Abstract
We evaluated the epigenetic contributions of repetitive DNA elements to human gene regulation. Human proximal promoter sequences show distinct distributions of transposable elements (TEs) and simple sequence repeats (SSRs). TEs are enriched distal from transcriptional start sites (TSSs) and their frequency decreases closer to TSSs being largely absent from the core promoter region. SSRs, on the other hand, are found at low frequency distal to the TSS and then increase in frequency starting ∼150bp upstream of the TSS. The peak of SSR density is centered around the -35bp position where the basal transcriptional machinery assembles. These trends in repetitive sequence distribution are strongly correlated, positively for TEs and negatively for SSRs, with relative nucleosome binding affinities along the promoters. Nucleosomes bind with highest probability distal from the TSS and the nucleosome binding affinity steadily decreases reaching its nadir just upstream of the TSS at the same point where SSR frequency is at its highest. Promoters that are enriched for TEs are more highly and broadly expressed, on average, than promoters that are devoid of TEs. In addition, promoters that have similar repetitive DNA profiles regulate genes that have more similar expression patterns and encode proteins with more similar functions than promoters that differ with respect to their repetitive DNA. Furthermore, distinct repetitive DNA promoter profiles are correlated with tissue-specific patterns of expression. These observations indicate that repetitive DNA elements mediate chromatin accessibility in proximal promoter regions and the repeat content of promoters is relevant to both gene expression and function.
INTRODUCTION
The prevalence of repetitive DNA sequences in mammalian genomes has been appreciated since the classic re-association kinetic (COT-curve) experiments of the late nineteen-sixties (Britten and Kohne, 1968). The completion of the human genome projects at the turn of the millennium further underscored the extent to which the human genome sequence is made up of repetitive DNA elements (Lander et al., 2001; Venter et al., 2001). There are several distinct categories of repetitive sequence elements in the human genome. Interspersed repeat sequences, also known as transposable elements (TEs), make up at least 45% of the euchromatic genome sequence, and novel human TE families continue to be discovered and characterized (Wang et al., 2005; Nishihara et al., 2006). Simple sequence repeats (SSRs) consist of tandem repeats of exact or nearly exact units of length k (k-mers), with k = 1-13 corresponding to microsatellites and k = 1-500 for minisatellites. Analysis of the human genome sequence showed that ∼3% of the euchromatic sequence was made up of SSRs, and both SSRs and TEs are thought to be far more abundant in heterochromatin. Segmental duplications of 1-200kb were initially shown to account for ∼3% of the human genome sequence (Lander et al., 2001), and more recent results reveal that copy number variants populate the genome to an even greater extent (Kidd et al., 2008).
The evolutionary significance and the functional role that repeated genomic elements, TEs in particular, play has long been a matter of speculation and inquiry. Once regarded as selfish, or parasitic, genomic elements with little or no phenotypic relevance (Doolittle and Sapienza, 1980; Orgel and Crick, 1980), it has since become apparent that TEs make substantial contributions to the structure, function and evolution of their host genomes (Kidwell and Lisch, 2001). Perhaps the most significant functional effect that TEs have had on their host genomes is manifest through the donation of regulatory sequences that control the expression of nearby genes (Feschotte, 2008). Studies of TE regulatory effects have focused, for the most part, on discrete well characterized regulatory elements such as transcription factor binding sites (Jordan et al., 2003; van de Lagemaat et al., 2003; Wang et al., 2007), enhancers (Bejerano et al., 2006) and alternative promoters (Dunn et al., 2003; Conley et al., 2008). A number of recent studies have also outlined the contributions of TEs to regulatory RNA genes (Smalheiser and Torvik, 2005; Borchert et al., 2006; Piriyapongsa and Jordan, 2007; Piriyapongsa et al., 2007). For this study, we sought to analyze the contribution of repetitive DNA to epigenetic aspects of gene regulation, specifically the relationship between repetitive DNA elements and the chromatin environment of human promoter sequences.
Genomic DNA in eukaryotes is wrapped around histone proteins and packaged into repeating subunits of chromatin called nucleosomes (Kornberg and Lorch, 1999). The importance of specific genomic sequences in determining the binding locations of nucleosomes has recently been confirmed (Segal et al., 2006). A number of factors point to a relationship between repetitive DNA elements, the local chromatin environment and epigenetic gene regulation. Densely compact heterochromatin is enriched for both TEs and SSRs in a number eukaryotic organisms (Dimitri and Junakovic, 1999). Heterochromatin functions to mitigate potentially deleterious effects associated with TEs by repressing both element transcription and ectopic recombination between dispersed element sequences (Grewal and Jia, 2007). In fact, it has been proposed that heterochromatin originally evolved to serve as a genome defense mechanism by silencing TEs (Henikoff and Matzke, 1997; Henikoff, 2000). In the plant Arabidopsis, de novo heterochromatin formation can be caused by insertions of TEs into euchromatin, and TEs are able to epigenetically silence genes when they are inserted nearby or inside them (Lippman et al., 2004). In other words, TEs have been shown to cause specific in situ changes in the chromatin environment that can spread locally and regulate gene expression in a way that is region-specific but sequence-independent (i.e. epigenetic).
The previously established connections between genome repeats, chromatin environment and gene regulation for model organisms, taken together with the repeat-rich nature of the human genome, suggest that repetitive sequence elements may play a role in regulating human gene expression by modulating the local chromatin environment. Specifically, we hypothesized that gene regulatory related differences in nucleosome binding at human promoter sequences are mediated in part by repetitive genomic elements. We evaluated the relationship between nucleosome binding, repetitive element promoter distributions and human gene expression to test this idea. Human proximal promoter sequences were characterized with respect to both their repetitive DNA architectures and predicted nucleosome binding affinities, and the repetitive DNA environment the promoters was considered with respect to patterns of gene expression.
MATERIALS AND METHODS
Promoter sequence analysis
Our analysis focused on proximal promoter sequence regions, which we define for a gene as ranging from -1kb at the 5′ end to the transcription start (TSS) at the 3′ end. We relied on the Database of Transcriptional Start Sites (DBTSS) to identify experimentally characterized TSS, based on aligned full-length cDNA sequences, in the human genome (Suzuki et al., 2002). These TSS were mapped to the March 2006 human genome reference sequence (NCBI Build 36.1) and used to extract 1kb proximal promoter sequences as described previously (Marino-Ramirez et al., 2004; Tharakaraman et al., 2005). This procedure was used to ensure analysis of the most accurate set of human proximal promoter sequences possible. For the additional three mammalian species analyzed — chimpanzee (Pan troglodytes), mouse (Mus musculus) and rat (Rattus norvegicus) — the locations of proximal promoter sequences were determined based on the 5′ most position of NCBI Refseq gene models (Pruitt et al., 2007). These positions were used to download 1kb proximal promoter sequences from the latest respective genome builds for each organism from the UCSC Genome Browser (Karolchik et al., 2003): chimpanzee n = 24,170, mouse n = 20,589 and rat n = 8,737.
The program RepeatMasker (Smit et al., 1996-2004) was used to detect and annotate repetitive elements in the proximal promoter sequences. RepeatMasker was run using 500bp of flanking sequence on either end of the proximal promoter regions analyzed to avoid edge effects in the detection of repeats. Repetitive elements detected by RepeatMasker were broken down into two main categories: interspersed repeats, also known as transposable elements (TEs), and simple sequence repeats (SSRs). SSRs may be annotated as low complexity sequences and correspond to runs of repeating k-mers where k = 1-13bp for microsatellites and k = 14-500 for minisatellites. TEs were further divided into specific classes: LINEs, SINEs, LTR and DNA as well as specific families L1 and Alu.
