Abstract
Wheat germ cell-free methods provide an important approach for the production of eukaryotic proteins. We have developed a protein expression vector for the TNT® SP6 High-Yield Wheat Germ Cell-Free (TNT WGCF) expression system (Promega) that is also compatible with our T7-based Escherichia coli intracellular expression vector pET15_NESG. This allows cloning of the same PCR product into either one of several pET_NESG vectors and this modified WGCF vector (pWGHisAmp) by In-Fusion LIC cloning (Zhu et al. in Biotechniques 43:354–359, 2007). Integration of these two vector systems allowed us to explore the efficacy of the TNT WGCF system by comparing the expression and solubility characteristics of 59 human protein constructs in both WGCF and pET15_NESG E. coli intracellular expression. While only 30% of these human proteins could be produced in soluble form using the pET15_NESG based system, some 70% could be produced in soluble form using the TNT WGCF system. This high success rate underscores the importance of eukaryotic expression host systems like the TNT WGCF system for eukaryotic protein production in a structural genomics sample production pipeline. To further demonstrate the value of this WGCF system in producing protein suitable for structural studies, we scaled up, purified, and analyzed by 2D NMR two 15N-, 13C-enriched human proteins. The results of this study indicate that the TNT WGCF system is a successful salvage pathway for producing samples of difficult-to-express small human proteins for NMR studies, providing an important complementary pathway for eukaryotic sample production in the NESG NMR structure production pipeline.
Electronic supplementary material
The online version of this article (doi:10.1007/s10969-010-9093-8) contains supplementary material, which is available to authorized users.
Keywords: Wheat germ cell free expression system, Micro cryo NMR probe, Structural genomics, Eukaryotic protein expression, Isotope enrichment
Introduction
Cell-free expression systems are capable of generating small quantities of soluble eukaryotic proteins in instances where traditional bacterial expression systems have failed [7, 17]. Cell-free systems can also accommodate a variety of isotopic enrichment schemes and are capable of routinely generating labeled protein in quantities sufficient for structural analysis by solution NMR [12, 13]. The Northeast Structural Genomics Consortium (NESG, www.nesg.org), a large-scale center funded by the National Institutes of General Medical Sciences (NIGMS), has to date created some 5,000 eukaryotic protein expression plasmids, mostly using modified pET vectors [pET_NESG vectors [1]] for cell-based production in E. coli. Although our success over the past 5 years in producing a specific protein is greatly enhanced by cloning multiple constructs of the same target, even using this strategy success rates in producing a soluble construct for a specific eukaryotic protein in our E. coli based expression system (26%) is significantly lower than the corresponding success rates for targets of prokaryotic origin (42%) (statistics posted at www.nesg.org). Within the NIGMS Protein Structure Initiative, the Center for Eukaryotic Structural Genomics (CESG) has pioneered the use of wheat germ cell-free systems for producing eukaryotic proteins for a wide range of applications (see for example [17, 18]). The wheat germ cell-free system is particularly advantageous for preparing samples for NMR studies, as it is relatively inexpensive to incorporate 15N and 13C isotopes when compared with other eukaryotic expression hosts. Here we describe progress in developing a cell-free protein expression pipeline that parallels the NESG HTP pET-based E. coli expression pipeline [1], allowing routine use of WGCF systems for salvage of certain eukaryotic proteins.
There are three widely utilized cell-free systems each based on cellular extracts [7]. These are derived from either E. coli, rabbit reticulocytes, or wheat germ. Such extracts contain all of the macromolecular machinery required for translation, such as ribosomes, elongation and termination factors, tRNAs, etc. E. coli cell-free expression systems have been used successfully in structural genomics [2, 4]; indeed the Riken Structural Genomics/Proteomics Initiative (RSGI) protein production pipeline is based on this type of cell-free translation. Unfortunately eukaryotic proteins often possess multiple domains, which tend to missfold in prokaryotic (e.g., E. coli) expression systems [16]. The rabbit reticulocyte system tends to have low efficiency, and hence not cost effective for producing the quantities of protein necessary for protein structure determination [7]. Wheat germ extracts, however, have been demonstrated to be well-suited for isotopic enrichment, and for cost-effective production of hundred microgram quantities of proteins sufficient for NMR studies [12, 13, 15, 17, 18].
With support from the Protein Structure Initiative of the NIGMS, the NESG has successfully constructed a protein structure production pipeline centered on an automated HTP multiplexed cloning platform [1]. The platform is fully integrated with a laboratory information management system and PCR primer/construct design tools [8]. To take full advantage of the NESG protein production infrastructure, we require a modified wheat germ cell-free (WGCF) vector that is compatible with the current cloning and expression procedures. In particular, we require a system that is compatible with the same PCR products used in our modified pET_NESG vectors (described in Acton et al. [1]).
Here we describe modifications to the Promega TNT® SP6 High-Yield Wheat Germ Cell-Free (TNT WGCF) expression system that allow utilization of the same PCR products generated for the NESG HTP pET-based cloning platform. Utilization of the TNT WGCF system avoids the cumbersome handling of mRNA, by coupling the processes of transcription and translation in one optimized extract [20]. This is achieved by the addition of SP6 RNA polymerase to the WGCF reaction and the inclusion of an SP6 promoter to the template DNA. Other commercially available systems require the separate generation of mRNA template. This finite mRNA supply is then added to the WGCF protein translation reaction. By contrast, the Promega TNT® requires only the addition of DNA template for protein expression.
