Abstract
Colony-stimulating factor 1 receptor (CSF1R) is expressed in monocytes/macrophages and dendritic cells. These cells play important roles in the innate immune response, which is regarded as an important aspect of asthma development. Genetic alterations in the CSF1R gene may contribute to the development of asthma. We investigated whether CSF1R gene polymorphisms were associated with the risk of asthma. Through direct DNA sequencing of the CSF1R gene, we identified 28 single nucleotide polymorphisms (SNPs) and genotyped them in 303 normal controls and 498 asthmatic patients. Expression of CSF1R protein and mRNA were measured on CD14-positive monocytes and neutrophils in peripheral blood of asthmatic patients using flow cytometry and real-time PCR. Among the 28 polymorphisms, two intronic polymorphism (+20511C>T and +22693T>C) were associated with the risk of asthma by logistic regression analysis. The frequencies of the minor allele at CSF1R +20511C>T and +22693T>C were higher in asthmatic subjects than in normal controls (4.6 vs. 7.7%, p = 0.001 in co-dominant and dominant models; 16.4 vs. 25.8%, p = 0.0006 in a recessive model). CSF1R mRNA levels in neutrophils of the asthmatic patients having the +22693CC allele were higher than in those having the +22693TT allele (p = 0.026). Asthmatic patients with the +22693CC allele also showed significantly higher CSF1R expression on CD14-positive monocytes and neutrophils than did those with the +22693TT allele (p = 0.045 and p = 0.044). The +20511C>T SNP had no association with CSF1R mRNA or protein expression. In conclusion, the minor allele at CSF1R +22693T>C may have a susceptibility effect in the development of asthma, via increased CSF1R protein and mRNA expression in inflammatory cells.
Electronic supplementary material
The online version of this article (doi:10.1007/s00439-010-0850-3) contains supplementary material, which is available to authorized users.
Introduction
Asthma and its phenotypes are complex traits induced by interactions between the surrounding environment and multiple disease susceptibility genetic factors (Sengler et al. 2002). Allergic asthma has been recognized as an antigen-dependent T-helper type 2 (Th2)-related disease with a cytokine response profile that includes interleukin (IL)-4, IL-5, and IL-13. These cytokines play important roles in the coordination and persistence of the airway inflammatory process in allergic asthma (Cohn et al. 2004).
Recently, the innate, non-antigen-dependent immune system has received as much attention in the pathogenesis of asthma as an antigen-dependent adaptive immune response because adaptive immune responses are dependent on activation of the innate system (Kanzler et al. 2007). Many innate immune receptors, such as the Toll-like receptors (TLR), NOD proteins, Dectin, CD14, and collectins, have been discovered (Beutler 2004). TLR2 is associated with increased allergic inflammation and airway hyper-reactivity in a murine allergic model (Redecke et al. 2004), whereas TLR4 ligands can decrease allergic responses (Velasco et al. 2005). In these processes, dendritic cells (DCs) play a central role in initiating and regulating the adaptive and innate immune responses (Iwasaki and Medzhitov 2004).
Colony-stimulating factor 1 (CSF1, also referred to as macrophage CSF or M-CSF) has been found to be an essential factor for the growth, survival, and differentiation of monocyte/macrophages (Stanley et al. 1997). Furthermore, CSF1 has important roles in DC differentiation and function. CSF1 upregulates human monocyte expression of the P2X7 extracellular ATP receptor (Zhang et al. 2005), which regulates DCs and macrophage inflammatory function, favoring the generation of cytokines that stimulate T helper 2 responses (la Sala et al. 2003).
CSF1 acts via specific binding to its high-affinity receptor CSF1R (CD115 antigen), encoded by the c-fms proto-oncogene. Upon binding, CSF1 induces CSF1R tyrosine phosphorylation, leading to the activation of Ras-ERK and PI3K and to the formation of DNA-binding complexes containing STAT-1, STAT-3, and STAT-5 (Hamilton 1997). The human CSF1R gene is located on chromosome 5q33–q35. Whole-genome analyses have shown that chromosome region 5q33–q35 contains a gene cluster of ADRB2 and IL17RB receptors, key molecules contributing to the development of asthma and atopy in several ethnic populations (Holberg et al. 2001; Ober et al. 1998; Xu et al. 2001; Yokouchi et al. 2000). Considering these biological effects of CSF1R and its linkage to chromosome 5q33–q35, genetic variants of CSF1R may be involved in asthma. However, no report has examined its association with asthma development. In an effort to discover polymorphism(s) in the CSF1R gene that may have effects in asthma and related phenotypes, we identified 28 SNPs of the CSF1R gene and genotyped them in 498 asthmatic patients and 306 normal controls.