Proximal promoter sequences, including 500bp flanks, were analyzed using the Nucleosome Prediction software developed by the Segal lab (Segal et al., 2006). This software was used to calculate the probability of each nucleotide being occupied by a nucleosome in all promoter sequences. These nucleosome occupancy probabilities are based on the periodicity of dinucleotides — AA/TT/TA — that are characteristic of genomic sequences that have been experimentally isolated as bound to nucleosomes. Predictions for the relative placement of nucleosomes along genomic sequence are further informed by a thermodynamic stability model. The nucleosome prediction model used in our analysis is based on experimentally characterized nucleosome bound sequences reported for chicken (Satchwell et al., 1986). The chicken model has been proven accurate when used on other vertebrate genomes (Segal et al., 2006). For sets of promoter sequences, nucleosome occupancy averages were calculated over each position of the 1kb proximal promoter regions and these average values were taken as the position-specific nucleosome binding affinities (nba) reported here.
Two sets of promoter sequence randomizations were done and position-specific nucleosome binding affinities were re-calculated on the randomized sequence sets. The first randomization consisted of randomly shuffling entire 1kb proximal promoter sequences. This has the effect of maintaining overall nucleotide composition of the promoter sequences while changing the dinucleotide composition as well as any regional nucleotide biases along the promoters. The second randomization procedure consisted on randomly shuffling non-overlapping 100bp windows along the promoter sequences in place. This has the effect of maintaining both overall and local nucleotide composition of the promoters while changing the dinucleotide composition.
Repeat-based promoter clustering
Human proximal promoter sequences were clustered solely based on their repetitive DNA architectures. To do this, we generated 1,000-unit vectors that represent the position-specific repeat content for each promoter sequence. A discrete value was assigned to each promoter sequence position (nucleotide) in the following manner:
Promoter sequence repeat vectors were then clustered using a combination of k-means clustering (k = 5, 10, 20) and Self Organized Mapping using the program Genesis (Sturn et al., 2002). We found that using k-means clustering with k = 5 followed by a Self Organized Map generated the most coherent clusters in terms of the repeat content of the vectors.
Gene Expression Analysis
We used version 2 of the Novartis mammalian gene expression atlas (GNF2), which provides replicate Affymetrix microarray data for 44,775 probes across 79 human tissues (Su et al., 2004). GNF2 expression data, in the form of Affymetrix signal intensity values, were obtained from the UCSC Table Browser (Karolchik et al., 2004), and Affymetrix probes were mapped to NCBI Refseq identifiers using the UCSC Table Browser tools. For each gene, the average, maximum and breadth of expression were computed across the 79 tissues in the GNF2 data set. Expression breadth is taken as the number of tissues where the gene has a signal intensity value > 350. Co-expression between gene pairs was measured by computing the Pearson correlation coefficient (r) between pairs of gene-specific expression signal intensity vectors:
where gi is the ith gene and tn is the expression level for that gene in the nth tissue.
For each repeat-specific promoter cluster, the average r-value for all pairwise comparisons between genes in the cluster was computed. In addition, the difference (diff) between the cluster-specific r-value averages (cluster-r) and all possible pairwise r-values between genes (all-r) was computed for each cluster:
The significance of these differences was computed using the normal deviate:
where sediff is the standard error of the difference.
Probabilistic analysis of promoter repeats
We used a probabilistic representation of the repeat content of the human proximal promoter sequence clusters in order to derive gene (promoter)-specific similarity scores that indicate the probability that any human gene (promoter) belongs to a specific repeat cluster. To do this, each proximal promoter sequence (1kb upstream of the TSS) in a cluster was divided into 20 non-overlapping windows of 50bp each. For each window (w), the probability (p) of the occurrence of a TE nucleotide, or SSR nucleotide or a non-repetitive (NR) nucleotide was calculated separately using the following formula:
where fb,w = counts of base b in window w and b represents counts of either TE nucleotides, or SSR nucleotides or non-repetitive nucleotides, N = number of sites in the window (50) and s(b) = a pseudocount function. The probabilities thus calculated for each window were averaged for all promoters in the cluster. This procedure was repeated to yield repetitive DNA probabilistic representation models for each of the six promoter clusters.
All the proximal promoter sequences analyzed were then scored against each of the six cluster-specific probabilistic models using a log-likelihood ratio approach illustrated as follows:
where fb = p,b,w, x 50, which is the model frequency used as background. Promoter-specific scores (S) were then computed as the sum of log-likelihood ratios over the 20 windows of 50bp each:
Using this method, we scored all genes (promoters) against each of the six cluster models to generate six cluster-specific gene (promoter) score vectors. This modeling and scoring method is a modification of the approach used to score sequence motifs, such as transcription factor binding sites, based on motif-characteristic position-weight matrices (Wasserman and Sandelin, 2004).
In order to relate promoter sequence repetitive DNA architecture to tissue-specific gene expression, the gene (promoter)-specific probabilistic repeat cluster scores were correlated with tissue-specific gene expression signal intensity values for each of the 79 tissues in GNF. This was repeated with gene (promoter)-specific scores assigned to each gene for each of the six repeat clusters. For example, for the cluster1 (c1) versus tissue1 (t1) comparison:
where gi is the ith gene, S is the score for the cluster1 model and e is the expression level for that gene in tissue1. In other words, each gene analyzed is assigned a repeat probability score for each of the six clusters, and these six sets of repeat probability promoter scores are individually correlated with the GNF2 tissue-specific expression values for the genes. This procedure resulted in a 6 × 79 matrix of correlation values.
Gene Ontology (GO) analysis
GO annotation terms (Ashburner et al., 2000) for human genes were obtained from the Gene Ontology Annotation database (http://www.ebi.ac.uk/GOA/). GO terms were further mapped to higher level GO slim categories. Expected versus observed frequencies of GO slim terms were compared using χ2 tests for each promoter repeat cluster, as well as for the combined TE− and TE+ groups, in order to look for over-represented GO slim categories. The pairwise similarity between GO terms was computed using modified semantic similarity method (Lord et al., 2003; Azuaje et al., 2005) as described previously (Marino-Ramirez et al., 2006; Tsaparas et al., 2006). The GO similarity difference (GOdiff) was calculated between the average pairwise similarity for GO terms from pairs of genes within TE groups (e.g. TE+) and the average pairwise GO similarity for all possible pairs of genes:
The significance of the difference was measured using the normal deviate as described for the gene expression analysis.
Statistical Analysis
Standard statistical tests were used to compare population means for pairwise (Student’s t-test) and for multiple comparisons (ANOVA), to correlated vectors of nucleosome binding affinities, TE and SSR densities, expression and promoter score values (Pearson correlation coefficient), to control for the confounding effects of multiple variables on correlation values obtained (partial correlation) and to evaluate the difference between observed and expected GO terms (χ2) (Zar, 1999).
