cells transfected with a NeoR plasmid and A3A-expression or control plasmids. (c) Quantification of the data in b and two additional experiments. The amount of A3A-expression plasmid was decreased as indicated. Inset: immunoblot of A3A-GFP and A3A-E72A-GFP and the same membrane stained with Ponceau S. (d) The number of colony forming cells in transfected cell populations in b and c was determined by diluting and plating cells into drug-free medium. (e) The proliferative capacity of transfected cells. (f) The percentage of apoptotic cells in transfected cell populations. Hydrogen peroxide (H2O2) was used as a cytotoxic control. (g) Transient expression of a GFP reporter plasmid in HEK-293 cells. (h) pDNA persistence in HeLa cells transfected with A3A or A3A-E72A was determined by quantitative PCR on DNA recovered 48 h post-transfection. For each pDNA, the A3A-E72A data were normalized to one. No t’fection: non-transfected controls. In c, e, g, and h, the mean and s.d. of three replica experiments is shown.
A3A expression resulted in a dose-dependent decrease in gene transfer efficiency in comparison to either control condition (Figs. 2b,c). This effect was potent: transfecting as little as 16 ng of A3A expression plasmid decreased the frequency of G418R colonies to 40% of controls (Fig. 2c). Similar data were obtained with HeLa cells or with a more efficient DNA transposon-mediated gene transfer system (Supplementary Fig. 5 and see below for a comparison with other human A3s). A3A expression did not decrease the number of colony-forming cells (assessed by plating in drug-free medium), nor was there any indication of altered proliferation or increased apoptosis in A3A-expressing cells (Figs. 2d-f). Identical results were obtained in experiments with transfection efficiencies ranging from 25-75%.
A3A glutamate 72 is part of a conserved zinc-coordinating catalytic motif, H-x-E-x28-C-x2-4-C, in which x is a non-conserved amino acid35,36. A3A-E72A is catalytically defective in vitro, in agreement with prior studies25,26 (Supplementary Fig. 4). The fact that this mutant did not decrease the efficiency of stable gene transfer suggested that this effect requires the DNA cytidine deaminase activity of A3A (addressed further below).
APOBEC3A inhibits transient gene expression
As a second measure of foreign DNA stability, we asked whether A3A affects transient gene expression. GFP-encoding plasmid DNA was transfected into HEK-293 cells and GFP fluorescence was monitored by flow cytometry (Fig. 2g). After 2 days of incubation, the higher A3A expression levels caused a small decrease in GFP fluorescence. By 5 days, all A3A levels caused significant reductions in GFP fluorescence, and a clear dose response was apparent. The rate of GFP fluorescence decay was also accelerated in A3A-expressing cells compared to control cells (~4% decline in GFP-positive cells per day versus 2% in the control cells). Thus, A3A expression caused a more rapid disappearance of transient reporter gene expression.
APOBEC3A destabilizes foreign plasmid DNA
Because reduced gene transfer efficiency and diminished reporter gene expression could result from a reduction in either the quantity or the integrity of the transfected DNA itself, we used quantitative PCR to directly measure plasmid DNA levels. These experiments revealed a 60% reduction in plasmid DNA levels in A3A expressing cells in comparison to control cells two days after transfection (Fig. 2h). It is noteworthy that this is likely an underestimate of the potency of A3A, because transfected DNA may resist DNase digestion (done prior to cell lysis to remove free DNA) or become sequestered in an A3A-impermeable compartment (e.g., endosomes). Nevertheless, these data showed that A3A compromises the physical integrity of foreign plasmid DNA, and that this effect requires the conserved catalytic glutamate.
APOBEC3A deaminates foreign DNA, creating UDG substrates
If A3A indeed triggers the clearance of foreign DNA by a mechanism involving DNA deamination, then, in addition to a requirement for catalytic activity, two major predictions follow. First, the immediate products of deamination, DNA uracils, might be substrates for excision by cellular uracil DNA glycosylases. A likely candidate is the major cellular uracil DNA glycosylase, UNG2, which functions normally in error-free excision repair of genomic uracils but also processes these lesions during error-prone antibody gene diversification events16,37 (Supplementary Table 1). Second, since DNA uridines base pair like thymidines and template the insertion of adenosines, they ought to amplify by PCR to result in C/G-to-T/A transition mutations. Furthermore, if A3A was the source of these mutations, then the mutated cytidines should be biased toward 5′-TC and 5′-CC dinucleotides, which are the reported targets for A3A deamination25,27,38,39 (Supplementary Table 2).
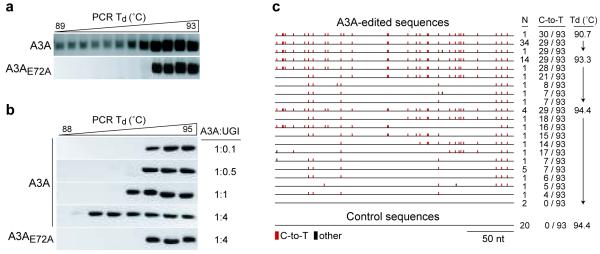
To test these predictions, we used a uracil DNA glycosylase inhibitor (UGI) to inhibit UNG2 and a technique called differential DNA denaturation (3D)-PCR to recover edited intermediates40,41. If UNG2 excises A3A-generated DNA uracils, then inhibiting it should cause an accumulation of uridine-containing intermediates. 3D-PCR is based on the fact that DNA with fewer inter-strand hydrogen bonds will amplify preferentially at lower denaturation temperatures (e.g., duplex DNA with G/U mismatches or A/T rich DNA). The combined use of these techniques allows for the detection of potentially short-lived uridine-containing DNA intermediates.
