Abstract
Deletion of glycine N-methyltransferase (GNMT) in mice, the main gene involved in liver S-adenosylmethionine (SAMe) catabolism, leads to the hepatic accumulation of this molecule and the development of fatty liver and fibrosis. To demonstrate that the excess of hepatic SAMe is the main agent contributing to liver disease in GNMT-KO mice, we treated 1.5-month old GNMT-KO mice for 6 weeks with nicotinamide (NAM), a substrate of the enzyme NAM N-methyltransferase. NAM administration markedly reduced hepatic SAMe content, prevented DNA-hypermethylation and normalized the expression of critical genes involved in fatty acid metabolism, oxidative stress, inflammation, cell proliferation, and apoptosis. More important, NAM treatment prevented the development of fatty liver and fibrosis in GNMT-KO mice. Because GNMT expression is down-regulated in patients with cirrhosis and there are subjects with GNMT mutations who have spontaneous liver disease, the clinical implication of the present findings is obvious at least with respect to these latter individuals. Especially since NAM has been used for many years to treat a broad spectrum of diseases including pellagra and diabetes without significant side effects, it should be considered in subjects with GNMT mutations.
Conclusions
These results indicate that the anomalous accumulation of SAMe in GNMT-KO mice can be corrected by NAM treatment leading to the normalization of the expression of many genes involved in fatty acid metabolism, oxidative stress, inflammation, cell proliferation and apoptosis, and to the reversion of the appearance of the pathologic phenotype.
Keywords: S-Adenosylmethionine, DNA methylation, Ras signaling, JAK/STAT signaling, hepatocytes
Introduction
Expression of glycine N-methyltransferase (GNMT) is predominant in hepatocytes, where it comprises about 1% of the total soluble protein, but is also found in other tissues such as pancreas and prostate (1). GNMT catalyzes the conversion of glycine into sarcosine (methyl-glycine), which is then oxidized to regenerate glycine (Figure 1). The function of this futile cycle is to catabolize excess S-adenosylmethionine (SAMe) synthesized by the liver after an increase in methionine concentration (i.e. after a protein-rich meal) to maintain a constant SAMe/S-adenosylhomocysteine (SAH) ratio and avoid aberrant methylation reactions (1,2). Accordingly, individuals with GNMT mutations that lead to inactive forms of the enzyme have elevated blood levels of methionine and SAMe, but the concentration of total homocysteine (the product of SAH hydrolysis) is normal (3,4). GNMT-knockout mice (GNMT-KO) recapitulate the situation observed in individuals with mutations of the GNMT gene (5,6) and have elevated methionine and SAMe both in serum and liver. These findings indicate that the hepatic reduction in total transmethylation flux caused by the absence of GNMT cannot be compensated by other methyltransferases that are abundant in the liver, such as guanidinoacetate N-methyltransferase, phosphatidylethanolamine N-methyltransferase or nicotinamide N-methyltransferase (NNMT), and that this situation leads to the accumulation of hepatic SAMe and increased transport of this molecule to the blood.
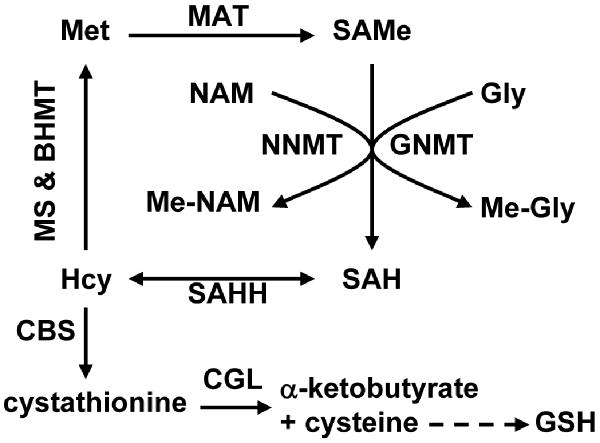
Figure 1.
Schematic representation of hepatic methionine metabolism. The first steps in hepatic methionine (Met) metabolism are conversion to S-adenosylmethionine (SAMe), through a reaction catalyzed by methionine adenosyltransferase (MAT), and transfer of the methyl group of SAMe to numerous methyl acceptors with formation of S-adenosylhomocysteine (SAH). Although there are a large number of SAMe-dependent methyltransferases, the reaction that quantitatively contributes most to the transmethylation flux is the methylation of glycine by glycine N-methyltransferase (GNMT, a reaction activated by SAMe) to form N-methyl-glycine (Me-Gly) (1,2). Accordingly, the liver of GNMT-KO mice shows a marked increase in the concentration of SAMe and aberrant methylation reactions of DNA and proteins (6). Nicotinamide (NAM) administration may be used to reduce the elevated levels of hepatic SAMe in GNMT-KO mice. NAM is methylated to form N-methyl-NAM (Me-NAM) by the enzyme NAM N-methyltransferase (NNMT). SAH is subsequently hydrolyzed to homocysteine (Hcy) by SAH hydrolase (SAHH), which is an important metabolic hub. Hcy can be remethylated to regenerate Met, via either methionine synthase (MS, a pathway inhibited by SAMe) and betaine homocysteine methyltransferase (BHMT), or used for the synthesis of cysteine and α-ketobutyrate as result of its transsulfuration. The transsulfuration pathway involves two enzymes: cystathionine β–synthase (CBS, which is activated by SAMe) and cystathionine γ-lyase (CGL). Cysteine is then utilized for the synthesis of glutathione (GSH), while α-ketobutyrate penetrates the mitochondria where is further metabolized. This coordinated modulation by SAMe of the flux of Hcy through the remethylation and transsulfuration pathways maximizes the production of cysteine, and consequently of GSH, after a methionine load minimizing the regeneration of this amino acid.