RESULTS AND DISCUSSION
Repetitive DNA and nucleosome binding affinity
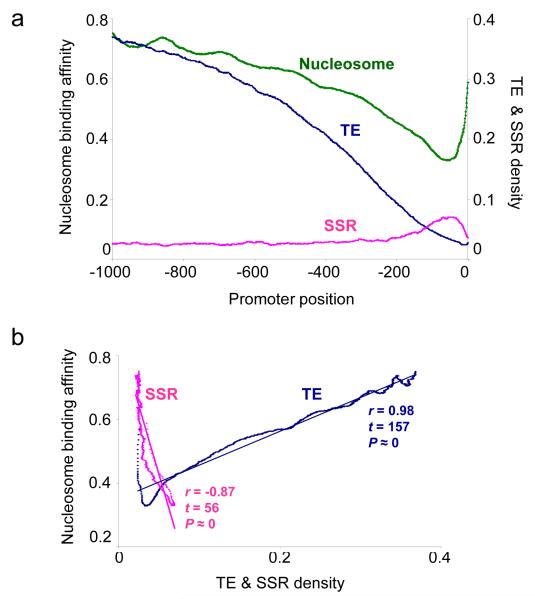
Experimentally characterized human gene proximal promoter sequences (n = 7,913) were taken from the Database of Transcriptional Start Sites (DBTSS) (Suzuki et al., 2002) and analyzed with respect to their repetitive DNA content and nucleosome binding affinities. The locations of repetitive DNA elements along promoter sequences were determined by the RepeatMasker program and nucleosome binding affinities were predicted using the method of Segal et al. (Segal et al., 2006). Two classes of repetitive DNA were analyzed separately: interspersed repeats, also known as transposable elements (TEs) and simple sequence repeats (SSRs), which are made up of runs of exact or nearly exact repeating k-mers. For each promoter position, from 1kb upstream to the transcriptional start site (TSS), the average TE and SSR densities over all promoter sequences were calculated as the fraction of sequences for which that position was occupied by a TE or SSR. Average nucleosome binding affinities across promoter positions were calculated as the fraction of sequences for which a given position was predicted to be occupied (bound) by a nucleosome. Average nucleosome binding affinities and the average TE density follow parallel trends along the proximal promoter regions (Figure 1a). Nucleosomes bind more tightly and TEs are found more frequently distal to the TSS, whereas nucleosomes bind promoter sequences most proximal to the TSS with lower affinity and TEs are rarely found close to the TSS. SSRs show a distinctly different trend with a higher density close to the TSS that corresponds to the decrease in nucleosome binding affinity. The SSR density matches the nucleosome binding even more closely than the TE density just upstream of the TSS. Nucleosome binding affinities decrease steadily from distal regions until ∼35bp upstream of the TSS, then the nucleosome binding affinity increases towards the TSS. Similarly, the SSR density increases to the same point and then drops off as the nucleosome binding affinity increases (Figure 1a). This core promoter region where nucleosome binding affinity is at its lowest and SSR density is at its highest corresponds to the location where the basal transcriptional machinery assembles, and RNA polymerase II binds, to initiate transcription.
Figure 1. Repetitive DNA density and nucleosome binding affinity along human proximal promoter sequences.
a) Average nucleosome binding affinities (green line, values on left y-axis) along with average TE densities (blue line, values on right y-axis) and average SSR densities (pink line, values on right y-axis) over 7,913 human proximal promoter sequences are plotted over each promoter position starting from -1,000bp upstream and progressing to the transcriptional start site (TSS at position 0). b) Linear trends and correlations relating position-specific nucleosome binding affinities (y-axis) to TE (blue) and SSR (pink) densities (x-axis) are shown. Statistical significance levels of the r-values are based on the Student’s t-distribution with df = n-2 = 998 where t = r*sqrt((n-2)/(1−r2)).
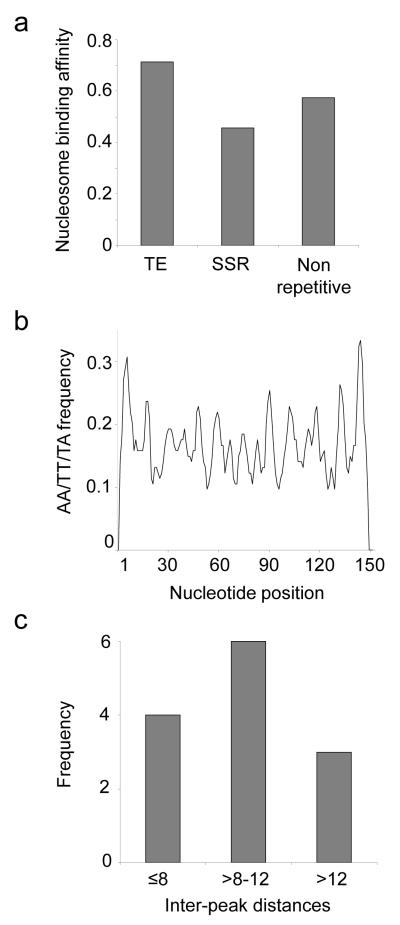
The correlations between nucleosome binding affinities with TE and SSR densities along human proximal promoter regions are robust and highly statistically significant (Figure 1b). Previously, we observed that nucleotide composition changes markedly along human proximal promoter sequences with an increase in CpG frequency close to the TSS (Marino-Ramirez et al., 2004), while the nucleosome binding prediction method we employed in this analysis relies on the periodicity of AT-rich dinucleotides (Segal et al., 2006). Thus, it is possible that the high (low) nucleosome binding affinity of TE (SSR) sequences in proximal promoter regions is a corollary effect of local differences in nucleotide composition. We attempted to control for this possibility in several ways. First of all, average nucleosome binding affinities were computed for all TE, SSR and non-repetitive sequences irrespective of their locations along proximal promoter regions. On average, TE sequences bind nucleosomes most tightly, followed by non-repetitive DNA and SSRs, which have the lowest nucleosome affinities (Figure 2a); all differences are highly statistically significant (ANOVA F = 4.5e11, P ≅ 0).
Figure 2. Nucleosome binding properties for repetitive versus non-repetitive DNA.
a) Average predicted nucleosome binding affinities are shown for TE, SSR and non-repetitive human promoter sequences. b) Periodicity of the nucleosome binding (wrapping) characteristic dinucleotides AA/TT/TA are shown for 39 experimentally characterized nucleosome bound TE sequences from chicken. c) Histogram showing the inter-peak distances for AA/TT/TA dinucleotides.
In addition to the binding affinity observations that are based on the nucleosome prediction software, we also analyzed the nucleosome wrapping characteristic AA/TT/TA dinucleotide frequencies along experimentally characterized nucleosome bound sequences from chicken (Satchwell et al., 1986) that we identified as being derived from TEs (n = 39). The chicken TE sequences show the characteristic AA/TT/TA dinucleotide periodicity expected of nucleosome bound sequences (Figure 2b); in fact, the average distance between dinucleotide peaks for these TE sequences is ∼10.3bp, which is close to the expected distance of 10.2bp corresponding to one turn of the DNA helix (Figure 2c). This is significant because DNA sequences are thought to wrap around nucleosomes by bending sharply at each repeating turn of the DNA helix, and this sharp bending is facilitated by the specific AA/TT/TA dinucleotides (Widom, 2001).
We also attempted to control for nucleotide composition effects by randomizing promoter sequences and re-calculating nucleosome binding affinities. First, entire 1kb promoter sequences were randomized and nucleosome binding affinities re-calculated. This control procedure has the effect of eliminating both native dinucleotide occurrences and local nucleotide composition biases. The average nucleotide binding affinity for such randomized promoter sequences (nba = 0.16) is ∼3x lower than seen for the observed promoter sequences (nba = 0.49), and the difference between random and observed affinities is highly significant (t = 23, P = 5.3e-100). In addition to differences in the magnitude of the nucleosome binding affinities, the relative affinity trends along the promoters were compared for the random versus observed sets. Partial correlation was used to control for the effects of the random sequences on the observed relationship between nucleosome binding affinity with TE and SSR densities along proximal promoters. The positive (negative) correlations between nucleosome binding for TE (SSR) do not change when the correlations between random sequences and nucleosome binding along the promoters are accounted for [rnba·TE|random1 = 0.99 and rnba·SSR|random1 = -0.85].