UGI-expressing HEK-293T cells were co-transfected with a reporter plasmid and A3A or A3A-E72A expression plasmids. Two days later, transfected DNA was recovered from cells and analyzed by 3D-PCR. At the highest denaturation temperatures, PCR products were apparent from both A3A and A3A-E72A expressing cells (Fig. 3a). However, at lower denaturation temperatures, PCR products were only obtained from A3A expressing cells. UNG2-proficient (UGI negative) cells were transfected similarly and PCR products were not detected at lower denaturation temperatures (data not shown). These results indicated that A3A edits transfected plasmid DNA and that UNG2 processes the edited molecules.
Figure 3.
APOBEC3A deaminates transfected plasmid DNA and generates lesions for uracil DNA glycosylase. (a) Agarose gel analysis of 3D-PCR products from stable UGI-expressing HEK-293T cells. Cells were transfected with plasmids encoding A3A or A3A-E72A and a mCherry (pTre2-mCherry) reporter construct. Total DNA was recovered 48 h post-transfection and analyzed by 3D-PCR at the indicated denaturation temperatures (Td). (b) Agarose gel analysis of 3D-PCR products from HEK-293 cells transiently transfected with increasing amounts of UGI plasmid, a GFP reporter construct (pEGFP-N3), and a plasmid encoding A3A or A3A-E72A. Total DNA was recovered 48 h post-transfection and analyzed by 3D-PCR at the indicated Td ranges. (c) Summary of the plasmid DNA sequences (nucleotides 1170-1426 of pEGFP-N3) recovered from the experiment in b. C/G-to-T/A hypermutations are indicated as red tics along the consensus sequence, and all other base substitutions as black tics. The control sequences were obtained from cells expressing A3A-E72A. The number of times each sequence was recovered, the number of C-to-T conversions in each sequence (out of 93 total cytidines), and the PCR Td used to amplify the populations of molecules from which the sequences are derived are indicated.
To confirm that UNG2 excises uracil from DNA molecules edited by A3A, we transfected HEK-293 cells with a reporter plasmid, A3A or A3A-E72A expression plasmid, and a range of UGI expression plasmid. In support of the prior experiments, higher doses of UGI (greater UNG2 inhibition) led to an increased abundance of lower denaturation temperature PCR products (Fig. 3b). As controls, UNG2 inhibition was alone insufficient to enable the recovery of low denaturation temperature PCR amplicons and, again, A3A catalytic activity was required.
To unambiguously determine whether the low denaturation temperature amplicons corresponded to A3A-edited molecules, we cloned and sequenced a number of amplicons from several independent PCRs (Fig. 3c). These analyses revealed high levels of C/G-to-T/A transition mutations in molecules recovered from A3A expressing cells. DNA cytidines within a 5′-TC dinucleotide context was preferred, consistent with prior studies25,27,38,39 (see below and Supplementary Table 2). Mutations were even apparent in the majority of molecules amplified at the highest denaturation temperature (94.4°C), indicating that selective amplification by 3D-PCR was not required to detect edited sequences. In contrast, no mutations were found in molecules recovered from A3A-E72A expressing cells. Overall, these data demonstrated that A3A deaminates foreign DNA in human cells and that the resulting uridines are substrates for UNG2-mediated excision.
Extensive foreign DNA editing occurs in human monocytes
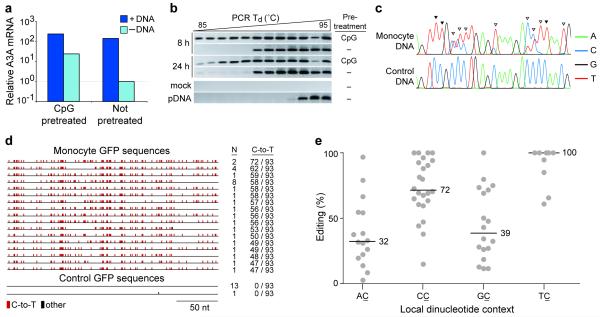
We next wanted to assess the impact of foreign DNA editing in A3A′s normal physiological context, i.e. in primary human phagocytes. We took advantage of the fact that a 100-fold difference in A3A expression exists between CpG-treated and mock-treated PBMCs (Fig. 1c). These two cell populations were prepared and transfected with GFP-encoding plasmid DNA or mock-transfected with buffer alone. We found that the introduction of plasmid DNA into cells caused A3A to be induced, regardless of whether the cells had been pre-treated with CpG DNA (Fig. 4a). These data are consistent with prior studies showing that the introduction of double-stranded DNA into leukocytes triggers strong TLR-independent IFN responses42,43.
Figure 4.
