Abstract
The transition from transcription initiation to elongation at the HIV-1 promoter is controlled by Tat, which recruits P-TEFb to TAR RNA to phosphorylate RNA polymerase II. It has long been unclear why the HIV-1 promoter is incompetent for elongation. We report that P-TEFb is recruited to the promoter in a catalytically inactive state bound to the inhibitory 7SK snRNP, thereby preventing elongation. It also has long been believed that TAR functions to recruit Tat to the promoter, but we find that Tat is recruited to the DNA template before TAR is synthesized. We propose that TAR binds Tat and P-TEFb as it emerges on the nascent transcript, competitively displacing the inhibitory 7SK snRNP and activating the P-TEFb kinase. Recruitment of an inhibitory snRNP complex at an early stage in the transcription cycle provides a new paradigm for controlling gene expression with a non-coding RNA.
RNA polymerase II (Pol II) activity can be regulated at several steps of the transcription cycle, including pre-initiation, DNA melting, initiation, promoter clearance, and elongation1. One well-studied case of regulation at the elongation step involves the HIV-1 Tat protein, which relieves a block to elongation at the viral promoter2–4 and functions after recruitment of the basal transcription machinery5. Without Tat, Pol II is stalled soon after initiation and is inefficiently converted to the processive form6,7, a step requiring the recruitment of positive transcription elongation factor b (P-TEFb) to a promoter-proximal RNA element (TAR) on the nascent HIV-1 pre-mRNA8. P-TEFb is composed of cyclinT1 (CycT1) and Cdk98–10 and is used at many promoters, including HIV-1, to phosphorylate Ser2 residues in the C-terminal domain (CTD) of Pol II11–13, thereby converting a non-phosphorylated form (Pol IIa) to a hyper-phosphorylated form (Pol IIo) that engages in productive elongation14–16
P-TEFb catalytic activity is tightly controlled in cells by switching the equilibrium between two states, an active P-TEFb form, which can be recruited to chromatin by interacting with Brd417,18, and an inactive ribonucleoprotein form containing the 7SK snRNP, composed of 7SK snRNA, Hexim1, and Larp7/Pip7S, in addition to the CycT1 and Cdk9 subunits10,19. Even though P-TEFb can be recruited to chromatin via Brd4, there is evidence that Tat can directly recruit P-TEFb to the HIV-1 promoter independently of Brd418. The inactive P-TEFb form also can also associate with a methylphosphate capping enzyme (Mepce), whose activity stabilizes the 7SK snRNA20.
Here we report the unexpected findings that Tat assembles into a complex with P-TEFb in its inactive 7SK snRNP form, and that this complex is recruited to the HIV-1 promoter before transcription initiation, in a TAR-independent manner. Once transcription begins, the nascent TAR hairpin is synthesized and is required to displace the inhibitory 7SK snRNP complex and activate the P-TEFb kinase, which occurs when Tat and CycT1 bind to TAR. Recruitment of the inhibitory 7SK snRNP complex at an early stage of the viral transcription cycle explains why P-TEFb is inactive at the viral promoter in the absence of Tat and establishes a new way to control the switch between transcription initiation and elongation.
RESULTS
P-TEFb/7SK snRNP assembles into Pol II complexes and Tat stimulates Cdk9 activity in vitro
We recently showed that P-TEFb (CycT1:Cdk9) is found not only in the previously described small and large P-TEFb complexes8,10,17,18, but also in purified Pol II complexes along with Tat21, like the Cdk7 and Cdk8 kinases22. We now have found that CycT1 and Cdk9 assemble into Pol II complexes even without Tat, along with non-phosphorylated Pol II (but not elongating Pol IIo), TBP, and splicing factors, and surprisingly, the 7SK snRNP (Hexim1, Larp7 and 7SK snRNA) (Fig. 1a). All 7SK snRNP components remained bound to Pol II complexes when purified further by gel filtration, yielding a single peak of 2–4 MDa (Fig. 1b), as previously reported22. P-TEFb immunoprecipitated from these Pol II complexes was catalytically inactive on a CTD substrate but, remarkably, the Cdk9 kinase was activated after incubating with GST-Tat (Fig. 1b), but not with a non-functional Tat mutant (data not shown). This result suggested that Tat may promote dissociation of the 7SK snRNP from P-TEFb preassembled with Pol II, as it does with the inactive complex in cells23,24. Indeed, the stimulation of P-TEFb catalytic activity correlates well with the displacement of Hexim1 and Larp7 from Pol II complexes, while CycT1 and Cdk9 remain bound (Fig. 1c).
Figure 1.
Inactive P-TEFb assembles into Pol II complexes and Tat activates its catalytic activity. (a) Partial composition of Pol II complexes purified on a TFIIS affinity resin22 was assessed by comparing Western blots of proteins and levels of 7SK snRNA eluted from a GST-TFIIS column to those from HeLa whole cell extracts (WCE) and a GST control column. The TFIIS used was an N-terminal fragment that possesses the same activity as full-length TFIIS22. The purified coomassie-stained recombinant GST and GST-TFIIS proteins used for the columns are shown on the left. The arrow indicates the position of full-length CycT1. (b) GST-TFIIS eluted complexes were loaded onto a Sepharose CL-2B gel filtration column and the single eluted peak was immunoprecipitated (IP) by either anti-Cdk9 or mock (normal rabbit IgG) antibodies. Both precipitates, either incubated or not with purified GST-Tat, were used in kinase assays using a GST-CTD substrate and analyzed by Western blot with the H5 antibody, which recognizes Pol IIo. (c) Pol II complexes were purified on a TFIIS affinity resin, incubated with GST or GST-Tat in vitro, immunoprecipitated with anti-Cdk9 antibody, and the presence of P-TEFb (CycT1 and Cdk9) and 7SK snRNP (Hexim1 and Larp7) was monitored by Western blot.
Assembly of Tat and P-TEFb/7SK snRNP at the HIV-1 promoter in vivo
Given that Tat and P-TEFb/7SK snRNP associate with Pol II and that Tat remodels the 7SK snRNP complex, we next examined Tat, P-TEFb/7SK snRNP, and co-factor occupancy at the HIV-1 promoter by chromatin immunoprecipitation (ChIP), utilizing eight PCR primer pairs (Fig. 2a) in a stable reporter cell line, with or without transfected Flag-tagged Tat (Supplementary Fig. 1 online). This high-resolution ChIP protocol provides a reasonably comprehensive view of factor occupancy in and around the HIV-1 promoter (Fig. 2a,b), extending previous studies of co-factor recruitment5,25,26, and for the first time, permits us to locate Tat. The ChIP signal for Tat was undetectable in mock transfections, or in cells transfected with a non-functional Tat mutant (Supplementary Fig. 2), but showed a high signal-to-noise at specific regions when transfected with optimal levels of Tat (Fig. 2b). As anticipated, Tat occupies the transcribed region where TAR is located (+103) but, remarkably, was present at almost equivalent levels in the core promoter (−75) (lower by a factor of 1.5). Tat also was seen in the downstream transcribing region, consistent with previous reports that Tat travels with the elongating Pol II27. High binding specificity is indicated by the absence of signal in the far upstream and downstream regions, −845 and +4490 respectively, and the relatively low signal at −352.
