Abstract
Late leaf spot (LLS) and rust are two major foliar diseases of groundnut (Arachis hypogaea L.) that often occur together leading to 50–70% yield loss in the crop. A total of 268 recombinant inbred lines of a mapping population TAG 24 × GPBD 4 segregating for LLS and rust were used to undertake quantitative trait locus (QTL) analysis. Phenotyping of the population was carried out under artificial disease epiphytotics. Positive correlations between different stages, high to very high heritability and independent nature of inheritance between both the diseases were observed. Parental genotypes were screened with 1,089 simple sequence repeat (SSR) markers, of which 67 (6.15%) were found polymorphic. Segregation data obtained for these markers facilitated development of partial linkage map (14 linkage groups) with 56 SSR loci. Composite interval mapping (CIM) undertaken on genotyping and phenotyping data yielded 11 QTLs for LLS (explaining 1.70–6.50% phenotypic variation) in three environments and 12 QTLs for rust (explaining 1.70–55.20% phenotypic variation). Interestingly a major QTL associated with rust (QTLrust01), contributing 6.90–55.20% variation, was identified by both CIM and single marker analysis (SMA). A candidate SSR marker (IPAHM 103) linked with this QTL was validated using a wide range of resistant/susceptible breeding lines as well as progeny lines of another mapping population (TG 26 × GPBD 4). Therefore, this marker should be useful for introgressing the major QTL for rust in desired lines/varieties of groundnut through marker-assisted backcrossing.
Electronic supplementary material
The online version of this article (doi:10.1007/s00122-010-1366-x) contains supplementary material, which is available to authorized users.
Introduction
Groundnut (Arachis hypogaea L.), also known as peanut, is an important oilseed crop in tropical and subtropical regions of the world. It is grown in six continents but mainly in Asia, Africa and America in over 100 countries with a world production of 37.10 m tons from an area of 23.11 m ha (FAO 2007). The low productivity of the crop in India and several African countries is ascribed to many biotic and abiotic stresses in the cultivation of the crop. Among the biotic stresses, two fungal diseases namely late leaf spot (LLS) caused by Phaeoisariopsis personata [(Berk. and Curt.) Deighton] and rust caused by Puccinia arachidis Speg. are widespread and economically most important. These diseases often occur together and cause yield loss up to 50–70% in the crop (Subrahmanyam et al. 1984). Besides adversely affecting the yield and quality of pod, it also affects the yield and quality of haulm. Though several effective fungicides are available to control the diseases but host-controlled resistance is considered the best strategy to surmount additional cost of production and hazardous effect of fungicides on the soil and environment.
Genetic studies on LLS and rust resistance suggest that resistance to these fungal diseases is complex and polygenic in nature and probably controlled by several recessive genes (Sharief et al. 1978; Nevill 1982; Green and Wynne 1986, 1987; Motagi 2001; Dwivedi et al. 2002). Furthermore, additive genetic variance seems to contribute predominantly to the resistance (Kornegay et al. 1980; Hamid et al. 1981; Anderson et al. 1986; Jogloy et al. 1987). Groundnut breeders across the world have developed superior varieties resistant to LLS and/or rust. However, co-occurrence of these two diseases and defoliating, partial and polygenic nature of LLS makes the identification of resistant and susceptible lines cumbersome through conventional screening techniques (see Leal-Bertioli et al. 2009).
Recent years have seen significant progress in the area of crop genomics applied to breeding (see Varshney et al. 2005). Several studies have demonstrated the utility of molecular markers and marker-assisted selection (MAS) to improve the efficiency of conventional breeding especially in the case of low heritable traits, where phenotypic selection is difficult, expensive, lack accuracy or precision (see Varshney et al. 2006). Among different types of marker systems, microsatellites or simple sequence repeat (SSR) markers, considered as markers of choice for breeding applications, have been extensively used in several crop species (see Gupta and Varshney 2000). In case of groundnut also, a large number of SSR markers have been developed by several groups during last 10 years (e.g., Hopkins et al. 1999; He et al. 2003; Ferguson et al. 2004; Moretzsohn et al. 2005; Mace et al. 2007; Cuc et al. 2008; Gautami et al. 2009). Development of more SSR markers is underway in groundnut from BAC (bacterial artificial chromosome)—end sequences (DR Cook, pers. commun; DJ Bertioli, pers. commun.) and transcript sequence data using next generation sequencing technologies (SJ Knapp, pers. commun.). However, genome mapping and trait mapping is still in its infancy in groundnut. For instance, SSR-based genetic map based on recombinant inbred line (RIL) mapping population of cultivated groundnut has been developed recently (Varshney et al. 2009) and some trait mapping studies have been conducted (Herselman et al. 2004; Varshney et al. 2009). One of the main bottlenecks for slow progress in molecular mapping in groundnut is low level of genetic diversity present in the germplasm of cultivated groundnut and non-availability of critical mass (large number of markers) and adequate molecular markers (e.g., single nucleotide polymorphisms, SNPs).
In view of above, the present study was undertaken to construct a genetic map and to identify the QTLs for LLS and rust in cultivated groundnut by using a recombinant inbred line (RIL) mapping population (TAG 24 × GPBD 4). Furthermore, validation of one SSR marker (IPAHM 103) associated using a major QTL for rust was also undertaken with several breeding lines and varieties which demonstrated the utility of this marker to breed rust resistant groundnut varieties through marker assisted selection.
Materials and methods
Mapping population
The mapping population comprised of 268 RILs derived from the cross TAG 24 × GPBD 4. TAG 24 is a popular high yielding cultivar in India but susceptible to both diseases (Patil et al. 1995), while GPBD 4, the leading groundnut variety in Karnataka and other southern states of India (Gowda et al. 2002), is high yielding and resistant to both diseases. The mapping population was developed by employing single seed descent (SSD) method from F2. The F6 progenies were used for phenotyping and F7 progenies for genotyping.
For validating the utility of a candidate marker IPAHM 103, a set of 46 highly diverse germplasm lines (10 susceptible cultivars, 13 hybrid derivatives from NcAc (North Carolina Accessions) genotypes, 11 interspecific derivatives, 8 mutant lines, 2 South American landraces and 2 advanced breeding lines) were used (ESM 1).
Phenotyping
Phenotyping of mapping population along with the parental genotypes for LLS was carried out in three environments, viz, Rainy 2004 (EL-I), 2005 (EL-II) and 2006 (EL-III) at Dharwad, India. Phenotyping of mapping population for rust was conducted at Dharwad in five environments, viz, Rainy 2004 (ER-I), Rainy 2005 (ER-II), Post-rainy 2007 (ER-III), Rainy 2007 (ER-IV) and Rainy 2008 (ER-V).
The inoculums for LLS and rust were produced and maintained separately on TMV-2 (a highly susceptible variety to LLS) and mutant 28-2 (resistant to LLS but highly susceptible to rust), respectively. The LLS conidia and rust urediniospores were isolated by soaking and rubbing infected leaves in water for 30 min and used for inoculation. RILs were sown in randomized block design (RBD) with two replications at Dharwad. Seeds of these RILs were treated with seed protectant before sowing. Ten seeds from each RIL were sown in 1 m rows with 30 cm and 10 cm inter- and intra-row spacing, respectively. Both the parental genotypes (TAG 24 and GPBD 4) were also sown after every 50 rows as controls.
Artificial disease epiphytotics were created in separate screening experiments for the two diseases using “spreader row technique”. Two more genotypes namely TMV 2 and mutant 28-2 were used as spreader rows for LLS and rust, respectively. Spreader rows were sown at every tenth row as well as border around the field to maintain the effective inoculum load. At 35 days after sowing, plants were inoculated uniformly in the evening with LLS/rust for a week. The inoculum containing 20,000 conidia/urediniospores per ml water was mixed with Tween 80 (0.2 ml/1,000 ml of water) as a mild surfactant and atomized on the plants using a Knapsack sprayer. High humidity was maintained by irrigating the field in the night with sprinkler or furrow irrigation. Additional inoculum was provided by placing pots containing diseased plants at every 50 rows. The non-targeted disease, i.e., rust/LLS in LLS and rust experiments, was controlled by spraying fungicide carbendazim (bavistin) 1 g l−1 and tridemorph (calixin) 1 ml l−1, respectively.
Disease scoring
Disease scoring for LLS was done at 70 days (LLS-I) and 90 days (LLS-II) after sowing, and for rust was scored at 70 days (LR-I), 80 days (LR-II), 90 days (LR-III), 105 days (LR-IV), 113 days (LR-V) and 120 days (LR-VI) after sowing in different seasons by using a modified 9-point scale (Subbarao et al. 1990). Components of resistance to rust, viz., incubation period (IP), latent period (LP) and infection type (IT) were also assessed in the greenhouse.