The marked steatosis and fibrosis observed in GNMT-KO mice (6) suggests that GNMT prevents these two processes both directly, by avoiding the excessive accumulation of SAMe in the hepatocytes, and indirectly, by preventing the activation of stellate cells through an as yet unidentified mechanism. The development of hepatocellular carcinoma (HCC) in GNMT-KO mice (6-8) suggests also that GNMT prevents carcinogenesis directly by avoiding the hypermethylation of DNA and histones of specific and critical carcinogenesis pathways such as the Ras and JAK/STAT inhibitors SOCS1-3, CIS, and RASSF1, RASFF4 (6). GNMT is silenced in human HCC and down-regulated in the livers of patients at risk of developing HCC such as in those with hepatitis C virus-and alcohol-induced cirrhosis (9,10). The identification of several individuals with mutations of GNMT as having mild to moderate liver disease with elevated serum aminotransferases (3,4) further suggests that GNMT plays a critical function in human liver health and its silencing can lead to disease.
Using the yeast two hybrid strategy, Rual et al (11) found interactions between GNMT and a variety of proteins including arrestin 3 and beta arrestin 1, two proteins involved in the regulation of G protein-coupled receptors and MAPK. This finding raises the question of whether the effect of GNMT silencing on the development of fatty liver, fibrosis and carcinogenesis is triggered only by the increase in cellular SAMe or involves the interaction of GNMT with other proteins. Here we show that treatment of GNMT-KO mice with nicotinamide (NAM), a substrate of NNMT, an enzyme mainly expressed in the liver that removes NAM by converting it to N-methyl-NAM (12), leads to the normalization of hepatic SAMe content and prevention of fatty liver and fibrosis formation.
Experimental Procedures
Experimental Design
Wild-type (WT) and GNMT-KO mice were fed a standard diet (Harland Teklad irradiated mouse diet 2014, Madison, WI) and housed in a temperature-controlled animal facility with 12 hours light-dark cycles. One-and-a-half month old GNMT-KO (n=20) and WT (n=5) mice were treated for 6 weeks with NAM (50 μM dissolved in the drinking water, which was replaced weekly, SIGMA-Aldrich) before sacrifice. For control groups, we used WT (n=15) and GNMT-KO mice (n=10) of the same age. At the time of sacrifice, livers were rapidly split into several pieces, some were snap frozen for subsequent RNA or protein extraction, and others were formalin fixed for histology and immunohistochemistry. Serum samples were also collected for determination of alanine aminotransferase (ALT) and aspartate aminotransferase (AST) activity. Animals were treated humanely and all procedures were in compliance with our institutions' guidelines for the use of laboratory animals.
Histology and Immunohistochemistry
Sections from formalin-fixed liver tissue were stained with hematoxylin and eosin or with Sirius red for collagen visualization. For α-SMA immunostaining and apoptosis detection, frozen liver tissue sections were fixed with 4% paraformaldehyde for 15 min at room temperature, followed by treatment with 3% hydrogen peroxide in methanol for 10 minutes. The sections were then incubated with 150 mM sodium citrate for 2 min followed by washes in PBS. For α-SMA immunolabeling, anti-α-SMA Cy3-conjugated antibody (SIGMA) was applied overnight at 4°C. For apoptosis detection, FITC-conjugated TUNEL enzyme was applied overnight at 4°C (in situ cell death detection kit, Roche). Washing in ultrapure H2O and then in PBS terminated the reaction. Nuclei were then labeled with Hoechst and the cover slips mounted in Citifluor mounting medium.
Quantitative Real-Time PCR
Total RNA was isolated using the RNeasy Mini Kit (Qiagen) including DNase treatment on column. Total RNA (1.5 μg) was retrotranscribed with Super Script III (Invitrogen) in the presence of random primers and oligo dT following manufacturer's instructions. Real-time PCR was performed using the BioRad iCycler thermalcycler. Five μl of a 1/20 dilution were used in each PCR reaction in a total reaction volume of 30 μl using iQ SYBR Green Super Mix (BioRad) and all reactions were performed in duplicate. PCR was carried out with the primers described in Table 1 (supplementary material). After checking specificity of the PCR products with the melting curve, Ct values were extrapolated to a standard curve performed simultaneously with the samples and data was then normalized to GAPDH expression. Validation of GAPDH as a housekeeping gene for normalizing RNA expression was carried out after measuring the expression of mRNA encoding GAPDH, actin, and ARP using quantitative real-time PCR in matched NAM treated and non-treated tissues obtained from 16 liver samples. GAPDH expression was no significantly different between these two groups.
Protein Isolation and Western Blot
Tissue was homogenized in lysis buffer (10 mM Tris/HCl pH 7.6. 5 mM EDTA, 50 mM NaCl, 1% Triton X–100, complete protease inhibitor cocktail, and 50 mM NaF). Samples were centrifuged (15,000g, 1 hour, 4°C) and supernatants were collected. Protein lysates (30 μg, determined by BCA) were electrophoresed on SDS-polyacrylamide gels and transferred onto nitrocellulose membranes. After blocking, membranes were incubated overnight at 4°C with specific antibodies against RASSF1A from Biosciences (San Diego, CA), pSTAT3 (Tyr705), pJAK2 (Tyr1007/1008), ERK1/2, pERK1/2 (Thr202/Tyr204) and pRAF1 (Ser338) from Cell Signaling Technology (Danvers, MA), STAT3, Ki-67 and cyclin D1 from Santa Cruz Biotechnology (Santa Cruz, CA), phospho-histone 3 (Ser10) from Millipore (Billerica, MA), and SOCS1 from Novus Biologicals (Littleton, CO). This was followed by 1-hour incubation with goat anti-mouse (Santa Cruz Biotechnology) or goat anti-rabbit (Biorad, UK) secondary antibodies conjugated to horseradish peroxidase. Immunoreactive proteins were detected by Western lightning Chemiluminescence Reagent (PerkinElmer LAS, INC. Boston).