Figure 2.
Distribution of Tat and cofactors at the HIV-1 promoter and dependence on TAR. (a) Schematic of the HIV-1 LTR promoter-luciferase (FFL) reporter integrated into a HeLa cell line, showing the locations of the upstream region (−845), enhancer elements (−352), core promoter containing Sp1 and TATA-boxes (−75), transcription start site (TSS; +1), TAR element, FFL coding region, stop codon, and polyadenylation signal (p(A)). The locations of eight amplicons used in PCR quantification of ChIP-enriched DNA are shown. Numbers indicate the positions of the central base pair of each amplicon relative to the TSS. (b) ChIP assays were performed with protein extracts from the reporter cell line 48 hr after a mock (grey bars) or Flag-tagged Tat transfection (black bars) using the antibodies indicated. Values represent the percentage of input DNA immunoprecipitated and are the average of four independent PCRs from two separate immunoprecipitations from two independent cell cultures. All standard deviations are <15%. (c) ChIP assays were performed as in panel b but using extracts pre-treated with RNase A. (d) ChIP assays were performed with protein extracts obtained from a HeLa HIV-1 LTRΔTAR-FFL cell line 48 hr after a mock (grey bars) or Flag-tagged Tat transfection (black bars) using the antibodies indicated.
Other factors also show tight binding distributions. In the absence of Tat, the basal transcription factors Sp1 and TBP were recruited exclusively to the core promoter (−75, Fig. 2b), where their binding sites are known to reside28,29 (Supplementary Fig. 3). Occupancy of both Sp1 and TBP was stimulated modestly (increased by a factor of 2 to 2.5) by Tat, consistent with its previously reported effects on basal factor recruitment5 and with our observed location of Tat in the same −75 region. Because very little Tat was observed in the upstream enhancer region (−352), a region also unnecessary for Tat activation in transcription reporter assays (Supplementary Fig. 3), it appears that Tat is recruited to and functions at the core promoter and downstream elements and not at the distal enhancer.
In the absence of Tat, both P-TEFb subunits (CycT1 and Cdk9) were found at moderate levels predominantly in the core promoter region (−75) (Fig. 2b). Interestingly, like P-TEFb, the 7SK snRNP components Hexim1 and Larp7 also were observed predominantly in the core promoter, consistent with their co-purification in Pol II complexes (Fig. 1a, b). Because Larp7 is an integral component of the 7SK snRNP, and because depletion of Larp7 leads to degradation of 7SK snRNA30, we infer that 7SK snRNA also is recruited to the HIV-1 promoter (see below). Upon Tat activation, CycT1 and Cdk9 occupancy increased by a factor of 2 to 3 at −75 and ~11 at +103 (Fig. 2b), implying that Tat further enhances P-TEFb recruitment at or near the transcription start site (TSS), and also increased P-TEFb occupancy by a factor of 6 to 22 through the transcribing unit, as expected given that P-TEFb also is associated with elongating Pol II complexes31–34. Interestingly, while occupancy of the P-TEFb subunits increases upon Tat activation, an especially large decrease in the 7SK snRNP components is observed concomitantly in the +103 region (lower by a factor of ~12 for Larp7 and ~15 for Hexim1) but not at the core promoter (lower by a factor of ~1.3 for Larp7 and ~2 for Hexim1). These results are consistent with a model in which Tat binding to the nascent RNA is coupled to the dissociation of 7SK snRNP and activation of the P-TEFb kinase (see below).
P-TEFb can be recruited to chromatin through interaction with Brd417,18, which is capable of stimulating HIV-1 transcription in a Tat-independent manner18. In our ChIP assays, Brd4 was constitutively present throughout the entire promoter and transcribing region and its profile was unchanged by Tat (Fig. 2b), in agreement with reports suggesting that the interaction between Brd4 and P-TEFb is not essential for Tat activation18.
Previous observations that Tat and P-TEFb interact with Pol II and HIV-1 preinitiation complexes (PICs)31,32,34,35 raised the possibilities that they assemble with a promoter-paused Pol II, like some pre-mRNA processing factors36, and may not be recruited to the HIV-1 promoter directly through TAR recognition. Therefore, we analyzed total Pol II and CTD phospho-isoforms by ChIP (Fig. 2b). In the absence of Tat, total Pol II was localized almost exclusively to the core promoter region (−75), and its occupancy rose by a factor of ~2 upon Tat activation and by a factor of 8 to 24 throughout the transcribing region (Fig. 2b). No Pol II was detected at the upstream enhancer region (−352), suggesting that chromatin looping between enhancer elements and the core promoter is not a major feature of Tat activation. Using Pol II CTD-specific antibodies to phosphorylated Ser5 (initiating) or Ser2 (elongating) Pol II, we observed exclusively the S5P-CTD form, but not the S2P-CTD, at the core promoter without Tat (Fig. 2b). Upon Tat activation, the level of S5P-CTD at the promoter increased by a factor of 5 to 6, suggesting that preloaded Pol II is not completely phosphorylated on Ser5-CTD residues before Tat activation, in agreement with a role of Tat in enhancing Ser5 phosphorylation in reconstituted transcription complexes in vitro33. The levels of both the S5P-CTD and S2P-CTD forms increased drastically (by a factor of 10 to 100) throughout the transcribing unit, coincident with enhanced recruitment of P-TEFb to the promoter and release of the 7SK snRNP (Fig. 2b). The patterns of Pol II accumulation at the core promoter are consistent with a paused Pol II model7, where Pol II pauses near the promoter before TAR is fully transcribed6 and allows Tat to operate at post-initiation steps.
