Abstract
Mutations in the two breast cancer susceptibility genes BRCA1 and BRCA2 are associated with increased risk of breast and ovarian cancer. Patients with mutations in both genes are rarely reported and often involve Ashkenazi founder mutations. Here we report the first identification of a Danish breast and ovarian cancer family heterozygote for mutations in the BRCA1 and BRCA2 genes. The BRCA1 nucleotide 5215G > A/c.5096G > A mutation results in the missense mutation Arg1699Gln, while the BRCA2 nucleotide 859 + 4A > G/c.631 + 4A > G is novel. Exon trapping experiments and reverse transcriptase (RT)–PCR analysis revealed that the BRCA2 mutation results in skipping of exon 7, thereby introducing a frameshift and a premature stop codon. We therefore classify the mutation as disease causing. Since the BRCA1 Arg1699Gln mutation is also suggested to be disease-causing, we consider this family double heterozygote for BRCA1 and BRCA2 mutations.
Keywords: BRCA1, BRCA2, Breast cancer, Double heterozygote, Ovarian cancer, Mutation
Introduction
Germ-line mutations in BRCA1 (MIM# 113705) and BRCA2 (MIM# 600185) confer a high risk of breast and ovarian cancer. A disease causing mutation will result in 60–80% risk of developing breast cancer for a female carrier, while the risk of getting ovarian cancer is 20–40% or 10–20% with a BRCA1 or BRCA2 mutation, respectively [reviewed in [1]]. Most mutations in BRCA1 and BRCA2 have been listed in Breast Cancer Information Core (BIC) [2], and includes pathogenic mutations such as frameshift, nonsense, and splicing mutations, of which several have been identified in Danish breast and/or ovarian cancer patients [3–6]. Moreover, several founder mutations have been identified in a variety of different populations, including Ashkenazi Jews, Icelanders and Greenlandic Inuits [7–9]. A large number of BRCA1/BRCA2 missense, silent and intron mutations are of unknown significance. To enable the clinicians to give the affected patients satisfactory advice about preventive care, assessment of these mutations of unknown significance by functional studies are advised. Intron variants can be analysed by RT–PCR or by examining the splicing pattern in vitro using a mini gene system (reviewed in [10]). Here we report the first identification of a Danish family with germ-line mutations in both the BRCA1 and BRCA2 genes. The BRCA1 nucleotide 5215G > A/c.5096G > A mutation results in the missense mutation Arg1699Gln, while the BRCA2 nucleotide 859 + 4A > G/c.631 + 4A > G mutation is novel. Functional characterization of the BRCA2 nucleotide 859 + 4A > G/c.631 + 4A > G mutation by exon trapping experiments and RT–PCR analysis, revealed that the mutation results in skipping of exon 7 and leads to incorporation of 18 out of frame amino acids followed by a stop codon. We therefore classify the mutation as disease causing. Since the BRCA1 Arg1699Gln mutation is also suggested to be disease-causing, we consider this family double heterozygote for BRCA1 and BRCA2 mutations.
Materials and methods
Patients
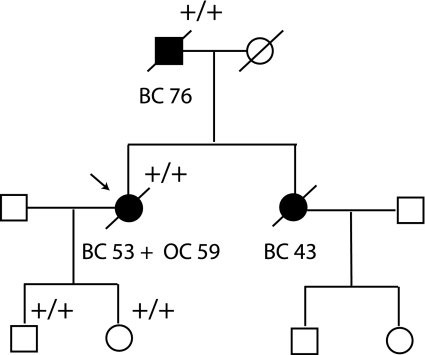
A 59 year-old woman with breast cancer (53) and ovarian cancer (59) was referred to genetic counselling. The family had two other cases of breast cancer (Fig. 1). Following verbal and written consent, blood samples were collected from the proband for mutation screening and after renewed consent a third blood sample was collected for RNA analysis. The family history was verified using the registry of the DBCG (Danish Breast Cancer Cooperative Group), hospital medical records and pathology reports, and genetic counselling was provided for family members.
Fig. 1.