A second randomization procedure was done to account for local differences in nucleotide composition along proximal promoter sequences. In this case, sequences were randomized within non-overlapping 100bp windows along the promoters. This had the effect of eliminating native dinucleotide occurrences while maintaining local nucleotide composition biases. As with the complete sequence randomization procedure, the locally randomized sequences have significantly lower nucleosome binding affinities (nba = 0.23) than the observed sequences (nba = 0.49), and this 2.1x difference is highly statistically significant (t = 17, P = 5.0e-55). Clearly, local nucleotide composition alone can not explain the observed nucleosome binding affinities. However, the relative trends in nucleosome binding show different local nucleotide composition effects for TEs versus SSRs. The partial correlation controlling for the effects of local nucleotide composition on the relationship between TE density and nucleosome binding eliminates the positive correlation seen across the entire promoter for the observed data [rnba·TE|random2 = -0.14]. This suggests that local nucleotide composition bias influences the decreasing trend in nucleosome binding affinities along proximal promoters irrespective of TE density. Interestingly, this same mitigating effect of local nucleotide composition is not seen for the relationship between SSRs and nucleosome binding [rnba·SSR|random2 = -0.53]. This suggested the possibility that most of the local nucleotide composition bias effect on the relationship between TEs and nucleosome binding may be confined to the region closest to the TSS where TEs are largely absent and SSRs are at their most dense (Figure 1a). Indeed, when partial correlation controlling for local nucleotide bias is done excluding 150bp upstream of the TSS, the positive correlation between TEs and nucleosome binding affinity remains [-1000 to -150 rnba·TE|random2 = 0.76]. In other words, positive TE effects on nucleosome binding are most evident away from the TSS, while the SSRs that inhibit nucleosome binding act closest to the TSS.
Taken together, these data suggest the intriguing possibility that the human genome utilizes repetitive DNA content along promoter regions to tune nucleosome binding in such a way as to facilitate maximum access of the basal transcriptional machinery just upstream of TSS. Furthermore, different classes of repeats play distinct roles in this process; TEs bind nucleosomes tightly yielding compact less accessible DNA, while SSRs extrude nucleosomes creating a relatively open chromatin environment.
Cross-species comparison
In addition to the human genome analysis, the relationship between nucleosome binding and repetitive DNA content of proximal promoter regions was evaluated for four additional mammalian species with complete genome sequences available: chimpanzee (Pan troglodytes), mouse (Mus musculus) and rat (Rattus norvegicus). For these species, NCBI Refseq gene models were used to define TSS and proximal promoter regions, while TE and SSR repeats and nucleosome binding were analyzed as was done for the human genome. The trends observed for human are highly similar to those seen for the other mammalian species (Supplementary Figure 1). In chimpanzee, mouse and rat, nucleosome binding affinities decrease steadily along the proximal promoter region until the core promoter, <50bp from the TSS, where nucleosome binding begins to increase. For these three species, TE density drops precipitously and steadily along the proximal promoter while SSR density increases sharply at first in the core promoter near the TSS and then drops off again as nucleosome binding affinity increases. Thus, repeat- rich mammalian genomes appear to use repetitive DNA elements to tune nucleosome binding and core promoter accessibility in similar ways. The conservation of the relationship between repetitive DNA and nucleosome biding in core promoters of several mammalian species suggests that this mechanism may have evolved early in the mammalian radiation as repetitive elements were proliferating within genomes. However, many of the repetitive elements that yield these patterns evolve rapidly and are lineage-specific. Accordingly, there may be an ongoing dynamic between repeat generation by mutation and/or transposition followed by selection based on the promoter location of the repeat and specific requirements for chromatin accessibility. For TEs in particular, this could simply mean that the elements are eliminated from core promoter regions close to the TSS by purifying selection. Indeed, negative selection against TE insertions closest to TSS would seem to be the easiest way to explain the observed pattern of TE density (Figure 1a and Supplementary Figure 1). However, our analysis of gene expression data, described in following sections, suggests that this is not the case. SSRs, on the other hand, appear to be favored in core promoter regions.
TE-specific effects on nucleosome binding affinity
The Repbase library of repetitive DNA elements used by the program RepeatMasker can be used to annotate TEs into different classes and families (Jurka et al., 2005; Kapitonov and Jurka, 2008). Using this approach, human TE sequences were divided into LINEs, (L1 and other LINES), SINEs (Alu and other SINEs), LTR retrotransposons, and DNA transposons to determine if different classes (families) of elements show differential nucleosome binding affinities (Table 1). In general, LINEs, LTR retrotransposons and DNA transposons have higher affinities for nucleosomes compared to SINEs. Specifically, L1 elements exhibit the highest nucleosome binding affinities while Alu elements display the lowest affinity for nucleosomes. All differences are statistically significant (Table 1, ANOVA).
Table 1.
Average* nucleosome binding affinities for TE classes (families)
| TE class (family)a | Avg nba ± s.e.b |
|---|---|
| L1 | 0.849 ± 6.8e-4 |
| LINE other | 0.805 ± 7.6e-4 |
| Alu | 0.510 ± 5.2e-4 |
| SINE other | 0.789 ± 7.0e-4 |
| LTR | 0.807 ± 7.9e-4 |
| DNA | 0.802 ± 9.8e-4 |
TEs are broken down by class (family) using RepeatMasker. The L1 and Alu families are considered separately from all other LINEs and SINEs respectively. All LTR and DNA elements are considered together as classes.
Average nucleotide binding affinities ± standard errors.
All differences are statistically significant (ANOVA F = 2.8e4, P≅0).
The differences in nucleosome binding affinities between L1 and Alu are consistent with their respective nucleotide compositions and perhaps also relevant to their genomic distributions. L1 elements, and LINEs in general, are more AT-rich than Alus (SINEs), and AT-rich sequences are more likely to bind nucleosomes tightly as discussed previously. L1 elements are also biased towards intergenic regions in their distribution, while Alu elements are found primarily in gene rich regions. In fact, it has been shown that Alus are preferentially retained in GC- and gene-rich regions of the genome, and this has been taken to suggest that they may be selectively fixed therein by virtue of some gene-related function that they play (Lander et al., 2001). Our data showing lower nucleosome binding for Alu elements suggests that they may be retained in gene regions by virtue of their ability to maintain a relatively open chromatin environment. Conversely, L1 elements may help to maintain compact chromatin structures characteristic of intergenic regions.
Promoter repeat architecture and gene expression levels
In light of the observed relationship between repetitive DNA elements and nucleosome binding, we used the repetitive DNA content of proximal promoter regions to group human genes into related clusters. The gene expression and functional properties of the clusters were then compared to their characteristic repeat architectures. To cluster human genes using their promoter repeat distributions, proximal promoter sequences were represented as 1,000-unit vectors with each position in a sequence-specific vector receiving a score indicating whether that particular nucleotide is part of a TE, SSR or non-repetitive sequence. These gene-specific promoter repeat vectors were then compared using a distance metric and clustered as described (Materials and Methods). This approach ensured that the clusters reflect both the abundance, or lack thereof, and the location of distinct repetitive DNA elements in human promoter sequences. In other words, this scheme relates human genes solely by virtue of their promoter repeat distributions.