The association of RNA-binding proteins with DNA observed in ChIP assays sometimes relies on an RNA bridge37. Thus, we next performed ChIP experiments on RNase treated extracts (Fig. 2c) to determine if Tat and P-TEFb/7SK snRNP recruitment to the promoter depend on RNA (TAR or other RNAs). RNase treatment reduced Tat occupancy at the core promoter (−75) by a factor of ~2.5 but much more dramatically where TAR is positioned (+103) (lower by a factor of ~18) (Fig. 2c), likely reflecting loss of binding to the nascent TAR, while Sp1, TBP, Brd4, and Pol II occupancy were nearly unchanged (compare Fig. 2b to 2c). In marked contrast, CycT1, Cdk9, Hexim1, and Larp7 occupancy were drastically reduced by RNase treatment (lower by a factor of 2 to 16), suggesting that all may assemble as part of the entire 7SK snRNP complex. Unexpectedly, the RNAse treatment caused only a small reduction in the levels of S2P-CTD Pol II throughout the transcribing region despite the nearly total loss of CycT1 and Cdk9, suggesting that P-TEFb may still somehow associate with elongating Pol II in a manner that is affected by RNA (Fig. 2c). The Tat AD alone, lacking the RNA-binding domain, also was found at the core promoter (lower by a factor of 3 compared to Tat occupancy), and even in the absence of TAR (Supplementary Fig. 4), suggesting that Tat can assemble at the promoter through protein-protein interactions alone, although full occupancy requires protein-RNA contacts as well.
Recruitment of Tat and P-TEFb/7SK snRNP to the HIV-1 promoter does not require TAR
Because we observed Tat and P-TEFb/7SK snRNP complexes in regions of the HIV-1 promoter upstream of the TSS, we wished to determine if TAR was necessary for their recruitment. We constructed a stable reporter cell line containing a TAR deletion and monitored factor occupancy with or without transfected Flag-tagged Tat (Fig. 2d). The results are particularly enlightening. First, Tat was still loaded at the promoter region (−75), showing a reduction in occupancy by a factor of only 2 when compared with TAR, whereas occupancy at +103 was virtually lost (lower by a factor of ~27). Second, CycT1, Cdk9, Hexim1, and Larp7, along with the basal transcription factors Sp1 and TBP, all were still loaded at the core promoter (−75) without TAR (compare Fig. 2d to 2b), demonstrating that TAR is not simply a binding site for Tat:P-TEFb recruitment. Third, Hexim1 and Larp7 were no longer displaced at +103 upon Tat activation, suggesting that the interaction of Tat:P-TEFb with TAR at the viral promoter is required to eject these subunits and presumably activate Cdk9. Finally, CycT1 and Cdk9 were no longer found throughout the transcribing region, nor was Pol II CTD Ser2 phosphorylation, providing compelling evidence that Hexim1 and Larp7 are not displaced from the viral promoter without TAR, and thus Cdk9 is not activated (Fig. 2d).
Tat assembles with 7SK snRNP in vivo
Previous studies have shown that Tat releases Hexim1 from inactive P-TEFb/7SK snRNP complexes23,24,30, and may do so in part by binding to a stem-loop near the 5’ end of 7SK snRNA where Hexim1 also binds30,38, thus competing with Hexim1 for CycT1-binding, as previously shown in vitro39. Given that Tat and 7SK snRNP, including Hexim1, are recruited to the HIV-1 promoter, we wished to further assess which interactions occur between Tat and the 7SK snRNP prior to promoter binding. We performed standard immunoprecipitation experiments with Flag-tagged Tat expressed stably in a 293 cell line, and also performed crosslinking conditions similar to those used for ChIP to trap potentially less stable or transient complexes37 with P-TEFb/7SK snRNP (Fig. 3). In the absence of crosslinking, CycT1, Cdk9, and Larp7 were detected along with Tat by Western blot and 7SK snRNA by RT-PCR (Fig. 3, lane 4). Interestingly, in the formaldehyde-crosslinked sample, we observed the same components but in addition also found Hexim1 and about 7-fold more 7SK snRNA, but no detectable U6 snRNA (Fig. 3, lane 5). No bound proteins were observed in control 293 cells not expressing Tat (Fig. 3, lane 6), and the crosslinking step did not affect the levels of input protein (Fig. 3, lanes 1–3). These results suggest that Tat and P-TEFb/7SK snRNP may form pre-assembled complexes before being recruited to the HIV-1 promoter.
Figure 3.
Tat assembles with the 7SK snRNP in vivo. RNA immunoprecipitation (RIP) was performed using antibodies to Flag-tagged Tat. Western blots (upper panels) indicate that components of P-TEFb (CycT1 and Cdk9) and 7SK snRNP (Larp7 and Hexim1) form a complex with Tat. The two lower panels utilized RT-PCR to detect 7SK and U6 snRNAs.
Determinants for P-TEFb/7SK snRNP recruitment to the HIV-1 promoter
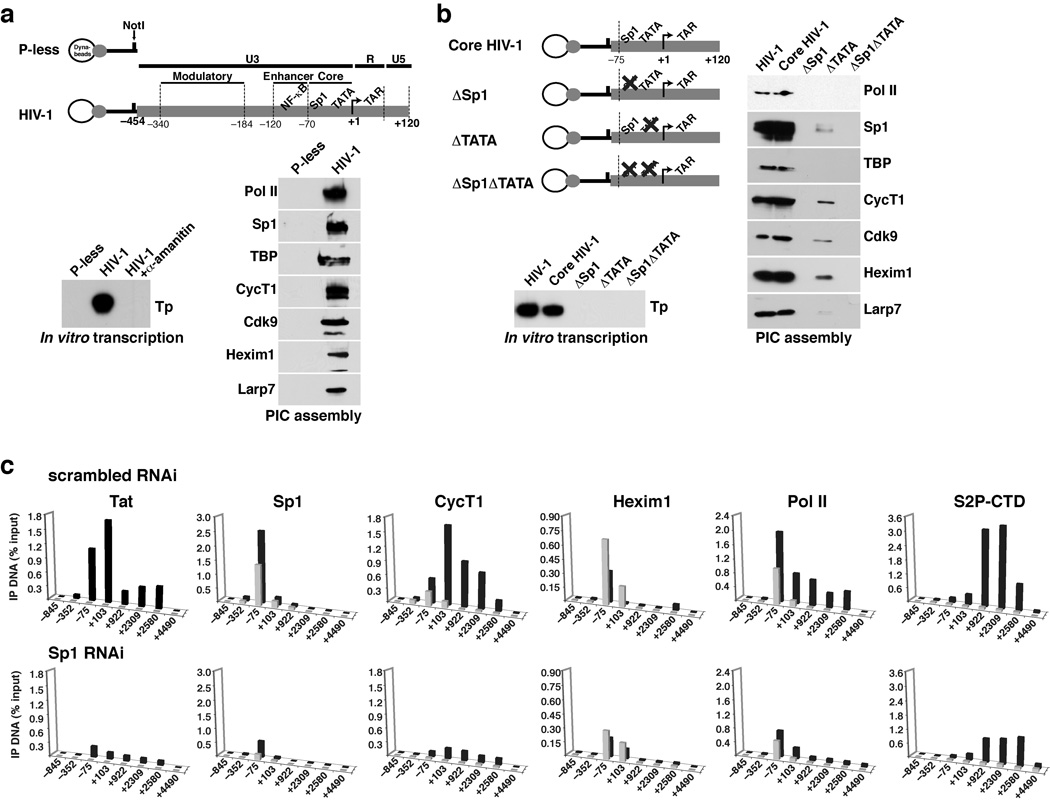
To provide direct evidence that P-TEFb/7SK snRNP complexes occupy the HIV-1 promoter, we purified transcription PICs using biotinylated templates and streptavidin-coated magnetic beads (Fig. 4a, top panel). Indeed, all components of the inactive P-TEFb complex were associated with the HIV-1 template, along with basal transcription factors (Sp1 and TBP), but not with a control promoterless template (Fig. 4a, bottom panel). The activity of the purified HIV-1 PICs was measured by in vitro transcription and primer extension and yielded the expected transcription product (Tp), which was inhibited by α-amanitin (Fig. 4a, bottom panel), confirming that the purified PICs were competent for Pol II transcription.