Promega provides a number of vectors that are compatible with the TNT WGCF system. In order to make the cloning and protein purification compatible with the current NESG platform, the pTSHQn vector (based on the Promega Riboprobe® System Vector pSP64) was subjected to several modifications. We describe the new vector, pWGHisAmp and demonstrate its use with the TNT WGCF system in obtaining good expression and solubility for eukaryotic protein targets. Using the NESG protein production platform, a set of 59 human protein constructs were then cloned and expressed in the TNT WGCF system and, in parallel, in our intracellular pET15_NESG E. coli system. To gauge the feasibility of using such samples for protein NMR structure applications, two of these human proteins were also isotope enriched and subsequently analyzed by 2D NMR using a micro cryo NMR probe. The TNT WGCF approach represents a viable salvage pathway of the NESG structure production pipeline for eukaryotic protein targets exhibiting poor expression and/or solubility in intracellular E. coli host systems.
Materials and methods
Creation of a WGCF expression vector compatible with NESG HTP cloning pipeline
The pTSHQn (KanR) vector (Promega) was modified to allow cloning of the same PCR product into the modified WGCF expression vector or our pET15_NESG vector for T7-based E. coli intracelluar expression. pET15_NESG produces proteins with a short N-terminal non-cleavable hexaHis tag (MGHHHHHHSH). The backbone of pTSHQn was PCR amplified with the primers WG-F (5′-GTGATGGTGATGGCCCATGGCGAATTCTCCTTATTCTATAG-3′) and WG-R (5′-GAGATCCGGCTGCTAAGGATCCTCTAGAGTCGACCTGC-3′). Following PCR, the reaction mixture was treated with DpnI to destroy the pTSHQn template and subjected to agarose gel electrophoresis. The amplified backbone of pTSHQn was excised from the gel and purified. The pET15_NESG linker, including the coding region for a hexaHis-tag, was amplified by PCR from the NESG-modified pET15_NESG plasmid [1] with the primers pET15_NESG linkerF (5′-ATGGGCCATCACCATCACCA-3′) and pET15_NESG linkerR (5′-TTAGCAGCCGGATCTCGAG-3′). The amplified pET15_NESG linker was also gel purified and was then subcloned into the amplified pTSHQn backbone by the In-FusionTM (Takara) ligation independent cloning method [21]. Transformation into E. coli XL-10 gold cells (Stratagene) was followed by selection on LB/Kan medium. The resulting recombinant clones were screened by PCR, and a correct clone, designated pWGHisKan, was validated by DNA sequencing.
To replace the kanamycin resistance gene of pWGHisKan with an ampicillin resistance gene, required for further standardization with the NESG cloning pipeline, pWGHisKan was PCR amplified excluding the kanamycin resistance gene using the primers WGHisF (5′-AACCATTACGTAGAAAGCCAGTCCGCAG-3′) and WGHisR (5′-TTGGTAATTCGAAATGACCGACCAAGCG-3′). The ampicillin resistance gene from pET15_NESG was also amplified utilizing the primers Amp-F (5′-TTCTACGTAATGGTTTCTTAGACGTCAGG-3′) and Amp-R (5′-ATTTCGAATTACCAATGCTTAATCAGTGAG-3′). The two PCR products were gel purified, subjected to In-FusionTM LIC, transformed into XL-10 gold cells, and selected on LB/Amp medium. Finally, the resulting pET-compatible WGCF expression plasmid, designated pWGHisAmp, was sequenced verified. The resulting expression vector provides proteins with an N-terminal non-cleavable hexaHis tag (MGHHHHHHSH), identical to the N-terminal tag provided by the pET15_NESG expression vector.
Cloning of target genes
The NESG has constructed a Human Cancer Pathway Interaction Network (HCPIN), providing structure–function annotations of key proteins associated with human cancer and developmental biology [9]. The long-range goal of the HCPIN project is to provide a comprehensive 3D structure–function database for human-cancer-associated proteins and protein complexes in the context of functional networks, using both experimental structures and high quality homology models (i.e., using protein templates with >80% sequence identity). Coding regions for 59 human protein targets or domains selected mostly from the HCPIN target list were cloned into both the pWGHisAmp vector for WGCF expression, and into the pET15_NESG vector for E. coli intracellular expression. Primer sequences were generated automatically using the Primer, Primer program [8]. The F-primers are 5′-ACCATCACAGCCAT plus gene specific sequences and the R-primers are 5′-GCAGCCGGATCTCGAGCTA plus gene specific sequences. PCR products were cloned into NdeI and XhoI digested pWGHisAmp vector by In-FusionTM LIC and transformed into XL-10 gold cells. Positive clones were screened first by colony PCR and then confirmed by DNA sequencing. The naming convention for NESG target protein id’s is described elsewhere [19]; the corresponding protein sequences are available on the HCPIN web site (http://nesg.org:9090/HCPIN/index.jsp).
T7-based E. coli intracellular protein expression
pET15_NESG plasmids were transformed into BL21(DE3)pMgK cells and expression analysis performed following standard NESG pipeline methods, as described elsewhere [1].