Marker genotyping
DNA of parents and RILs (F7) was isolated using SIGMA® GenElute Plant Genomic DNA Kit. DNA quality was checked and quantified on 0.8% agarose gel with known concentrations of uncut lambda DNA standard. Initially the parents, TAG 24 and GPBD 4, were screened for polymorphism with a 1089 SSR markers, compiled from several sources, as mentioned in Varshney et al. (2009). Polymorphic SSR markers identified between these genotypes, subsequently were used for genotyping the mapping population.
Polymerase chain reactions (PCRs) were performed as given in Varshney et al. (2009) with some modifications: 5 μl of reaction volume (instead of 10 μl) containing reduced amount of other PCR components was used. Another modification was in the amplification profile, as higher annealing temperature (65°C instead of 60°C) was used to increase the specificity of amplicons.
As in Varshney et al. (2009), the PCR products were tested on 1.2% agarose gel to check for amplification and, subsequently depending on the use of normal or fluorescent dyes labeled primers, PCR amplicons were separated using polyacrylamide gel electrophoresis (PAGE) or capillary electrophoresis. Allele sizing of the electrophoresis data was carried out using Genescan 3.1 software (Applied Biosystems, USA) and Genotyper 3.1 (Applied Biosystems, USA).
Statistical analysis
Phenotypic data
The analysis of variance (ANOVA) at different stages of disease scoring for LLS and rust was performed to test the significance of differences between RILs. To assess and quantify the genetic variability among the RILs, phenotypic coefficient of variance (PCV) and heritability in the broad sense (h2b.s) were estimated. Correlation coefficient (r) among the different stages of LLS, rust and LLS with rust was also estimated. All necessary computation for the field trials was performed with the software package GenStat 10th edition (Payne et al. 2007).
Linkage map construction
Marker genotyping data obtained on the mapping population were used for the linkage analysis using MAPMAKER/EXP V 3.0 (Lander et al. 1987; Lincoln et al. 1992). A minimum LOD score of 3.0 and maximum recombination fraction (θ) of 0.5 were set as threshold values for linkage group determination by using “Make Chromosome” command and a set of anchor markers was identified from Varshney et al. (2009). The most likely marker order within each linkage group was estimated by using three point analyses (“three point” command). Marker orders were confirmed by comparing the Log-likelihood of the possible orders using multipoint analysis (“compare” command). The “try” command was used to determine the exact position of the new marker orders. The new marker orders were again confirmed with the “compare” commands. Finally, the marker order as per linkage group was calculated using RECORD program (van Os et al. 2005). Recombination fraction was converted into map distances in centiMorgans (cM) using Kosambi mapping function (Kosambi 1944). The inter-marker distances calculated from MAPMAKER were used to construct linkage map by using MAPCHART version 2.2 (Voorrips 2006).
QTL analysis
All necessary computation for QTL mapping and estimation of their additive effects was performed with the software package PLABQTL version 1.1 (Utz and Melchinger 1996). The method of CIM with cofactors (Zeng 1994; Jansen and Stam 1994) was used for the detection and mapping of QTLs. The genotypic data of 56 mapped markers and mean phenotypic data of 268 RILs were used for CIM QTL analysis. Cofactors were selected using stepwise regression (Miller 1990) with an F to enter and F to delete value of 3.5. A LOD threshold of 2.5 was chosen to declare a putative QTL as significant. Estimates of the additive effect of each detected QTL, the total LOD score, the total proportion of phenotypic variation explained (PVE) by all the detected QTLs were obtained by fitting a multiple linear regression model that simultaneously included all the detected QTLs for the traits in question.
For analyzing QTL × environment (QTL × E) interactions in case of LLS, phenotyping data for LLS-I and LLS-II scored in all three environments (Rainy 2004, Rainy 2005 and Rainy 2006) were averaged and analyzed together with mapping data for 56 mapped SSR markers. Because of rust scoring at different stages in different environments, QTL × E interaction could not be analyzed.
Single marker analysis (SMA), which is based on simple linear regression method (Haley and Knott 1992), was also performed to tag and confirm potential SSR markers linked to the trait.
Results
Phenotyping of mapping population
A set of 268 RILs derived from a susceptible female parent TAG 24 and highly resistant male parent GPBD 4 for LLS and rust, along with the parental genotypes, was phenotyped for LLS in three environments, viz., Rainy 2004 (EL-I), 2005 (EL-II) and 2006 (EL-III) at Dharwad and for rust in five environments, viz., Rainy 2004 (ER-I), Rainy 2005 (ER-II), Post-rainy 2007 (ER-III), Rainy 2007 (ER-IV) and Rainy 2008 (ER-V). The mean disease score of parent GPBD 4 for LLS (1.00–3.50) and rust (2.00–4.00) showed consistently lower disease incidence than TAG 24 (LLS: 4.75–9.00 and rust: 3.71–8.00) at all the scoring stages and environments (Table 1). Phenotypic data on 268 RILs for LLS and rust showed near normal distribution for both diseases, but slightly skewed toward susceptibility to LLS and rust at later stages, i.e., LLS-II, LR-III, LR-V and LR-VI. The majority of RILs showed variation for both diseases, within the parental limits. A few RILs exhibited transgressive variation for susceptible reaction. Analysis of variance on these phenotyping data revealed significant differences between recombinant inbred lines in the reaction to LLS and rust.
Table 1.
Mean of parents and recombinant inbred lines (RILs) and estimates of phenotypic coefficient of variation (PCV) and broad sense heritability (h2b.s) for LLS and rust in TAG 24 × GPBD 4 mapping population
| Environments/traitsa | Scoring stagea | Mean | PCV (%) | h2b.s (%) | ||
|---|---|---|---|---|---|---|
| TAG 24 | GPBD 4 | RILs ± SE | ||||
| LLS | ||||||
| EL-I | LLS-I | 6.75 | 1.75 | 4.49 ± 0.71 | 31.00 | 48.50 |
| LLS-II | 9.00 | 3.00 | 6.95 ± 0.86 | 22.84 | 40.87 | |
| EL-II | LLS-I | 7.00 | 1.75 | 5.38 ± 0.51 | 25.82 | 73.53 |
| LLS-II | 7.75 | 2.00 | 6.38 ± 0.40 | 21.71 | 82.81 | |
| EL-III | LLS-I | 4.75 | 1.00 | 3.14 ± 0.49 | 33.55 | 55.40 |
| LLS-II | 8.50 | 3.50 | 6.14 ± 0.35 | 23.56 | 64.70 | |
| Rust | ||||||
| ER-I | LR-III | 7.00 | 3.00 | 4.61 ± 0.72 | 38.61 | 67.0 |
| ER-II | LR-I | 3.71 | 2.00 | 3.09 ± 0.37 | 36.24 | 77.0 |
| LR-III | 4.50 | 3.00 | 4.13 ± 0.56 | 43.09 | 80.0 | |
| ER-III | LR-IV | 5.00 | 2.25 | 3.55 ± 0.51 | 36.61 | 69.0 |
| LR-V | 5.50 | 3.35 | 4.92 ± 0.63 | 33.33 | 70.0 | |
| LR-VI | 7.00 | 3.50 | 5.62 ± 0.73 | 59.58 | 68.0 | |
| ER-IV EI | LR-II | 6.00 | 3.00 | 5.07 ± 0.69 | 44.58 | 81.48 |
| LR-III | 8.00 | 4.00 | 6.30 ± 0.68 | 35.24 | 81.16 | |
| ER-IV EII | LR-I | 4.80 | 2.60 | 3.36 ± 0.50 | 26.53 | 37.10 |
| LR-II | 5.60 | 3.40 | 4.31 ± 0.45 | 21.51 | 52.60 | |
| LR-III | 6.93 | 3.50 | 5.34 ± 0.57 | 21.02 | 47.61 | |
| ER-V | IP | 9.00 | 18.00 | 10.42 ± 0.94 | 15.83 | 34.0 |
| LP | 11.00 | 25.00 | 15.91 ± 1.46 | 26.02 | 75.0 | |
| IT | 1.00 | 4.00 | 2.55 ± 0.28 | 47.84 | 89.0 | |
aEnvironments and stages are abbreviated for LLS as EL-I Rainy 2004, EL-II Rainy 2005, EL-III Rainy 2006, LLS-I 70 days to score, LLS-II 90 days to score, and for rust as ER-I Rainy 2004, ER-II Rainy 2005, ER-III Post-rainy 2007, ER-IV Rainy 2007, ER-V Rainy 2008, EI experiment I, EII experiment II, IP incubation period in (days), LP latent period (in days), IT infection type (in days)
The phenotypic coefficient of variation (PCV) estimates for LLS were high at LLS-I compared to LLS-II (Table 1). High PCV (21.71–33.55%) at different stages and environments revealed substantial variation for LLS in the RIL population. The heritability ranged from 40.87 to 82.81%. It was moderate to high with very high heritability observed in the EL-II at both stages of LLS. The PCV for rust was high at later (LR-II and LR-III) stages. Moderate to high PCV (15.83–59.58%) was observed at different stages of rust. High to very high heritability (34.0–89.0%) was observed in different screening environments (Table 1).