Figure 4.
7SK snRNP and Tat recruitment to HIV-1 PICs are Sp1-dependent. (a) Promoterless and full-length LTR templates were immobilized to streptavidin-coated magnetic beads through a biotin moiety at the 5’ end, and a NotI site was used to cleave the promoter and elute proteins from the beads, which were then analyzed by Western blot. The activity of the purified HIV-1 PICs was measured by in vitro transcription and primer extension and yielded the expected transcription product (Tp), and was inhibited by α-amanitin treatment. (b) The immobilized full-length HIV-1 LTR, core, and mutant templates shown were incubated with HeLa nuclear extracts and washed, and proteins were eluted with NotI and identified by Western blot. In vitro transcription of the PICs produced the correct Tp. (c) HeLa HIV-1 LTR reporter cells were transfected with a control scrambled siRNA or a siRNA against Sp1, resulting in >80% depletion of Sp1 and decrease in reporter activity (Supplementary Fig. 5), and ChIP assays were performed using the antibodies indicated. Values represent the percentage of input DNA immunoprecipitated and are the average of four independent PCRs from two separate immunoprecipitations from two independent cell cultures. All standard deviations are <15%
The ChIP occupancy profile of Tat, basal transcription factors, and P-TEFb/7SK snRNP components (Fig. 2) indicated that the core promoter region is important for their recruitment. To test whether the core promoter is sufficient for transcription complex assembly in vitro and transcription activation, we synthesized a minimal template containing the Sp1, TATA, initiator, and TAR elements (base pairs −105 to +120) but lacking the 5’ enhancer region (Core HIV-1, Fig. 4b). Indeed, PIC assembly with the core template was identical to the full-length promoter (Fig. 4b), as was Tat activation in transcription reporter assays (Supplementary Fig. 3). Deleting the Sp1 or TATA elements reduced or eliminated all interactions, basal transcription, and Tat activation (Fig 4b, Supplementary Fig. 3), suggesting that the basal machinery, Sp1 as suggested previously29,40,41, and P-TEFb/7SK snRNP, all contribute to forming a Tat-responsive complex.
We further assessed the in vivo relevance of Sp1 to Tat assembly at the promoter by depleting Sp1 with RNAi and monitoring factor occupancy by ChIP. Sp1 occupancy at the HIV-1 promoter decreased by a factor of ~8 (Fig. 4c) and Tat activity in transcription reporter assays decreased by a factor of ~10 (Supplementary Fig. 5). Furthermore, Sp1 knockdown almost completely eliminated Tat occupancy at the promoter, strongly decreased CycT1 and Hexim1 occupancy, and reduced the levels of Pol II CTD Ser2 phosphorylation (Fig. 4c), suggesting that Sp1 may participate in P-TEFb/7SK snRNP interactions at the viral promoter in vivo. Indeed, previous data suggest that Sp1 and P-TEFb interact in vivo and that the interaction activates HIV-1 transcription29,41,42.
DISCUSSION
Current models of HIV-1 transcription activation propose that Tat and P-TEFb are recruited to the promoter by binding to TAR on the nascent transcript. However, here we provide evidence that Tat and P-TEFb, in complex with the inhibitory 7SK snRNP, are loaded into HIV-1 PICs prior to transcription initiation and in the absence of TAR (Figs. 2 and 4). Further, we provide evidence that Tat can displace the 7SK snRNP to activate the Cdk9 kinase (Fig. 1), consistent with previous reports that Tat releases Hexim1 from the large and inactive P-TEFb complexes23,24,30, and importantly, that this activation step occurs as TAR emerges on the transcript (Fig. 2) from a promoter-paused Pol II7. These results lead to the following two-step model for Tat activation (Fig. 5a): First, the catalytically inactive P-TEFb/7SK snRNP complex is recruited to the promoter with Tat, most likely as a pre-assembled complex (Fig. 3) and through interactions with Sp1 or other basal transcription factors, and remains bound in promoter-paused transcription complexes (Figs. 4 and 5a). Second, the inhibitory 7SK snRNP is released by competitive binding of the Tat:P-TEFb complex to TAR as it is transcribed, thereby activating Cdk9 and Pol II CTD phosphorylation (Fig. 5a). Importantly, Tat and P-TEFb/7SK snRNP complexes are recruited to PICs even in a TAR-deleted promoter (Fig. 2d), and in the absence of transcription (Fig. 4a), providing direct evidence that TAR does not function solely as a recruitment site for Tat and CycT1. Even though Tat assembles into PICs in the TAR-deleted promoter, it does not activate Cdk9 and therefore the transcription complexes remain incompetent for elongation (Fig. 5b). In this revised model, TAR provides the essential function of transferring Tat and CycT1 to the nascent RNA, displacing the inhibitory 7SK snRNP in the process (Fig. 5a).
Figure 5.