Expression screening a WGCF small-scale batch reaction with FluoroTect™ detection
Cell-free protein synthesis screening was performed using TNT® SP6 High-Yield WGCF (Promega) and FluoroTect™ (Promega) in vitro fluorophore labeling of target proteins. DNA was purified using Qiaprep 96 Turbo (Qiagen). For each reaction, 14 μL of TNT WGCF lysate (Promega) was mixed with 2 μg of plasmids, 0.5 μL of FluoroTect™ (Promega), and nuclease-free H2O to a final volume of 20 μL. These batch reactions were incubated in microfuge tubes at room temperature for 2 h followed by addition of 1 μL of RNase A (Qiagen) to each reaction, and incubated at 37 °C for 10 min to hydrolyze unincorporated fluorophore-labeled tRNA. Total protein solubility was determined by SDS PAGE analysis of 1 μL of the total reaction compared with the 1 μL of the soluble protein supernatant resulting from centrifugation at 3,000×g at 4 °C for 10 min. Detection of FluoroTect™ labeled proteins was accomplished directly by scanning the gels with a laser-based fluorescent gel scanner (Typhoon 8600, GE Healthcare), with excitation of 488 nm and emission of 532 nm.
Protein production scale-up using WGCF dialysis reaction
Select targets identified as well-expressed and soluble in the HTP FluoroTect screen were subsequently scaled up using a dialysis process. For each 100 μl small-scale dialysis reaction, 60 μL of wheat germ lysate (TNT WGCF) was mixed with 20 μg of expression plasmid purified using a Nucleobond Xtra Maxi Plus kit (Macherey–Nagel GmbH & Co. KG). Nuclease-free H2O was added to obtain the final reaction volume. The reactions were set up in microfuge tubes, transferred into GeBaFlex-tube mini dialysis cups (Gen Bio-Applications Ltd), and inserted into a 2 mL microfuge tube containing 1 mL of dialysis buffer. Dialysis buffer [20] consists of 12 mM HEPES, pH 7.6, 0.5 mM spermidine, 5 mM DTT, 80 μM amino acids, 100 mM KOAc, 1.2 mM ATP, 0.1 mM GTP, 10 mM CP, and 1.5 mM Mg(OAc)2. Reactions were incubated at room temperature for 24 h and then at 4° C for 1–2 days before protein expression analysis.
Isotope enrichment
For isotope enrichment, 2–3 mL dialysis reactions were carried out using amino-acid-depleted WGCF lysate and 15N-, 13C enriched amino acids. In these reactions, 1.2–1.8 mL of TNT WGCF lysate minus amino acids (not yet commercially available) was mixed with 400–600 μg of purified plasmid [prepared using a Nucleobond Xtra Maxi Plus kit (Macherey–Nagel GmbH & Co. KG], and 1 mM (or 100 μM) 15N-, 13C-enriched amino acids (Cambridge Isotopes Laboratories). Nuclease-free H2O was added to make up the final reaction volume. 15N-, 13C-enriched HR3597B was prepared in two 3 mL dialysis reactions containing 100 μM 15N-, 13C-enriched amino acids; 15N, 13C-enriched human ubiquitin was prepared in a 2 mL dialysis reaction containing 1 mM 15N-, 13C-labeled amino acids. The reactions were incubated in 3 mL Slide-A-Lyzer dialysis cassettes (Pierce) against 100 mL dialysis buffer containing 15N-,13C-labeled amino acids (same concentration) at room temperature with gentle stirring for 24 h, and then transferred to 4° C for 1–2 days before protein expression analysis.
Protein purification
To quantitate protein synthesis in the 100 μL dialysis reaction, purification was performed using MagneHis (Promega) magnetic beads. The reaction mixture was transferred to a 1.5 mL microfuge tube and mixed with 350 μL of Binding Buffer A (50 mM HEPES, pH 7.5, 500 μM NaCl and 20 mM imidazole), followed by centrifugation (10,000×g for 10 min). The supernatant was collected, and 150 μL was then transferred to a new microfuge tube containing 30 μL of magnetic beads. Following mixing and incubation at room temperature, the magnetic beads were isolated using a 96-Well Magnet Type A (Qiagen), the remaining solution was discarded. Using the same beads this process was repeated for additional 150 μL aliquots of supernatant. The beads were washed three times with 150 μL of binding buffer using the 96-Well Magnet Type A to capture the magnetic beads. Bound proteins were eluted in 50 μL of Elution Buffer (50 mM Tris, pH 7.5, 500 mM NaCl, 500 mM imidazole) and the results visualized by SDS–PAGE analysis.
Mini columns with 0.5–1 mL of Ni–NTA resin (Qiagen) were used to purify the 15N-, 13C-enriched proteins, synthesized with larger scale dialysis reactions. Briefly, 5 volumes of Binding Buffer B (50 mM Tris, pH 7.5, 500 mM NaCl, 40 mM imidazole) were added to the reaction mixtures, followed by centrifugation at 10,000×g for 10 min. The supernatant was loaded onto a Ni–NTA column and washed with 10 volumes of Binding Buffer. The bound proteins were eluted with Elution Buffer and analyzed by SDS PAGE.