Family pedigree. Breast and ovarian cancer are indicated as well as the age at diagnosis. Diagonal slash indicates deceased, while the proband is indicated with an arrow. Mutation positive individuals are indicated with +. +/+ indicates individuals with both mutations
BRCA1 and BRCA2 screening
Genomic DNA was purified from whole blood using the QIAamp DNA mini kit (Qiagen), and from paraffin-embedded tissue using the Chemagic DNA tissue kit (Chemagen) according to the manufacturer’s instructions. BRCA1 and BRCA2 screening was performed as recently described [4]. Sequence variations were verified in a new blood sample. The BRCA1 mutation is numbered according to GenBank accession number U14680, in which A in the AUG start codon has number 120, while the BRCA2 mutation is numbered according to GenBank accession number NM_000059, in which A in the AUG start codon has number 229. Moreover, the BRCA1 and BRCA2 mutations are numbered according to GenBank accession number NC_00017.10 and NC_000013.9 using the guidelines from the Human Genetic Variation Society.
In silico analyse
In silico prediction of the functional consequence of the missense variants was performed using SIFT (Sorting Intolerant From Tolerant): http://blocks.fhcrc.org/sift/SIFT.html [11] Polyphen: http://coot.embl.de/PolyPhen/ [12] and PMut: http://mmb2.pcb.ub.es:8080/PMut/ [13]. The following three splice site prediction programmes were used to predict the effect of the BRCA2 IVS7 + 4A > G mutation on the efficiency of splicing: www.fruitfly.org/seq_tools/splice.html [14], www.cbs.dtu.dk/services/NetGene2 [15] and www.umd.be/SSF [16]. The genomic sequence spanning the BRCA2 nucleotide 859 + 4 A > G/c.631 + 4 A > G mutation was submitted according to the guidelines of each programme and default settings were used in all predictions.
Exon trapping analysis
pSPL3-BRCA2-wild type plasmid and pSPL3-BRCA2-mutant plasmid, containing BRCA2 exon 7 and flanking intron sequences, was constructed by cloning genomic DNA from the proband using the following primers BRCA2-F, 5′-GATCACCTCGAGCGTTATACCTTTGCCCTGAGATTTA -3′ and BRCA2-R 5′-GATCACCTGCAGTTTAATGCCCCAATTACCAC -3′ into the pSPL3 splicing vector as recently described [5]. All constructs were verified by sequencing. COS7 cells were transfected using Fugene 6 (Roche) with either pSPL3-BRCA2-wild-type or pSPL3-BRCA2-mutant plasmids, RNA was purified and RT–PCR was performed as recently described [5].
RNA analysis
A fresh blood sample was obtained from the patient. Total cellular RNA was isolated with Trizol (Invitrogen) according to the manufacturer’s instructions. For reverse transcription–PCR (RT–PCR), cDNA was synthesized using random hexamer primers and M-MuLV reverse transcriptase (New England Biolabs) as described by the supplier. The cDNA was amplified with the BRCA2 specific primers 5′-CAAATTAGACTTAGGAAGGAATGTTCC-3′ and 5′-CATGAGGAAATACAGTTTCAGATGC-3′. The samples were separated by agarose gel electrophoresis and visualized by ethidium bromide staining. Finally, the bands were purified, cloned into pCR-Blunt II-TOPO (Invitrogen) and sequenced using an ABI3730 DNA analyzer (Applied Biosystems).
Results
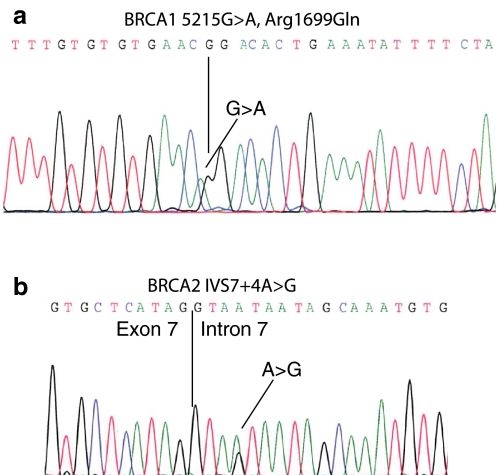
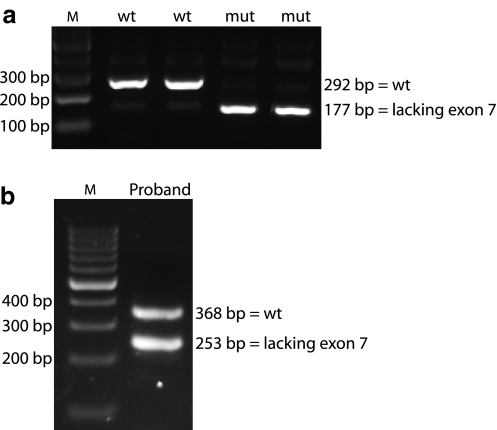