Materials and methods
Subjects
Subjects were recruited from the Genome Research Center for Allergy and Respiratory Diseases at Soonchunhyang University, Bucheon, Seoul, and Chunan Hospital, Korea. All patients were diagnosed by a physician and met the definition of asthma in the Global Initiative for Asthma (GINA) guidelines (Bateman et al. 2008). All patients had a history of dyspnea and wheezing during the previous 12 months, plus one of the following: (1) >15% increase in FEV1 or >12% increase plus 200 mL following inhalation of a short-acting bronchodilator, (2) <10 mg/mL PC20 methacholine, and (3) >20% increase in FEV1 following 2 weeks of treatment with systemic or inhaled steroids and long-acting bronchodilators. The normal subjects were recruited from the patients’ spouses and members of the general population who had no respiratory symptoms and had an FEV1 > 75% of the predicted value, PC20 methacholine >10 mg/mL, and normal findings on a plain chest X-ray. Twenty-four common inhalant allergens [e.g., dust mites (Dermatophagoides farinae and D. pteronyssinus), aspergillus, cat fur, dog fur, cockroaches, grasses, trees, and ragweed pollens] were used for a skin prick test. Atopy was defined as having a wheal reaction greater than or equal to histamine or 3 mm in diameter. Total IgE was measured using the Fluoroenzymeimmunoassay (Phadia AB, Uppsala, Sweden). Written informed consent was obtained from all subjects. Study protocols were approved by the Institutional Review Board of Soonchunhyang University Hospital.
Sequence analysis of human CSF1R
We sequenced CSF1R gene, including a promoter region (1.5 kb), to discover single nucleotide polymorphisms (SNPs) in DNA samples from 24 Koreans using the BigDye Terminator (v3.1) Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) and an ABI PRISM 3700 DNA analyzer (Applied Biosystems). The primer sets used for amplification and sequencing were designed based on sequence information from GenBank (Ref. Genome seq.; NC_000005.8). Information regarding primers is given in Supplementary Table 1 (http://www.snp-genetics.com/reference/additional_list.asp).
SNP selection and genotyping
Nineteen SNPs discovered in the CSF1R gene and nine SNPs in the International HapMap database (http://hapmap.ncbi.nlm.nih.gov/) were selected for this study (Fig. 1b). These included three SNPs in the promoter (−27162C>T, −27068C>G, and −27022C>T), 21 SNPs in introns (−26465G>A, −26306G>A, +5815C>T, +6610A>G, +8498T>C, +9180C>T, +9219A>T, +11680G>T, +13198G>A, +14665G>A, +15629G>A, +18363C>T, +20511C>T, +21997C>T, +22693T>C, +23443G>A, +25268C>T, +25995C>G, +28801C>G, +29446G>A, and +30473C>A), three SNPs in exons (+5648C>T, +8313C>T, and +15859A>G), and one SNP in the 3′-UTR (+33128A>G). The SNPs were genotyped at a multiplex level using Illumina’s Golden Gate genotyping system (Oliphant et al. 2002), and data quality was assessed using duplicate DNA (n = 10). The genotype quality score for keeping data was set to 0.25. SNPs that could not satisfy the following criteria were excluded: (1) a minimum call rate of 90%, (2) no duplicate error, and (3) Hardy–Weinberg equilibrium greater than p > 0.001. Twenty-eight SNPs were successfully genotyped. Information about the primers is provided in Supplementary Table 1.
Fig. 1.
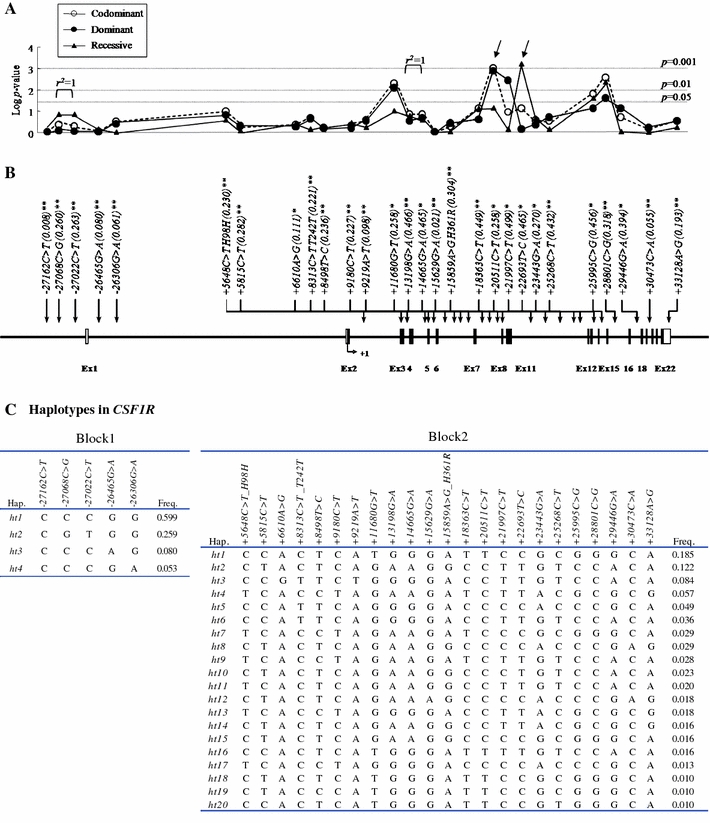
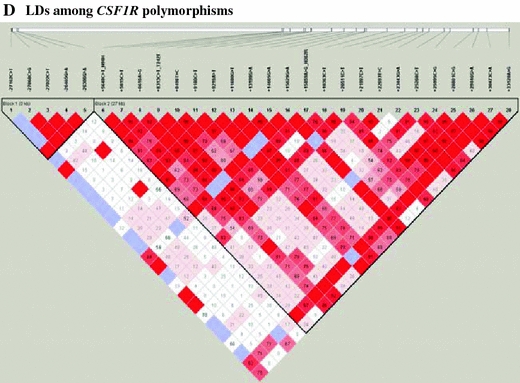
Gene map, p value, haplotype list, and linkage disequilibrium of SNPs in the CSF1R gene. a Log p values for the association analysis of SNPs with asthma risk. b Gene map and locations of the SNPs in the CSF1R gene at chromosome 5q33–q35 (60 kb). The first base of the translation start site is denoted as nucleotide +1. The black and white blocks represent the untranslated regions and coding regions of CSF1R, respectively. *polymorphisms selected for large-scale genotyping, considering allele frequencies, locations, and LD after discovery. **SNPs selected in the HapMap database. p values of each SNP were demonstrated in three different models (co-dominant, dominant, and recessive). c Haplotypes of CSF1R. d Linkage disequilibrium coefficient (|D′| and r 2) among CSF1R SNPs