APOBEC3A mutates foreign DNA in primary human cells. (a) A3A expression by qRT-PCR in CpG-pretreated or untreated monocytes transfected with plasmid DNA (+DNA) or with buffer alone (−DNA) at 8 h post-transfection. Levels are relative to those in untreated, mock-transfected cells. (b) 3D-PCR products from the experiment in a. Total DNA was isolated and a portion of the GFP gene was amplified using a gradient of PCR denaturation temperatures (Td). DNA extracted from mock-transfected cells and the mock DNA mixed with input plasmid were subjected to the same PCR scheme as controls (labeled “mock” and “pDNA”, respectively). (c) Representative chromatograms of PCR products from DNA transfected into monocytes or control reactions (nucleotides 1289-1314 of pEGFP-N3). Cytidines that have been edited in all or a fraction of molecules are indicated with filled or open arrowheads, respectively. (d) Plasmid DNA recovered from monocytes was subjected to 3D-PCR, cloned, and sequenced. C/G-to-T/A mutations are indicated as red tics; other base substitutions as black tics. The number of times each sequence was recovered and the number of C-to-T conversions in each molecule are indicated. (e) A quantification of the DNA editing site dinucleotide contexts detected by directly sequencing 3D-PCR amplicons (as in c). The editing percentages were grouped by dinucleotide context and the median percentage for each group is indicated. For instance, 5 of the 9 cytidines within 5′-TC dinucleotides were edited in 100% of the molecules amplified (i.e., only a T peak was evident in the chromatogram).
Next, to determine whether endogenous A3A mutates transfected plasmid DNA, we recovered DNA from cells at 8 and 24 h post-transfection and used 3D-PCR to amplify a region of the GFP gene. For a control template, we mixed GFP plasmid with DNA prepared from mock-transfected cells. This control only amplified at the highest denaturation temperatures (Fig. 4b, pDNA). In contrast, we were able to detect plasmid DNA amplification at lower denaturation temperatures using the DNA recovered from transfected monocytes. Cloning and sequencing these PCR products showed that they resulted from amplification of extensively edited plasmid DNA, with some molecules having as many as 72/93 cytidines edited in the 257 nucleotide amplicon (Figs. 4c,d).
We also noted that considerably more amplification occurred at lower denaturation temperatures using DNA recovered from the CpG-pretreated cells than from the non-pretreated cells at 8 h post-transfection (Fig. 4b). This suggested that the plasmid DNA had been more extensively mutated in the cells that were expressing higher levels of A3A at the time of transfection. At 24 h post-transfection, weak bands appeared at the lowest denaturation temperatures in the mock-pretreated cells, likely corresponding to mutations catalyzed by A3A that had been induced by plasmid DNA transfection itself (Fig. 4b).
In addition to cloning and sequencing individual molecules, we also directly sequenced populations of PCR products (Figs. 4c and Supplementary Fig. 6). Remarkably, these analyses revealed evidence for C-to-U conversion of 70/72 cytidines (97%) within a 200 nucleotide region of the GFP gene (i.e., these cytidines had been edited in at least some of the molecules amplified; Supplementary Fig. 6). A strong bias toward deamination at 5′-TC and 5′-CC dinucleotides was observed, fully consistent with catalysis by A3A (Fig. 4e and Supplementary Table 2; compare with prior reports25,27,38,39). There was a high degree of correspondence between the most frequently edited sites in these primary cell experiments and those observed in plasmid DNA recovered from A3A-expressing HEK-293 cells (Figs. 3, 4, and Supplementary Fig. 6). We noted that the editing appeared to be strand-specific (Figs. 3c and 4d). However, additional experiments using partly degenerate primer sets (with G’s replaced by R’s) revealed that both DNA strands were edited (data not shown, but see Online Methods for assay details).
Interestingly, in contrast to experiments in HEK-293 cells, we detected hyper-edited DNA molecules recovered from primary monocytes without inhibiting UNG2. This suggested that editing is more extensive in primary monocytes (consistent with the dramatic induction of A3A), but many other alternatives such as lower UNG2 activity may also be tenable. Essentially identical results were obtained using PBMCs or CD14-enriched phagocytes from three independent donors and analyzed with additional sets of PCR primers (e.g., Supplementary Fig. 7 and data not shown). These results clearly demonstrated that foreign DNA is hypermutated extensively in primary human cells. The main enzyme responsible is likely A3A, because its expression is highly induced (making it the best expressed A3 family member), the kinetics of induction correlate with the appearance of low denaturation temperature 3D-PCR amplicons, and the primary cell foreign DNA hypermutation patterns are similar to those observed in cell culture experiments with only A3A present25,27,38,39 (Figs. 1, 3, 4 and Supplementary Table 2). RNAi experiments intended to knock-down A3A were not successful, likely because the A3A induction was too strong (both the siRNA itself and the foreign DNA are immunostimulatory; data not shown and Fig. 4a).
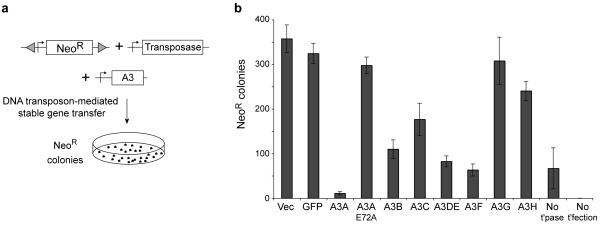
Foreign DNA restriction by multiple human APOBEC3s
Humans have six APOBEC3 proteins in addition to A3A, and most of these have much broader expression profiles32,44-46. To test whether any of these proteins are capable of inhibiting foreign DNA, we used an efficient DNA transfer system based on the cut-and-paste transposon Sleeping Beauty47 (Fig. 5a). Here, stable gene transfer results from both random and transposase-mediated integration events. As expected, A3A nearly eliminated DNA transfer, and restriction depended upon catalytic activity (Fig. 5b). Several other human A3 proteins also inhibited DNA transfer but to different degrees: A3B, A3DE and A3F were most potent, A3C and A3H showed intermediate levels, and A3G had essentially no activity. These restriction profiles contrast with those for HIV-1, with A3G and A3F being the most potent inhibitors18,19 (Supplementary Table 1 and Supplementary Fig. 1). Similar results were obtained in stable gene transfer experiments analogous to those described in Fig. 2 (data not shown). Taken together with the aforementioned results, it is possible that A3C and A3F make minor contributions to foreign DNA editing in primary human phagocytes and, further, that multiple A3s may contribute to foreign DNA restriction in other cell types (Supplementary Table 2).