The proband was referred to genetic counselling, since she was diagnosed with breast cancer at the age of 53 and ovarian cancer at the age of 59. Her deceased father and deceased sister were diagnosed with breast cancer at the age of 76 and 43, respectively (Fig. 1). Blood samples were collected, genomic DNA was purified and the entire coding region and the exon–intron boundaries of BRCA1 and BRCA2 were screened by dHPLC and sequencing. Moreover, the DNA was examined for large genomic rearrangements by MLPA analysis. The analysis identified a BRCA1 nucleotide 5215G > A/c.5096G > A mutation (Fig. 2a), which introduces a missense mutation changing amino acid 1699 from an arginine to a glutamine. Moreover, a novel BRCA2 nucleotide 859 + 4A > G/c.631 + 4A > G mutation of unknown significance were identified (Fig. 2b). Both mutations were also identified in the proband’s two children. Moreover, analysis of genomic DNA from paraffin-embedded cancer tissue from the probands deceased father revealed that both mutations were inherited from him (Fig. 1). To indicate whether the mutations could be disease-causing, three different protein prediction programmes were used to predict the functional consequence of the BRCA1 Arg1699Gln mutation [11–13], while three different splice site prediction programmes were used to predict the effect of the BRCA2 nucleotide 859 + 4A > G/c.631 + 4A > G mutation on splicing of exon 7 [14–16]. All three protein prediction programmes estimated the BRCA1 Arg1699Gln mutation to be pathogenic. Moreover, two of the splice site programmes predicted the BRCA2 nucleotide 859 + 4A > G/c.631 + 4A > G mutation to have an effect on splicing of exon 7, while the third one (NetGene2) was unable to find both the wild type and the mutated splice site. It was therefore decided to analyse the BRCA2 nucleotide 859 + 4A > G/c.631 + 4A > G mutation by functional analysis using exon trapping. A fragment including BRCA2 exon 7 containing either the wild-type or the nucleotide 859 + 4A > G/c.631 + 4A > G mutation, respectively, 148 bp of intron 6 (IVS6), and 344 bp of intron 7 (IVS7) was cloned into the minigene vector pSPL3. The wild-type and mutant constructs was then transfected into COS-7 cells (in duplicates) and after 48 h mRNA was purified and examined by RT–PCR in duplicate on a 2% agarose gel (Fig. 3a). The normal wild-type BRCA2 exon 7 generated one transcript comprising the expected 292 bp and a very faint band of 177 bp, while the BRCA2 exon 7 nucleotide 859 + 4 A > G/c.631 + 4 A > G mutant yielded one faint band of 292 bp and a very strong band of 177 bp, respectively. Sequencing revealed that the 292 bp band contained exon 7, while the 177 bp band lacked exon 7. Finally, RT–PCR analysis using mRNA purified from whole blood was performed. The PCR products were analysed on a 1% agarose gel (Fig. 3b). Two PCR products were amplified from the patient—a 368 bp band and a smaller 253 bp band. Cloning and sequence analysis revealed that the 368 bp band contained exon 7, while the 253 bp band lacked exon 7 (data not shown).
Fig. 2.
Identification of the BRCA1 nucleotide 5215G > A/c.5096G > A mutation, and the BRCA2 nucleotide 859 + 4A > G/c.631 + 4A > G mutation. DNA was purified from the patient and the BRCA1 and BRCA2 genes were amplified using intronic primer pairs flanking each exon. The PCR products were pre-screened by dHPLC (denaturing high performance liquid chromatography) and sequenced. The analysis revealed a a BRCA1 nucleotide 5215G > A/c.5096G > A mutation and a b BRCA2 nucleotide 859 + 4A > G/c.631 + 4A > G mutation
Fig. 3.