Buffer exchange and sample concentration
Target-protein containing fractions were pooled and exchanged into 1× NMR Buffer (20 mM MES, pH 6.5, 200 mM NaCl, 10 mM DTT and 1× Roche Protease Inhibitor Cocktail). In brief, 1× NMR Buffer was added to the eluate (4 mL final volume), mixed and transferred to a 4 mL Amicon Ultra centrifugal filter device (5 kDa MWCO). This was subsequently followed by centrifugation at 3,000×g at 4°C for 40 min. The flow through was discarded and 1× NMR Buffer was added to the sample to 4 mL and again centrifuged at 3,000×g at 4°C for 50 min. Remaining sample (about 150 μL) was then transferred to a new 1.5 mL Amicon Ultra centrifugal filter device (5 kDa MWCO) and mixed with 300 μL of 1× NMR Buffer and centrifuged at 10,000×g for 40 min at 4° C. The flow through solution was discarded and 400 μL of 1× NMR buffer added to the remaining material (~25 μL). The sample was again subjected to centrifugation at 10,000×g for 40 min at 4° C. Then another 400 μL of 1× NMR Buffer was added and the sample subjected to a final centrifugation as previously described. 2H2O and 2, 2-dimethyl-2-silapentane-5-sulfonic acid (DSS) were added at 10 and 1%, for locking and referencing, respectively. Following buffer exchange into 1× NMR Buffer and the addition of 2H2O and DSS, samples we’re concentrated down to a volume of about 38 μL for NMR measurements.
Labeling efficiency analysis
An aliquot of each purified 15N-, 13C-enriched protein was run on a 4–12% NuPage SDS–PAGE gel (Invitrogen). Target bands (about 1–2 μg) were excised and submitted to the Biological Mass Spectrometry Facility of the Center for Advanced Technology and Medicine. In-gel tryptic digestions were performed and analyzed on a 4800 MALDI-TOF/TOF Analyzer (Applied Biosystems). Theoretical average masses, both labeled and unlabeled, were calculated for several 15N-,13C-enriched tryptic peptides and compared to the average masses observed by mass spectrometry. Labeling efficiency was calculated from the ratio of observed to calculated differences from theoretical unlabeled average mass.
2D NMR analysis
2D 15N-1H HSQC spectra were aquired for the WGCF-derived proteins ubiquitin (35 μL, ~0.3 mM) and HR3597B (35 μL at ~0.4 mM) in NMR buffer. In both cases, the NMR data was collected using the following typical parameters: 64 scans with 64 t1 (15N-dimension) increments and 2048 t2 (1H-dimension) increments using a 1 s recycle delay. Total data collection time in each case was 2 h and 40 min. Data were collected at a temperature of 25° C using a Bruker 600 MHz spectrometer fitted with a 1.7 mM micro-cryoprobe. Chemical shifts were referenced to internal DSS.
Results
Vector engineering
To date the NESG has created some 5,000 pET-based expression constructs for eukaryotic protein, including expression vectors for 2,963 constructs corresponding to 1,743 human proteins (statistics available on line at www.nesg.org). These E. coli intracellular expression constructs consist of domains and full-length proteins cloned into one of three NESG-modified pET vectors [1]. In 2006, the NESG adopted the In-FusionTM (Takara) Ligation Independent Cloning (LIC) method to meet most of its cloning needs. Briefly, LIC cloning, by strand displacement and resection by the InFusion enzyme [14], requires that the target PCR product share 15 nucleotides complimentary to the site of vector linearization.
The WGCF expression vector pTSHQn from Promega, contains an N-terminal metal-affinity purification tag sequence MASSHQHQHQHQHQAIA (HQ tag) and a kanamycin (kanr) resistance gene for selection. This affinity tag and selection agent differ from those currently employed by the NESG. The pTSHQn vector was accordingly modified to make it more compatible with the existing protein production platform and PCR products. First, pWGHisKan was created by replacing the N-terminal HQ tag and ORF of pTSHQn with a pET15_NESG linker sequence and MGHHHHHHSH non-cleavable hexaHis tag. This change allows the same PCR product to be cloned into both the pET15_NESG vector for bacterial intracellular expression or into pWGHisKan for cell-free expression. Secondly, the kanamycin resistance gene of pWGHisKan was replaced with an ampicillin resistance gene to generate a new fully compatible vector called pWGHisAmp. These changes are depicted graphically in Fig. 1. Proteins produced by either intracellular bacterial expression or WGCF will have the same protein sequence and affinity tag, thereby allowing for parallel cloning and expression screening. However, proteins produced with the WGCF system may exhibit advantageous differences in folding and aggregation properties.
Fig. 1.