Detailed analysis of these data sets showed highly significant and positive correlation between stages in each of the environment for LLS (r = 0.37–0.87; P < 0.01) and rust (r = 0.37–0.98; P < 0.01). The correlations were high even across the environments in LLS, but negative correlation was observed between LLS and rust (data not shown).
Molecular marker and linkage map analysis
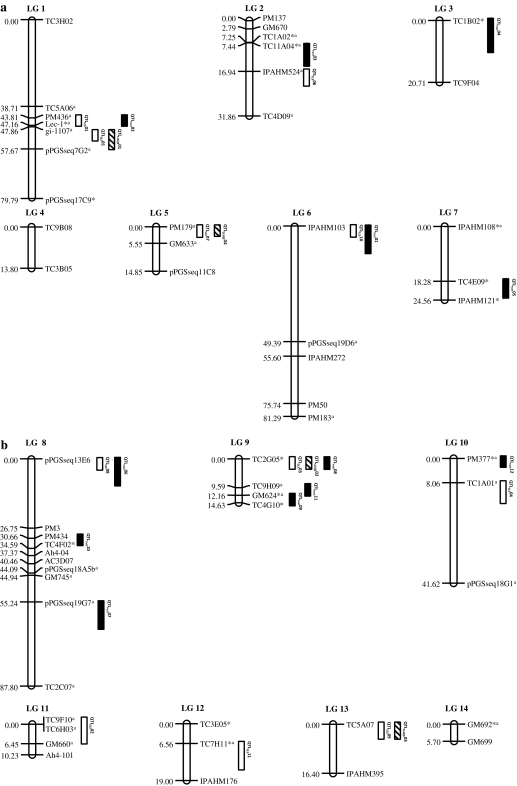
A total of 1089 SSR markers, as mentioned in Varshney et al. (2009), were screened on two parental genotypes of the mapping population of which 907 markers showed high quality amplification in both genotypes. Of these 907 markers, only 67 SSR markers showed polymorphism between parental genotypes. Subsequently, genotyping data were obtained on the complete set of 268 RILs for 67 markers. Chi-square (χ2) test was performed on the genotypic data to test the null hypothesis for the expected 1:1 Mendelian segregation on all the scored markers. Of these, 20 markers (29.85%) showed segregation distortion (SD). Due to the limited number of polymorphic markers, the distorted markers were also used for linkage map construction. Linkage map analysis of 67 markers assigned a total of 56 markers to 14 linkage groups (LGs) spanning 462.24 cM with an average marker interval of 8.25 cM (Fig. 1). Eleven SSR markers, however, remained unlinked. The number of markers mapped per linkage group ranged from two (LG 3, LG 4, LG 13 and LG 14) to ten (LG 8). The lengths of linkage groups ranged from 5.70 cM (LG 14) to 87.80 cM (LG 8).
Fig. 1.
Genetic linkage map based on TAG 24 × GPBD 4 population showing QTL positions for LLS and rust. Seven linkage groups each have been shown in a (LG 1–LG 7) and b (LG 8–LG 14). Asterisk represents the markers showing segregation distortion. Distorted markers are indicated with suffix “a” indicating markers from tetraploid reference map (Varshney et al. 2009). Numbers on the left of each linkage group are Kosambi map distances. On the right hand side of the linkage groups, QTLs for LLS and rust, as mentioned in Tables 2, 3 and 4, have been shown using the following boxes: Open rectangle indicates QTL for LLS; rectangle with diagonal lines indicates QTL across the environment for LLS; closed rectangle indicates QTL for rust
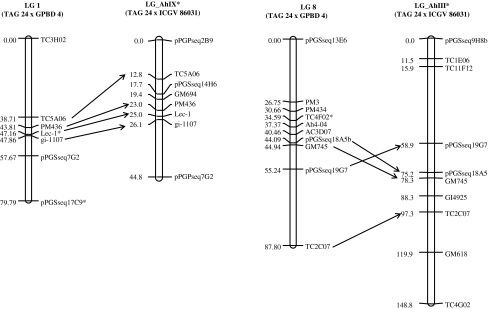
The linkage map obtained in the present study was also compared with the reference map for cultivated groundnut species developed based on mapping population TAG 24 × ICGV 86031 (Varshney et al. 2009). Comparison of these two maps showed 28 common markers on 13 linkage groups. The order of these common markers is congruent in the majority of the cases, except for four markers that were mapped on different linkage groups (Fig. 2; ESM 2).
Fig. 2.
Comparison of two linkage groups of the developed map with the tetraploid reference map (Varshney et al. 2009). LG1 and LG 8 of TAG 24 × GPBD 4 aligned through common markers with LG_AhIX and LG_AhIII of TAG 24 × ICGV86031, respectively. These common markers in both linkage maps are in the same order
Identification of QTLs for LLS resistance
Mapping data for all 56 SSR markers assigned to the genetic map were analyzed together with phenotypic data obtained at both stages (LLS-I and LLS-II) in all three environments (Rainy 2004, Rainy 2005 and Rainy 2006) by using PLABQTL version 1.1 (Utz and Melchinger 1996). As a result, a total of 11 QTLs were detected on ten linkage groups at LLS-I and LLS-II stages in all three environments (Table 2). However, all these QTLs were minor as all of them explained <10% PV (Collard et al. 2005).
Table 2.
Features of QTLs for late leaf spot (LLS) identified in TAG 24 × GPBD 4 population
| QTL | Marker intervalA | LG | Position (cM) | LOD | Phenotypic variation (R2P, %) | Additive effect |
|---|---|---|---|---|---|---|
| QTLLLS01 | PM436-Lec-1ab | 1 | 44–46 | 2.66–3.32 | 3.70-6.50 | −0.241 to −0.342 |
| QTLLLS02 | TC9F10-GM660ab | 11 | 0 | 2.65–2.80 | 3.10-4.40 | −0.260 to −0.208 |
| QTLLLS03 | TC2G05-TC9H09bdef | 9 | 0 | 2.65–5.11 | 3.80-4.80 | −0.213 to −0.273 |
| QTLLLS04 | TC1A01-pPGSseq18G1b | 10 | 12 | 5.11 | 1.80 | −0.193 |
| QTLLLS05 | gi-1107-pPGSseq7G2 cd | 1 | 48–50 | 2.89–2.96 | 1.70-2.90 | −0.163 to −0.223 |
| QTLLLS06 | IPAHM524-TC4D09 cd | 2 | 18 | 4.70–6.03 | 2.90-4.40 | −0.231 to −282 |
| QTLLLS07 | PM179-GM633cdf | 5 | 0–4 | 2.64–6.03 | 3.00-4.80 | −0.219 to −286 |
| QTLLLS08 | pPGSseq13E6-PM3c | 8 | 2 | 2.61 | 4.90 | −0.325 |
| QTLLLS09 | TC5A07-IPAHM395d | 13 | 0 | 5.68 | 4.30 | −0.256 |
| QTLLLS10 | IPAHM103-pPGSseq19D6f | 6 | 0 | 2.95 | 2.60 | 0.209 |
| QTLLLS11 | TC7H11-IPAHM176f | 12 | 18 | 2.79 | 2.00 | −0.183 |
ASuperscripts on group of markers associated with QTLs represent environment and stages as follows: aLLS-I EL-I; bLLS-II EL-I; cLLS-I EL-II; dLLS-II EL-II; eLLS-I EL-III; fLLS-II EL-III; details about these abbreviations are given in “Material and methods” as well as in Table 1
Four QTLs were detected for LLS-I (resistance values scored at 70 days after sowing) and LLS-II (resistance values were scored at 90 days after sowing) stages in the EL-I environment (Rainy 2004) on linkage groups LG1, LG9, LG10 and LG11. Of these, two QTLs (QTLLLS01, QTLLLS02) were found to be common at both the stages (LLS-I and LLS-II). However, these QTLs showed 3.70 and 3.10% PVE in LLS-I and 6.50 and 4.40% PVE in LLS-II. In the EL-II environment (Rainy 2005), a total of six QTLs were detected for both stages (LLS-I and LLS-II) on linkage groups LG1, LG2, LG5, LG8, LG9 and LG13 with PVE ranging from 1.70 to 4.90%. Three QTLs (QTLLLS05, QTLLLS06, QTLLLS07) were co-localized at both the stages having 1.70–2.90, 2.90–4.40 and 3.00–4.80% PVE. In the case of the third environment (EL-III), QTL analysis revealed one QTL (QTLLLS03) for LLS-I and accounted for 3.90% PVE. Four QTLs (QTLLLS03, QTLLLS07, QTLLLS10, QTLLLS11) were detected for LLS-II and revealed 2.00–4.30% PVE. One QTL (QTLLLS03) was co-mapped in LLS-I and LLS-II with 3.90–4.30% PVE. Interestingly, a QTL (QTLLLS03) was found for LLS-II in all the three screening environments (EL-I, EL-II and EL-III). This QTL was detected at LOD values ranging from 2.65 to 5.11 with 3.80–4.80% PVE. The resistant alleles for all QTLs, except one detected at LLS-II (QTLLLS10), were contributed by the resistant parent, GPBD 4.