Figure 5.
Foreign DNA restriction by multiple human APOBEC3 proteins. (a) Schematic of the DNA transposon-mediated stable gene transfer experiments. (b) A histogram showing the mean and s.d. of three independent transposon-mediated stable gene transfer experiments done in the presence of the indicated A3 expression constructs. Controls included the number of NeoR colonies resulting from random integration (no transposase; No t’pase) and from no transfection (no foreign DNA; No t’fectection). Flow cytometry confirmed that all of the A3-GFP constructs were expressed similarly (data not shown).
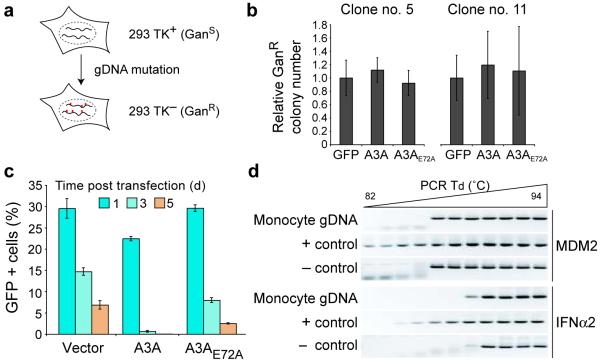
Mutation of nuclear genomic DNA is undetectable
The extraordinary level of A3A-dependent foreign DNA mutation prompted us to ask whether genomic DNA may also be susceptible to attack. We first established stable HEK-293 clones expressing a HSV thymidine kinase (TK) reporter gene (Fig. 6a and Online Methods). TK-positive cells are sensitive to gancyclovir (GanS), and genetic inactivation of TK confers resistance to this drug (GanR). This sensitive assay was used to ask whether transient A3A expression caused an elevated genomic mutation frequency. Although the level of A3A expressed in these experiments caused a strong reduction in foreign gene expression, it had no effect on the frequency of GanR mutation (Figs. 6b,c). Similar results were obtained for an independently established cell line with TK integrated elsewhere in the genome (Fig. 6b).
Figure 6.
Lack of detectable genomic DNA mutation in cells that restrict foreign DNA. (a) Schematic of the genomic TK mutation assay. (b) Two independent clonal HEK-293 cell lines harboring integrated TK genes were transfected with the indicated expression plasmids and selected with gancyclovir-containing medium. The relative mean number of GanR (TK−) colonies and s.d. from duplicate or quadruplicate experiments is shown. (c) GFP (or A3-GFP) expression in transfected HEK-293-TK cells (clone #5 in b). (d) 3D-PCR results with primers specific to MDM2 or IFNA2 genomic loci (as described in Fig. 4b). For a positive control (+), genomic DNA was heat denatured and incubated with purified A3A prior to PCR. For a negative control (−), total DNA recovered from untransfected HEK-293 cells was subjected to the same PCR amplification procedures.
To generalize these findings to primary human cells, we used 3D-PCR to try to detect genomic DNA mutation in monocytes expressing high levels of A3A (i.e., in cells where foreign DNA is edited; Fig. 4). Two distinct genomic loci, IFNA2 and MDM2, were analyzed, and no evidence for genomic hypermutation was observed (Fig. 6d). The 3D-PCR profiles were indistinguishable for DNA derived from A3A-induced cells and control DNA templates. As a positive control, the DNA recovered from monocytes was heat denatured and treated with recombinant A3A in vitro and, as expected, amplification occurred at much lower denaturation temperatures. It is also notable that, as anticipated, each 3D-PCR amplicon has a unique amplification threshold that correlates with its overall G/C content (MDM2: 38%; IFNα2: 49%; GFP: 59%).
Both TK mutation and 3D-PCR datasets were consistent with control experiments showing that neither A3A transient transfection into HEK-293 or HeLa cells causes overt signs of cell cycle arrest, growth defects, or apoptosis (Figs. 2d-f and data not shown). Thus, using several measures, we did not observe genomic DNA editing in the same cells in which foreign DNA mutation is readily detectable. We conclude that, in contrast to foreign DNA, endogenous nuclear DNA is much less susceptible and possibly even completely resistant to A3A-dependent DNA deamination.
DISCUSSION
The recognition and clearance of foreign intracellular DNA has been postulated to be a fundamental arm of the innate immune response42,43,48. Although both TLR-dependent and - independent pathways sense foreign DNA and trigger robust IFN production, little is known about the downstream effector proteins, some of which presumably must function to mediate DNA clearance. Here, we show that several A3 proteins, particularly A3A, are such foreign DNA restriction factors. A3A is expressed predominantly in phagocytic cells and it is strongly IFN-inducible, consistent with a front-line role in foreign DNA restriction. A3A is also induced by CpG single-stranded DNA or double-stranded plasmid DNA, presumably through the induction of IFN by the TLR9 pathway or the TLR-independent DNA sensing pathway(s), respectively. A3A reduces foreign DNA stability and integrity by three measures: stable gene transfer, transient gene expression, and DNA persistence. The extraordinary levels of foreign DNA C/G-to-T/A hypermutation, a requirement for the A3A active site glutamate, and the strong influence of UGI combine to demonstrate that foreign DNA restriction is a deamination- and uracil excision-dependent process (Supplementary Table 1).