Vector engineering schema illustrating the creation of the plasmid pWGHisAmp from pTSHQn (Promega), and the NESG modified pET15 vector. PCR and In-FushionTM ligation independent cloning methods were used to replace the ORF and HQ metal affinity tag in pTSHQn with the pET15 linker region and 6xHis metal affinity tag to generate pWGHisKan. This was subsequently followed by replacement of the kanamycin resistance gene in pWGHisKan with the pET15 ampicillin resistance gene to generate the final vector pWGHisAmp. PCR derived products can now be cloned into either the NESG modified pET15 bacterial expression plasmid or into the pWGHisAmp plasmid for wheat germ cell-free expression
Screening of human protein expression with the modified WGCF system and comparison with in-cell expression in E. coli
A set of 58 non-structured protein targets (see supplementary data) were selected from the Human Cancer Pathway Protein Interaction Network (HCPIN) [9], along with full length human ubiquitin (sp id: P62988). These targets were cloned into our pET15_NESG and pWGHisAmp vectors. Expression and solubility in the pET15_NESG system was assayed following transformation into BL21(DE3)pMgK E.coli cells. Expression was ranked on a relative scale from 0 to 5, where 5 indicates the highest levels of expression observed for that system. The degree to which the total protein expressed remained soluble following lysate centrifugation was also represented on a relative scale of 0–5, where 5 is indicates >90% of the expressed protein is in the soluble fraction.
In this work, we used the FluroTect™ GreenLys technology [6, 11] to introduce fluorescent-labeled Lys residues at certain AAA codon sites, allowing visualization of target protein even in the presence of high levels of background proteins from the WGCF system itself. The FluoroTect™ GreenLys in vitro Translation Labeling System allows the fluorescent labeling of translation products through the use of a modified charged lysine tRNA labeled with the fluorophore BODIPY®-FL. Using this system, fluorescently labeled lysine residues are incorporated into nascent proteins during translation. The fluorescent lysine is added to the translation reaction as a charged fluorescent lysine-tRNA complex (FluoroTect™GreenLys tRNA) rather than a free amino acid. Although only a fraction of the few Lys residues in each protein molecule are modified, it cannot be excluded that the FluoroTect™ tag affects (negatively or positively) the observed solubility of the targeted protein. In addition, the apparent expression level (i.e., fluorescence intensity) depends on the number of Lys residues that have been substituted with a ε-labeled lysine via the modified tRNA, and can only be compared qualitatively with other methods of protein visualization such as Coomassie Blue dye binding. Further, the fluorescence detection technology is also considerably more sensitive than the aforementioned dye binding. This raises the possibility that solubility may only appear greater as the protein levels detected are below that favoring aggregation. Despite these important caveats, the FluoroTect™ labeling approach is an excellent first pass screening method for assessing expression and solubility of proteins produced in the WGCF system at relatively low levels compared to background proteins. These targets can then be scaled up without FluoroTect™ labeling.
E. coli intracellular protein expression and solubility levels were scored for each protein on a scale of 0–5 for expression (E) in the total cell extract; and on a scale of 0–5 for solubility (S) of the expressed protein in the soluble extract (S) based on Commassie Blue stain binding using SDS–PAGE. In our hands, proteins with a product E × S (ES score) >12 can be scaled up to produce purified proteins at levels of >20 mg/L. WGCF expression results were scored in a similar way, using FluoroTect™ labeling with SDS–PAGE gels. These FluoroTect™ ES scores are not directly comparable to the ES scores for E. coli intracellular expression. In our experience, WGCF FluoroTect™ES scores ≥8 can be scaled up (as described below) to provide 50–100 μg yields of protein, sufficient for NMR analysis using microprobe NMR technologies that have been described elsewhere [3].
E. coli intracellular expression results were grouped into three classes: proteins with poor expression (E ≤ 1), proteins with promising expression (E ≥ 2) but poor solubility (S ≤ 1), and proteins with both promising expression (E ≥ 2) and solubility (S ≥ 2). In Table 1, Poor expression and solubility in the cell-based system are indicated with a “−” symbol, promising expression and solubility with a “+” symbol.
Table 1.
Expression and solubility screening results for E. coli intracellular and Promega TNT WGCF expression
| E. coli | Total constructs | WGCF ES ≥ 8 | % salvage (%) | |
|---|---|---|---|---|
| Exp. | Sol. | |||
| − | − | 34 | 19 | 46 |
| + | − | 9 | 6 | 67 |
| + | + | 16 | 15 | 94 |
| 59 | 40 | 68 | ||
The scoring protocol is explained in the text
An SDS–PAGE gel showing results for some of the 59 WGCF screening reactions is presented in Fig. 2. Overall 40 of the 59 human proteins tested (~70%) were expressed with ES values greater than 8 in the WGCF system (Table 1). More importantly, of 43 human proteins which could not be produced in the E. coli intracellular system (34 with low or no expression and 9 that were expressed but not soluble), 25 were expressed and soluble with ES values ≥8 in the WGCF system (sufficient for NMR analysis). This corresponds to a salvage rate of ~60% (25/43).
Fig. 2.