For analyzing QTL × E interactions, data from individual environment and means across the environments (EL-I, EL-II and EL-III) for LLS-I and LLS-II were used with mapping data and a significant QTL × E interaction was observed at LLS-I (3.19; P < 0.01) and LLS-II (8.09; P < 0.01) stages of scoring (data not shown). While three QTLs were observed for averaged data of LLS-I at LOD value ranging from 2.50 to 3.51 with 1.70 to 2.20% PVE, 4 QTLs were observed for averaged data for LLS-II. However, three QTLs (QTLLLSQE01, QTLLLSQE02, QTLLLSQE03) identified in LLS-II were co-mapped with the same QTLs identified for LLS-I. Here again, the favorable alleles for resistance to LLS in all QTLs came from the resistant parent GPBD 4 (Table 3).
Table 3.
Identification of QTLs across environments for LLS in TAG 24 × GPBD 4 population
| Scoring stage | QTL | LG | Marker interval | Position (cM) | LOD value | Phenotypic variation (R2P, %) | Additive effect |
|---|---|---|---|---|---|---|---|
| LLS-I | QTLLLSQE01 | 1 | gi-1107-pPGSseq7G2 | 56 | 3.47 | 1.70 | −0.134 |
| QTLLLSQE02 | 9 | TC2G05-TC9H09 | 0 | 3.51 | 2.20 | −0.145 | |
| QTLLLSQE03 | 13 | TC5A07-IPAHM395 | 0 | 2.50 | 1.90 | −0.132 | |
| LLS-II | QTLLLSQE01 | 1 | gi1107-pPGSseq7G2 | 54 | 4.04 | 3.20 | −0.209 |
| QTLLLSQE04 | 5 | PM179-GM633 | 2 | 2.74 | 2.60 | −0.176 | |
| QTLLLSQE02 | 9 | TC2G05-TC9H09 | 0 | 5.22 | 5.70 | −0.258 | |
| QTLLLSQE03 | 13 | TC5A07-IPAHM395 | 0 | 2.68 | 2.10 | −0.155 |
−ve sign indicates that favorable allele has come from resistant parent GPBD 4
+ve sign indicates that favorable allele has come from susceptible parent TAG 24
Identification of QTLs for rust
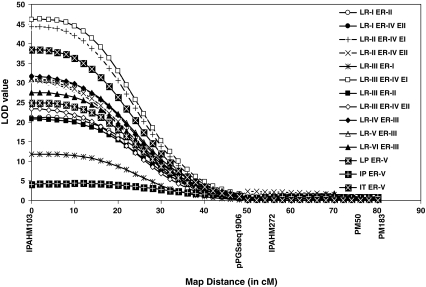
QTL analysis for rust data for a total of six stages, i.e., 70 days (LR-I), 80 days (LR-II), 90 days (LR-III), 105 days (LR-IV), 113 days (LR-V) and 120 days (LR-VI) after sowing collected under five different environments, i.e., Rainy 2004 (ER-I), Rainy 2005 (ER-II), Post-rainy 2007 (ER-III), Rainy 2007 (ER-IV) and Rainy 2008 (ER-V) revealed a total of 12 putative QTLs on 8 different linkage groups (Table 4). Of these, 11 QTLs had small effects with PVE ranging from 1.70 to 7.00% on seven different linkage groups. However, one major QTL (QTLrust01) was detected at an LOD score ranging from 4.35 to 44.32 on linkage group 6 contributing 6.90–55.20% PVE (Fig. 3).
Table 4.
Features of QTLs for rust identified in the TAG 24 × GPBD 4 population
| QTL | Marker intervalA | LG | Position (cM) | LOD | Phenotypic variation (R2P, %) | Additive effectB |
|---|---|---|---|---|---|---|
| QTLrust01 | IPAHM103-pPGSseq19D6abcdefghijklmn | 6 | 0–12 | 4.35–44.32 | 6.90–55.20 | 0.522 to −1.718 |
| QTLrust02 | PM436-Lec-1bc | 1 | 46 | 3.22–3.51 | 2.20–4.50 | 0.143–0.323 |
| QTLrust03 | TC11A04-IPAHM524b | 2 | 16 | 3.51 | 2.10 | 0.148 |
| QTLrust04 | TC1B02-TC9F04bcdefjkl | 3 | 0–14 | 2.86–4.91 | 1.70–5.20 | 0.143–0.328 |
| QTLrust05 | TC4E09-IPAHM121i | 7 | 24 | 2.59 | 2.60 | −0.105 |
| QTLrust06 | pPGSseq13E6-PM3j | 8 | 20 | 4.24 | 4.90 | −0.372 |
| QTLrust07 | pPGSseq19G7-TC2C07i | 8 | 76 | 3.15 | 2.00 | 0.120 |
| QTLrust08 | TC2G05-TC9H09i | 9 | 2 | 3.09 | 2.30 | 0.145 |
| QTLrust09 | GM624-TC4G10dei | 9 | 14 | 2.94–3.87 | 2.80–7.00 | −0.208 to −0.247 |
| QTLrust10 | PM434-TC4F02j | 8 | 4 | 3.16 | 6.80 | 0.355 |
| QTLrust11 | TC9H09-GM624k | 9 | 12 | 2.80 | 6.00 | −0.199 |
| QTLrust12 | PM377-TC1A01l | 10 | 0 | 2.53 | 3.90 | 0.259 |
ASuperscripts on group of markers associated with QTLs represent environment and stages as follows: aLR-III ER-I; bLR-I ER-II; cLR-III ER-II; dLR-IV ER-III; eLR-V ER-III; fLR-VI ER-III; gLR-II ER-IV EI; hLR-III ER-IV EI; iLR-I ER-IV EII; jLR-II ER-IV EII; kLR-III ER-IV EII; lIP ER-V; mLP ER-V; nIT ER-V; details of these abbreviations are given in “Materials and methods”as well as Table 1
B−ve sign indicates that the favorable allele has come from resistant parent GPBD 4 except for IP and LP; +ve sign indicates that the favorable allele has come from the susceptible parent TAG 24
Fig. 3.
A snapshot showing the occurrence of a major QTL (QTLrust01) on LG 6 for rust based on phenotyping data obtained for different stages of infection under five environments. Different environments and stages of phenotyping in the figure have been abbreviated as follows. Environments: ER-I Rainy 2004, ER-II Rainy 2005, ER-III Post-rainy 2007, ER-IV Rainy 2007, ER-V Rainy 2008. Stages: LR-I phenotype scored at 70 days after sowing (DAS), LR-II phenotype scored at 80 DAS, LR-III phenotype scored at 90 DAS, LR-IV phenotype scored at 105 DAS, LR-V phenotype scored at 113 DAS, LR-VI phenotype scored at 120 DAS, IP incubation period, LP latent period, IT infection type. X-axis shows the linkage group 6 (LG 6) with relative position of different markers and Y-axis shows the LOD value for which QTL for rust has been detected for the above-mentioned environment/stage
In case of Rainy 2004 environment (ER-I), one QTL (QTLrust01) was detected for LR-III with 18.40% PVE. Similarly in the case of Rainy 2005 environment (ER-II), four QTLs were detected for LR-I and LR-III with 2.10–30.50% PVE. Among them, three QTLs (QTLrust01, QTLrust02 and QTLrust04) were common between LR-I and LR-III stages of rust. In case of Post-rainy 2007 environment (ER-III), three QTLs were identified for LR-IV, LR-V and LR-VI with 2.80–36.80% PVE. Interestingly, two QTLs (QTLrust01, QTLrust04,) were co-mapped in all three different stages with PVE ranging from 33.80–36.80% and 3.10–5.20%. In Rainy 2007 environment (ER-IV), nine QTLs were identified for LR-I, LR-II and LR-III stages of rust with 1.70–55.20% PVE. One QTL (QTLrust01) was co-mapped with LR-II and LR-III of EI accounted for 52.70–55.20% PVE and all three stages of EII contributed 35.20–43.50% PVE. In case of Rainy 2008 (ER-V) season, three QTLs (QTLrust01, QTLrust04, QTLrust12) were identified with PVE ranging from 2.70 to 48.00% and one QTL (QTLrust01) was present in the three components of rust. The resistant allele is contributed by the resistant parent GPBD 4 in five QTLs (QTLrust01, QTLrust05, QTLrust06, QTLrust09 and QTLrust11) and TAG 24 for seven QTLs (QTLrust02, QTLrust03, QTLrust04, QTLrust07, QTLrust08, QTLrust10 and QTLrust12).