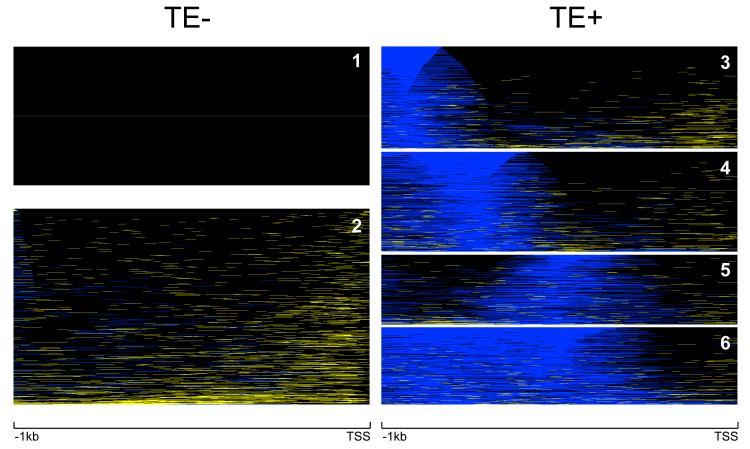
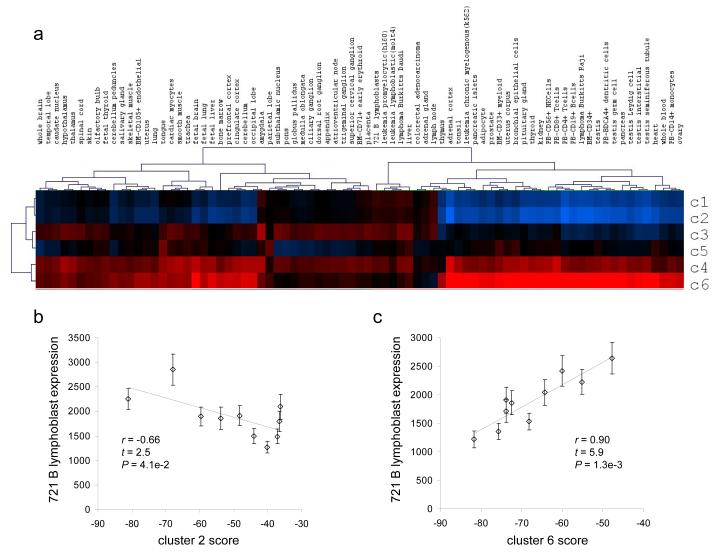
We obtained six repeat-specific clusters of human genes in this way (Figure 3), each cluster representing a distinct overall pattern of TE and/or SSR content and distribution. Two of these clusters (c1 & c2, TE−) consist of genes that are largely devoid of TEs, while four consist of genes with increasing TE densities (c3 — c6, TE+). c1 does not contain any repetitive DNA, while c2 is enriched in SSR sequences and has very low TE content. c3 — c6 have progressively more TE content with locations shifting slightly towards the TSS.
Figure 3. Clusters of human proximal promoters based on their repetitive DNA sequence distributions.
Proximal promoter sequences are represented left-to-right from position -1,000bp upstream to the transcriptional start site (TSS). Promoter sequences are color coded according to their repeat element distributions. Individual promoter nucleotide positions occupied by TEs are shown in blue, SSR positions are shown in yellow and non-repetitive positions are shown in black. The vertical size of the clusters corresponds to the number of sequences in each cluster. There are two (c1 & c2) clusters that contain promoters largely devoid of TE sequences (TE−), and the promoter sequences of the remaining four clusters (TE+, c3 — c6) contain increasing numbers of TEs.
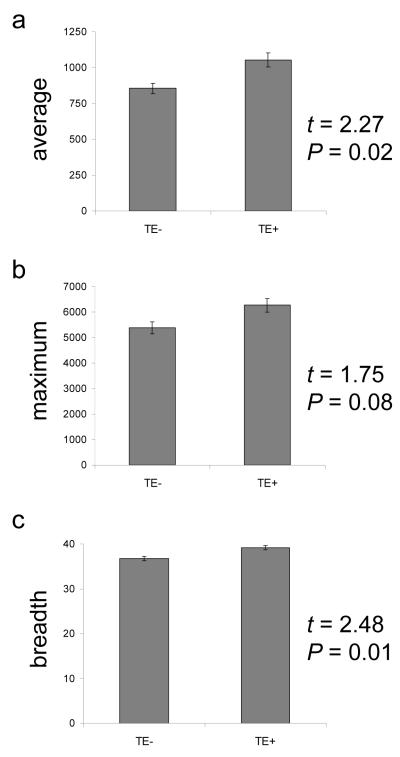
The gene expression properties of the human genes in these clusters were analyzed using version 2 of the Novartis mammalian gene expression atlas (GNF2) (Su et al., 2004). This data set consists of Affymetrix microarray experiments, performed in replicate, on 79 different human tissue (cell) samples. For each human gene, over 79 tissues, we computed the average expression level, maximum expression level and breadth of expression as described (Materials and Methods); cluster-specific averages for each of these parameters were then compared (Figure 4). We were surprised to find that clusters that contain TEs (c3 — c6, TE+) have higher average, maximum and breadth of expression than clusters that are largely devoid of TEs (c1 & c2, TE−). Gene expression levels are known to correlate with a number of measures of gene ‘importance’ such as sequence and phylogenetic conservation, fitness effects, numbers of protein interactions etc (Duret and Mouchiroud, 2000; Pal et al., 2001; Krylov et al., 2003; Zhang and Li, 2004; Wolf et al., 2006). In other words, genes that are more highly and broadly expressed are under greater purifying selection than genes with lower expression levels. If TEs are eliminated from proximal promoter sequences by purifying selection, then one may expect that TE+ promoters would have lower, and not higher as we observe, levels of gene expression than TE− promoters. In other words, our analysis of repeat cluster gene expression levels argues against the straightforward interpretation that the paucity of TEs in proximal promoter sequences, and their decreasing frequency closer to TSS, is a result of purifying selection against disruptive insertions in core promoters.
Figure 4. Gene expression comparison for TE− versus TE+ promoter clusters.
Human gene expression data are from the Novartis mammalian gene expression atlas version 2 (GNF2). a) Average level of expression, (b) maximum level of expression and (c) breadth of expression across 79 human tissues (cells) are compared for genes that have TE− versus TE+ promoter sequences. Statistical significance levels are based on the Student’s t-test.
On the other hand, one may expect that genes with more restricted and more tightly regulated expression, such as developmental genes, would have more TE sensitive promoters than genes that are highly and broadly expressed. In fact, developmental genes are known to have promoters that are largely devoid of TEs (Simons et al., 2006; Simons et al., 2007). This may reflect that fact that such genes are more finely and tightly regulated and accordingly contain more complex promoters with higher numbers of cis-regulatory elements. If this is indeed the case, then the paucity of TEs in proximal promoter regions may still be explained, to some extent, by purifying selection against disruptive insertions. Discrimination between these two hypotheses regarding the selective elimination, or lack thereof, of proximal promoter TE sequences awaits further analysis.
Promoter repeat architecture and tissue-specific gene co-expression
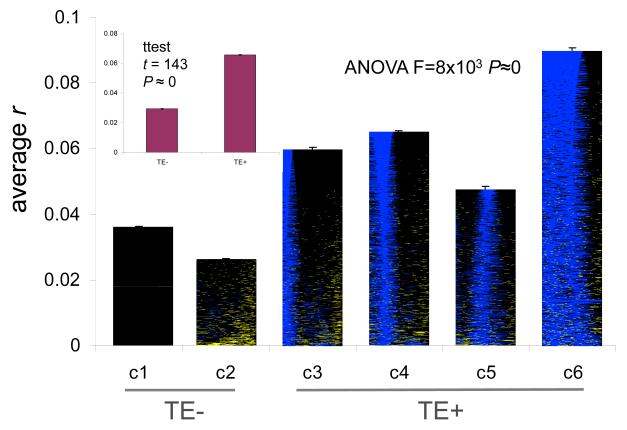
In addition to analyzing repeat cluster gene expression levels, we also evaluated the relationship between the tissue-specific expression patterns of genes across the 79 tissues from GNF2 and their promoter repeat content. To do this, gene-specific vectors of expression levels across tissues were compared using the Pearson correlation coefficient (r); positive values of r indicate gene pairs that are co-expressed across tissues. For each cluster, average r-values were computed based on all pairwise comparisons within the cluster (Figure 5). Higher average r-values are associated with increasing TE promoter content of the clusters. For instance, there is a positive (R = 0.77), albeit marginally significant (z = 1.72, P = 0.1), rank correlation between cluster TE content and co-expression. In addition, all four TE+ clusters have greater average co-expression than either of the TE− clusters, and the average r-value for TE+ clusters together is significantly greater than seen for the combined TE− clusters (Figure 5).