Measurement of CSF1R protein expression on CD14-positive monocytes and neutrophils of asthmatic patients
Neutrophils and mononuclear cells (PBMC) were purified from the heparinized peripheral blood of asthmatic patients using a Percoll gradient (1.115, 1.100, 1.090, and 1.070). Mouse anti-human M-CSF R mAb (R&D Systems, Minneapolis, USA) was incubated with 5 × 105 PBMC or neutrophils, followed by treatment with 50 μg of PE-conjugated goat anti-mouse IgG (BD Biosciences, San Jose, CA, USA) for CSF1R staining. Additionally, PBMC was incubated with FITC-conjugated mouse anti-human CD14 (BD Biosciences). Stained cells were analyzed using a flow cytometer (FACScan, Becton-Dickinson, Mountain View, CA, USA). For isotype-matched controls, PE-conjugated mouse IgG1 (R&D Systems) and FITC-conjugated mouse IgG2a kappa (BD Biosciences) were used, with the same concentration of each antibody tested.
RNA extraction and real-time PCR of CSF1R m-RNA
Total RNA was isolated from neutrophils using a modified guanidium thiocyanate–phenol–chloroform extraction method (Schreiber et al. 1989). We quantified RNA and reverse transcribed cDNA from 3 μg of total RNA. DNase I (10,000 U/mL; Stratagene, La Jolla, CA, USA)-treated RNA was reverse-transcribed by incubating with 0.5 mM dNTP, 2.5 mM MgCl2, 5 mM DTT, 1 μL of random hexamer (50 μg/μL), and SuperScript II RT (200 unit/μL, Life Technologies, Grand Island, NY, USA) at 42°C for 50 min, and heat-inactivated at 70°C for 15 min.
Specific primer pairs for RT-PCR were as follows: β-actin forward: 5′-GGACTTCGAGCAAGAGATGG-3′; β-actin reverse: 5′-AGCACTGTGTTGGCGTACAG-3′ and CSF1R forward: 5′-CACCTTCACCCTCTCTCTGC-3′; CSF1R reverse: 5′-AGCATCTTCACAGCCACCTT-3′. Amplification was performed for 30 cycles (one cycle: 30 s at 94°C, 30 s at 55°C, and 40 s at 72°C) with initial denaturation at 94°C for 5 min and a final extension at 72°C for 10 min. The size and amount of the PCR products generated were determined by agarose gel electrophoresis in the presence of ethidium bromide and analyzed with the Kodak EDAS 1D analysis package. The cDNA fragment of the CSF1R gene contained from exon10 to exon12. cDNA was aliquoted into tubes containing specific primer pairs for human PGK1 (phosphoglycerate kinase1), human β-actin, and CSF1R genes for amplification (191, 233, and 263 bp fragments, respectively) using RT-PCR. Nucleotide sequences of the primers were as follows: PGK1 forward: 5′-CCTGGGCGGAGCTAAAGTTG-3′; PGK1 reverse: 5′-TCTCAGCTTTGGACATTAGGTCT-3′; β-actin forward: 5′-GGACTTCGAGCAAGAGATGG-3′; β-actin reverse: 5′-AGCACTGTGTTGGCGTACAG-3′; CSF1R forward: 5′-CAGAGCCTGCTGACTGTTGA-3′; CSF1R reverse: 5′-TTGCCCTCATAGCTCTCGAT-3. The primers of PGK1 and β-actin were prepared using the Primerbank web server (http://pga.mgh.harvard.edu/primerbank/). RT-PCR was performed with the StepOne™ Real-Time PCR System (Applied Biosystems, CA, USA). The reactions were prepared with 20 μL PCR mixture consisting of 10 μL master mix (iQ SYBR Green Supermix; Bio-Rad, Hercules, CA, USA), 1 μg/μL cDNA template, and 1 μL each of the CSF1R primer pair. Reactions were denatured at 95°C for 10 min and amplified for 40 cycles at 95°C for 15 s, 57°C for 32 s, and 72°C for 30 s. A melting curve for all products was obtained immediately after amplification by increasing temperature in 0.3°C increments from 60°C for 85 cycles of 15 s each. The threshold cycle (Ct) of CSF1R or endogenous reference gene was defined as the fractional cycle number at which the fluorescence of PCR products passed the fixed threshold. Using the 2−ΔΔCt method, the data are presented as the fold change in gene expression, normalized to an endogenous reference gene (PGK1 and β-actin) and relative to a control (+22693 TT and +22693 CC genotype of CSF1R). For the control sample, ΔΔCt equals zero and 20 = 1, so that the fold change in gene expression relative to the control = 1, by definition (Livak and Schmittgen 2001). For the other samples, evaluation of 2−ΔΔCt indicates the fold change in gene expression relative to the control. Fold differences in CSF1R mRNA expression in each patient were also calculated by comparing 2−ΔΔCt.