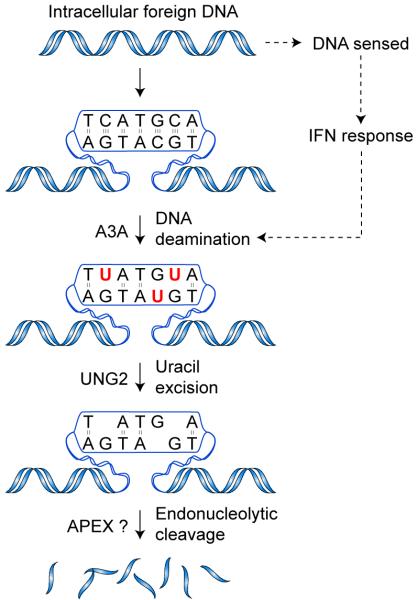
Our data indicate a model in which foreign DNA clearance is initiated by A3A-dependent deamination of DNA cytidines to uridines (Fig. 7). The subcellular location in which this occurs is presently unclear. Additionally, it is possible that the naked, double-stranded plasmid DNA substrates used in our experiments are rendered transiently single-stranded by cellular enzymes such as a polymerase or helicase. Regardless, the uridine lesions in foreign DNA are subsequently converted by the cellular uracil DNA glycosylase UNG2 to nuclease-susceptible abasic sites. It is likely that these abasic sites are processed by the endonuclease APEX, which cleaves the DNA phosphodiester backbone37, but many other cellular nucleases such as TREX1 could also be involved49. Additionally, DNase1 and 2 may contribute to foreign DNA clearance, but it is notable that these enzymes are ubiquitously expressed and not considered part of the IFN response48,50.
Figure 7.
A model for foreign DNA restriction. Foreign DNA enters the cell by escaping from an endosomal or phagosomal compartment, by infection, or by other means. TLR-dependent or - independent DNA sensing initiates signaling cascades that result in production of IFN, which in turn induces A3A expression. A3A engages the foreign DNA, deaminating multiple cytidines in a molecule. The resulting uracils are excised by UNG2, creating nuclease-sensitive abasic sites. Cleavage of the backbone by APEX1 or other nucleases results in fragmentation and degradation of the foreign DNA.
Prior to this study, physiologically relevant DNA deamination substrates were thought to be limited to antibody gene DNA and to viral and retroelement single-strand DNA replication intermediates (Supplementary Table 1). Here, we demonstrate that naked double-stranded plasmid DNA from E. coli is subject to restriction by A3A and other human APOBEC3 proteins. It is reasonable to assume that B-form, double-stranded DNA from other types of bacteria (i.e., all other bacterial DNA) will be similarly restricted. Restriction-susceptible DNA may also originate from other microbes (fungi or viruses; foreign DNA) and/or from apoptotic or necrotic cells (self DNA). Indeed, DNA released from apoptotic cells has been shown to trigger an IFN response51. However, it is also possible that most restriction susceptible viruses will have evolved a counter-defense mechanism such as lentivirus Vif or foamy virus Bet18,19. Further experiments with natural and synthetic DNA substrates will shed light on the types and characteristics of foreign DNA subject to A3-mediated restriction.
Our studies raise important questions about how A3 proteins discriminate between foreign and self-DNA. It is possible that the foreign DNA restriction mechanism is compartmentalized to help mitigate the obvious risk to the normal genomic DNA of the cell. However, epitope-tagged versions of A3A distribute cell-wide and penetrate the nuclear compartment25 (data not shown), and yet they do not elevate genomic mutation frequencies (Fig. 6). Therefore, presumably at least one additional mechanism exists to ensure the integrity of cellular genomic DNA. The issue of target specificity may be analogous to prokaryotic restriction and modification systems, in which self-DNA is protected from restriction by nucleic acid modifications (usually methylation). Analogous DNA modifications or epigenetic markings may serve to protect genomic DNA from A3-mediated DNA editing. Perhaps the relevant imprints are removed from the DNA of dying cells. Again, experiments with synthetic DNA substrates are likely to be informative.
An ancestral AID gene duplicated and diverged to root the APOBEC3 locus, which has expanded significantly in several mammalian lineages including that defined by modern primates36,52. It is therefore not surprising that the foreign DNA restriction mechanism described here shares some features with the mechanism of AID-mediated antibody gene diversification and others with the mechanisms of APOBEC3-mediated retroelement restriction (Supplementary Table 1). However, the foreign DNA restriction mechanism described here is clearly distinct, targeting exogenous double-stranded DNA (not chromosomal or replication-associated substrates), requiring both DNA deamination and uracil excision, and occurring in different cell types. The A3A-dependent foreign DNA restriction mechanism described here appears to be the result of evolutionary and functional specialization. The fact that other A3s are to varying extents also capable of foreign DNA restriction, yet much less IFN responsive, suggests that the mechanism described here may be both constitutively operative in most cells (a housekeeping function) and strongly inducible in specific cells such as phagocytes that frequently encounter foreign DNA. Along these lines, it is notable that mouse APOBEC3 is induced in dendritic cells by DNA transfection or CpG DNA incubation43. Further studies with a variety of cell types and tissues from humans, mice, and other species will help address these important regulatory and evolutionary points. Nevertheless, despite many new questions, our current studies establish foreign DNA deamination as a new fundamental effector function within the broader innate immune response.