Proposed model of HIV-1 transcription activation by Tat. (a) Tat assembles into complexes with P-TEFb (CycT1 and Cdk9) and the 7SK snRNP (Hexim1, Larp7, and 7SK snRNA). This Tat-7SK snRNP complex is recruited to HIV-1 PICs containing the Pol IIa form and the basal transcription machinery (e.g. Sp1, TBP), among other possible promoter-specific factors, and remains bound in the paused state (Paused complex). As transcription proceeds and TAR is synthesized, Tat facilitates the transfer of P-TEFb to the nascent RNA site. We propose that this Tat-TAR binding event competitively displaces 7SK snRNP and activates Cdk9 to phosphorylate Ser2 residues in the CTD (P) and assemble competent transcription elongation complexes containing a Pol IIo form. Hexim1 may dissociate from Larp7/7SK snRNA complexes, as it is not stably bound30, and may be replaced by hnRNP proteins in a transcription-dependent manner59,60. (b) In the absence of TAR, Tat and P-TEFb do not transfer to the nascent RNA and evict the 7SK snRNP, preventing Ser2-CTD phosphorylation (P) and formation of elongation complexes.
Several previous observations are consistent with this basic model of Tat activation: (i) P-TEFb has been found in HIV-1 PICs in the absence of Tat and remains associated with productive elongation complexes31–34,43; (ii) Tat has been found in complexes with Pol II and HIV-1 PICs prior to synthesis of TAR21,35,44; and Hexim1 over-expression inhibits Tat activation45. The finding that Tat is present in PICs is consistent with the observation that it stimulates HIV-1 transcription complex assembly5, perhaps by enhancing P-TEFb recruitment or stabilizing PICs at the promoter (Figs. 2 and 4).
In our model, P-TEFb is transferred to the nascent RNA co-transcriptionally soon after TAR is synthesized, allowing Tat and CycT1 to bind and competitively displace the 7SK snRNP. We envision that the role of TAR in ejecting the inhibitory 7SK snRNP subunits ensures that Cdk9 becomes activated on the nascent RNA and effectively times the switch between initiation and elongation. The precise nature of the nascent RNA appears unimportant for triggering Cdk9 activation, as the Tat-TAR interaction can be functionally replaced by heterologous RNA-protein interactions when the Tat activation domain is fused to the cognate RNA-binding domain46–49. Therefore, it is not TAR per se but rather the protein-RNA binding step that dissociates 7SK snRNP and activates the P-TEFb kinase. In contrast, the Tat activation domain is indispensable for activation, consistent with the observation that it is the minimal domain required to displace Hexim1 from P-TEFb by competing for the same interaction surface on CycT1, and to activate Cdk910,23,39. Even when the Tat activation domain alone is recruited to the HIV-1 promoter (Supplementary Fig. 4), it does not activate transcription, consistent with the idea that Tat does not dissociate the 7SK snRNP without the TAR binding step (Figs. 2 and 5). In agreement with the proposed co-transcriptional role for TAR, our ChIP data show that Tat cannot displace Hexim1 from the 7SK snRNP or increase Pol II Ser2 CTD-phosphorylation when TAR is deleted (Fig. 2b,d), although recombinant Tat can do so at high concentrations in vitro in the absence of template DNA or TAR (Fig. 1c). We speculate that an additional step in which Tat does not activate the P-TEFb kinase until TAR is synthesized is not recapitulated under these in vitro conditions.
It is interesting that both Tat and Hexim1 are able to bind to a 5’ region of 7SK snRNA in vitro30,38, that Hexim1 possesses an arginine-rich similar to the RNA-binding of Tat10, and that both compete for the same interaction surface on CycT123,39,45. These results provide evidence for molecular mimicry between the viral and host protein-RNA complexes (Tat-TAR and Hexim1-7SK snRNA) and suggest a competition model in which Tat takes the place of Hexim1 during the formation of transcription elongation complexes (Fig. 5), consistent with the observation that Tat displaces Hexim1 from the 7SK snRNP. Our findings also help resolve two apparent paradoxes. First, drugs that impair Pol II activity, like actinomycin D, activate the HIV-1 promoter50; and intriguingly it stimulates the release of Hexim1 from the HIV-1 promoter mimicking Tat, albeit not as efficiently (Supplementary Fig. 6). Second, drugs that reduce the levels of the inactive P-TEFb complex correspondingly reduce HIV-1 replication rates51.
There are striking similarities between Tat:P-TEFb loading at the HIV-1 promoter and transfer to the nascent pre-mRNA with co-transcriptional processes in which some pre-mRNA processing factors, such as the capping machinery and splicing factors, initially are loaded on a promoter-paused Pol II complex and later transferred to the pre-mRNA36,52,53. The proposed co-transcriptional mechanism helps solve a longstanding puzzle as to why HIV-1 evolved the use of an RNA site, in this case TAR, to regulate transcription. In our model, TAR does not function like a simple enhancer to recruit Tat to the nascent RNA, but rather allows an RNA-binding step to time the transition into productive elongation. This resembles the stimulatory effect of splicing factors on transcription elongation54. It remains to be determined if cellular promoters regulate P-TEFb activity by related mechanisms, but it is interesting that the 3’ UTR of HIC mRNA activates HIV-1 transcription elongation in a P-TEFb-dependent manner by displacing 7SK snRNA through a yet uncovered mechanism55. It is further possible that inactive P-TEFb complexes are assembled at cellular promoters and become activated by mechanisms that do not require a TAR-like RNA element. Indeed, Gal4-Tat and Gal4-P-TEFb fusions can activate transcription of an HIV-1 promoter containing several Gal4 DNA-binding sites, although multiple sites are required and activation is weaker than with TAR (5,56; D’Orso, unpublished data), implying that the normal pathway of activation may not be fully recapitulated by artificial tethering of transcription factors or co-activators. Similarly, recruiting a Gal4-P-TEFb fusion to the hsp70 promoter in Drosophila activates transcription in the absence of heat shock, but to a lower level than when activated by heat shock factor-157.
HIV-1 transcription activation by Tat provides yet another example in which a virus exploits a cellular control mechanism, in this case the coupling of transcription with pre-mRNA processing53 through the use of a regulatory non-coding RNA. It will be interesting to test if acetylation of the Tat activation domain, which enhances formation of the elongation complex on TAR49, facilitates Tat-7SK snRNP assembly or the eviction of 7SK snRNP from the HIV-1 promoter. Understanding how Tat-7SK snRNP assembly/disassembly may be coordinated with the activity of the 7SK snRNA capping enzyme Mepce20,30, with the activities of other elongation factors, such as DSIF and NELF, or with histone modifications at the HIV-1 promoter6,26 also requires further investigation. Finally, as mentioned above, it seems unlikely that the assembly of inactive P-TEFb/7SK snRNP complexes at the HIV-1 promoter is unique, raising the question of whether other cellular genes utilize similar mechanisms to control the switch between initiation and elongation.