Statistical analyses
Linkage disequilibria (LD) between loci were measured as Lewontin’s D′ (|D′|) and r 2 . Haplotypes of each individual were inferred using the algorithm (PHASE, version 2.0), developed by Stephens et al. (2001). The associations of CSF1R genotypes and haplotypes with the risk of asthma were analyzed by logistic analysis, controlling for age (continuous value), gender (male = 0, female = 1), smoking status (non-smoker = 0, ex-smoker = 1, smoker = 2) and atopy status (non-atopy = 0, atopy = 1) as co-variables. Statistical analyses were performed using the SAS (v9.1; SAS Institute, Cary, NC, USA) and SPSS software (v11; SPSS, Inc., Chicago, IL, USA). p values of <0.05 were deemed to indicate statistical significance. The effective number of independent marker loci in CSF1R was calculated to correct for multiple testing using SNPSpD (http://genepi.qimr.edu.au/general/daleN/SNPSpD/) based on the spectral decomposition (SpD) of matrices of pair-wise LD between SNPs (Nyholt 2004). The number of independent marker loci in CSF1R was calculated as 4.5008 and 20.0301 in CSF1R-ht-block1 and CSF1R-ht-block2. The final number of independent marker loci in CSF1R was 24.531. In vitro analysis, differences between independent groups and samples were evaluated using a non-parametric Kruskal–Wallis H test for continuous data. When significant differences were found, a Mann–Whitney U test was performed to detect differences between pairs of samples. Statistical significance was defined as p < 0.05.
Results
Characteristics of the study subjects
The clinical characteristics of the study subjects are summarized in Table 1. Significant differences were found in mean age and prevalence of smoking between the normal controls and asthmatic patients (p = 0.001). FVC, FEV1, and PC20 values were significantly lower in the asthmatic subjects than in the normal controls (p < 0.001). IgE level, atopy prevalence, and eosinophil number were significantly higher in the asthmatic patients (p < 0.001).
Table 1.
Clinical profile and laboratory findings of the study subjects
| Normal controls | Asthmatics | |
|---|---|---|
| Number | 303 | 498 |
| Age (year, range) | 48 (8–83) | 43 (11–87)* |
| Onset of age (year, range) | 41 (1–75) | |
| Percentage of males | 44.3 | 45.1 |
| Current/ex-smoker (%) | 17.5/11.8 | 17/20* |
| FVC (% predicted) | 93.7 ± 12.4 | 82.9 ± 18.1** |
| FEV1 (% predicted) | 103.09 ± 14.77 | 79.33 ± 22.18** |
| PC20 methacholine (mg/ml) | 24.29 ± 2.47 | 4.02 ± 6.17** |
| Total IgE (IU/ml) | 115.9 ± 10.8 | 420.0 ± 55.1** |
| Peripheral blood eosinophil (%) | 2.5 ± 0.1 | 5.6 ± 0.25** |
| Atopy (%) | 33.1 | 62.5** |
Values are mean ± SE
* p = 0.001 and ** p < 0.001 are obtained using t test or χ2 test between asthmatics and normal controls
Association of SNPs in CSF1R with asthma risk
Pair-wise comparisons of the SNPs revealed two sets of absolute LDs (|D′| = 1 and r 2 = 1: −27068C>G, −27022C>T; +13198G>A, +14665G>A); several complete LDs (|D′| = 1 and r 2 ≠ 1) were also found (Fig. 1d). Haplotypes of CSF1R were constructed using the PHASE software, and eight major haplotypes with over 5% of MAF were used for further statistical analyses (Fig. 1c). The genotype distributions of the 28 SNPs were in Hardy–Weinberg equilibrium for all subjects (p > 0.05, Supplementary Table 2). The frequency of each of the 28 polymorphisms and eight haplotypes was compared between the asthmatic participants and normal controls using logistic regression. The allele frequencies in the asthmatic and normal control subjects are shown in Supplemental Table 3, and the p values of each SNP are shown in Fig. 1a. Among the 28 polymorphisms tested, two intronic polymorphisms (+20511C>T and +22693T>C) were associated with asthma risk (Fig. 1a; Table 2; p = 0.001 in co-dominant and dominant models and p = 0.0006 in a recessive model). After correcting for multiple comparisons, the difference remained significant in the two CSF1R SNPs (p = 0.02 and 0.01, respectively). Frequencies of the T alleles at CSF1R +20511C>T in the asthmatic group were higher than in the normal control group (49.5 vs. 36.7%, OR = 1.55, p = 0.001). In the analysis of the recessive model, the frequency of the C allele at CSF1R +22693T>C in the asthmatic group was higher than in the normal control group (25.8 vs. 16.4%, OR = 1.99, p = 0.0006). The data suggest that the T alleles at CSF1R +20511C>T and the C allele at CSF1R +22693T>C have a genetic susceptibility effect on the development of asthma.