Validation of a candidate marker: IPAHM 103 for molecular breeding of rust
As IPAHM 103 marker was found associated with the major QTL (QTLrust01), identified through CIM analysis, contributing up to 55.20% PV, this marker seems to be a candidate marker for deployment in MAS for molecular breeding, though one of two fragments amplified by this marker behaves as a dominant marker. Therefore, the potential and utility of this candidate marker IPAHM 103 was further validated using a set of resistant and susceptible germplasm lines with different genetic background and another mapping population (TG 26 × GPBD 4) segregating for rust resistance. In this direction, a set of 46 genotypes included released varieties, hybrid derivatives from NcAc (North Carolina Accessions), interspecific derivatives, mutant lines, cultivars from South American landraces and advanced breeding lines were tested. Phenotyping data for rust resistance were already available for these lines. Allelic data for the marker IPAHM 103 were obtained on these genotypes in this study. Based on the phenotypic and genotypic data, distinct marker profiles for these candidate markers were observed for resistant and susceptible lines for rust except in the case of six genotypes (VR5, MN 1-28, Girnar 1, ICGV 86590, ICGV 87921, ICGV 86156) (ESM 1).
A new mapping population (TG 26 × GPBD 4) that segregates for rust was also used for validation of the candidate marker. Allelic data were obtained for 53 markers including IPAHM 103 for all 146 RILs of this mapping population. Phenotyping data for all these 146 RILs were already available. Genotyping data and phenotyping data were subjected to CIM and SMA analyses. In these analyses, the marker IPAHM 103 was found as the nearest marker for the QTL that contributes 24.10–48.90% PV in CIM analysis and 27.98–51.96% PV in SMA. Single marker analysis and CIM yielded similar results in both TAG 24 × GPBD 4 and TG 26 × GPBD 4 populations (Table 5).
Table 5.
Estimates of phenotypic variation for IPAHM 103 using single marker analysis (SMA) and composite interval mapping (CIM) analysis in TAG 24 × GPBD 4 and TG 26 × GPBD 4 mapping populations
| Environmenta | Stages of Scoring | SMA (R2P, %) | CIM (R2P, %) |
|---|---|---|---|
| Mapping population-TAG 24 × GPBD 4 | |||
| ER-I | LR-III | 19.11 | 18.40 |
| ER-II | LR-I | 28.07 | 30.50 |
| LR-III | 28.35 | 28.70 | |
| ER-III | LR-IV | 39.46 | 36.60 |
| LR-V | 39.45 | 36.80 | |
| LR-VI | 34.04 | 33.80 | |
| ER-IV EI | LR-II | 53.95 | 52.50 |
| LR-III | 54.42 | 55.20 | |
| ER-IV EII | LR-I | 36.59 | 39.10 |
| LR-II | 40.58 | 43.50 | |
| LR-III | 33.80 | 35.20 | |
| ER-V | IP | 7.91 | 6.90 |
| LP | 34.88 | 34.70 | |
| IT | 48.91 | 48.00 | |
| Mapping population-TG 26 × GPBD 4 | |||
| ER-II | LR-I | 32.79 | 32.30 |
| LR-III | 27.98 | 28.90 | |
| ER-III | LR-I | 33.01 | 32.70 |
| LR-III | 37.49 | 36.70 | |
| LR-IV | 32.64 | 31.10 | |
| ER-IV EI | LR-I | 24.86 | 24.10 |
| LR-III | 38.50 | 35.80 | |
| LR-IV | 36.86 | 35.10 | |
| ER-IV EII | LR-I | 49.39 | 46.10 |
| LR-III | 51.96 | 48.90 | |
aAbbreviation for different environments (ER) are given in “Materials and methods” as well as in Table 1; EI experiment I, EII experiment II, IP incubation period, LP latent period, IT infection type
Discussion
QTL detection mainly depends on the biometrical methods to analyze, the type of mapping population, marker density, population size and heritability of traits (Melchinger 1998). QTL analysis in groundnut was mostly thwarted in past because of the less number of SSR markers available in the public domain and lack of suitable mapping population with sufficient molecular and trait diversity. However, recent efforts at the international level have facilitated development of a large number of SSR markers. Similarly, a few groups have developed some good mapping populations. As a result, linkage mapping and QTL detection have now become possible in cultivated groundnut (Varshney et al. 2009).
Identification of resistant breeding material to major foliar diseases is one of the challenging objectives of groundnut breeders. The simultaneous occurrence of LLS and rust, and the dominating and defoliating nature of LLS make the visual selection for rust resistance very difficult. Mapping of resistance genes to these diseases, therefore, is very important because resistance is quantitative in nature and governed by recessive genes. Therefore, the present study was conducted to determine the location and effects of QTLs for LLS and rust and identify the diagnostic marker(s) for deployment in breeding.
Phenotypic evaluation
The mapping population consisting of 268 RILs exhibited significant variation in the resistance to LLS and rust. The magnitude of variation was moderate to high as revealed by phenotypic coefficient of variation and with high to very high heritable variation. High positive correlation between disease scores at different stages and across environments revealed consistency in disease development in different lines. The near normal to normal distribution revealed the quantitative nature of resistance. Precise phenotyping of traits is one of the paramount factors in QTL detection strategies. Since resistance to LLS and rust is complex with several components of resistance (Nevill 1982; Green and Wynne 1986, 1987; Motagi 2001; Dwivedi et al. 2002), the power of QTL detection could be increased by phenotyping the mapping population for the components of resistance such as: incubation period, latent period, lesion size and lesion on the main stem for LLS; number of pustule and pustule diameter for rust.
Polymorphism assessment and segregation distortion
Although a variety of marker systems, e.g., RFLP (Halward et al. 1991) and RAPDs (Kochert et al. 1991), were tested for DNA polymorphism in cultivated groundnut lines, a very low level of polymorphism was observed that was mainly ascribed to the origin of cultivated groundnut by a single event of polyploidization and further isolation from wild relatives (Halward et al. 1991; Young et al. 1996). SSR markers, due to their multi-allelic nature on the other hand showed a relatively better polymorphism in cultivated groundnut (He et al. 2003, 2005; Mace et al. 2006; Nimmakayala et al. 2007; Hong et al. 2008; Varshney et al. 2009). However, even by using a large number (1,089) of SSR markers, only 6.15% markers showed polymorphism between the parents, TAG 24 and GPBD 4. The low level of polymorphisms obtained in this set of genotypes was not unexpected as other diversity and mapping studies showed similar kind of results (Moretzsohn et al. 2004, 2005; Ferguson et al. 2004; Varma et al. 2005, Varshney et al. 2009). To achieve higher number of marker polymorphism between parental genotypes for developing good genetic maps, it would be desirable to use either more appropriate marker systems such as SNPs or genetically diverse genotypes for developing mapping population (see Paterson et al. 2004).
Out of 67 markers for which genotyping data were obtained, 20 (29.8%) showed SD that was relatively less as compared to earlier mapping studies such as Burow et al. 2001 (68%), Moretzsohn et al. 2005 (51%) and Varshney et al. 2009 (35%). In general, higher SD is obtained in the mapping populations developed from highly diverse genotypes with less genome similarities, e.g., cultivated × wild/synthetic genotypes. Therefore, relatively less SD can be attributed to low diversity nature of the parental genotypes (TAG 24 and GPBD 4) or to the use of a large size of the mapping population (268 RILs) employed in the present study. However, it is important to note that the use of distorted markers may affect the estimation of map distances and the order of markers.