Exon trapping and RT–PCR analysis. a COS-7 cells were transfected with pSPL3-BRCA2-exon 7 wild-type or pSPL3-BRCA2-exon 7 mutant plasmids in duplicates. Total RNA was isolated, RT–PCR analysis was performed and the PCR products were resolved on a 2% agarose gel. The 292 bp product corresponds to inclusion of exon 7 (unaltered splicing), while the 172 bp product corresponds to the exclusion of exon 7. The sizes of the DNA marker (M) are indicated to the right. b RT–PCR was performed on RNA purified from whole blood. The cDNA was amplified with specific BRCA2 primers. The sample was separated by agarose gel electrophoresis and visualized by ethidium bromide staining. Two RT–PCR products (368 and 253 bp) were obtained from the patient (Lane 2). The PCR products were cloned and sequence analysis revealed that the 253 bp band lacked exon 7 (data not shown)
Discussion
Patients double heterozygote for pathogenic mutations in BRCA1 and BRCA2 are rarely observed, and most reports involve the three Ashkenazi Jewish founder mutations BRCA1 nucleotide 185delAG and nucleotide 5382insC and BRCA2 nucleotide 6174delT (reviewed in [17]) [18–20]. However, four reports describe other BRCA1 and BRCA2 mutations, including a Dutch study describing two patients with a Dutch BRCA1 founder mutation and two different BRCA2 frameshift mutations [17], an Australian study describing one patient with a BRCA1 frameshift mutation and a BRCA2 splice site mutation [21], while a Korean study reported two cases of double heterozygosity for frameshift or nonsense mutations in the BRCA1 and BRCA2 genes [22]. Finally, a Spanish family with a pathogenic BRCA1 Ala1708Glu missense mutation and a BRCA2 frameshift mutation has been identified [23].
In this study, we have identified a Danish breast and ovarian cancer family with germ-line mutations in the BRCA1 and BRCA2 genes. Interestingly, the proband, the probands father, and the probands two children have inherited both mutations. The BRCA1 mutation changes the amino acid residue argenine on position 1699 to a glutamine. Arg1699 is highly conserved from man to plants [24]. It resides in the BRCT domain of BRCA1, which is involved in protein–protein interaction (reviewed in [25]). Crystal structure of the BRCA1 BRCT domain has shown that Arg1699 participates in the salt bridge with Asp1840 and Glu1836 [26]. Moreover, the Arg1699Gln mutation changes the amino acid residue from basic to polar, and in silico analysis using the SIFT, Polyphen or PMut software all regarded the BRCA1 Arg1699Gln mutation as disease-causing. Finally, we have only observed the mutation in breast and/or ovarian cancer families, and not in 200 healthy controls. Still, there have previously been inconclusive reports regarding the Arg1699Gln. Some in silico based studies have not been able to classify the mutation and group it as a variant of unknown significance [27, 28]. In contrast, other studies consider it to be deleterious using in silico analysis [29, 30]. Functional studies have shown that the BRCA1 Arg1699Gln mutation reduces the transcriptional activity of BRCA1 [31]. Moreover a loss in phosphospecificity of BRCA1 Gln1699 in its binding to BACH1—compared to BRCA1 Arg1699—has been observed [32]. Finally, the Arg1699Gln mutation appears to be defective in nuclear foci formation [33]. Moreover it should be noted that another substitution on this position (Arg1699Trp) is considered deleterious based on both functional and co-segregation studies [27]. Together, these data indicate that the BRCA1 Arg1699Gln mutation could be disease-causing, although it may have a lower penetrance.
The BRCA2 nucleotide 859 + 4A > G/c.631 + 4A > G mutation has not previously been reported in the BIC database or in the literature. Several other mutations have been identified in this area, including nucleotide 859 + 2T > G/c.631 + 2T > G, which disrupt the splice site consensus sequence and induce skipping of exon 7 [34], nucleotide 859G > A/c.631G > A, which also has been shown to cause skipping of exon 7 [35], as well as nucleotide 859 + 1G > A/c.631 + 1G > A [2]. Analysis of the BRCA2 nucleotide 859 + 4A > G/c.631 + 4A > G mutation using splice site prediction programmes suggested that the mutation affected the exon 7 splice doner site. Therefore we performed exon trapping experiments and RT–PCR analysis on RNA purified from whole blood. The analysis revealed that the mutation resulted in skipping of exon 7, which introduces a premature stop codon after incorporation of 18 out of frame amino acids. We therefore classify this mutation as disease-causing. Interestingly, even though the family is double heterozygote for mutations in BRCA1 and BRCA2, the affected family members do not have breast and/or ovarian cancer at a younger age than individuals with single BRCA1 or BRCA2 deleterious mutations. This is in agreement with other studies showing that double heterozygosity does not seem to lead to a more severe phenotype [17].
In summary, we here report the first Danish case of a patient who is double heterozygote for BRCA1 and BRCA2 mutations. Genetic screening is now offered to all family members.