Table 2.
Genotype distribution of CSF1R polymorphisms in asthmatics (BA) and normal subjects (NC) of the study subjects
| Loci | Position | rsSNP | Subject | N (%) | Co-dominant | Dominant | Recessive | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C/C | C/R | R/R | OR(95% CI) | p | p corr | OR(95% CI) | p | p corr | OR(95% CI) | p | p corr | ||||
| +20511C>T | Intron11 | rs216144 | NC | 166 (63.4) | 84 (32.1) | 12 (4.6) | 1.55 (1.19–2.00) | 0.001 | 0.02 | 1.67 (1.22–2.28) | 0.001 | 0.03 | 1.85 (0.94–3.65) | 0.07 | 1 |
| BA | 236 (50.5) | 195 (41.8) | 36 (7.7) | ||||||||||||
| +22693T>C | Intron11 | rs216140 | NC | 74 (28.2) | 145 (55.3) | 43 (16.4) | 1.22 (0.98–1.51) | 0.08 | 1 | 0.94 (0.67–1.32) | 0.71 | 1 | 1.99 (1.34–2.95) | 0.0006 | 0.01 |
| BA | 136 (29) | 212 (45.2) | 121 (25.8) | ||||||||||||
* Controlled for age (continuous value), sex (male = 0, female = 1), atopy status (non-atopy = 0, atopy = 1), and smoking status (non-smoker = 0, ex-smoker = 1, smoker = 2) as co-variables. The effective number of independent marker loci in CSF1R was calculated to correct for multiple testing, using the software SNPSpD (http://genepi.qimr.edu.au/general/daleN/SNPSpD) (Nyholt 2004). The number of independent marker loci in CSF1R was calculated as 4.5008 and 20.0301 in ht-block1 and ht-block2
Flow cytometric analysis for CSF1R protein expression on peripheral blood CD14-positive monocytes and neutrophils from asthmatic subjects
The CSF1R expression was measured on peripheral blood neutrophils and CD14-positive monocytes obtained from asthmatic patients by flow cytometry (Fig. 2). The CD14+ monocytes from asthmatic patients with the C alleles at +22693T>C showed significantly higher CSF1R expression than those with the TT alleles at +22693T>C (46.33 ± 4.62 vs. 83.52 ± 12.64, p = 0.045; Fig. 2e). The expression showed a similar trend in neutrophils. The level of CSF1R expression was significantly increased on neutrophils having the +22693 CC alleles versus those having +22693 T alleles (2.42 ± 0.3 vs. 5.62 ± 1.75, p = 0.044; Fig. 2f). In contrast, CSF1R expression was not different according to the genotypes of CSF1R +20511C>T (data not shown).
Fig. 2.