Global DNA Methylation
Global DNA methylation was assessed by two different methods. In the first method, 5-methyl-cytosine (5mC) genomic content was determined by high-performance capillary electrophoresis (13). In brief, genomic DNA samples were boiled, treated with nuclease P1 (Sigma) for 16 hours at 37°C and with alkaline phosphatase (Sigma) for an additional 2 hours at 37°C. After hydrolysis, total cytosine and 5mC content were measured by capillary electrophoresis using a P/ACE MDQ system (Beckman Coulter). Relative 5mC content was expressed as the percentage of total cytosine content (methylated and nonmethylated). Each sample was assayed in triplicate. The second method used to assess global DNA methylation is based in the use of HpaII methylation-sensitive restriction endonucleases that leaves a 5′-guanine overhang after DNA cleavage, with subsequent single nucleotide extension with radiolabeled [3H]dCTP (14). The extent of [3H]dCTP incorporation after restriction enzyme treatment is directly proportional to the number of unmethylated (cleavage) CpG sites.
Hepatic SAMe, SAH and Glutathione Levels
Liver specimens were homogenized in 0.4 M perchloric acid on ice for 5 min and centrifuged at 1,000g for 15 min at 4°C. The aqueous layer was quantitatively removed, neutralized with 3M KOH and centrifuged at 3,000g for 10 min at 4°C. SAMe and SAH concentrations were determined by LC/MS using a Waters ACQUITY-UPLC system coupled to a Waters Micromass LCT Premier Mass Spectrometer equipped with a Lockspray ionization source as described previously (15). The amount of glutathione (GSH) was determined using the Sigma GSH kit.
Statistical Analysis
Student t test was used to evaluate statistical significance. Values of P < 0.05 were considered statistically significant.
Results
Nicotinamide Treatment Reduces Hepatic SAMe Content in GNMT Knockout Mice
Our previous studies showed that SAMe levels of both liver (5) and serum (6) of GNMT-KO mice are much higher than in WT animals. This is accompanied by development of liver injury and eventually by development of HCC (9). In order to prove that the pathologic phenotype is a result of the elevated levels of SAMe in the liver we sought to reduce the elevated levels by administration of NAM and see whether this would reverse the appearance of the pathologic phenotype. The enzyme, NNMT, uses SAMe to form N-methyl-NAM which is excreted in the urine (Figure 1). In order to verify this hypothesis, NAM was added to the drinking water of 1.5-month old GNMT-KO and WT mice for 6 weeks and at the end of this period the hepatic SAMe content was determined. As previously demonstrated (5), SAMe content in the livers of 3-month old GNMT-KO animals was about 40-fold higher than in WT animals (Table 2). As hypothesized, the livers of NAM-treated GNMT-KO animals exhibited markedly lower SAMe levels than untreated GNMT-KO mice. The administration of NAM to WT animals had no significant effect on hepatic SAMe content. This result is compatible with the function of GNMT as a SAMe buffer. SAMe is an allosteric regulator of GNMT (1). Accordingly, when the hepatic content of SAMe increases, as a result of its augmented synthesis or reduced catabolism, GNMT activity is stimulated; and when the content of SAMe diminishes, as a result of a decrease in its synthesis or increased consumption, GNMT activity is reduced. The amount of hepatic SAH in GNMT-KO mice was similar to that of WT animals (Table 2). However, in the livers of NAM-treated GNMT-KO mice SAH content was about 1.7-fold higher compared to the untreated animals. The administration of NAM to WT animals had no significant effect on hepatic SAH content. It is remarkable that the levels of hepatic GSH are similar in the WT and GNMT-KO animals in spite of the significant reduction in transmethylation reactions. This is probably due to the activation by SAMe of cystathionine β-synthase (CBS), the first enzyme linking homocysteine with GSH synthesis, and also to the inhibition by SAMe of homocysteine remethylation (2) (Figure 1). In WT and GNMT-KO mice, NAM administration had no significant effect on hepatic GSH content (Table 2).
Table 2.
Effect of nicotinamide (NAM) administration on hepatic S-adenosylmethionine (SAMe), S-adenosylhomocysteine (SAH) and glutathione (GSH) content in wild-type (WT) and GNMT-KO mice.
| WT | GNMT-KO | WT + NAM | GNMT-KO + NAM | |
|---|---|---|---|---|
| SAMe nmol/g tissue |
87.8 ± 8.9 | 3,245 ± 134* | 63.8 ± 14.4 | 94.4 ± 9.7** |
| SAH nmol/g tissue |
41.9 ± 6.5 | 36.3 ± 8.6 | 38.3 ± 6.9 | 62.2 ± 13.4** |
| GSH μmol/g tissue |
13.8 ± 0.8 | 14.4 ± 0.3 | 12.4 ± 0.2 | 14.5 ± 0.2 |
P <0.05, GNMT-KO versus WT mice;
P <0.05, GNMT-KO versus GNMT-KO + NAM mice. Results represent the mean ± SEM of quintuplicate experiments.
Nicotinamide Treatment Prevents Liver Injury, Steatosis and Fibrosis in GNMT Knockout Mice
Next, we determined the levels of serum aminotransferases in NAM-treated GNMT-KO mice. We have previously demonstrated that both serum ALT and AST are increased in GNMT-KO mice compared with WT animals (6). We observed that in NAM-treated GNMT-KO mice the serum aminotransferases were significantly reduced when compared with untreated GNMT-KO animals (AST activity was 55.3 ± 4.1 U/l in WT mice, 266.3 ± 27 U/L in GNMT-KO animals, and 61.8 ± 9.5 U/l in NAM-treated GNMT-KO; and ALT activity was 24.5 ± 3 U/l in WT mice, 177.8 ± 10.4 U/L in GNMT-KO animals, and 35.6 ± 1.4 U/l in NAM-treated GNMT-KO; n = 5, P < 0.05 NAM-treated GNMT-KO versus untreated GNMT-KO for both aminotransferases).