Total expression (A) and solubility (B) characteristic of 14 human targets, expressed in wheat germ small-scale “batch” reactions, with FluoroTect™ detection and excitation at 532 and 488 nm, respectively. Expression and solubility characteristics corresponding to in-cell E. coli expressed protein are indicated above the gel (“−” corresponding to poor expression or solubility and “+” indicating promising expression or solubility). The identities by NESG target ID of the proteins in the order listed are as follows: HR2951, HR2928, HR3111, HR2930, HR2934, HR2887, HR2965, HR2924, HR2889, HR2891, HR3007, HR3057, HR3051 and HR2886 (the corresponding protein sequences can be accessed on line at http://nmr.cabm.rutgers.edu:9090/PLIMS/targetSummary.jsf)
Scale up using dialysis reaction with isotopic enrichment
Consistent with recent work from Promega [20], we observe that the protein yield obtained using the simple batch reaction used above for screening is lower than what can be obtained using a dialysis reaction. Several proteins showing good WGCF expression and solubility in the screening reactions were selected for scale up using the dialysis method. Small scale dialysis reactions (usually 100 μL) were performed before scale-up with labeled amino acids, so as to validate which proteins could be successfully produced prior to isotopic enrichment. Yield at this scale is generally not detectable by SDS–PAGE gel analysis of the whole WGCF reaction mixture without FluoroTect™ labeling. Therefore, it is necessary to perform IMAC purification followed by SDS–PAGE gel analysis. Several proteins, including some proteins which failed to be produced by E. coli intracellular expression, were randomly selected to test on small scale dialysis reaction. After course purification by MagneHis (Promega), all of the selected target proteins were demonstrated to be produced with a yield of about 2–6 μg per 100 μL dialysis reaction (data not shown).
To test whether the proteins produced by the WGCF system are amenable for structural analysis by NMR, two human proteins (HR3597B and human ubiquitin) were produced with larger scale dialysis reactions containing 15N-, 13C-labeled amino acids. HR3597B, a putative zinc-binding protein, showed very low expression in the E. coli intracellular expression system.
15N-,13C-enriched human ubiquitin was produced using a single 2 mL dialysis reaction containing 1 mM labeled amino acids; HR3597B was produced using two separate 3 mL dialysis reactions (total 6 mL) with 100 μM labeled amino acids. Samples were purified by Ni–NTA IMAC, as described in “Methods and materials”, and buffer exchanged to 35–40 μL of NMR Buffer. Protein yield and purity was estimated from the SDS–PAGE data in Fig. 3. About 1 μL of the 15N-, 13C-enriched ubiquitin NMR buffer exchanged preparation was run in lane 1 of panel 3A. Exactly 10 μL of the 15N-, 13C-enriched HR3597B Ni–NTA-purified sample, and 1 μL of the NMR buffer exchanged preparation were run in lanes 1 and 2, respectively of panel 3B. Lysozyme standards at various concentrations were utilized to estimate protein yield. Approximately 120 μg of 15N-, 13C-enriched ubiquitin was produced from a 2 mL dialysis reaction (60 μg protein/mL dialysis reaction) and about 200 μg of 15N-,13C-enriched HR3597B from a 6 mL dialysis reaction (30 μg protein/mL dialysis reaction). Following buffer exchange into NMR Buffer, final concentrations were estimated as 0.3 mM for ubiquitin and 0.4 mM for HR3597B. HR3597B has an ES value of 9 in the FluoroTect™ screening assay, demonstrating that an ES value of 8 is a reasonable threshold for determining which targets may be scaled up for subsequent structural analysis. Contaminating proteins with non-engineered Ni-affinity such as those found at ~50 kDa are from the WGCF lysate itself, and are not isotope-enriched. Therefore, they are not observed in isotope-filtered 2D [1H-15N]–HSQC and related triple resonance NMR spectra. Although these proteins do not appear to interfere with NMR studies we are exploring alternative purification methods (HaloTag, Promega; Strep-TagII, Novagen). We have found the former method removes the contaminating bands and that the HSQC spectra of the HaloTag purified HR3597B did not differ significantly from the Ni–NTA purified protein. Although this indicates the contaminating proteins are innocuous, we are in the process of developing alternate purification strategies to allow for production of more homogenous WGCF protein samples.
Fig. 3.
SDS–PAGE analysis of purified double-labeled protein produced by larger-scale dialysis reaction. Lane M in panels A and B correspond to Precision Plus Protein Marker (Bio-rad). In panel A, lane 1 was loaded 1 μL of purified 15N-,13C-enriched human ubiquitin in NMR buffer (40 μL preparative volume). In panel B lane 1 was loaded 10 μL of Purified HR3597B in elution buffer (1,900 μL preparative volume) and in lane 2, 1 μL of purified HR3597B in NMR buffer (40 μL preparative volume). Lanes 2–5 of panel A contain 0.5, 1.0, 2.0 and 3.0 μg of lysozyme and Lanes 3–5 of panel B contain 0.3, 1.0 and 2.0 μg of lysozyme
Labeling efficiency
MALDI-TOF mass spectroscopy was employed to examine the masses of four tryptic peptides of 15N-, 13C-enriched ubiquitin produced in the presence of 0.1 and 1.0 mM 15N-, 13C-labeled amino acids. Using 1.0 mM concentrations of labeled amino acids in the cell free expression media, typical labeling efficiencies are generally >90% (Table 2). We observe a 1.3-fold enhancement in labeling efficiencies with the 1.0 mM labeled amino acid supplemented dialysis mixture relative to the reaction using 0.1 mM concentration (Table 2). It is not known if the same enhancement in labeling efficiency could be optimized with intermediate concentrations of labeled amino acids (e.g. 0.5 mM) in the reaction mixture. Future work will be directed at this question.
Table 2.