Genetic map and its comparison with the reference map
Due to low level of polymorphism, as mentioned earlier, there has been slow progress in developing genetic maps in groundnut and especially in cultivated groundnut. The present study presents a genetic map with 56 SSR marker loci and 14 linkage groups (LGs) that span 462.24 cM with an average marker interval of 8.25 cM. The map coverage is much lower than diploid maps developed by Moretzsohn et al. 2005 (1,230.89 cM) and Gobbi et al. 2006 (754.8 cM), but the tetraploid maps, as compared to diploid maps, are of direct significance to genetic improvement of cultivated groundnut. Therefore, this map was compared with other tetraploid maps and, in terms of marker density, the present map was found to be superior to the AFLP map of Herselman et al. 2004 (139.4 cM; 5 LGs), but less dense than the RFLP map of Burow et al. 2001 (2,210 cM; 23 LGs); Hong et al. 2008 (679 cM, 20 LGs) and Varshney et al. 2009 (1,270.5 cM, 22 LGs). It is also important to mention that although some genetic maps were developed in past, these were not used to identify QTLs associated with biotic stresses.
The developed map in this study based on the RIL population (TAG 24 × GPBD 4) was compared in detail with the SSR-based genetic map developed based on the RIL population (TAG 24 × ICGV 86031) of Varshney et al. (2009). Comparison of these maps revealed only 28 common markers on these two maps. In general, a good congruence was observed in marker orders on these maps. For instance, LG_AhIX (TAG 24 × ICGV 86031) with LG 1 (TAG 24 × GPBD 4), LG_AhIV with LG 2 and LG_AhIII with LG 8 showed good congruence (Fig. 2; ESM 2). These analyses indicate the possibility of preparing a combined (consensus) genetic linkage map of groundnut with higher marker density, if both the maps are saturated with a sufficient number of common markers.
QTLs for LLS and leaf rust
As the expression of quantitative traits is influenced by environment, QTLs identified for the trait may vary for different environments in which the trait is phenotyped. The present study revealed 11 QTLs in the individual environment and four QTLs across environments associated with resistance to LLS. However, all these QTLs were minor as they contributed 1.70–6.50% of the phenotypic variation. Similarly, in case of rust, a total of 12 QTLs were detected and the majority of them were minor QTLs (1.70–7.00% PVE). However, one major QTL (QTLrust01) contributed a considerable amount of variation toward the total PVE (6.90–55.20%). The results of the present study reconfirms that the genetics of resistance to rust and rust components are complex and controlled by both major and minor QTLs (Motagi 2001; Dwivedi et al. 2002). However, it was intriguing to find a major QTL (QTLrust01), which explained as large as 55.20% of PVE and/or detected at high LOD scores (up to 44.32) and was also consistent over environments. The presence of major QTL accompanied by minor QTLs appears to be a common phenomenon in disease resistance studies (George et al. 2003; Nair et al. 2005; Welter et al. 2007; Shuancang et al. 2009).
As mentioned above, the majority of QTLs identified were minor QTLs that were prone to inconsistency. In addition to the environmental conditions that vary according to seasons, size of the population (Miklas et al. 2001), low density of marker loci, incomplete marker genotyping data and genotyping/phenotyping errors may be attributed for the appearance of small effect QTLs. Such inconsistencies have been found in other studies, which report identification of small and medium effect QTLs, specific to the screening environment (Ender and Kelly 2005).
In some cases, co-mapping of a few QTLs was observed. For instance, six QTLs were co-mapped for LLS-I and LLS-II in one or the other environments (QTLLLS01, QTLLLS02, QTLLLS03, QTLLLS05, QTLLLS06, QTLLLS07, 1.70–6.50% PVE) and one QTL (QTLLLS03, 3.80–4.80% PVE) was observed across the environments. In the case of rust, four QTLs were co-mapped at one or the other stages, but two QTLs (QTLrust01, QTLrust04) were consistently present in all the three different stages and in the majority of the environments. These consistent QTLs can be a future target for MAS and candidate gene approach, but map saturation can enhance the magnitude of effects of these QTLs. Further, such QTLs can be validated and used in marker-assisted breeding.
Parallelism in the QTLs conferring resistance to LLS and rust
Although no phenotypic correlation was found between LLS and rust phenotyping data, QTL analysis revealed four common QTLs (QTLLLS01 and QTLrust10, QTLLLS03 and QTLrust08, QTLLLS08 and QTLrust06, QTLLLS10 and QTLrust01) conferring resistance to both diseases. It is noteworthy here that these are mainly small effects QTLs, except for QTLrust01 that contributes 1.70–7.00% PVE. It is quite possible that saturation of map can yield common QTL with large effect that may be of interest to breeders.
Classical genetic analyses, conducted in the past on LLS and rust, indicated multiple recessive genes governing resistance in LLS (Sharief et al. 1978; Nevill 1982) and as few recessive genes confer rust resistance (Kalekar et al. 1984; Knauft 1987; Paramasivam et al. 1990). In the present investigation, near normal to normal phenotypic distribution of RILs and many small effects QTLs (1.70–6.50%) identified for LLS indicate governance of LLS resistance by many and small effect loci. Similarly in the case of rust, normal phenotypic distribution of RILs, small effect QTLs and one major QTL (QTLrust01) contributing as much as 55.20% PV indicate the possibility of oligogenes, as well as additional minor genes, controlling the resistance or environmental variation, affecting disease development. Hence, the results of the present study corroborate that the genetics of these traits is complex in nature and is controlled by few major and large number of minor genes.
Validation of IPAHM 103 marker allele with respect to resistance
The present study was undertaken to identify the QTLs for LLS and rust resistance, so that molecular markers associated with such QTLs can be used in MAS to accelerate disease resistance breeding. Although several QTLs were identified in this study, only one major QTL (QTLrust01) was identified for resistance to rust that can be used in MAS. Therefore, validation of the marker IAPHM 103 was undertaken in different genetic background with an objective to test the reliability of the marker. For this purpose, a variety of highly diverse germplasm lines derived from multi-crosses and multi-parent were used to check the presence of resistance allele in resistant genotypes and vice versa. IPAHM 103 was strongly associated with resistance in the majority of cases (hybrid derivatives from NcAcs, interspecific derivatives, mutant lines) except in few (VR 5, MN 1-28, Girnar 1, ICGV 86590, ICGV 87921, ICGV 86156) germplasm lines. Intriguingly, the appearance and disappearance of one of two fragments amplified by the marker IPAHM 103 with resistance and susceptibility indicate the possibility of association of the marker with the resistance gene itself. For instance, VL 1 and VL 2 (resistance to rust) are mutants derived from Dharwad Early Runner (DER, susceptible to rust). When VL 1 was mutated using ethyl methane sulfonate (EMS), M 28-2, VB 9, Mutant 110 and Mutant 45 were obtained but with susceptible rust reaction (ESM 1). Similarly, the presence and absence of the candidate fragment amplified by IPAHM 103 marker were observed with respect to rust reaction in DER based mutants.
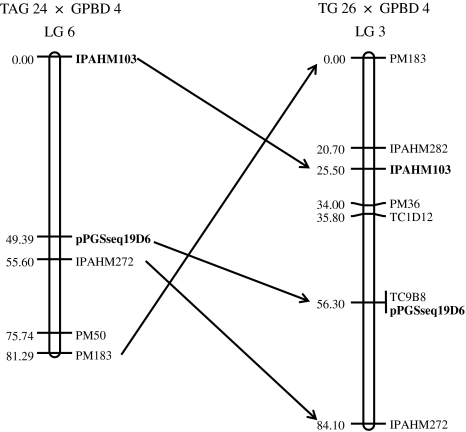
Validation of the nearest marker IPAHM 103 in germplasm lines of different genetic background and mapping population indicated that it could be directly used for marker-assisted breeding for rust resistance in groundnut. QTL analysis provides putative location of potential genomic regions. Therefore, further efforts in fine mapping and map-based cloning of this locus are necessary to study candidate resistance genes and cellular pathways involved in resistance. A major QTL (QTLrust01) for resistance to rust was mapped on LG6, which was further compared with the map derived from another mapping population (TG 26 × GPBD 4). The order and orientation of anchor markers were congruent, but some more additional markers were added between IPAHM 103 and pPGSseq19D6 (Fig. 4).
Fig. 4.