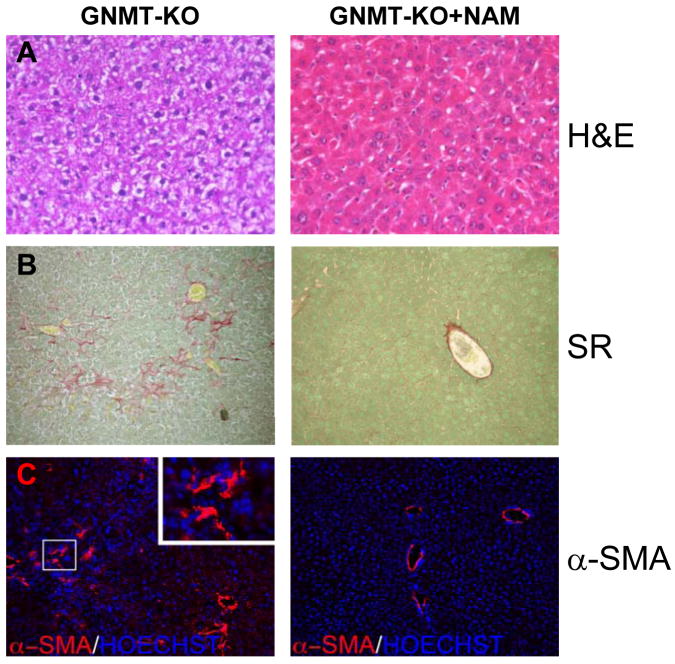
Furthermore, histological examination revealed that the livers of 3-month old GNMT-KO mice treated with NAM lacked signs of steatosis or fibrosis. As previously reported (6), 3-months old GNMT-KO mice exhibited extensive accumulation of liver fat, evident on hematoxylin and eosin staining, and fibrosis, demonstrated by Sirius red staining and α-SMA immunohistochemistry (Figure 2). In contrast, NAM-treated GNMT-KO mice showed no signs of either steatosis or fibrosis (Figure 2). Liver histology was normal in NAM-treated WT animals.
Figure 2.
Hepatic steatosis and fibrosis in GNMT-KO mice after NAM administration. (A) Livers of 3-month old GNMT-KO mice treated with NAM during the last 6 weeks lacked signs of hepatic steatosis (white droplets) compared to GNMT-KO livers. NAM administration in GNMT-KO mice prevented also (B) collagen deposition (Sirius red staining) and (C) α-SMA expression, both indicative of liver fibrosis. At least 10 animals per group were examined (H&E, hematoxylin and eosin staining; SR, Sirius red staining; and α-SMA, α-smooth muscle actin). Original magnification 40× (Zeiss A×10 microscope).
Nicotinamide Treatment Normalizes Hepatic Gene and Protein Expression in GNMT Knockout Mice and Prevents Apoptosis
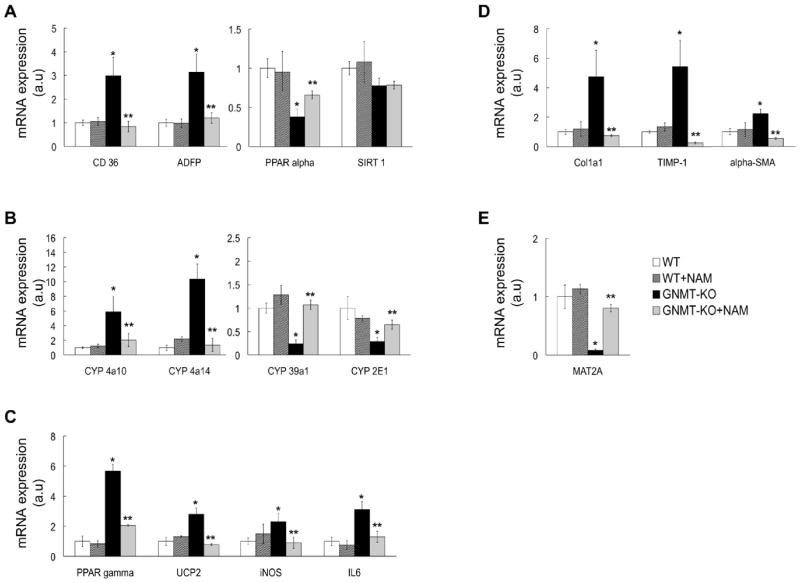
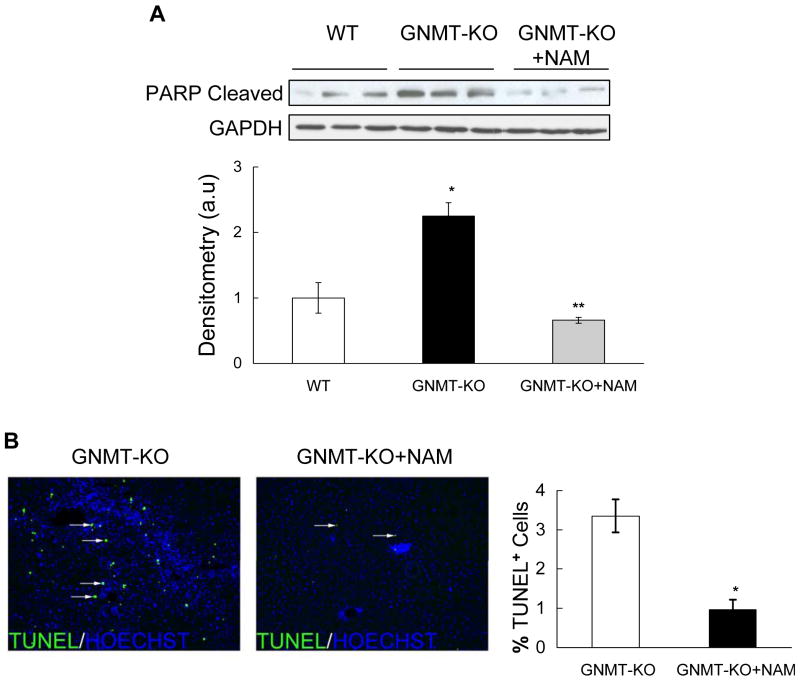
Consistent with the high SAMe levels, the liver expression of methionine adenosyltransferase 2A (MAT2A), a gene whose expression is inhibited by SAMe (16), was markedly reduced in GNMT-KO mice but normal in NAM-treated KO animals (Figure 3e). Similarly, the livers of 3-months old GNMT-KO mice showed marked alterations in the expression of critical genes involved in lipid metabolism (CD36, ADFP, PPARα and PPARγ), oxidative stress and inflammation (CYP2E1, CYP39A1, CYP4A10, CYP4A14, UCP2, PPARγ, IL6 and iNOS), and extracellular matrix regulation (COL1A1, TIMP-1, α-SMA); and the treatment of GNMT-KO mice with NAM prevented completely (CD36, ADFP, CYP4A10, CYP4A14, CYP39A1, UCP2, IL6, iNOS, COL1A1, α-SMA) or largely (PPARα, PPARγ, CYP2E1, TIMP-1) the abnormal expression of these genes in the liver (Figure 3a-e). NAM administration to WT mice had no significant effect on the expression of these genes (Figure 3a-e), indicating that the effect of NAM on gene expression in GNMT-KO mice is mediated by its capacity to reduce hepatic SAMe content. The hepatic expression of SIRT1, a NAD+-dependent protein deacetylase that is an important regulator of energy metabolism modulating many aspects of glucose and lipid homeostasis (17), was similar in WT and GNMT-KO mice and was not modified by NAM administration (Figure 3a). Lastly, the livers of 3-months old GNMT-KO mice showed marked apoptosis as demonstrated by PARP cleavage and TUNEL immunostaining, which was also prevented in NAM-treated GNMT-KO animals (Figure 4a,b).