METHODS
HIV-1 LTR reporter HeLa cell lines
To generate cell lines with an integrated reporter for ChIP analyses, we transfected HeLa cells with Ssp1-linearized pcDNA3.1 bearing the HIV-1 LTR reporters using Lipofectamine2000. We selected clones for more than four weeks in DMEM–10% FBS with 750 µg/ml of geneticin. We analyzed forty clones transfected with the wild-type reporter by transcription activation assays with HIV-1 Tat and selected several with reproducible activity levels. For ChIP analyses, we chose a single active clone containing 14 integrants and responsive to Tat. To generate the HIV-1 LTRΔTARFFL-containing cell line, we selected 32 clones and chose one containing 15 integrants that was responsive to TNF-µ for the ChIP assays21. The number of integration sites was determined by quantitative PCR21 with PCR primers shown in Supplementary Methods online (Supplementary Table 1 online).
ChIP
We transfected reporter cell lines with Tat plasmids by calcium phosphate, incubated for 36 hr, and washed in PBS. We crosslinked the chromatin with 1% formaldehyde for 5–15 min at room temperature and stopped the reaction by adding glycine (to 150 mM). We washed cells with PBS, harvested in RIPA buffer, and sonicated samples to generate ~150–bp DNA fragments. For immunoprecipitation, we pre-cleared 1 mg of protein extract for 1 hr with 50 µl of protein A- or G-dynabeads before adding 2–10 µg of antibody (Supplementary Table 2) to a fresh tube containing dynabeads. We incubated reaction mixtures overnight at 4°C with 50 µl of protein A/G-dynabeads pre-blocked with 0.5 mg ml−1 bovine serum albumin and 0.1 mg ml−1 of salmon sperm DNA. We washed the beads twice with RIPA buffer, four times with ChIP wash buffer (100 mM Tris-HCl pH 8.5, 500 mM LiCl, 1% (v/v) Nonidet P-40, 1% (w/v) deoxycholic acid), twice with RIPA buffer, and twice with 1× Tris-EDTA. We eluted immunocomplexes for 15 min at 65°C with 1% (w/v) SDS, and reversed the crosslinking by adjusting the solution to 200 mM NaCl and incubating for 16 hr at 65°C. We used a fraction of the DNA as the template in PCR reactions, which were performed in the exponential range of amplification (24 to 32 cycles, depending on the primer combination and antibody used). We electrophoresed amplification products (~70 bp) in 2.5% (w/v) agarose gels and visualized with ethidium bromide. PCR primer sequences are shown in Supplementary Table 1. Quantification of PCR products was performed with Scion Image1.63 software using known input DNA standards.
In vivo RNA Immunoprecipitation (RIP)
We crosslinked cells with formaldehyde to stably trap RNA-protein complexes formed in vivo37. Briefly, we crosslinked 293 T-Rex Tat cells in 0.1% (v/v) formaldehyde for 8 min at room temperature; washed and resuspended cell pellets in 50 mM Tris (pH 7.4), 1% (v/v) NP-40, 0.5% (w/v) sodium deoxycholate, 0.05% (w/v) SDS, 2 mM MgCl2, and 150 mM NaCl; and sonicated three times for 20 sec each on a Branson Sonifier 450 with microtip. We incubated extracts for 3 hr at 4°C with protein G dynabeads pre-bound with mouse anti-Flag or mouse anti-GFP antibody as controls. We washed bound samples four times for 10 min in binding buffer supplemented with 1 M urea and 0.5 M NaCl and then resuspended in 50 mM Tris pH 6.3, 5 mM EDTA, 1% (w/v) SDS, and 10 mM DTT. We reversed crosslinks by heating for 45 min at 70°C, purified RNA with TRIzol reagent, and performed first-strand cDNA synthesis using random decamers. We detected 7SK and U6 snRNAs (Supplementary Table 3) by RT-PCR.
RNAi
For Sp1 knockdown, we plated HeLa cells to a density of 5×105 cells per well in 6-well plates and analyzed by Western blot 72 hr after transfecting Sp1 siRNA (sc-29487) or scrambled siRNA (sc-37007) (Santa Cruz). For RNAi-ChIP, we plated cells in 15 cm2 dishes, transfected with the siRNA for 72 hr, and then re-transfected with a Tat-Flag (or empty vector) for 36 hr before harvesting for ChIP assays.
Purification of Pol II complexes
We used a TFIIS affinity resin to purify Pol II22,58. To prepare the resin, we lysed E. coli BL21 DE3 cells expressing GST or GST-TFIIS(N) (a TFIIS N terminal fragment22) by sonication in TBS buffer containing 150 mM NaCl and protease inhibitors (2 mM PMSF and 1 mM EDTA) on ice. We clarified and treated supernatants with 1% (v/v) Triton X-100 for 10 min, centrifuged, and bound to a glutathione-agarose resin. We prepared HeLa whole cell extracts (WCE) as described22, and purified Pol II complexes eluted from the TFIIS resin further by gel filtration on a Sepharose CL-2B column in 20 mM Tris-HCl pH 7.9, 0.1 mM DTT, 0.1 mM EDTA, 20% glycerol, and 150 mM NaCl. We analyzed composition by Western blotting with a panel of antibodies and by RT-PCR of TRIZol-purified RNAs to detect 7SK RNA with primers shown in Supplementary Table 3. We immunoprecipitated P-TEFb from the Pol II preparation using an anti-Cdk9 antibody and used it for two experiments: kinase activity (Fig. 1b) and disassembly of 7SK snRNP in vitro (Fig. 1c). To measure CTD kinase activity, we incubated 50 ng of GST-CTD and 100 µM ATP in the absence or presence of GST-Tat (100 ng) in 50 mM Tris-HCl pH 7.5, 1.5 mM DTT, 5 mM MnCl2, and 4 mM MgCl2 for 60 min at 30°C. We detected phosphorylated GST-CTD (CTD IIo form) by Western blotting with a Ser2P-CTD antibody. GST-Tat (residues 1–72) was expressed from a pGEX2T vector and purified on a glutathione-agarose column. To test the role of GST-Tat on the integrity of the 7SK snRNP complex in vitro, we purified Pol II complexes on a GST-TFIIS column, incubated for 30 min with GST or GST-Tat, immunoprecipitated Cdk9 as in Fig. 1b, and analyzed bound factors by Western blot. To monitor transcription, we prepared nuclear extracts49 and performed transcription reactions using immobilized linear templates and primer extension.