Acknowledgments
We thank Peter B. Nielsen and Martin Skygge for technical assistance. This study was supported by the Neye Foundation and the Danish Cancer Society.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- BIC
Breast cancer information core
- DBCG
Danish breast cancer cooperative group
- RT–PCR
Reverse transcriptase–PCR
References
- 1.Nathanson KL, Wooster R, Weber BL. Breast cancer genetics: what we know and what we need. Nat Med. 2001;7:552–556. doi: 10.1038/87876. [DOI] [PubMed] [Google Scholar]
- 2.http://www.research.nhgri.nih.gov/bic/
- 3.Thomassen M, Hansen TV, Borg A, et al. BRCA1 and BRCA2 mutations in Danish families with hereditary breast and/or ovarian cancer. Acta Oncol. 2008;47:772–777. doi: 10.1080/02841860802004974. [DOI] [PubMed] [Google Scholar]
- 4.Hansen TV, Bisgaard ML, Jonson L, et al. Novel de novo BRCA2 mutation in a patient with a family history of breast cancer. BMC Med Genet. 2008;9:58. doi: 10.1186/1471-2350-9-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hansen TV, Steffensen AY, Jonson L, et al. The silent mutation nucleotide 744 G – > A, Lys172Lys, in exon 6 of BRCA2 results in exon skipping. Breast Cancer Res Treat. 2010;119:547–550. doi: 10.1007/s10549-009-0359-4. [DOI] [PubMed] [Google Scholar]
- 6.Hansen TO, Jonson L, Albrechtsen A, et al. Large BRCA1 and BRCA2 genomic rearrangements in Danish high risk breast-ovarian cancer families. Breast Cancer Res Treat. 2009;115:315–323. doi: 10.1007/s10549-008-0088-0. [DOI] [PubMed] [Google Scholar]
- 7.Neuhausen S, Gilewski T, Norton L, et al. Recurrent BRCA2 6174delT mutations in Ashkenazi Jewish women affected by breast cancer. Nat Genet. 1996;13:126–128. doi: 10.1038/ng0596-126. [DOI] [PubMed] [Google Scholar]
- 8.Thorlacius S, Olafsdottir G, Tryggvadottir L, et al. A single BRCA2 mutation in male and female breast cancer families from Iceland with varied cancer phenotypes. Nat Genet. 1996;13:117–119. doi: 10.1038/ng0596-117. [DOI] [PubMed] [Google Scholar]
- 9.Hansen TV, Ejlertsen B, Albrechtsen A, et al. A common Greenlandic Inuit BRCA1 RING domain founder mutation. Breast Cancer Res Treat. 2009;115:69–76. doi: 10.1007/s10549-008-0060-z. [DOI] [PubMed] [Google Scholar]
- 10.Baralle D, Baralle M. Splicing in action: assessing disease causing sequence changes. J Med Genet. 2005;42:737–748. doi: 10.1136/jmg.2004.029538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferrer-Costa C, Gelpi JL, Zamakola L, et al. PMUT: a web-based tool for the annotation of pathological mutations on proteins. Bioinformatics. 2005;21:3176–3178. doi: 10.1093/bioinformatics/bti486. [DOI] [PubMed] [Google Scholar]
- 14.Reese MG, Eeckman FH, Kulp D, et al. Improved splice site detection in Genie. J Comput Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 15.Hebsgaard SM, Korning PG, Tolstrup N, et al. Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res. 1996;24:3439–3452. doi: 10.1093/nar/24.17.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeo G. Burge CB: maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 17.Leegte B, van der Hout AH, Deffenbaugh AM, et al. Phenotypic expression of double heterozygosity for BRCA1 and BRCA2 germline mutations. J Med Genet. 2005;42:e20. doi: 10.1136/jmg.2004.027243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedman E, Bar-Sade Bruchim R, Kruglikova A, et al. Double heterozygotes for the Ashkenazi founder mutations in BRCA1 and BRCA2 genes. Am J Hum Genet. 1998;63:1224–1227. doi: 10.1086/302040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bell DW, Erban J, Sgroi DC, et al. Selective loss of heterozygosity in multiple breast cancers from a carrier of mutations in both BRCA1 and BRCA2. Cancer Res. 2002;62:2741–2743. [PubMed] [Google Scholar]