Figure 3.
Gene expression in GNMT-KO mice after NAM administration. NAM administration in GNMT-KO prevented alterations in the expression of critical genes involved in (A) lipid metabolism (CD36, ADFP, PPARα and SIRT1), (B) oxidative stress (CYP4A10, CYP4A14, CYP39A1, and CYP2E1), (C) inflammation (PPARγ, UCP2, iNOS, and IL6), (D) extracellular matrix regulation (COL1A1, TIMP-1 and α-SMA) and (E) MAT2A. Gene expression was assessed by real time PCR using specific primers and values were normalized with GAPDH mRNA expression. Each bar represents the mean ± SEM of at least quintuplicate experiments. Results are expressed in arbitrary units (a.u). *P <0.05, GNMT-KO versus WT mice; **P <0.05, GNMT-KO versus GNMT-KO + NAM mice.
Figure 4.
Hepatic apoptosis in GNMT-KO mice after NAM administration. (A) Upper: Western blot of liver extracts from WT, GNMT-KO and GNMT-KO mice after NAM administration, incubated with the indicated antibodies. Data are representative of an experiment performed three times. Bottom: A graphical representation (mean ± SEM) of the densitometric changes of PARP cleavage expressed in arbitrary units (a.u). (*P <0.05, GNMT-KO versus WT mice; **P <0.05, GNMT-KO versus GNMT-KO+NAM mice). (B) Livers of 3-month old GNMT-KO mice treated with NAM showed a decrease in TUNEL positive cells compared to GNMT-KO livers. The number of TUNEL-positive cells were counted and expressed as the percentage of Hoechst nuclei per field in liver specimens from GNMT-KO mice and GNMT-KO mice treated with NAM. Data (mean ± SEM) are the average of five experiments performed independently. *P <0.05, GNMT-KO versus GNMT-KO + NAM mice.
Nicotinamide Treatment Prevents DNA Hypermethylation and the Activation of Ras and JAK/STAT Pathways in GNMT Knockout Mice
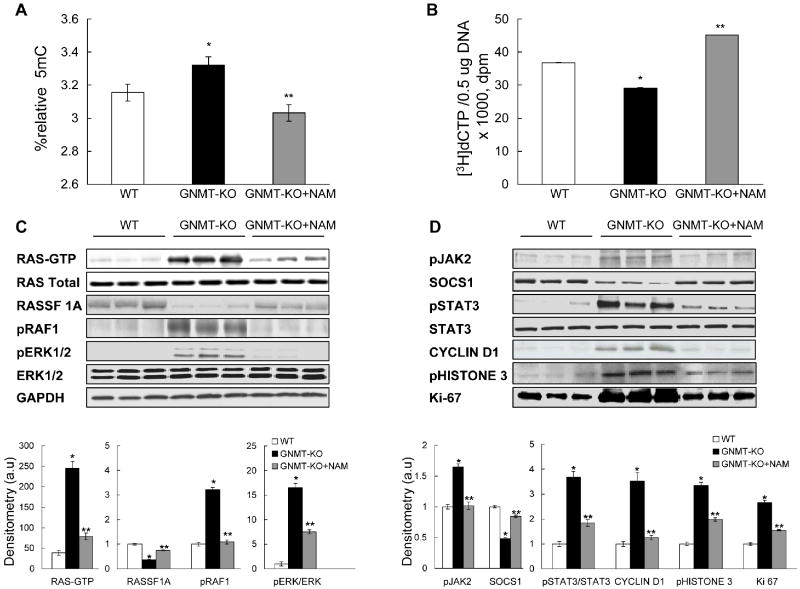
We have previously reported the existence of global DNA hypermethylation in the livers of GNMT-KO mice (6). Global DNA methylation, assayed both by the quantification of 5mC groups (Figure 5a) and by the measurement of the number of unmethylated CpG sites (Figure 5b), was significantly reduced in the livers of GNMT-KO animals treated with NAM. This agrees with the observation that NAM administration induces a marked reduction of hepatic SAMe content and also an increase in SAH content.
Figure 5.