Comparative mapping of IPAHM 103, diagnostic marker for rust QTL (QTLrust01) and other linked markers between TAG 24 × GPBD 4 and TG 26 × GPBD 4 mapping populations. Three markers namely IPAHM 103, pPGSseq19D6 and IPAHM 272 show collinearity between two maps. Furthermore, three additional markers, namely PM36, TC1D12 and TC9B8 have been integrated in the QTL (QTLrust01) region in TG 26 × GPBD 4 mapping population
Conclusions
The present study yielded partial linkage map of groundnut and QTLs for LLS and rust. Though QTLs for LLS are minor (<10% PVE), a major QTL explaining the quantum of phenotypic variation as much as 55.20% for the QTL of rust has been reported here. Thus, the present report of a QTL is the first of its kind. However, further studies are needed to narrow down the marker interval of the identified QTL by saturating the genetic map using more polymorphic markers.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
Authors are thankful to the National Fund of the Basic and Strategic Research in Agriculture (NFBSRA) of the Indian Council of Agriculture (ICAR-RKV, MVCG), Department of Biotechnology (DBT) of the Government of India (MVCG, RKV) and the Tropical Legume I of Generation Challenge Programme (RKV) for financial support to undertake this study. Thanks are also due to Dr T. Nepolean of the Indian Agricultural Research Institute (IARI), New Delhi, Dr P. L. Kulwal of Mahatma Phule Krishi Vidyapeeth (MPKV), Rahuri and the two anonymous reviewers for their valuable suggestions.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Anderson WF, Wynne JC, Green CC. Potential for incorporation of early and late leaf spot resistance in peanut. Plant Breed. 1986;97:163–170. doi: 10.1111/j.1439-0523.1986.tb01047.x. [DOI] [Google Scholar]
- Burow MD, Simpson CE, Starr JL, Paterson AH. Transmission genetics of chromatin from a synthetic amphidiploid to cultivated peanut (Arachis hypogaea L.): broadening the gene pool of a monophyletic polyploid species. Genetics. 2001;159:823–837. doi: 10.1093/genetics/159.2.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collard BCY, Jahufer MZZ, Brouwer JB, Pang ECK. An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: the basic concepts. Euphytica. 2005;142:169–196. doi: 10.1007/s10681-005-1681-5. [DOI] [Google Scholar]
- Cuc LM, Mace ES, Crouch JH, Quang VD, Long TD, Varshney RK. Isolation and characterization of novel microsatellite markers and their application for diversity assessment in cultivated groundnut (Arachis hypogaea) BMC Plant Biol. 2008;8:55–65. doi: 10.1186/1471-2229-8-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi SL, Pande S, Rao JN, Nigam SN. Components of resistance to late leaf spot and rust among interspecific derivatives and their significance in a foliar disease resistance breeding in groundnut (Arachis hypogaea L.) Euphytica. 2002;125:81–88. doi: 10.1023/A:1015707301659. [DOI] [Google Scholar]
- Ender M, Kelly JD. Identification of QTL associated with white mold resistance in common bean. Crop Sci. 2005;45:2482–2490. doi: 10.2135/cropsci2005.0064. [DOI] [Google Scholar]
- FAO (2007) Food and Agricultural Organization of the United Nation, FAO Statistical Database (http://faostat.fao.org/faostat/collections?subset=agriculture)
- Ferguson ME, Burow MD, Schulze SR, Bramel PJ, Paterson AH, Kresovich S, Mitchell S. Microsatellite identification and characterization in peanut (A. hypogaea L.) Theor Appl Genet. 2004;108:1064–1070. doi: 10.1007/s00122-003-1535-2. [DOI] [PubMed] [Google Scholar]
- Gautami B, Ravi K, Lakshmi Narasu M, Hoisington DA, Varshney RK (2009) Novel set of groundnut SSRs for genetic diversity and interspecific transferability. Int J Integr Biol (IJIB) 7:100–106
- George ML, Prasanna BM, Rathore RS, Setty TA, Kasim F, Azrai M, Vasal S, Balla O, Hautea D, Canama A, Regalado E, Vargas M, Khairallah M, Jeffers D, Hoisington D. Identification of QTLs conferring resistance to downy mildews of maize in Asia. Theor Appl Genet. 2003;107:544–551. doi: 10.1007/s00122-003-1280-6. [DOI] [PubMed] [Google Scholar]
- Gobbi A, Teixeira C, Moretzsohn M, Guimaraes P, Leal-Bertioli S, Bertioli D, Lopes CR, Gimenes M (2006) Development of a-linkage map to species of B genome related to the peanut (Arachis hypogaea–AABB). In: XIV International Plant and Animal Genome conference, San Diego, CA, USA, pp 679
- Gowda MVC, Motagi BN, Naidu GK, Diddimani SB, Sheshagiri R. GPBD 4: A Spanish bunch groundnut genotype resistant to rust and late leaf spot. Int Arachis Newsl. 2002;22:29–32. [Google Scholar]
- Green CC, Wynne JC. Diallel and generation means analyses for the components of resistance to Cercospora arachidicola in peanut. Theor Appl Genet. 1986;73:228–235. doi: 10.1007/BF00289279. [DOI] [PubMed] [Google Scholar]
- Green CC, Wynne JC. Genetic variability and heritability for resistance to early leaf spot in four crosses of Virginia-type peanut. Crop Sci. 1987;27:18–21. doi: 10.2135/cropsci1987.0011183X002700010005x. [DOI] [Google Scholar]
- Gupta PK, Varshney RK. The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat. Euphytica. 2000;113:163–185. doi: 10.1023/A:1003910819967. [DOI] [Google Scholar]
- Haley CS, Knott SA. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity. 1992;69:315–324. doi: 10.1038/hdy.1992.131. [DOI] [PubMed] [Google Scholar]
- Halward TM, Stalker HT, Larue EA, Kochert G. Genetic variation detectable with molecular markers among unadapted germplasm resources of cultivated peanut and related wild species. Genome. 1991;34:1013–1020. [Google Scholar]
- Hamid MA, Isleib TG, Wynne JC, Green CC. Combining ability analysis of Cercospora leaf spot resistance and agronomic traits in Arachis hypogaea L. Oleagineux. 1981;36:605–612. [Google Scholar]
- He G, Meng R, Newman M, Gao G, Pittman RN, Prakash CS. Microsatellites as DNA markers in cultivated peanut (Arachis hypogaea L.) BMC Plant Biol. 2003;3:3–9. doi: 10.1186/1471-2229-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G, Meng R, Gao H, Guo B, Gao G, Newman M, Pittman RN, Prakash CS. Simple sequence repeat markers for botanical varieties of cultivated peanut (Arachis hypogaea L.) Euphytica. 2005;142:131–136. doi: 10.1007/s10681-005-1043-3. [DOI] [Google Scholar]
- Herselman L, Thwaites R, Kimmins FM, Courtois B, Van Der Merwe PJA, Seal SE. Identification and mapping of AFLP markers linked to peanut (Arachis hypogaea L.) resistance to the aphid vector of groundnut rosette disease. Theor Appl Genet. 2004;109:1426–1433. doi: 10.1007/s00122-004-1756-z. [DOI] [PubMed] [Google Scholar]
- Hong YB, Liang XQ, Chen XP, Liu HY, Zhou GY, Li SX, Wen SJ. Construction of genetic linkage map based on SSR markers in peanut (Arachis hypogaea L.) Agric Sci China. 2008;7:915–921. [Google Scholar]
- Hopkins MS, Casa AM, Wang T, Mitchell SE, Dean RE, Kochert GD, Kresovich S. Discovery and characterization of polymorphic simple sequence repeats (SSRs) in peanut. Crop Sci. 1999;39:1243–1247. doi: 10.2135/cropsci1999.0011183X003900040047x. [DOI] [Google Scholar]
- Jansen RC, Stam P. High resolution of quantitative traits into multiple loci via interval mapping. Genetics. 1994;136:1445–1447. doi: 10.1093/genetics/136.4.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jogloy S, Wynne JC, Beute MK. Inheritance of late leaf spot resistance and agronomic traits in peanut. Peanut Sci. 1987;14:86–90. doi: 10.3146/i0095-3679-14-2-9. [DOI] [Google Scholar]
- Kalekar AR, Patil BC, Deokar AB. Inheritance of resistance to rust in groundnut. Madras Agric J. 1984;71:125–126. [Google Scholar]
- Knauft DA (1987) Inheritance of rust resistance in groundnut. In: McDonald D, Subrahmanyam P, Wightman JA, Mertin JV (eds) Groundnut rust disease. In: Proceedings of discussion group meeting, International Crop Research Institute for the Semi Arid Tropics. Patancheru, India, pp 183–187