Finally, the existence of a foreign DNA restriction system in human cells and possibly in other vertebrates has major implications. First, for the basic biomedical sciences, it is likely that transient or stable gene delivery will be influenced by the A3 repertoire of a cell. For instance, the T cell line CEM and its derivative CEM-SS are differentially amenable to stable gene transfection, with the latter A3-deficient line more efficient at stably incorporating foreign DNA (data not shown). We suggest that the difficulty in transfecting many cell lines and primary cells may be due significantly to their expressed A3 repertoire. Second, for the applied biomedical sciences, such as non-viral gene therapy, it is possible that endogenous A3 proteins undermine gene transfer efficiency and/or mutate therapeutically intended DNA. Third, the efficacy of DNA-based vaccines or adjuvants could be dramatically impacted by the foreign DNA restriction mechanism described here. Future studies defining the range of cell types and species in which this foreign DNA defense operates will undoubtedly serve to broaden these implications.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/nsmb/.
Supplementary Material
ACKNOWLEDGEMENTS
We thank J. Albin, J. Hultquist, D. Kaufman, L. Lackey, and D. Trono for thoughtful feedback, P. Hackett and S. McIvor (Univ. of MN) for SB reagents, H. Bull (Univ. Saskatchewan) for help with antibody production, B. Thielen and J. Lingappa for sharing in vitro DNA deaminase assay protocols, and M. Cornwell and J. Valesano for technical assistance, B. Cullen (Duke Univ.), R. Tsien (UCSD), and J. DiNoia (Institut de recherches cliniques de Montréal) for plasmid constructs, and J. Hanten (3M) for reagents and helpful discussions in the early stages of these studies. M.D.S. was supported in part by a 3M Graduate Fellowship and a Cancer Biology Training Grant (CA009138). M.B.B. was supported in part by the Children’s Cancer Research Fund, Minneapolis, MN. This work was supported by grants from the National Institutes of Health, GM090437 and AI064046.
Appendix
ONLINE METHODS
DNA constructs
An A3A cDNA (NM_145699) was obtained from B. Cullen22. The construction of A3A-expressing derivatives and all other constructs is in the full methods online.
Cell culture
Cell lines were maintained in 10% FBS- and pen/strep-supplemented DMEM at 37°C and 5% CO2. Cells were transfected with TransIT-LT1 (Mirus Bio Corporation). Cell viability was monitored by propidium iodide staining and flow cytometry, and proliferation by Cell Titer 96 aqueous reagent (Promega).
HEK-293T clones stably expressing uracil DNA glycosylase were created by transfecting cells with pEF-UGI and screening puromycin resistant clones for UNG2 inhibition40. Clonal HEK-293 cell lines harboring a stably integrated TK-neo cassette were generated by transfecting cells with pCMV-SB-10 and pT2-TK-neo and screening G418 resistant clones for sensitivity to gancyclovir.
Primary cell experiments
Blood was obtained from healthy donors (Memorial Blood Centers, Minneapolis, MN). PBMCs were isolated by density gradient centrifugation using Ficoll Paque Plus (GE Healthcare Life Sciences). Monocytes were enriched by negative selection of non-monocytes using magnetic separation (MACS separation, Miltenyi Biotec) or centrifugation (RosetteSep, Stem Cell Technologies). CD14+ cells were stained with CD14-FITC (Miltenyi Biotec). Some experiments included 2 ng/ml universal type 1 interferon (R&D Systems) or 3 μM CpG oligonucleotide.
Immunoblotting
Cellular proteins were extracted with 25 mM HEPES (pH 7.4), 10% glycerol, 150 mM NaCl, 0.5% Triton X-100, 1 mM EDTA, 1 mM MgCl2, 1 mM ZnCl2, and protease inhibitors and clarified by centrifugation (10 minutes at 20,800g at 4°C). 15 μg were fractionated by SDS-PAGE, transferred to PVDF (Millipore), and probed with an anti-A3A polyclonal antiserum, an anti-GFP monoclonal antibody (Clontech), or an anti-eEF1alpha monoclonal antibody (Upstate). The anti-A3A polyclonal serum was generated by immunizing a rabbit with a peptide corresponding to A3A residues 171-199 (CPFQPWDGLEEHSQALSGRLRAILQNQGN) mixed with TiterMax Gold adjuvant (Sigma). Primary antibodies were detected with fluorescently labeled secondary antibodies and imaging (LI-COR Biosciences).
Quantitative reverse-transcription PCR assays
cDNA was made from total RNA (Qiagen) by reverse transcription with random hexamers and AMV reverse transcriptase (Roche) and used to template qPCR reactions (Lightcycler 480, Roche). Data were normalized to the geometric mean of at least two of the following three reference genes: TBP, RPL13A, or HPRT. Supplementary Table 3 lists all primer and probe sequences.
Gene transfer experiments
250,000 HEK-293 or HeLa cells were plated into 6-well plates. After 24 h of incubation, the cells were transfected with pcDNA3.1 (NeoR, Invitrogen) and pEF-A3A-GFP or pEF-A3A-E72A-GFP. Two days post transfection, 100,000 cells were plated into 10cm dishes in media containing 1 mg per ml G418 (Cellgro). Colony forming efficiency was monitored simultaneously by plating serial cell dilutions into drug-free media. After 12-14 days of additional incubation, colonies were fixed and stained with crystal violet. Sleeping Beauty DNA transposon-mediated gene transfer assays were described47.