Figure 5. Gene co-expression for repeat-specific proximal promoter clusters.
Average pairwise Pearson correlation coefficients (r) for gene expression across 79 human tissues are shown for clusters 1-6 (see Figure 3) as well as for the TE− versus TE+ clusters (inset). Statistical significance levels are based on ANOVA for multiple comparisons and on the Student’s t-test for the TE− versus TE+ comparison.
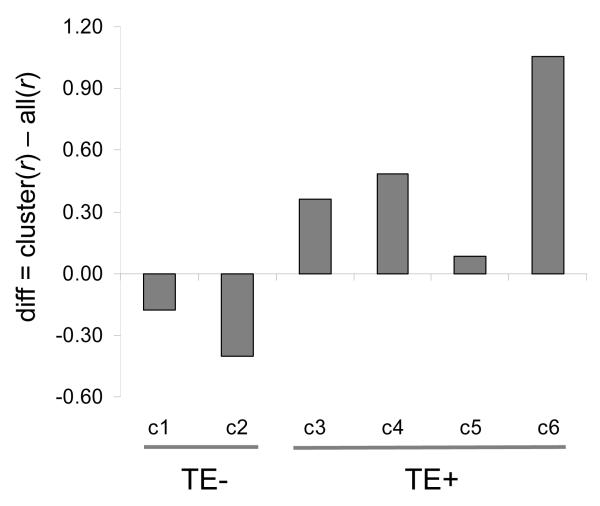
The possibility of gene co-regulation within repeat clusters was also evaluated by taking the difference between the average r-value for all pairwise comparisons within clusters to average pairwise r-value for all gene comparisons (Materials and Methods). If genes within clusters are co-regulated, then the value of this difference should be positive, whereas no co-regulation will yield a negative difference value. The TE− clusters 1 & 2 have negative difference values indicating that genes with no TEs in their promoters are less co-expressed with other genes possessing a similar lack of repeats than they are with all genes. On the other hand, the TE+ clusters 3-6 all have positive difference values further demonstrating that genes with similar repetitive DNA profiles in their promoters are more closely co-expressed than are random pairs of genes. The difference values for each cluster are statistically significant (7.3>z>100.6, 1.4e-13<P<0).
Taken together, these observations on gene co-expression also argue against the notion that TE insertions in proximal promoter sequences are basically disruptive or deleterious, since the presence of similar TE promoter distributions implies a higher level of gene co-regulation than the absence of TEs does. This is not to say that the majority of de novo TE insertions in and around functional promoter sequences are not deleterious, clearly they are. However, the repeat sequences that have been fixed in proximal promoter sequences do appear to make functionally relevant contributions to chromatin accessibility and help to regulate levels and specific patterns of gene expression.
Probabilistic analysis of promoters and gene expression
Given the relationship between gene expression and the repetitive DNA architecture of human promoters we observed, we wanted to further evaluate the propensity of human genes to be expressed in specific tissues based on the repetitive DNA content of their promoters. To do this, we used a probabilistic representation of cluster-specific promoter architectures together with the GNF2 expression data. This involved partitioning 1kb proximal promoter sequences into 20 non-overlapping windows of 50bp each, and for a given cluster, representing the probability of observing TE, SSR or non-repetitive nucleotides in each window (Materials and Methods). The probabilistic representation of promoter repeat architectures we employed is mathematically analogous to the probabilistic representations of position weight matrices (PWMs) used to summarize position-specific residue frequencies among collections of sequence motifs such as transcription factor biding sites (Wasserman and Sandelin, 2004). Accordingly, promoter repeat profiles can be represented as sequence logos showing the probability and distribution for sites of different repeat classes (Supplementary Figure 2). The cluster-specific promoter repeat profiles can then be used to score individual promoter sequences just as PWM representations can be used to score putative motif sequences. Connecting these cluster- and position-specific promoter repeat profiles to tissue-specific gene expression profiles was done in a way that is similar to the methodology used to connect the presence of transcription factor binding site motifs to specific gene expression patterns (Conlon et al., 2003).
For each of the 79 tissues in GNF2, each promoter sequence was given six cluster-specific scores, and for each cluster, the gene-specific scores were correlated with the tissue-specific gene expression levels (Materials and Methods). This resulted in a 6-by-79 matrix of cluster-by-tissue correlations (Figure 7). The TE+ clusters 4 & 6 show particularly high correlations with a number of tissues, such as B lymphoblasts (Figure 7b and 7c), whereas the TE− clusters 1 & 2 show low correlations with the same tissues and lower correlations overall. This indicates that certain repeat-rich promoter architectures play a role in driving tissue-specific expression, while repeat poor promoters have less coherent regulatory properties. In addition, the differences in promoter score-expression level correlations across tissues and between clusters indicate that different repeat contexts are likely to have tissue-specific regulatory functions. Hierarchical clustering of the tissues and the clusters, according to the promoter score-expression level correlations, groups related tissues together including reproductive tissues, immune related cells and cancer samples (Figure 7a). This indicates that TE-rich promoters may help to regulate genes that function specifically in these tissues further underscoring the biological significance of promoter sequence repetitive DNA profiles.
Figure 7. Promoter repetitive DNA architecture and tissue-specific gene expression.
Probabilistic models were used to represent the repetitive DNA architectures of each repeat-specific cluster (see Figure 3 and Supplementary Figure 2). Cluster-specific probabilistic models were used to score individual promoter sequences in terms of how closely they resemble a given cluster (Materials and Methods). Vectors of cluster-specific gene scores were correlated with vectors of gene expression values specific human tissues. a) A heat map illustrating the relative correlation values between gene (promoter)-specific scores for each cluster and tissue-specific gene expression values for the 79 tissues in the Novartis gene expression atlas version 2 (GNF2). Relatively high (positive) correlations between gene-cluster scores and gene expression levels are shown in red and low (negative) correlations are shown in blue. Two specific examples of such correlations are shown in panels b & c. b) Gene (promoter)-specific scores based on the probabilistic model for cluster 2 are negatively correlated with gene expression levels in a B lymphoblast cell line. c) Gene (promoter)-specific scores based on the probabilistic model for cluster 6 are positively correlated with gene expression levels in a B lymphoblast cell line. In other words, genes with repetitive DNA promoter profiles that most closely resemble cluster 6 are more highly expressed in the B lymphoblast cell line, whereas genes with repetitive DNA promoter profiles that resemble cluster 2 have lower levels of B lymphoblast expression.
Gene Ontology analysis
Having established a connection between repetitive DNA promoter architectures and gene regulation, we wondered whether genes with similar promoter repeat distributions encoded proteins with related functions. In order to test this, we used analysis of Gene Ontology (GO) terms for genes within and between the TE− versus the TE+ repeat-specific promoter clusters (Figure 3). A modified version of the GO semantic similarity measure (Lord et al., 2003; Azuaje et al., 2005) was used to compare the similarities between GO terms within clusters versus the background GO similarity among all pairs of genes. As described previously (Marino-Ramirez et al., 2006; Tsaparas et al., 2006), the GO semantic similarity approach measures the pairwise similarity between annotation terms along the GO directed acyclic graph in order to evaluate the functional similarity between pairs of genes. For TE− and TE+ genes, the GO similarity difference (GOdiff) is equal to the average GO similarity for all gene pairs within clusters minus the average GO similarity for all possible gene pairs (Materials and Methods). Negative values of GOdiff indicate that gene pairs are more similar within clusters than for all possible pairs. Both the TE− and TE+ gene sets encode proteins that are significantly more functionally similar than the background comparison set [TE− = -3.4e-3, z = 34, P≅0; TE+ = -7.9e-3, z = 11, P = 4.8e-3]. However, within the TE+ clusters, pairs of genes encode proteins that are significantly more functionally similar, on average, than the pairs of genes found within the TE− clusters (t = 5.8, P = 6.4e-9). This is consistent with the stronger signal of gene co-regulation seen for clusters of promoter sequences that are enriched for TEs and underscores the potential biological significance of repeat-rich promoter sequences in the human genome.