- Kochert G, Halward T, Branch WD, Simpson CE. RFLP variability in peanut (Arachis hypogaea L.) cultivars and wild species. Theor Appl Genet. 1991;81:565–570. doi: 10.1007/BF00226719. [DOI] [PubMed] [Google Scholar]
- Kornegay JL, Beute MK, Wynne JC. Inheritance of resistance to Cercospora arachidicola and Cercosporidium personatum in six Viginia-type peanut lines. Peanut Sci. 1980;7:4–9. doi: 10.3146/i0095-3679-7-1-2. [DOI] [Google Scholar]
- Kosambi DD. The estimation of map distances from recombination values. Ann Eugen. 1944;12:172–175. [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburn L. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Leal–Bertioli SCM, Jose ACVF, Alves-Freitas DMT, Moretzsohn MC, Guimaraes PM, Nielen S, Vidigal BS, Pereira RW, Pike J, Favero AP, Parniske M, Varshney RK, Bertioli DJ. Identification of candidate genome regions controlling disease resistance in Arachis. BMC Plant Biol. 2009;9:112–123. doi: 10.1186/1471-2229-9-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln S, Daly M, Lander E (1992) Construction of genetics maps with MAPMAKER/EXP 3.0, 3rd edn. Whitehead Institute Technical Report
- Mace ES, Phong DT, Upadhyaya HD, Chandra S, Crouch JH. SSR analysis of cultivated groundnut (Arachis hypogaea L.) germplasm resistant to rust and late leaf spot diseases. Euphytica. 2006;152:317–330. doi: 10.1007/s10681-006-9218-0. [DOI] [Google Scholar]
- Mace ES, Varshney RK, Mahalakshmi V, Seetha K, Gafoor A, Leeladevi Y, Crouch JH. In silico development of simple sequence repeat markers within the aeshynomenoid/dalbergoid and genistoid clades of the Leguminosae family and their transferability to Arachis hypogaea, groundnut. Plant Sci. 2007;174:51–60. doi: 10.1016/j.plantsci.2007.09.014. [DOI] [Google Scholar]
- Melchinger AE (1998) Advances in the analysis of data on quantitative trait loci. In: Proceedings of 2nd International Crop Science Congress, New Delhi, India
- Miklas PN, Johnson W, Delorme R, Gepts P. QTL conditioning physiological resistance and avoidance to white mold in dry bean. Crop Sci. 2001;41:309–315. doi: 10.2135/cropsci2001.412309x. [DOI] [Google Scholar]
- Miller AJ (1990) Subset selection in regression. Chapman and Hall, London, p 288
- Moretzsohn MC, Hopkins MS, Mitchell SE, Kresovich S, Valls JFM, Ferreira ME. Genetic diversity of peanut (Arachis hypogaea L.) and its wild relatives based on the analysis of hypervariable regions of the genome. BMC Plant Biol. 2004;4:11. doi: 10.1186/1471-2229-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretzsohn MC, Leoi L, Proite K, Guimara PM, Leal-Bertioli SCM, Gimenes MA, Martins WS, Valls JFM, Grattapaglia D, Bertioli DAJ. Microsatellite-based gene-rich linkage map for the AA genome of Arachis (Fabaceae) Theor Appl Genet. 2005;111:1060–1071. doi: 10.1007/s00122-005-0028-x. [DOI] [PubMed] [Google Scholar]
- Motagi (2001) Genetic analysis of resistance to late leaf spot and rust vis-à-vis productivity in groundnut (Arachis hypogaea L.). Dissertation, University of Agriculture Sciences, Dharwad, India
- Nair SK, Prasanna BM, Garg A, Rathore RS, Setty TA, Singh NN. Identification and validation of QTLs conferring resistance to sorghum downy mildew (Peronosclerospora sorghi) and Rajasthan downy mildew (P. heteropogoni) in maize. Theor Appl Genet. 2005;110:1384–1392. doi: 10.1007/s00122-005-1936-5. [DOI] [PubMed] [Google Scholar]
- Nevill DJ. Inheritance of resistance to Cercosporidium personatum in groundnuts: a genetic model and its implications for selection. Oleagineux. 1982;37:355–362. [Google Scholar]
- Nimmakayala P, Jeong J, Asturi SR, Thomason Y, Tallury S, Reddy UK (2007) Genetic diversity of peanut (Arachis hypogaea L.) and its wild relatives based on AFLP and microsatellite polymorphisms. In: XV International Plant and Animal Genomes Conference. San Diego, CA, USA, pp 427
- Paramasivam K, Jayasekhar M, Rajsekharan R, Veerbadhiran P. Inheritance of rust resistance in groundnut (A. hypogaea L.) Madras Agric J. 1990;77:50–52. [Google Scholar]
- Paterson AH, Stalker HT, Gallo-Meagher M, Burrow MD, Dwivedi SL, Crouch JH, Mace ES. Genomics and genetic enhancement of peanut. In: Wilson RF, Stalker HT, Brummer EC, editors. Legume crop genomics. Champaign: AOCS Press; 2004. pp. 97–109. [Google Scholar]
- Patil SH, Kale DM, Deshmukh SN, Fulzele GR, Weginwar BG. Semi dwarf, early maturing and high yielding new groundnut variety, TAG 24. J Oilseed Res. 1995;12:254–257. [Google Scholar]
- Payne RW, Murray DA, Harding SA, Baird DB, Soutar DM. GenStat for windows (10th edition) introduction. Hemel Hempstead: VSN International; 2007. [Google Scholar]
- Sharief Y, Rawlings JO, Gregory WC. Estimates of leaf spot resistance in three interspecific hybrids of Arachis. Euphytica. 1978;27:741–751. doi: 10.1007/BF00023710. [DOI] [Google Scholar]
- Subbarao PV, Subrahmanyam P, Reddy PM (1990) A modified nine point disease scale for assessment of rust and late leaf spot of groundnut. In: Second International Congress of French Phytopathological Society. 28–30 November 1990, Montpellier, France, pp 25
- Subrahmanyam P, Williams JH, Mcdonald D, Gibbons RW. The influence of foliar diseases and their control by selective fungicides on a range of groundnut (Arachis hypogaea L.) genotypes. Ann Appl Biol. 1984;104:467–476. doi: 10.1111/j.1744-7348.1984.tb03029.x. [DOI] [Google Scholar]
- Utz HF, Melchinger AE (1996) PLABQTL: A program for composite interval mapping of QTL. J Agric Genomics 2:1–5
- van Os H, Stam P, Visser RGF, van ECK HJ. RECORD: a novel method for ordering loci on a genetic linkage map. Theor Appl Genet. 2005;112:30–40. doi: 10.1007/s00122-005-0097-x. [DOI] [PubMed] [Google Scholar]
- Varma TSN, Dwivedi SL, Pande S, Gowda MVC. SSR markers associated with resistance to rust (Puccinia arachidis Speg.) in groundnut (Arachis hypogaea L.) Sabro J Breed Genet. 2005;37:107–119. [Google Scholar]
- Varshney RK, Graner A, Sorrells ME. Genomics assisted breeding for crop improvement. Trends Plant Sci. 2005;10:621–630. doi: 10.1016/j.tplants.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Varshney RK, Hoisington DA, Tyagi AK. Advances in cereal genomics and applications in crop breeding. Trends Biotechnol. 2006;24:490–499. doi: 10.1016/j.tibtech.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Varshney RK, Bertioli DJ, Moretzsohn MC, Vadez V, Krishnamurthy L, Aruna R, Nigam SN, Moss BJ, Seetha K, Ravi K, He G, Knapp SJ, Hoisington DA. The first SSR-based genetic linkage map for cultivated groundnut (Arachis hypogaea L.) Theor Appl Genet. 2009;118:729–739. doi: 10.1007/s00122-008-0933-x. [DOI] [PubMed] [Google Scholar]
- Voorrips RE (2006) MapChart 2.2: software for the graphical presentation of linkage maps and QTLs. Plant Research International, Wageningen, The Netherlands
- Welter LJ, Go¨ktu¨rk-Baydar N, Akkurt M, Maul E, Eibach R, Topfer R, Zyprian EM. Genetic mapping and localization of quantitative trait loci affecting fungal disease resistance and leaf morphology in grapevine (Vitis vinifera L.) Mol Breed. 2007;20:359–374. doi: 10.1007/s11032-007-9097-7. [DOI] [Google Scholar]
- Young ND, Weeden NF, Kochert G. Genome mapping in legumes (Family Fabaceae) In: Paterson AH, Landes RG, Austin TX, editors. Genome mapping in plants. Austin: Landes Company; 1996. pp. 211–227. [Google Scholar]
- Yu S, Zhang F, Yu R, Zou Y, Qi J, Zhao X, Yu Y, Zhang D, Li L. Genetic mapping and localization of a major QTL for seedling resistance to downy mildew in Chinese cabbage (Brassica rapa ssp. pekinensis) Mol Breed. 2009;23:573–590. doi: 10.1007/s11032-009-9257-z. [DOI] [Google Scholar]
- Zeng ZB. Precision mapping of quantitative trait loci. Genetics. 1994;136:1457–1468. doi: 10.1093/genetics/136.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.