Labeling efficiency of ubiqutin produced using the modified Promega TNT WGCF system with two different concentrations of labeled amino acids in the reaction mixture
| Peptide sequence | Position | 0.1 mM labeled aa’s (%) | 1 mM labeled aa’s (%) |
|---|---|---|---|
| EGIPPDQQR | 44–52 | 68.9 | 93.7 |
| ESTLHLVLR | 74–82 | 73.4 | 96.4 |
| IQDKEGIPPDQQR | 40–52 | 66.6 | 94.0 |
| TITLEVEPSDTIENVK | 22–37 | 77.2 | 93.2 |
2D NMR analysis
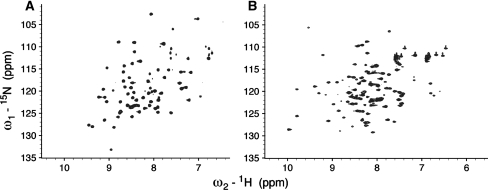
The 2D HSQC spectrum collected on 15N-,13C-enriched ubiquitin (shown in Fig. 4) is well dispersed, with good signal-to-noise, and superposes well with the 2D HSQC spectrum of a 15N-,13C-enriched ubiquitin sample produced by conventional in-cell pET vector system (not shown). The 2D HSQC spectra collected on 15N-, 13C-enriched HR3597B is also of promising quality, sufficient to demonstrate the value of the WGCF system. Secondary structure prediction (SSpro, Ver.4.5) [5] identifies regular secondary structure (helix or beta) for only 35 of 100 amino acids in HR3597B. Also, a consensus of 9 predictors of disorder, determined with the aid of the NESG Dismeta server (http://www-nmr.cabm.rutgers.edu/bioinformatics/disorder/), indicates disorder for ~30 amino acids, and suggests that this protein may require Zn as a structural co-factor. Accordingly, we judge the spectral quality to be sufficient to demonstrate the feasibility of isotope-enrichment using the TNT WGCF expression system.
Fig. 4.
Micro cryo NMR probe 15N-1H 2D HSQC spectra at 25 °C of A human ubiquitin and B NESG human protein target HR3597B produced with 15N,13C-enrichment using the Promega TNT® WGCF system. Spectra were obtained using a 600 MHz 1.7 mm Bruker micro cryo NMR probe using 100–200 μg of protein sample each
Discussion
WGCF provides an important approach for producing eukaryotic proteins, and is especially valuable for producing hundred microgram quantities of isotope-enriched protein samples for NMR studies [12, 13, 15, 18]. We have previously demonstrated that good quality 3D structures of small (<10 kDa) proteins can be determined by NMR using <100 μg quantities of protein samples with micro NMR probes [3]. These technologies are enhanced by the recent introduction of Bruker micro cryo NMR probes, like the one used in this work. Thus, there is a natural synergy between WGCF protein expression and micro cryo NMR probe technologies. In addition, WGCF systems can accommodate selective labeling strategies for single proteins—something that cannot easily be achieved in whole cell systems. This includes incorporation of SAIL (“Stereo Array Isotope Labeling”) amino acids, which are valuable for NMR studies of larger proteins [10].
In this work, modifications were made to pTSHQn, a TNT® SP6 High-Yield WGCF expression system vector, to enable parallel implementations of WGCF and in-cell pET vector cloning strategies. Using this system we have assessed expression and solubility for 59 NESG human protein targets, ~60% of which could not be expressed in a soluble form in our E. coli intracellular pET system. Forty of these human proteins were produced at levels sufficient to be considered for NMR studies (>100 μg) using the TNT WGCF system; 25 of these could not be produced in soluble form using the cell-based E. coli system. Modifications were made in the TNT WGCF system by depletion of amino-acids from the lysate, to allow incorporation of labeled amino acids. Human ubiquitin and NESG human protein target HR3597B were 15N-, 13C-enriched, and purified using a modified TNT WGCF medium. Each reaction produced ~30–60 μg of labeled protein per mL of WGCF dialysis reaction volume. In this work, TNT WGCF expression and micro cryo NMR probe technology were combined for the first time, and promising HSQC results were obtained for the two test proteins, suggesting that the structural analysis by NMR of such WGCF produced proteins will become routine in the not-so-distant future.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank Colleen Ciccosanti, James Hartnett, Janet Huang, Li-Chung Ma, Melissa Maglaqui, Dongyan Wang, and Rong Xiao for helpful discussions. We also thank Haiyan Zheng, Caifeng Zhao, Meiqian Qian and Peter Lobel of the Biological Mass Spectrometry Facility at CABM for MALDI-TOF analysis of isotope enriched proteins. This work was supported by grant U54-GM074958 from the Protein Structure Initiative of National Institute of General Medical Sciences.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Acton TB, Gunsalus KC, Xiao R, Ma LC, Aramini J, et al. Robotic cloning and protein production platform of the Northeast Structural Genomics Consortium. Methods Enzymol. 2005;394:210–243. doi: 10.1016/S0076-6879(05)94008-1. [DOI] [PubMed] [Google Scholar]