Global DNA methylation and activation of Ras and JAK/STAT pathways in GNMT-KO mice liver after NAM administration. (A, B) Quantification of global DNA methylation in genomic DNA samples isolated from livers of WT, GNMT-KO mice, and GNMT-KO mice treated with NAM. In (A), total cytosine and 5-methylcytosine (5mC) content was determined in genomic DNA, and relative 5mC content, expressed as the percentage of total cytosine content (methylated and non-methylated), was determined. Data represent means ± SEM from 5 animals per subgroup. In (B), the DNA methylation status was determined by measuring the number of DNA unmethylated CpG sites using the cytosine extension assay. Two micrograms of DNA per sample were subjected to restriction digestion with the HpaII endonuclease. Lower incorporation of [3H]dCTP means a higher DNA methylation status. Data represent means ± SEM from 5 animals per subgroup. *P <0.05, GNMT-KO versus WT mice; **P <0.05, GNMT-KO versus GNMT-KO + NAM mice. (C and D) Western blot analysis of activated Ras (Ras-GTP), the RAS inhibitor RASSF1A, the Ras downstream effectors, pRAF and pERK1/2, activated JAK2 (pJAK2), the JAK inhibitor SOCS1, and the JAK downstream effectors pSTAT3, cyclin D1, pHistone 3 and Ki67. The downregulation of RASSF1A and SOCS1 expression in GNMT-KO mice compared to WT mice was prevented by NAM administration. The induction of Ras-GTP, pRAF1, pERK1/2, pJAK2, pSTAT3, cyclin D1 and of the mitotic markers pHistone 3 and Ki67 in GNMT-KO livers was prevented or markedly reduced by NAM administration in GNMT-KO mice as compared to untreated knockout animals. Bottom: a graphical representation (mean ± SEM) of the densitometric changes of the mentioned proteins expressed in arbitrary units (a.u). *P <0.05, GNMT-KO versus WT mice; **P <0.05, GNMT-KO versus GNMT-KO+NAM mice.
Next, we examined the Ras and JAK/STAT signaling pathways. We have previously shown the persistent activation of the Ras and JAK/STAT signaling pathways via suppression of Ras and JAK/STAT inhibitors such as RASSF1A and SOCS1 in GNMT-KO mice liver (6). Here we observed that NAM administration to GNMT-KO mice prevented the hepatic suppression of RASSF1A and SOCS1 protein expression (Figure 5c,d). Concomitant with this normalization of RASSF1A protein expression, we observed that the livers of NAM-treated GNMT-KO mice exhibited markedly lower expression of Ras and downstream effectors of Ras involved in cell proliferation and survival, including pRAF1 and pERK1/2, than untreated knockout animals (Figure 5c). Ras activity, assessed by immunoprecipitation with anti-pan Ras antibody and probed with anti-RAF-1 antibody, was markedly increased in GNMT-KO mice liver but much less elevated in NAM-treated GNMT-KO mice (Figure 5c). Similarly, the pERK1/2 content was increased more than 15-fold in GNMT-KO mice liver and only 7-fold in NAM-treatment knockout animals (Figure 5c). The levels of pRAF1 were elevated in GNMT-KO mice as compared to WT animals but similar in WT and NAM-treated GNMT-KO mice (Figure 5c). Similarly, concurrent with the normalization of SOCS1 protein expression, we observed that while the liver protein levels of pSTAT3 and of the downstream mitotic markers effectors pHistone 3 and Ki-67 were significantly elevated in both GNMT-KO mice groups as compared to WT animals, the induction in the NAM-treated group was significantly lower than in the untreated group (Figure 5d). The protein levels of activated JAK2 tyrosine kinase (pJAK2) and cyclin D1 were elevated in GNMT-KO mice as compared to WT animals but similar in WT and NAM-treated GNMT-KO mice (Figure 5d).
Discussion
SAMe is synthesized by methionine adenosyltransferase (MAT). In mammals there are three isoforms of MAT (MATI, MATII and MATIII) that are encoded by two genes (MAT1A and MAT2A). MATI and MATIII are tetrameric and dimeric forms, respectively, of the same subunit (α1) encoded by MAT1A, whereas the MATII isoform is a tetramer of a different subunit (α2) encoded by MAT2A. Adult differentiated liver expresses predominantly MAT1A, whereas extrahepatic tissues and fetal liver express MAT2A (2,18). The prevalent liver form, MATIII, has lower affinity for its substrates, is activated by methionine and has higher Vmax, contrasting with the other two enzymes (2,18). Based on the differential properties of hepatic MAT isoforms, it has been postulated that MATIII is the truly liver-specific isoform (2). Under normal conditions MATI would, as MATII outside the liver, synthesize most SAMe required by the hepatic cells. However, after an increase in methionine concentration, i.e. after a protein-rich meal, conversion to the high activity MATIII would occur and methionine excess will be eliminated (2). This will lead to an accumulation of SAMe and to the activation of GNMT, the main enzyme involved in hepatic SAMe catabolism (Figure 1) (1). Consequently, the excess of SAMe will be eliminated and converted to homocysteine via SAH. Once formed, the excess of homocysteine will be used for methionine regeneration or utilized for the synthesis of cysteine and α-ketobutyrate as result of its transsulfuration (2,18). Cysteine is then utilized for the synthesis of GSH as well as other sulfur containing molecules such as taurine, while α-ketobutyrate penetrates the mitochondria where it is further metabolized.
Consistent with this model, MAT1A-KO animals, despite over-expressing MAT2A in the liver (19), have high blood methionine and reduced hepatic SAMe, whereas GNMT-KO mice show increased liver SAMe (6). GNMT-KO mice, which are neither obese nor diabetic, spontaneously develop fatty liver and fibrosis three months after birth, and about five months later develop HCC (6). In GNMT-KO mice, the expression of hepatic MAT2A was down regulated but its levels was normal in NAM-treated animals. This finding is consistent with the changes observed in hepatic SAM content, since MAT2A expression is inhibited when the concentration of SAMe increases (16). Three-month old GNMT-KO mice showed an induction of CD36, a fatty acid translocase whose elevated expression is sufficient to increase hepatic fatty acid uptake (20) and of ADFP, a lipid droplet protein whose deficiency confers resistance to fatty liver development (21), as well as an increase in the expression of PPARγ, a transcription factor whose over-expression in the liver induces steatosis (22), and a reduction in the expression of hepatic PPARα, a lipid-activated transcription factor primarily expressed in the liver, in which it has been shown to activate β-oxidation of fatty acids (23). Related to this, we have previously observed that the activation of AMP-activated protein kinase (AMPK), a main regulator of cellular energy stores that activates fatty acid oxidation, is blocked in GNMT-KO mice due to the inhibitory effect that the accumulation of SAMe exerts on this pathway (24, 25).