Supplementary Material
ACKNOWLEDGEMENTS
We thank K. Yamamoto, P. Walter, C. Guthrie, R. Andino, J. Gross and N. Krogan for comments. We are grateful to J. Greenblatt (University of Toronto) for reagents, members of the Frankel and Yamamoto lab for discussions, and the reviewers for their insightful comments. This work was initially supported by an amfAR Mathilde Krim fellowship in Basic Biomedical Research (106988-43-RFNT) and a NIH K99/R00 grant A112185 to I.D., and NIH grants AI29135 and P50 GM082250 (HARC Center) to A.D.F.
Footnotes
AUTHOR CONTRIBUTIONS
I.D. and A.D.F. designed research, I.D. performed research; I.D. and A.D.F. analyzed data and wrote the paper
REFERENCES
- 1.Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 2.Feinberg MB, Baltimore D, Frankel AD. The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcriptional elongation. Proc. Natl. Acad. Sci. U S A. 1991;88:4045–4049. doi: 10.1073/pnas.88.9.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laspia MF, Rice AP, Mathews MB. HIV-1 Tat protein increases transcriptional initiation and stabilizes elongation. Cell. 1989;59:283–292. doi: 10.1016/0092-8674(89)90290-0. [DOI] [PubMed] [Google Scholar]
- 4.Garber ME, et al. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 1998;12:3512–3527. doi: 10.1101/gad.12.22.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raha T, Cheng SW, Green MR. HIV-1 Tat stimulates transcription complex assembly through recruitment of TBP in the absence of TAFs. PLoS Biol. 2005;3:e44. doi: 10.1371/journal.pbio.0030044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Z, Klatt A, Gilmour DS, Henderson AJ. Negative elongation factor NELF represses human immunodeficiency virus transcription by pausing the RNA polymerase II complex. J. Biol. Chem. 2007;282:16981–16988. doi: 10.1074/jbc.M610688200. [DOI] [PubMed] [Google Scholar]
- 7.Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peterlin BM, Price DH. Controlling the Elongation Phase of Transcription with P-TEFb. Mol. Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 9.Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 10.Zhou Q, Yik JH. The Yin and Yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol. Mol. Biol. Rev. 2006;70:646–659. doi: 10.1128/MMBR.00011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim YK, Bourgeois CF, Isel C, Churcher MJ, Karn J. Phosphorylation of the RNA polymerase II carboxyl-terminal domain by CDK9 is directly responsible for human immunodeficiency virus type 1 Tat-activated transcriptional elongation. Mol. Cell. Biol. 2002;22:4622–4637. doi: 10.1128/MCB.22.13.4622-4637.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahn SH, Kim M, Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3' end processing. Mol. Cell. 2004;13:67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- 14.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 15.Gomes NP, et al. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 2006;20:601–612. doi: 10.1101/gad.1398206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol. Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jang MK, et al. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 18.Yang Z, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 19.Yik JH, et al. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell. 2003;12:971–982. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- 20.Jeronimo C, et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol. Cell. 2007;27:262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D'Orso I, Grunwell JR, Nakamura RL, Das C, Frankel AD. Targeting tat inhibitors in the assembly of human immunodeficiency virus type 1 transcription complexes. J. Virol. 2008;82:9492–9504. doi: 10.1128/JVI.00763-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan G, Aso T, Greenblatt J. Interaction of elongation factors TFIIS and elongin A with a human RNA polymerase II holoenzyme capable of promoter-specific initiation and responsive to transcriptional activators. J. Biol. Chem. 1997;272:24563–24571. doi: 10.1074/jbc.272.39.24563. [DOI] [PubMed] [Google Scholar]
- 23.Barboric M, et al. Tat competes with HEXIM1 to increase the active pool of P-TEFb for HIV-1 transcription. Nucleic Acids Res. 2007;35:2003–2012. doi: 10.1093/nar/gkm063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedore SC, et al. Manipulation of P-TEFb control machinery by HIV: recruitment of P-TEFb from the large form by Tat and binding of HEXIM1 to TAR. Nucleic Acids Res. 2007;35:4347–4358. doi: 10.1093/nar/gkm443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bres V, Gomes N, Pickle L, Jones KA. A human splicing factor, SKIP, associates with P-TEFb and enhances transcription elongation by HIV-1 Tat. Genes Dev. 2005;19:1211–1226. doi: 10.1101/gad.1291705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bres V, Yoshida T, Pickle L, Jones KA. SKIP interacts with c-Myc and Menin to promote HIV-1 Tat transactivation. Mol Cell. 2009;36:75–87. doi: 10.1016/j.molcel.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keen NJ, Gait MJ, Karn J. Human immunodeficiency virus type-1 Tat is an integral component of the activated transcription-elongation complex. Proc. Natl. Acad. Sci. U S A. 1996;93:2505–2510. doi: 10.1073/pnas.93.6.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berkhout B, Gatignol A, Rabson AB, Jeang KT. TAR-independent activation of the HIV-1 LTR: evidence that tat requires specific regions of the promoter. Cell. 1990;62:757–767. doi: 10.1016/0092-8674(90)90120-4. [DOI] [PubMed] [Google Scholar]
- 29.Kamine J, Subramanian T, Chinnadurai G. Sp1-dependent activation of a synthetic promoter by human immunodeficiency virus type 1 Tat protein. Proc. Natl. Acad. Sci. U S A. 1991;88:8510–8514. doi: 10.1073/pnas.88.19.8510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krueger BJ, et al. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 2008;36:2219–2229. doi: 10.1093/nar/gkn061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Isel C, Karn J. Direct evidence that HIV-1 Tat stimulates RNA polymerase II carboxyl-terminal domain hyperphosphorylation during transcriptional elongation. J. Mol. Biol. 1999;290:929–941. doi: 10.1006/jmbi.1999.2933. [DOI] [PubMed] [Google Scholar]
- 32.Ping YH, Rana TM. Tat-associated kinase (P-TEFb): a component of transcription preinitiation and elongation complexes. J. Biol. Chem. 1999;274:7399–7404. doi: 10.1074/jbc.274.11.7399. [DOI] [PubMed] [Google Scholar]