Given the functional coherence of repeat-specific clusters demonstrated by the GO similarity analysis, we wanted to evaluate whether certain GO functional categories are over-represented within specific clusters. To do this, we traced the GO terms represented in the dataset to GO slim terms (Table 2). GO slim categories provide a higher level view of more granular individual GO annotations in order to provide an overview of the kinds of functions that may be over-represented in different groups. The observed counts of GO slim categories for each of the six repeat-specific clusters, as well as for the combined TE− and TE+, groups were compared to their expected values based on the background GO slim frequencies across all clusters to look for over-represented terms. Genes in the electron transport, cytoplasm, catalytic activity and oxidoreductase activity categories were found to be over-represented in TE+ clusters and accordingly under-represented in the TE− clusters, whereas genes in cell communication, multicellular organismal development, regulation of biological process and transcription regulator activity categories are over-represented in TE− clusters and under represented in TE+ clusters. Evaluation of over-represented GO terms in individual clusters reveals coherence across the three categories of GO terms: molecular function, cellular component and biological process. For instance, the TE+ cluster 5 has an over-represented receptor and transporter activities in the molecular function category that agree with the cell surface cellular component term and the response to stimulus biological process term. The over-represented catalytic activity molecular process term for the most TE-rich cluster 6 corresponds to a cytoplasmic cellular component term along with metabolic and biosynthetic biological process terms. In a general sense, the coherence of GO functional annotations within repeat-specific clusters and the differences between clusters are consistent with biological significance of the regulatory differences seen for these clusters.
Table 2.
| Groupb | Molecular Functionc | Cellular Componentc | Biological Processc |
|---|---|---|---|
|
| |||
| TE− | GO:0030528 : transcription regulator activity | - | GO:0007154 : cell communication |
| GO:0007275 : multicellular organismal development |
|||
| GO:0050789 : regulation of biological process |
|||
|
| |||
| TE+ | GO:0003824 : catalytic activity | GO:0005737 : cytoplasm | GO:0006810:transport |
| GO:0016491 : oxidoreductase activity | GO:0007154:cell communication | ||
|
| |||
| C1 | GO:0005198 : structural molecule activity | - | - |
|
| |||
| C2 | GO:0016301 : kinase activity | - | GO:0007154 : cell communication |
| GO:0016491 : oxidoreductase activity | GO:0007275 : multicellular organismal development |
||
| GO:0030528 : transcription regulator activity | |||
| GO:0007610 : behavior | |||
| GO:0030154 : cell differentiation | |||
| GO:0050789 : regulation of biological process |
|||
|
| |||
| C3 | - | - | - |
|
| |||
| C4 | GO:0003824 : catalytic activity | GO:0005737 : cytoplasm | GO:0006944 : membrane fusion |
| GO:0009056 : catabolic process | |||
|
| |||
| C5 | GO:0004872 : receptor activity | GO:0009986 : cell surface | GO:0050896 : response to stimulus |
| GO:0005215 : transporter activity | |||
| GO:0022857 : transmembrane transporter activity |
|||
|
| |||
| C6 | GO:0003824 : catalytic activity | GO:0005622 : intracellular | GO:0008152 : metabolic process |
| GO:0005737 : cytoplasm | GO:0009058 : biosynthetic process | ||
GO slim categories provide a high level view of GO functions and subsume a number of lower (more granular) GO functional annotation categories.
Repeat-specific clusters 1-6 along with the combined TE+ and TE− groups (see Figure 3).
GO functional annotation categories.
Statistical significance for over-represented terms was evaluated using with χ2 tests with at least χ2>4.2, P<0.04.
Conclusion
We have uncovered a connection between repetitive DNA sequences and nucleosome binding in human proximal promoter regions along with an influence of repetitive DNA promoter sequences on specific patterns of gene expression. Interestingly, different classes of repetitive elements function differently to mediate nucleosome binding; TEs bind nucleosomes tightly and are generally excluded from core promoter regions, while SSRs have a low affinity for nucleosomes and are enriched just upstream of TSSs. Thus, it appears that repetitive sequence elements are differentially utilized to tune the accessibility to promoter sequences by transcription factors, particularly the basal transcriptional machinery that assembles just upstream of the TSS, via changes in the local chromatin environment.
Supplementary Material
Supplementary Figure 1. Repetitive DNA density and nucleosome binding affinity along mammalian proximal promoter sequences. Average nucleosome binding affinities (green line, values on left y-axis) along with average TE densities (blue line, values on right y-axis) and average SSR densities (pink line, values on right y-axis) over species-specific sets of proximal promoter sequences are plotted over each promoter position starting from -1,000bp upstream and progressing to the transcriptional start site (TSS at position 0). Trends are shown for chimpanzee (a), mouse (b) and rat (c).
Supplementary Figure 2. Sequence logos representing promoter repeat architectures for six clusters (c1-c6) of human proximal promoter sequences. For each cluster, the probability of observing TE (blue ‘T’), SSR (yellow ‘S’) or non-repetitive (black ‘N’) sequence residues is represented along proximal promoter sequences from -1kb to the TSS (x-axis) by the relative bits of information (i.e. the height of residue) on the y-axis.
Figure 6. Differences in gene co-expression between cluster-specific gene pairs versus all possible pairs of genes.