SIRT1 is an important modulator of lipid homeostasis. Thus, it has been shown that SIRT1 transgenic mice are more resistant to develop fatty liver than WT animals in response to a high-fat diet (26), and that mice with a hepatocyte-specific deletion of SIRT1 are more prone to develop steatosis than WT animals when administered a high-fat diet (27), indicating that this protein deacetylase is relevant in preventing fatty liver. From this perspective, and since the expression of SIRT1 is not altered in GNMT-KO mice, it seems that this protein deacetylase is not playing a significant role in the generation of steatosis in this animal model. Moreover, the finding that the administration to GNMT-KO mice of NAM, an inhibitor of SIRT1 activity (28), prevented rather than aggravated the abnormal expression of CD36, ADFP, PARPα and PARPα further supports the conclusion that the development of steatosis in GNMT-KO mice is independent of SIRT1. This may be due to the low dose of NAM used in the present experiments (50 μM) as compared to the high concentration used to inhibit SIRT1 activity in culture cells (5 mM) (29).
One of the factors associated with the development and progression of steatosis is the oxidative stress originated by toxic lipid peroxidation catalyzed by cytochrome P4502E1 (CYP2E1), the main enzyme involved in NADPH-dependent reduction of oxygen leading to lipid peroxidation (30). The CYPs constitute a super-family of heme-containing microsomal mono-oxygenases that play a central role in the detoxification of xenobiotics, as well as in the metabolism of endogenous compounds including fatty acids. CYP2E1 expression and activity is up-regulated in SAMe-deficient, MAT1A-KO mice liver (31). In contrast, CYP2E1 as well as the expression of CYP39A1, an oxygenase catalyzing the rate-limiting step of bile acid synthesis (32), are reduced in GNMT-KO mice liver but the expression of two alternative fatty acid-hydroxylases (CYP4A10 and CYP4A14, the two major CYP4A genes) is markedly induced. It has been demonstrated that CYP4A enzymes are key intermediates of an adaptive response to perturbation of hepatic lipid metabolism. Thus, in CYP2E1-KO mice lipid peroxidation induced by the accumulation of hepatic fatty acids in response to a methyl-deficient diet is mediated by the up-regulation of CYP4A10 and CYP4A14 expression (33). SAMe is known to be an inhibitor of CYP2E1 activity (34) and, although the Ki is relatively high, it is likely that at the elevated concentration of SAMe present in GNMT-KO liver a direct effect of this molecule on CYP2E1 activity may be also responsible for the induction of CYP4A genes. Again, normalization of SAMe content in GNMT-KO by NAM treatment prevented the abnormal expression of CYP2E1, CYP39A1, CYP4A10 and CYP4A14.
Additionally, NAM treatment prevented the abnormal expression of critical genes involved in the generation of oxidative stress (UCP2, PPARγ, IL6 and iNOS) and liver fibrosis (COL1A1, TIMP-1 and α-SMA) as well as prevented apoptosis (determined both by PARP cleavage and TUNEL immunostaining). These findings agree with the observation that in whole blood stimulated with endotoxin NAM is an anti-inflammatory agent inhibiting PARP activation, iNOS expression as well as the stimulation of pro-inflammatory cytokines such as IL6 and iNOS (35). Whether in this experimental setting NAM also reduced cellular SAMe content is not known. Finally, NAM administration prevented global DNA hypermethylation, normalized the expression of RASSF1A and SOCS1 tumor suppressors, which are frequently inactivated by promoter methylation in human HCC (36), and inhibited or markedly reduced the activation of the Ras and JAK/STAT proliferative pathways in GNMT-KO mice, agreeing with the hypothesis that NAM exerts its therapeutic activity primarily via reduction of hepatic SAMe content.
Summing up, these results indicate that deletion of GNMT is associated with an increased expression of genes inducing steatosis (CD36, ADFP, PPARγ, CYP4A10, CYP4A14, UCP2) and also with a reduction in the expression of PPARα, a major activator of fatty acid oxidation, and that these changes are prevented by NAM administration. Moreover, these findings indicate that the hepatic reduction in total transmethylation flux caused by deletion of GNMT and the concomitant accumulation of SAMe can be compensated by NNMT if exogenous NAM is provided. Additionally, our results indicate that NAM administration to GNMT-KO mice prevents global DNA hypermethylation as well as the abnormal expression of numerous genes involved in fatty acid metabolism, oxidative stress, fibrosis, apoptosis and proliferation observed in untreated animals. More significant, NAM treatment not only normalized the expression of all these genes and proteins in GNMT-KO mice, but also prevented the development of fatty liver and fibrosis. The mechanism by which GNMT deletion leads to fibrosis is not known. Possibly, increased lipid accumulation and apoptosis in GNMT-KO hepatocytes may activate hepatic stellate cells (37,38), the central mediators of liver fibrogenesis. Accordingly, NAM may attenuate fibrogenesis by preventing hepatic fat accumulation and apoptosis via lowering SAMe content. At present, however, other alternatives as a direct effect of NAM on stellate cell activation cannot be excluded.
In summary, these results rule out the possibility that the interaction of GNMT with other target proteins, such as arrestin 3 and beta arrestin 1 (11), may play a critical role in the initiation of liver disease in GNMT-KO mice. Because GNMT expression is down-regulated in patients with cirrhosis and HCC, and there are subjects with GNMT mutations who have spontaneous liver disease (3,4,9,10), the clinical implications of the present findings is obvious at least with respect to these latter individuals. NAM has been used for many years to treat a broad spectrum of diseases including pellagra and diabetes without significant side effects (39,40). Therefore, our findings suggest individuals with GNMT mutations are likely to benefit from NAM treatment.