- 2.Aoki M, Matsuda T, Tomo Y, Miyata Y, Inoue M, Kigawa T, Yokoyama S. Automated system for high-throughput protein production using the dialysis cell-free method. Protein Expr Purif. 2009;68(2):128–136. doi: 10.1016/j.pep.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 3.Aramini JM, Rossi P, Anklin C, Xiao R, Montelione GT. Microgram-scale protein structure determination by NMR. Nat Methods. 2007;4(6):491–493. doi: 10.1038/nmeth1051. [DOI] [PubMed] [Google Scholar]
- 4.Busso D, Kim R, Kim SH. Using an Escherichia coli cell-free extract to screen for soluble expression of recombinant proteins. J Struct Funct Genomics. 2004;5(1–2):69–74. doi: 10.1023/B:JSFG.0000029197.44728.c5. [DOI] [PubMed] [Google Scholar]
- 5.Cheng J, Randall AZ, Sweredoski MJ, Baldi P (2005) SCRATCH: a protein structure and structural feature prediction server. Nucleic Acids Res 33(Web Server issue): W72–W76 [DOI] [PMC free article] [PubMed]
- 6.Coleman MA, Lao VH, Segelke BW, Beernink PT. High-throughput, fluorescence-based screening for soluble protein expression. J Proteome Res. 2004;3(5):1024–1032. doi: 10.1021/pr049912g. [DOI] [PubMed] [Google Scholar]
- 7.Endo Y, Sawasaki T. Cell-free expression systems for eukaryotic protein production. Curr Opin Biotechnol. 2006;17(4):373–380. doi: 10.1016/j.copbio.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 8.Everett JK, Acton TB, Montelione GT. Primer Prim’er: a web based server for automated primer design. J Struct Funct Genomics. 2004;5(1–2):13–21. doi: 10.1023/B:JSFG.0000029238.86387.90. [DOI] [PubMed] [Google Scholar]
- 9.Huang YJ, Hang D, Lu LJ, Tong L, Gerstein MB, et al. Targeting the human cancer pathway protein interaction network by structural genomics. Mol Cell Proteomics. 2008;7(10):2048–2060. doi: 10.1074/mcp.M700550-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kainosho M, Torizawa T, Iwashita Y, Terauchi T, Mei Ono A, et al. Optimal isotope labelling for NMR protein structure determinations. Nature. 2006;440(7080):52–57. doi: 10.1038/nature04525. [DOI] [PubMed] [Google Scholar]
- 11.Kobs G, Hurst R, Betz N, Godat B. FluoroTect™ GreenLys in vitro translation labeling system. Promega Notes. 2001;77:23–27. [Google Scholar]
- 12.Kohno T. Production of proteins for NMR studies using the wheat germ cell-free system. Methods Mol Biol. 2005;310:169–185. doi: 10.1007/978-1-59259-948-6_12. [DOI] [PubMed] [Google Scholar]
- 13.Kohno T, Endo Y. Production of protein for nuclear magnetic resonance study using the wheat germ cell-free system. Methods Mol Biol. 2007;375:257–272. doi: 10.1007/978-1-59745-388-2_13. [DOI] [PubMed] [Google Scholar]
- 14.Marsischky G, LaBaer J (2004) Many paths to many clones: a comparative look at high-throughput cloning methods. Genome Res 14(10B): 2020–2028 [DOI] [PubMed]
- 15.Morita EH, Shimizu M, Ogasawara T, Endo Y, Tanaka R, et al. A novel way of amino acid-specific assignment in (1)H-(15)N HSQC spectra with a wheat germ cell-free protein synthesis system. J Biomol NMR. 2004;30(1):37–45. doi: 10.1023/B:JNMR.0000042956.65678.b8. [DOI] [PubMed] [Google Scholar]
- 16.Netzer WJ, Hartl FU. Recombination of protein domains facilitated by co-translational folding in eukaryotes. Nature. 1997;388(6640):343–349. doi: 10.1038/41024. [DOI] [PubMed] [Google Scholar]
- 17.Vinarov DA, Loushin Newman CL, Markley JL. Wheat germ cell-free platform for eukaryotic protein production. FEBS J. 2006;273(18):4160–4169. doi: 10.1111/j.1742-4658.2006.05434.x. [DOI] [PubMed] [Google Scholar]
- 18.Vinarov DA, Lytle BL, Peterson FC, Tyler EM, Volkman BF, et al. Cell-free protein production and labeling protocol for NMR-based structural proteomics. Nat Methods. 2004;1(2):149–153. doi: 10.1038/nmeth716. [DOI] [PubMed] [Google Scholar]
- 19.Wunderlich Z, Acton TB, Liu J, Kornhaber G, Everett J, et al. The protein target list of the Northeast Structural Genomics Consortium. Proteins. 2004;56(2):181–187. doi: 10.1002/prot.20091. [DOI] [PubMed] [Google Scholar]
- 20.Zhao KQ, Hurst R, Slater MR, Bulleit RF. Functional protein expression from a DNA based wheat germ cell-free system. J Struct Funct Genomics. 2007;8(4):199–208. doi: 10.1007/s10969-007-9035-2. [DOI] [PubMed] [Google Scholar]
- 21.Zhu B, Cai G, Hall EO, Freeman GJ. In-fusion assembly: seamless engineering of multidomain fusion proteins, modular vectors, and mutations. Biotechniques. 2007;43(3):354–359. doi: 10.2144/000112536. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.