Plasmid DNA Q-PCR assays
HEK-293 or HeLa cells were transfected as above, and at the indicated times post transfection, cells were harvested and total DNA extracted (Qiagen). 50 ng total DNA was used as template for each qPCR reaction. Reactions were performed on an iCycler instrument using SYBR Green I (BioRad). Plasmid DNA levels were normalized to genomic DNA levels as measured by a qPCR assay specific for the beta actin locus. Primers are listed in Supplementary Table 3.
3D-PCR and DNA sequencing assays to detect hyper-editing in cell lines
HEK-293 cells, HEK-293T cells, or HEK-293T cells stably expressing UGI were transfected with reporter plasmids pTRE2-Δpuro-mCherry or pEGFP-N3, pcDNA3.1-UGI or pcDNA3.1, and pEF-A3A-GFP or pEF-A3A-E72A-GFP. 24 h post-transfection, cells were treated with DNaseI (Roche) to remove extracellular input plasmid. 24 h later, cells were harvested and total DNA extracted (Qiagen). 50 ng DNA was used as input to PCR using primers listed in Supplementary Table 4 and Taq DNA polymerase (Roche), which amplifies uracil-containing DNA (25 cycles: 94°C,15s; 50°C, 30s; and 68°C, 2min). 0.25 μl of this PCR was used as template for nested PCR with Phusion DNA polymerase (Finnzymes) and primers listed in Supplementary Table 4 (25 cycles: Td gradient as indicated; 52°C, 30s; and 72°C, 15s). PCR products were visualized on agarose gels.
Sequencing to detect edited DNA
PCR products were recovered from agarose gels (Qiagen). 20 ng PCR product was sequenced directly (Genewiz) or cloned, and sequenced. The amplicon contained nucleotides 1170-1426 of pEGFP-N3 (Genbank U57609.1). PCR product sequences were analyzed using phred software version 0.071220.b [www.phrap.org53]. During the course of these studies we observed that the level of A3A-induced deamination is so high that primer design is difficult. For instance, since almost every DNA cytidine is a potential A3A substrate, an ideal PCR primer set must be devoid of G’s. Since this is not feasible, we elected to minimize the number of G’s and, at unavoidable positions, have the primers synthesized with R’s (G or A). This approach enabled us to detect C/G-to-T/A transitions in both strands of plasmid substrates recovered from A3A-overexpressing HEK-293 cells or from A3A-induced (IFN or CpG-treated) CD14-positive monocytes (data not shown).
Primary cell transfection
Monocytes were incubated for 20-24 h with 3 μM CpG DNA or mock treated. 2×107 monocytes were transfected with 2 μg endotoxin-free pEGFP-N3 (Qiagen) using nucleofection (Amaxa). 3 h post-transfection, cells were treated with DNaseI to remove extracellular DNA. Cells were harvested periodically for RNA and protein preparation. Gene expression was monitored using qRT-PCR, and recovered DNA was analyzed as above.
TK selection experiments
The frequency of cells converting from TK+ to TK− was used as a measure of genomic DNA mutation. Inactivating mutations of the integrated TK gene confer GanR. TK-neo clones were transfected, as above, with A3A or control expression plasmids. Two days later, 1×106 cells were plated into Gan-containing (5 μM) medium. GanR colonies were allowed to grow for 12-14 days, and then stained with crystal violet.
Additional References for Online Methods
- 53.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
REFERENCES
- 1.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 2.Uematsu S, Akira S. Toll-like receptors and Type I interferons. J Biol Chem. 2007;282:15319–15323. doi: 10.1074/jbc.R700009200. [DOI] [PubMed] [Google Scholar]
- 3.Takaoka A, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 4.Roberts TL, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–1060. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 5.Burckstummer T, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 6.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hornung V, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 11.Van Damme N, et al. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe. 2008;3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts WK, Hovanessian A, Brown RE, Clemens MJ, Kerr IM. Interferon-mediated protein kinase and low-molecular-weight inhibitor of protein synthesis. Nature. 1976;264:477–480. doi: 10.1038/264477a0. [DOI] [PubMed] [Google Scholar]
- 13.Kerr IM, Brown RE, Hovanessian AG. Nature of inhibitor of cell-free protein synthesis formed in response to interferon and double-stranded RNA. Nature. 1977;268:540–542. doi: 10.1038/268540a0. [DOI] [PubMed] [Google Scholar]
- 14.Meurs E, et al. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell. 1990;62:379–390. doi: 10.1016/0092-8674(90)90374-n. [DOI] [PubMed] [Google Scholar]
- 15.Bass BL, Weintraub H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell. 1988;55:1089–1098. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- 16.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 17.Longerich S, Basu U, Alt F, Storb U. AID in somatic hypermutation and class switch recombination. Curr Opin Immunol. 2006;18:164–174. doi: 10.1016/j.coi.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 18.Chiu YL, Greene WC. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu Rev Immunol. 2008;26:317–353. doi: 10.1146/annurev.immunol.26.021607.090350. [DOI] [PubMed] [Google Scholar]