Average pairwise Pearson correlations (r) for gene expression across 79 human tissues were measured for all possible gene pairs and this value was subtracted from the average pairwise r-values for genes within each repeat-specific cluster (c1 — c6). A negative value indicates that genes within the cluster have less similar co-expression than background, whereas a positive value indicates that genes within a cluster are more highly co-expressed than expected.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuaje F, Wang H, Bodenreider O. Onotlogy-driven similarity approaches to supporting gene functional assessment. Proc ISMB SIG meeting on Bio-ontologies. 2005;2005:9–10. [Google Scholar]
- Bejerano G, Lowe CB, Ahituv N, King B, Siepel A, Salama SR, Rubin EM, Kent WJ, Haussler D. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature. 2006;441:87–90. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- Britten RJ, Kohne DE. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968;161:529–40. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- Conley AB, Piriyapongsa J, Jordan IK. Retroviral promoters in the human genome. Bioinformatics. 2008 doi: 10.1093/bioinformatics/btn243. in press. [DOI] [PubMed] [Google Scholar]
- Conlon EM, Liu XS, Lieb JD, Liu JS. Integrating regulatory motif discovery and genome-wide expression analysis. Proc Natl Acad Sci U S A. 2003;100:3339–44. doi: 10.1073/pnas.0630591100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitri P, Junakovic N. Revising the selfish DNA hypothesis: new evidence on accumulation of transposable elements in heterochromatin. Trends Genet. 1999;15:123–4. doi: 10.1016/s0168-9525(99)01711-4. [DOI] [PubMed] [Google Scholar]
- Doolittle WF, Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980;284:601–3. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- Dunn CA, Medstrand P, Mager DL. An endogenous retroviral long terminal repeat is the dominant promoter for human beta1,3-galactosyltransferase 5 in the colon. Proc Natl Acad Sci U S A. 2003;100:12841–6. doi: 10.1073/pnas.2134464100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D. Determinants of substitution rates in mammalian genes: expression pattern affects selection intensity but not mutation rate. Mol Biol Evol. 2000;17:68–74. doi: 10.1093/oxfordjournals.molbev.a026239. [DOI] [PubMed] [Google Scholar]
- Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Heterochromatin function in complex genomes. Biochim Biophys Acta. 2000;1470:O1–8. doi: 10.1016/s0304-419x(99)00034-7. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Matzke MA. Exploring and explaining epigenetic effects. Trends Genet. 1997;13:293–5. doi: 10.1016/s0168-9525(97)01219-5. [DOI] [PubMed] [Google Scholar]
- Jordan IK, Rogozin IB, Glazko GV, Koonin EV. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 2003;19:68–72. doi: 10.1016/s0168-9525(02)00006-9. [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–7. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat Rev Genet. 2008;9:411–2. doi: 10.1038/nrg2165-c1. author reply 414. [DOI] [PubMed] [Google Scholar]
- Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ, Weber RJ, Haussler D, Kent WJ. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–4. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–6. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, Haugen E, Zerr T, Yamada NA, Tsang P, Newman TL, Tuzun E, Cheng Z, Ebling HM, Tusneem N, David R, Gillett W, Phelps KA, Weaver M, Saranga D, Brand A, Tao W, Gustafson E, McKernan K, Chen L, Malig M, Smith JD, Korn JM, McCarroll SA, Altshuler DA, Peiffer DA, Dorschner M, Stamatoyannopoulos J, Schwartz D, Nickerson DA, Mullikin JC, Wilson RK, Bruhn L, Olson MV, Kaul R, Smith DR, Eichler EE. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell MG, Lisch DR. Perspective: transposable elements, parasitic DNA, and genome evolution. Evolution Int J Org Evolution. 2001;55:1–24. doi: 10.1111/j.0014-3820.2001.tb01268.x. [DOI] [PubMed] [Google Scholar]
- Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–94. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- Krylov DM, Wolf YI, Rogozin IB, Koonin EV. Gene loss, protein sequence divergence, gene dispensability, expression level, and interactivity are correlated in eukaryotic evolution. Genome Res. 2003;13:2229–35. doi: 10.1101/gr.1589103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lippman Z, Gendrel AV, Black M, Vaughn MW, Dedhia N, McCombie WR, Lavine K, Mittal V, May B, Kasschau KD, Carrington JC, Doerge RW, Colot V, Martienssen R. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–6. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- Lord PW, Stevens RD, Brass A, Goble CA. Investigating semantic similarity measures across the Gene Ontology: the relationship between sequence and annotation. Bioinformatics. 2003;19:1275–83. doi: 10.1093/bioinformatics/btg153. [DOI] [PubMed] [Google Scholar]
- Marino-Ramirez L, Bodenreider O, Kantz N, Jordan IK. Co-evolutionary Rates of Functionally Related Yeast Genes. Evol Bioinform Online. 2006;2:295–300. [PMC free article] [PubMed] [Google Scholar]
- Marino-Ramirez L, Spouge JL, Kanga GC, Landsman D. Statistical analysis of over-represented words in human promoter sequences. Nucleic Acids Res. 2004;32:949–58. doi: 10.1093/nar/gkh246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara H, Smit AF, Okada N. Functional noncoding sequences derived from SINEs in the mammalian genome. Genome Res. 2006;16:864–74. doi: 10.1101/gr.5255506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgel LE, Crick FH. Selfish DNA: the ultimate parasite. Nature. 1980;284:604–7. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- Pal C, Papp B, Hurst LD. Highly expressed genes in yeast evolve slowly. Genetics. 2001;158:927–31. doi: 10.1093/genetics/158.2.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piriyapongsa J, Jordan IK. A Family of Human MicroRNA Genes from Miniature Inverted-Repeat Transposable Elements. PLoS ONE. 2007;2:e203. doi: 10.1371/journal.pone.0000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piriyapongsa J, Marino-Ramirez L, Jordan IK. Origin and evolution of human microRNAs from transposable elements. Genetics. 2007;176:1323–37. doi: 10.1534/genetics.107.072553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–5. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satchwell SC, Drew HR, Travers AA. Sequence periodicities in chicken nucleosome core DNA. J Mol Biol. 1986;191:659–75. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–8. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons C, Makunin IV, Pheasant M, Mattick JS. Maintenance of transposon-free regions throughout vertebrate evolution. BMC Genomics. 2007;8:470. doi: 10.1186/1471-2164-8-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons C, Pheasant M, Makunin IV, Mattick JS. Transposon-free regions in mammalian genomes. Genome Res. 2006;16:164–72. doi: 10.1101/gr.4624306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR, Torvik VI. Mammalian microRNAs derived from genomic repeats. Trends Genet. 2005;21:322–6. doi: 10.1016/j.tig.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. RepeatMasker. 1996-2004 http://repeatmasker.org
- Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–8. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, Cooke MP, Walker JR, Hogenesch JB. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–7. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Yamashita R, Nakai K, Sugano S. DBTSS: DataBase of human Transcriptional Start Sites and full-length cDNAs. Nucleic Acids Res. 2002;30:328–31. doi: 10.1093/nar/30.1.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharakaraman K, Marino-Ramirez L, Sheetlin S, Landsman D, Spouge JL. Alignments anchored on genomic landmarks can aid in the identification of regulatory elements. Bioinformatics. 2005;21(Suppl 1):i440–8. doi: 10.1093/bioinformatics/bti1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaparas P, Marino-Ramirez L, Bodenreider O, Koonin EV, Jordan IK. Global similarity and local divergence in human and mouse gene co-expression networks. BMC Evol Biol. 2006;6:70. doi: 10.1186/1471-2148-6-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Lagemaat LN, Landry JR, Mager DL, Medstrand P. Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet. 2003;19:530–6. doi: 10.1016/j.tig.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Miklos G.L. Gabor, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, et al. The sequence of the human genome. Science. 2001;291:1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Wang H, Xing J, Grover D, Hedges DJ, Han K, Walker JA, Batzer MA. SVA elements: a hominid-specific retroposon family. J Mol Biol. 2005;354:994–1007. doi: 10.1016/j.jmb.2005.09.085. [DOI] [PubMed] [Google Scholar]
- Wang T, Zeng J, Lowe CB, Sellers RG, Salama SR, Yang M, Burgess SM, Brachmann RK, Haussler D. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc Natl Acad Sci U S A. 2007;104:18613–8. doi: 10.1073/pnas.0703637104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet. 2004;5:276–87. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
- Widom J. Role of DNA sequence in nucleosome stability and dynamics. Q Rev Biophys. 2001;34:269–324. doi: 10.1017/s0033583501003699. [DOI] [PubMed] [Google Scholar]
- Wolf YI, Carmel L, Koonin EV. Unifying measures of gene function and evolution. Proc Biol Sci. 2006;273:1507–15. doi: 10.1098/rspb.2006.3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zar JH. Biostatistical Analysis. Fourth ed Prentice-Hall; Upper Saddle River: 1999. [Google Scholar]
- Zhang L, Li WH. Mammalian housekeeping genes evolve more slowly than tissue-specific genes. Mol Biol Evol. 2004;21:236–9. doi: 10.1093/molbev/msh010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Repetitive DNA density and nucleosome binding affinity along mammalian proximal promoter sequences. Average nucleosome binding affinities (green line, values on left y-axis) along with average TE densities (blue line, values on right y-axis) and average SSR densities (pink line, values on right y-axis) over species-specific sets of proximal promoter sequences are plotted over each promoter position starting from -1,000bp upstream and progressing to the transcriptional start site (TSS at position 0). Trends are shown for chimpanzee (a), mouse (b) and rat (c).
Supplementary Figure 2. Sequence logos representing promoter repeat architectures for six clusters (c1-c6) of human proximal promoter sequences. For each cluster, the probability of observing TE (blue ‘T’), SSR (yellow ‘S’) or non-repetitive (black ‘N’) sequence residues is represented along proximal promoter sequences from -1kb to the TSS (x-axis) by the relative bits of information (i.e. the height of residue) on the y-axis.