- 20.Frank TS, Deffenbaugh AM, Reid JE, et al. Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: analysis of 10, 000 individuals. J Clin Oncol. 2002;20:1480–1490. doi: 10.1200/JCO.20.6.1480. [DOI] [PubMed] [Google Scholar]
- 21.Smith M, Fawcett S, Sigalas E, et al. Familial breast cancer: double heterozygosity for BRCA1 and BRCA2 mutations with differing phenotypes. Fam Cancer. 2008;7:119–124. doi: 10.1007/s10689-007-9154-8. [DOI] [PubMed] [Google Scholar]
- 22.Choi DH, Lee MH, Bale AE, et al. Incidence of BRCA1 and BRCA2 mutations in young Korean breast cancer patients. J Clin Oncol. 2004;22:1638–1645. doi: 10.1200/JCO.2004.04.179. [DOI] [PubMed] [Google Scholar]
- 23.Caldes T, de la Hoya M, Tosar A, et al. A breast cancer family from Spain with germline mutations in both the BRCA1 and BRCA2 genes. J Med Genet. 2002;39:e44. doi: 10.1136/jmg.39.8.e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giannini G, Capalbo C, Ottini L, et al. Clinical classification of BRCA1 DNA missense variants: H1686Q is a novel pathogenic mutation occurring in the ontogenetically invariant THV motif of the N-terminal BRCT domain. J Clin Oncol. 2008;26:4212–4214. doi: 10.1200/JCO.2008.18.2089. [DOI] [PubMed] [Google Scholar]
- 25.Glover JN. Insights into the molecular basis of human hereditary breast cancer from studies of the BRCA1 BRCT domain. Fam Cancer. 2006;5:89–93. doi: 10.1007/s10689-005-2579-z. [DOI] [PubMed] [Google Scholar]
- 26.Williams RS, Bernstein N, Lee MS, et al. Structural basis for phosphorylation-dependent signaling in the DNA-damage response. Biochem Cell Biol. 2005;83:721–727. doi: 10.1139/o05-153. [DOI] [PubMed] [Google Scholar]
- 27.Goldgar DE, Easton DF, Deffenbaugh AM, et al. Integrated evaluation of DNA sequence variants of unknown clinical significance: application to BRCA1 and BRCA2. Am J Hum Genet. 2004;75:535–544. doi: 10.1086/424388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chenevix-Trench G, Healey S, Lakhani S, et al. Genetic and histopathologic evaluation of BRCA1 and BRCA2 DNA sequence variants of unknown clinical significance. Cancer Res. 2006;66:2019–2027. doi: 10.1158/0008-5472.CAN-05-3546. [DOI] [PubMed] [Google Scholar]
- 29.Abkevich V, Zharkikh A, Deffenbaugh AM, et al. Analysis of missense variation in human BRCA1 in the context of interspecific sequence variation. J Med Genet. 2004;41:492–507. doi: 10.1136/jmg.2003.015867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomez Garcia EB, Oosterwijk JC, Timmermans M, et al. A method to assess the clinical significance of unclassified variants in the BRCA1 and BRCA2 genes based on cancer family history. Breast Cancer Res. 2009;11:R8. doi: 10.1186/bcr2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vallon-Christersson J, Cayanan C, Haraldsson K, et al. Functional analysis of BRCA1 C-terminal missense mutations identified in breast and ovarian cancer families. Hum Mol Genet. 2001;10:353–360. doi: 10.1093/hmg/10.4.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clapperton JA, Manke IA, Lowery DM, et al. Structure and mechanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer. Nat Struct Mol Biol. 2004;11:512–518. doi: 10.1038/nsmb775. [DOI] [PubMed] [Google Scholar]
- 33.Lovelock PK, Spurdle AB, Mok MT, et al. Identification of BRCA1 missense substitutions that confer partial functional activity: potential moderate risk variants? Breast Cancer Res. 2007;9:R82. doi: 10.1186/bcr1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pyne MT, Brothman AR, Ward B, et al. The BRCA2 genetic variant IVS7 + 2T– > G is a mutation. J Hum Genet. 2000;45:351–357. doi: 10.1007/s100380070007. [DOI] [PubMed] [Google Scholar]
- 35.Pensabene M, Spagnoletti I, Capuano I, et al. Two mutations of BRCA2 gene at exon and splicing site in a woman who underwent oncogenetic counseling. Ann Oncol. 2009;20:874–878. doi: 10.1093/annonc/mdn724. [DOI] [PubMed] [Google Scholar]