- 19.Malim MH, Emerman M. HIV-1 accessory proteins--ensuring viral survival in a hostile environment. Cell Host Microbe. 2008;3:388–398. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 20.Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol. 2004;14:1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 21.Bogerd HP, Wiegand HL, Doehle BP, Lueders KK, Cullen BR. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 2006;34:89–95. doi: 10.1093/nar/gkj416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bogerd HP, et al. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci U S A. 2006;103:8780–8785. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muckenfuss H, et al. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J Biol Chem. 2006;281:22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- 24.Chiu YL, et al. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc Natl Acad Sci U S A. 2006;103:15588–15593. doi: 10.1073/pnas.0604524103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H, et al. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr Biol. 2006;16:480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 26.Narvaiza I, et al. Deaminase-independent inhibition of parvoviruses by the APOBEC3A cytidine deaminase. PLoS Pathog. 2009;5:e1000439. doi: 10.1371/journal.ppat.1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vartanian JP, Guétard D, Henry M, Wain-Hobson S. Evidence for editing of human papillomavirus DNA by APOBEC3 in benign and precancerous lesions. Science. 2008;320:230–233. doi: 10.1126/science.1153201. [DOI] [PubMed] [Google Scholar]
- 28.Suspène R, Guétard D, Henry M, Sommer P, Wain-Hobson S, Vartanian JP. Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc Natl Acad Sci U S A. 2005;102:8321–8326. doi: 10.1073/pnas.0408223102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng G, et al. Myeloid differentiation and susceptibility to HIV-1 are linked to APOBEC3 expression. Blood. 2007 doi: 10.1182/blood-2006-10-051763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka Y, et al. Anti-viral protein APOBEC3G is induced by interferon-alpha stimulation in human hepatocytes. Biochem Biophys Res Commun. 2006;341:314–319. doi: 10.1016/j.bbrc.2005.12.192. [DOI] [PubMed] [Google Scholar]
- 31.Bonvin M, et al. Interferon-inducible expression of APOBEC3 editing enzymes in human hepatocytes and inhibition of hepatitis B virus replication. Hepatology. 2006;43:1364–1374. doi: 10.1002/hep.21187. [DOI] [PubMed] [Google Scholar]
- 32.Koning FA, Newman EN, Kim EY, Kunstman KJ, Wolinsky SM, Malim MH. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J Virol. 2009;83:9474–9485. doi: 10.1128/JVI.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng G, Lei KJ, Jin W, Greenwell-Wild T, Wahl SM. Induction of APOBEC3 family proteins, a defensive maneuver underlying interferon-induced anti-HIV-1 activity. J Exp Med. 2006;203:41–46. doi: 10.1084/jem.20051512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang FX, Huang J, Zhang H, Ma X, Zhang H. APOBEC3G upregulation by alpha interferon restricts human immunodeficiency virus type 1 infection in human peripheral plasmacytoid dendritic cells. J Gen Virol. 2008;89:722–730. doi: 10.1099/vir.0.83530-0. [DOI] [PubMed] [Google Scholar]
- 35.Conticello SG, Langlois MA, Yang Z, Neuberger MS. DNA deamination in immunity: AID in the context of its APOBEC relatives. Adv Immunol. 2007;94:37–73. doi: 10.1016/S0065-2776(06)94002-4. [DOI] [PubMed] [Google Scholar]
- 36.LaRue RS, et al. The artiodactyl APOBEC3 innate immune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC Mol Biol. 2008;9:104. doi: 10.1186/1471-2199-9-104. 120 pages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kavli B, Otterlei M, Slupphaug G, Krokan HE. Uracil in DNA--general mutagen, but normal intermediate in acquired immunity. DNA Repair (Amst) 2007;6:505–516. doi: 10.1016/j.dnarep.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 38.Aguiar RS, Lovsin N, Tanuri A, Peterlin BM. Vpr.A3A chimera inhibits HIV replication. J Biol Chem. 2008;283:2518–2525. doi: 10.1074/jbc.M706436200. [DOI] [PubMed] [Google Scholar]
- 39.Henry M, Guetard D, Suspene R, Rusniok C, Wain-Hobson S, Vartanian JP. Genetic editing of HBV DNA by monodomain human APOBEC3 cytidine deaminases and the recombinant nature of APOBEC3G. PloS one. 2009;4:e4277. doi: 10.1371/journal.pone.0004277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Di Noia J, Neuberger MS. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 2002;419:43–48. doi: 10.1038/nature00981. [DOI] [PubMed] [Google Scholar]
- 41.Suspène R, Henry M, Guillot S, Wain-Hobson S, Vartanian JP. Recovery of APOBEC3-edited human immunodeficiency virus G->A hypermutants by differential DNA denaturation PCR. J Gen Virol. 2005;86:125–129. doi: 10.1099/vir.0.80426-0. [DOI] [PubMed] [Google Scholar]
- 42.Ishii KJ, et al. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- 43.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 44.Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol Cell. 2002;10:1247–1253. doi: 10.1016/s1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- 45.Jarmuz A, et al. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- 46.Liddament MT, Brown WL, Schumacher AJ, Harris RS. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr Biol. 2004;14:1385–1391. doi: 10.1016/j.cub.2004.06.050. [DOI] [PubMed] [Google Scholar]
- 47.Ivics Z, Hackett PB, Plasterk RH, Izsvák Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 48.Ishii KJ, Akira S. Innate immune recognition of, and regulation by, DNA. Trends Immunol. 2006;27:525–532. doi: 10.1016/j.it.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 49.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kawane K, Nagata S. Nucleases in programmed cell death. Methods Enzymol. 2008;442:271–287. doi: 10.1016/S0076-6879(08)01414-6. [DOI] [PubMed] [Google Scholar]
- 51.Okabe Y, Kawane K, Akira S, Taniguchi T, Nagata S. Toll-like receptor-independent gene induction program activated by mammalian DNA escaped from apoptotic DNA degradation. J Exp Med. 2005;202:1333–1339. doi: 10.1084/jem.20051654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Conticello SG, Thomas CJ, Petersen-Mahrt S, Neuberger MS. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol. 2005;22:367–377. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.