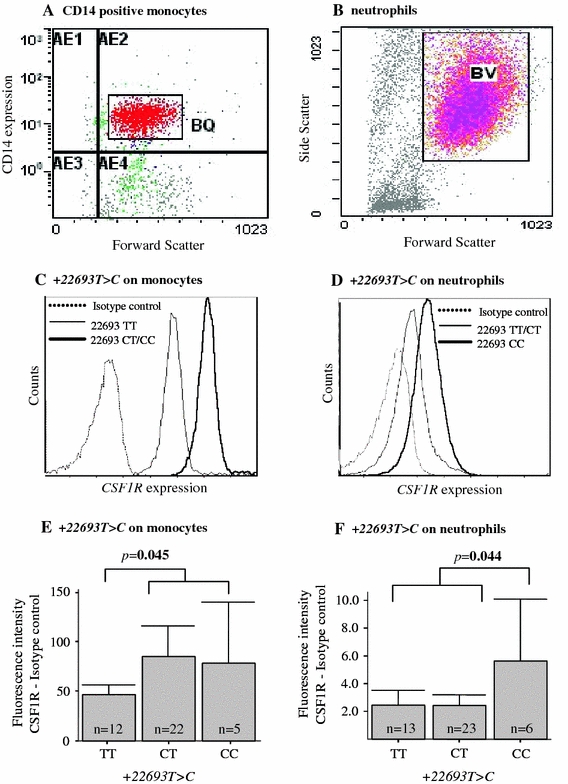
CSF1R protein expression on CD14+ monocytes and neutrophils in the peripheral blood of asthmatic subjects. a Gating for CD14+ among monocytes using an FITC-conjugated CD14 Ab. b Gating for neutrophils using side scatter and forward scatter. c, e Levels of CSF1R expression were compared between monocytes from asthmatic patients with the T or C alleles on CSF1R +22693T>C. d, f Levels of CSF1R expression were compared between neutrophils from asthmatic patients having the C allele at CSF1R +22693T>C and having the T allele at CSF1R +22693T>C
Comparison of CSF1R mRNA levels from its SNP subtypes
To investigate the genetic effect of the +22693 T>C polymorphism on mRNA expression of the CSF1R gene in neutrophils, CSF1R mRNA levels were measured. Because CSF1R +22693 T>C is located in intron 11, we amplified mRNA containing the CSF1R gene using primers to detect alternative splicing, as described in “Materials and methods” and Fig. 3a. To identify the RT-PCR products, sequences were examined using the BLAST search program after direct sequencing. The amplified RT-PCR fragment from exon 8 to exon 13 was 758 bp in length (Fig. 3b) and that of the RT-PCR product from exon 10 to exon 12 was 263 bp. Both of them shared 100% sequence identity with the human CSF1R gene. In the RT-PCR products, no alternative splicing was found in the common or rare homozygotes of +22693 T>C (Fig. 3b). Quantitation of mRNA expression in neutrophils from 12 asthmatics was performed by determining the concentration of the CSF1R RT-PCR products, corrected by that of β-actin and PGK1 (Fig. 3c). The expression level of total CSF1R mRNA in neutrophils was higher in patients having +22693 T alleles versus those having +22693 C alleles (Fig. 3d).
Fig. 3.
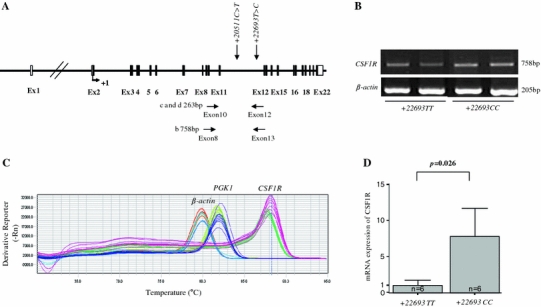
Comparison of CSF1R mRNA levels from its SNP subtypes. a Diagram of PCR primers. The RT-PCR product size from exon 8 to exon 13 is 758 bp. The RT-PCR product size from exon 10 to exon 12 is 263 bp. b Neutrophils containing T allele or C allele were harvested, and CSF1R mRNA was measured using RT-PCR. c Melt curve using RT-PCR is shown. d CSF1R mRNA was measured using RT-PCR. Data are expressed as the ratio of CSF1R mRNA over β-actin and PGK1 mRNA. The experiments were performed in duplicate
Discussion
In this study, we identified 28 genetic variants in the exons, introns, and flanking region of the CSF1R gene. Among them, we discovered that two polymorphisms in introns were associated with a risk of asthma development by logistic analysis. The rare allele frequencies of CSF1R +20511C>T and CSF1R +22693T>C were significantly higher in asthmatic patients versus normal controls, and this remained significant after correcting for multiple comparisons. This indicates that the rare alleles, CSF1R +20511C>T and CSF1R +22693T>C, in the intron may have some genetic susceptibility effect on the risk of asthma.
Genetic variants exert their effects via alteration of the gene product, the protein. The effects of CSF1 are mediated by the CSF1 receptor tyrosine kinase (CSF1R) through autophosphorylation of CSF1R and the subsequent phosphorylation of downstream molecules. Triggering this phosphorylation cascade increases gene transcription and protein translation and induces cytoskeletal remodeling by several signaling pathways, leading to the survival, proliferation, and differentiation of target cells (Pixley and Stanley 2004). Functional derangement of phosphorylation of CSF1R has been documented in pulmonary alveolar proteinosis (PAP). PAP is an autoimmune disorder in which autoantibodies interfere with signaling through the granulocyte–macrophage colony-stimulating factor receptor, leading to macrophage and neutrophil dysfunction (Juvet et al. 2008). Additionally, CSF1 is able to prime some innate immune responses by modulation of Toll-like receptors. Stimulation of monocytes and macrophages with CSF1 downregulates the expression of TLR1, TLR5, TLR2, TLR6, and TLR9 (O’Mahony et al. 2008; Sweet et al. 2002) and increases expression of CD14 (Ji et al. 2004). Thus, CSF1 enhances cytokine production in response to LPS, but suppresses the CpG DNA response (Sweet et al. 2002). These data suggest that CSF1R expression may be associated with various inflammatory responses, including the innate immune response, in asthma pathogenesis. However, no association between genetic variants in the CSF1R gene or protein has been documented in asthma.