- 33.Zhou M, et al. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol. Cell. Biol. 2000;20:5077–5086. doi: 10.1128/mcb.20.14.5077-5086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou C, Rana TM. A bimolecular mechanism of HIV-1 Tat protein interaction with RNA polymerase II transcription elongation complexes. J. Mol. Biol. 2002;320:925–942. doi: 10.1016/s0022-2836(02)00556-9. [DOI] [PubMed] [Google Scholar]
- 35.Garcia-Martinez LF, Ivanov D, Gaynor RB. Association of Tat with purified HIV-1 and HIV-2 transcription preinitiation complexes. J. Biol. Chem. 1997;272:6951–6958. doi: 10.1074/jbc.272.11.6951. [DOI] [PubMed] [Google Scholar]
- 36.Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat. Struct. Mol. Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA. Reversible crosslinking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods. 2002;26:182–190. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- 38.Czudnochowski N, Vollmuth F, Baumann S, Vogel-Bachmayr K, Geyer M. Specificity of Hexim1 and Hexim2 complex formation with cyclin T1/T2, importin alpha and 7SK snRNA. J. Mol. Biol. 2010;395:28–41. doi: 10.1016/j.jmb.2009.10.055. [DOI] [PubMed] [Google Scholar]
- 39.Schulte A, et al. Identification of a cyclin T-binding domain in Hexim1 and biochemical analysis of its binding competition with HIV-1 Tat. J. Biol. Chem. 2005;280:24968–24977. doi: 10.1074/jbc.M501431200. [DOI] [PubMed] [Google Scholar]
- 40.Berkhout B, Silverman RH, Jeang KT. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell. 1989;59:273–282. doi: 10.1016/0092-8674(89)90289-4. [DOI] [PubMed] [Google Scholar]
- 41.Kamine J, Chinnadurai G. Synergistic activation of the human immunodeficiency virus type 1 promoter by the viral Tat protein and cellular transcription factor Sp1. J. Virol. 1992;66:3932–3936. doi: 10.1128/jvi.66.6.3932-3936.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yedavalli VS, Benkirane M, Jeang KT. Tat and trans-activation-responsive (TAR) RNA-independent induction of HIV-1 long terminal repeat by human and murine cyclin T1 requires Sp1. J. Biol. Chem. 2003;278:6404–6410. doi: 10.1074/jbc.M209162200. [DOI] [PubMed] [Google Scholar]
- 43.Montanuy I, Torremocha R, Hernandez-Munain C, Sune C. Promoter influences transcription elongation: TATA-box element mediates the assembly of processive transcription complexes responsive to cyclin-dependent kinase 9. J. Biol. Chem. 2008;283:7368–7378. doi: 10.1074/jbc.M706243200. [DOI] [PubMed] [Google Scholar]
- 44.Cujec TP, et al. The human immunodeficiency virus transactivator Tat interacts with the RNA polymerase II holoenzyme. Mol. Cell. Biol. 1997;17:1817–1823. doi: 10.1128/mcb.17.4.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fraldi A, et al. Inhibition of Tat activity by the HEXIM1 protein. Retrovirology. 2005;2:42. doi: 10.1186/1742-4690-2-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Southgate C, Zapp ML, Green MR. Activation of transcription by HIV-1 Tat protein tethered to nascent RNA through another protein. Nature. 1990;345:640–642. doi: 10.1038/345640a0. [DOI] [PubMed] [Google Scholar]
- 47.Selby MJ, Peterlin BM. Trans-activation by HIV-1 Tat via a heterologous RNA binding protein. Cell. 1990;62:769–776. doi: 10.1016/0092-8674(90)90121-t. [DOI] [PubMed] [Google Scholar]
- 48.Smith CA, Calabro V, Frankel AD. An RNA-binding chameleon. Mol. Cell. 2000;6:1067–1076. doi: 10.1016/s1097-2765(00)00105-2. [DOI] [PubMed] [Google Scholar]
- 49.D'Orso I, Frankel AD. Tat acetylation modulates assembly of a viral-host RNA-protein transcription complex. Proc. Natl. Acad. Sci. U S A. 2009;106:3101–3106. doi: 10.1073/pnas.0900012106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Casse C, Giannoni F, Nguyen VT, Dubois MF, Bensaude O. The transcriptional inhibitors, actinomycin D and alpha-amanitin, activate the HIV-1 promoter and favor phosphorylation of the RNA polymerase II C-terminal domain. J. Biol. Chem. 1999;274:16097–16106. doi: 10.1074/jbc.274.23.16097. [DOI] [PubMed] [Google Scholar]
- 51.Biglione S, et al. Inhibition of HIV-1 replication by P-TEFb inhibitors DRB, seliciclib and flavopiridol correlates with release of free P-TEFb from the large, inactive form of the complex. Retrovirology. 2007;4:47. doi: 10.1186/1742-4690-4-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ujvari A, Luse DS. Newly Initiated RNA encounters a factor involved in splicing immediately upon emerging from within RNA polymerase II. J. Biol. Chem. 2004;279:49773–49779. doi: 10.1074/jbc.M409087200. [DOI] [PubMed] [Google Scholar]
- 53.Kornblihtt AR, de la Mata M, Fededa JP, Munoz MJ, Nogues G. Multiple links between transcription and splicing. RNA. 2004;10:1489–1498. doi: 10.1261/rna.7100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fong YW, Zhou Q. Stimulatory effect of splicing factors on transcriptional elongation. Nature. 2001;414:929–933. doi: 10.1038/414929a. [DOI] [PubMed] [Google Scholar]
- 55.Young TM, Tsai M, Tian B, Mathews MB, Pe'ery T. Cellular mRNA activates transcription elongation by displacing 7SK RNA. PLoS One. 2007;2:e1010. doi: 10.1371/journal.pone.0001010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Southgate CD, Green MR. The HIV-1 Tat protein activates transcription from an upstream DNA-binding site: implications for Tat function. Genes Dev. 1991;5:2496–2507. doi: 10.1101/gad.5.12b.2496. [DOI] [PubMed] [Google Scholar]
- 57.Lis JT, Mason P, Peng J, Price DH, Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 2000;14:792–803. [PMC free article] [PubMed] [Google Scholar]
- 58.Robert F, Blanchette M, Maes O, Chabot B, Coulombe B. A human RNA polymerase II-containing complex associated with factors necessary for spliceosome assembly. J. Biol. Chem. 2002;277:9302–9306. doi: 10.1074/jbc.M110516200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barrandon C, Bonnet F, Nguyen VT, Labas V, Bensaude O. The transcription-dependent dissociation of P-TEFb-HEXIM1-7SK RNA relies upon formation of hnRNP-7SK RNA complexes. Mol. Cell. Biol. 2007;27:6996–7006. doi: 10.1128/MCB.00975-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van Herreweghe E, et al. Dynamic remodelling of human 7SK snRNP controls the nuclear level of active P-TEFb. EMBO J. 2007;26:3570–3580. doi: 10.1038/sj.emboj.7601783. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.