The intronic SNPs of CSF1R gene in the present study may lead to differential production in the quantity of CSF1R protein or to structural variants. Thus, functional validation is necessary to confirm the genetic effect. To validate the genetic effect of CSF1R +20511C>T and CSF1R +22693T>C, the protein level on neutrophils and monocytes was analyzed by flow cytometry because these cells are known to express the receptor on their surface (Buckle et al. 1990). To separate monocytes clearly by flow cytometry, CD14+ cells were gated to eliminate other cells. Asthmatic patients having the rare alleles had significantly increased CSF1R expression on neutrophils and CD14+ monocytes compared with those with the common +22693T>C T alleles. These data indicate that the rare allele genotype increases protein expression of CSF1R gene, leading to asthma development via differential regulation of neutrophils, monocytes, and other immune cells, including dendritic cells, although we did not measure CSF1R protein expression on dendritic cells. Interestingly, CSF1R expression was independent from the CSF1R +20511C>T genotype.
It is well known that intronic SNPs can affect transcript processing through alternative splicing or producing RNA secondary structure (Chen and Stephan 2003). Because the CSF1R +22693T>C is located in the intron, we next checked the presence of alternatively spliced forms of CSF1R by RT-PCR.
No alternative splicing was found in the subjects having rare or common homozygotes. Thus, one of the explanations for the differential protein expression of the CSF1R gene may be different mRNA production, due to a change in RNA secondary structure according to genotype (Zuker 2003). The secondary structure containing the C allele of +22693T>C has a ΔG of −127.25 kcal/mol, and the T allele has a ΔG of −131.45 kcal/mol (Supplemental Fig. 1). These data suggest that this SNP may affect the secondary structure of the CSF1R gene. To validate the functional effect of the +22693 T>C on RNA processing, we measured mRNA levels using RT-PCR. We found that mRNA expression in neutrophils having the C alleles at +22693 C was significantly higher than in those having the T alleles at +22693. This indicates that the differential RNA production may be derived from a genetic effect of the SNP.
In summary, we genotyped human CSF1R gene polymorphisms and found that the rare allele CSF1R +22693T>C was associated with the risk of asthma via increasing CSF1R mRNA and protein expression on neutrophils and monocytes. This information on the genetic polymorphism of the CSF1R gene may be useful for improving asthma treatment and developing new strategies for controlling bronchial asthma.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The study was supported by the Korea Health 21 R&D Project, Ministry for Health, Welfare and Family Affairs, Republic of Korea (A010249), and samples were generously donated by the Collaborative Biobank of Korea at Soonchunhyang University Bucheon Hospital.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
E. K. Shin and S.-H. Lee contributed equally as the first author.
Contributor Information
Hyoung Doo Shin, Email: hdshin@sogang.ac.kr.
Choon-Sik Park, Phone: +82-32-6215105, FAX: +82-32-6215023, Email: mdcspark@unitel.co.kr.
References
- Bateman ED, Hurd SS, Barnes PJ, Bousquet J, Drazen JM, FitzGerald M, Gibson P, Ohta K, O’Byrne P, Pedersen SE, Pizzichini E, Sullivan SD, Wenzel SE, Zar HJ. Global strategy for asthma management and prevention: GINA executive summary. Eur Respir J. 2008;31:143–178. doi: 10.1183/09031936.00138707. [DOI] [PubMed] [Google Scholar]
- Beutler B. Innate immunity: an overview. Mol Immunol. 2004;40:845–859. doi: 10.1016/j.molimm.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Buckle AM, Jayaram Y, Hogg N. Colony-stimulating factors and interferon-gamma differentially affect cell surface molecules shared by monocytes and neutrophils. Clin Exp Immunol. 1990;81:339–345. doi: 10.1111/j.1365-2249.1990.tb03342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Stephan W. Compensatory evolution of a precursor messenger RNA secondary structure in the Drosophila melanogaster Adh gene. Proc Natl Acad Sci USA. 2003;100:11499–11504. doi: 10.1073/pnas.1932834100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn L, Elias JA, Chupp GL. Asthma: mechanisms of disease persistence and progression. Annu Rev Immunol. 2004;22:789–815. doi: 10.1146/annurev.immunol.22.012703.104716. [DOI] [PubMed] [Google Scholar]
- Hamilton JA. CSF-1 signal transduction. J Leukoc Biol. 1997;62:145–155. doi: 10.1002/jlb.62.2.145. [DOI] [PubMed] [Google Scholar]
- Holberg CJ, Halonen M, Solomon S, Graves PE, Baldini M, Erickson RP, Martinez FD. Factor analysis of asthma and atopy traits shows 2 major components, one of which is linked to markers on chromosome 5q. J Allergy Clin Immunol. 2001;108:772–780. doi: 10.1067/mai.2001.119158. [DOI] [PubMed] [Google Scholar]
- Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat Immunol. 2004;5:987–995. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- Ji XH, Yao T, Qin JC, Wang SK, Wang HJ, Yao K. Interaction between M-CSF and IL-10 on productions of IL-12 and IL-18 and expressions of CD14, CD23, and CD64 by human monocytes. Acta Pharmacol Sin. 2004;25:1361–1365. [PubMed] [Google Scholar]
- Juvet SC, Hwang D, Waddell TK, Downey GP. Rare lung disease II: pulmonary alveolar proteinosis. Can Respir J. 2008;15:203–210. doi: 10.1155/2008/528948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzler H, Barrat FJ, Hessel EM, Coffman RL. Therapeutic targeting of innate immunity with Toll-like receptor agonists and antagonists. Nat Med. 2007;13:552–559. doi: 10.1038/nm1589. [DOI] [PubMed] [Google Scholar]
- la Sala A, Ferrari D, Di Virgilio F, Idzko M, Norgauer J, Girolomoni G. Alerting and tuning the immune response by extracellular nucleotides. J Leukoc Biol. 2003;73:339–343. doi: 10.1189/jlb.0802418. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Mahony DS, Pham U, Iyer R, Hawn TR, Liles WC. Differential constitutive and cytokine-modulated expression of human Toll-like receptors in primary neutrophils, monocytes, and macrophages. Int J Med Sci. 2008;5:1–8. doi: 10.7150/ijms.5.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober C, Cox NJ, Abney M, Di Rienzo A, Lander ES, Changyaleket B, Gidley H, Kurtz B, Lee J, Nance M, Pettersson A, Prescott J, Richardson A, Schlenker E, Summerhill E, Willadsen S, Parry R. Genome-wide search for asthma susceptibility loci in a founder population. The Collaborative Study on the Genetics of Asthma. Hum Mol Genet. 1998;7:1393–1398. doi: 10.1093/hmg/7.9.1393. [DOI] [PubMed] [Google Scholar]
- Oliphant A, Barker DL, Stuelpnagel JR, Chee MS (2002) BeadArray technology: enabling an accurate, cost-effective approach to high-throughput genotyping. Biotechniques (Suppl):56–58, 60–61 [PubMed]
- Pixley FJ, Stanley ER. CSF-1 regulation of the wandering macrophage: complexity in action. Trends Cell Biol. 2004;14:628–638. doi: 10.1016/j.tcb.2004.09.016. [DOI] [PubMed] [Google Scholar]
- Redecke V, Hacker H, Datta SK, Fermin A, Pitha PM, Broide DH, Raz E. Cutting edge: activation of Toll-like receptor 2 induces a Th2 immune response and promotes experimental asthma. J Immunol. 2004;172:2739–2743. doi: 10.4049/jimmunol.172.5.2739. [DOI] [PubMed] [Google Scholar]
- Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengler C, Lau S, Wahn U, Nickel R. Interactions between genes and environmental factors in asthma and atopy: new developments. Respir Res. 2002;3:7. doi: 10.1186/rr179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley ER, Berg KL, Einstein DB, Lee PS, Pixley FJ, Wang Y, Yeung YG. Biology and action of colony-stimulating factor-1. Mol Reprod Dev. 1997;46:4–10. doi: 10.1002/(SICI)1098-2795(199701)46:1<4::AID-MRD2>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet MJ, Campbell CC, Sester DP, Xu D, McDonald RC, Stacey KJ, Hume DA, Liew FY. Colony-stimulating factor-1 suppresses responses to CpG DNA and expression of toll-like receptor 9 but enhances responses to lipopolysaccharide in murine macrophages. J Immunol. 2002;168:392–399. doi: 10.4049/jimmunol.168.1.392. [DOI] [PubMed] [Google Scholar]
- Velasco G, Campo M, Manrique OJ, Bellou A, He H, Arestides RS, Schaub B, Perkins DL, Finn PW. Toll-like receptor 4 or 2 agonists decrease allergic inflammation. Am J Respir Cell Mol Biol. 2005;32:218–224. doi: 10.1165/rcmb.2003-0435OC. [DOI] [PubMed] [Google Scholar]
- Xu J, Meyers DA, Ober C, Blumenthal MN, Mellen B, Barnes KC, King RA, Lester LA, Howard TD, Solway J, Langefeld CD, Beaty TH, Rich SS, Bleecker ER, Cox NJ. Genomewide screen and identification of gene-gene interactions for asthma-susceptibility loci in three U.S. populations: collaborative study on the genetics of asthma. Am J Hum Genet. 2001;68:1437–1446. doi: 10.1086/320589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokouchi Y, Nukaga Y, Shibasaki M, Noguchi E, Kimura K, Ito S, Nishihara M, Yamakawa-Kobayashi K, Takeda K, Imoto N, Ichikawa K, Matsui A, Hamaguchi H, Arinami T. Significant evidence for linkage of mite-sensitive childhood asthma to chromosome 5q31–q33 near the interleukin 12 B locus by a genome-wide search in Japanese families. Genomics. 2000;66:152–160. doi: 10.1006/geno.2000.6201. [DOI] [PubMed] [Google Scholar]
- Zhang XJ, Zheng GG, Ma XT, Lin YM, Song YH, Wu KF. Effects of various inducers on the expression of P2X7 receptor in human peripheral blood mononuclear cells. Sheng Li Xue Bao. 2005;57:193–198. [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