Supplementary Material
Acknowledgments
We thank Begoña Rodríguez for technical assistance.
Financial support: This work is supported by grants from NIH AT-1576 (to S.C.L., M.L.M-C. and J.M.M.), DK15289 and DK080010 (to C.W.), SAF 2008-04800, HEPADIP-EULSHM-CT-205 and ETORTEK-2008 (to J.M.M. and M.L.M.-C); Sanidad Gobierno Vasco 2008111015 (to M.L.M.-C); FISP107-1067 (to J.J. and ER-M). CIBERehd and CIBERdem are funded by the Instituto de Salud Carlos III.
List of abbreviations in alphabetical order
- ADFP
Adipose differentiation-related protein
- COL1A1
pro-α1-collagen type-I
- CBS
cystathionine β-synthase
- CYP2E1
cytochrome P4502E1
- CYP39A1
cytochrome P45039A1
- CYP4A10
cytochrome P4504A10
- CYP4A14
cytochrome P4504A14
- CD36
fatty acid translocase CD36
- GSH
glutathione
- GAPDH
glyceraldehyde phosphate dehydrogenase
- GNMT
glycine N-methyltransferase
- HCC
hepatocellular carcinoma
- iNOS
inducible nitric oxide synthase
- IL6
interleukin-6
- JAK
Janus kinase
- MAT
methionine adenosyltransferase
- 5mC
5-methyl-cytosine
- NAM
nicotinamide
- NNMT
nicotinamide N-methyltransferase
- PARP
poly (ADP-ribose) polymerase
- PPARα
peroxisome proliferator-activated receptor-α
- PPARγ
peroxisome proliferator-activated receptor-γ
- RASSF1A
Ras-association domain family/tumor suppressor-1A
- (SAMe)
S-adenosylmethionine
- α-SMA
α–smooth muscle actin
- SIRT1
sirtuin-1
- SOCS1
suppressor of cytokine signaling-1
- STAT3
signal transducer and activator of transcription 3
- TIMP1
TIMP tissue inhibitor of metalloproteinase-1
- UCP2
uncoupling protein-2
References
- 1.Luka Z, Mudd SH, Wagner C. Glycine N-methyltransferase and regulation of S-adenosylmethionine levels. J Biol Chem. 2009;284:22507–22511. doi: 10.1074/jbc.R109.019273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mato JM, Martínez-Chantar ML, Lu SC. Methionine metabolism and liver disease. Annu Rev Nutr. 2008;28:273–293. doi: 10.1146/annurev.nutr.28.061807.155438. [DOI] [PubMed] [Google Scholar]
- 3.Mudd SH, Cerone R, Schiaffino MC, Fantasia AR, Minniti G, Caruso U, et al. Glycine N-methyltransferase deficiency: a novel inborn error causing persistent isolated hypermethioninemia. J Inherit Metab Dis. 2001;24:448–464. doi: 10.1023/a:1010577512912. [DOI] [PubMed] [Google Scholar]
- 4.Augoustides-Savvopoulou P, Luka Z, Karyda S, Stabler SP, Allen RH, Patsiaoura K, et al. Glycine N-methyltransferase deficiency: a new patient with a novel mutation. J Inherit Metab Dis. 2003;26:745–759. doi: 10.1023/B:BOLI.0000009978.17777.33. [DOI] [PubMed] [Google Scholar]
- 5.Luka Z, Capdevila A, Mato JM, Wagner C. A glycine N-methyltransferase knockout mouse model for humans with deficiency of this enzyme. Transgenic Res. 2006;15:393–397. doi: 10.1007/s11248-006-0008-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez-Chantar ML, Vázquez-Chantada M, Ariz U, Martínez N, Varela M, Luka Z, et al. Loss of the glycine N-methyltransferase gene leads to steatosis and hepatocellular carcinoma in mice. Hepatology. 2008;47:1191–1199. doi: 10.1002/hep.22159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liao YJ, Liu SP, Lee CM, Yen CH, Chuang PC, Chen CY, et al. Characterization of a glycine N-methyltransferase gene knockout mouse model for hepatocellular carcinoma: Implications of the gender disparity in liver cancer susceptibility. Int J Cancer. 2009;124:816–826. doi: 10.1002/ijc.23979. [DOI] [PubMed] [Google Scholar]
- 8.Wagner C, Luka Z, Mato JM. Hepatocellular carcinoma in GNMT-/- mice. Toxicol Appl Pharmacol. 2009;237:246–247. doi: 10.1016/j.taap.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 9.Chen YM, Shiu JY, Tzeng SJ, Shih LS, Chen YJ, Lui WY, et al. Characterization of glycine N-methyltransferase gene expression in human hepatocellular carcinoma. Int J Cancer. 1998;75:787–793. doi: 10.1002/(sici)1097-0215(19980302)75:5<787::aid-ijc20>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 10.Avila MA, Berasain C, Torres L, Martín-Duce A, Corrales FJ, Yang H, et al. Reduced mRNA abundance of the main enzymes involved in methionine metabolism in human liver cirrhosis and hepatocellular carcinoma. J Hepatol. 2000;33:907–914. doi: 10.1016/s0168-8278(00)80122-1. [DOI] [PubMed] [Google Scholar]
- 11.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 12.Cantoni GL. Methylation of nicotinamide with soluble enzyme system from rat liver. J Biol Chem. 1951;189:203–216. [PubMed] [Google Scholar]
- 13.Fraga MF, Uriol E, Borja Diego L, Berdasco M, Esteller M, Cañal MJ, et al. High-performance capillary electrophoretic method for the quantification of 5-methyl 2′-deoxycytidine in genomic DNA: application to plant, animal and human cancer tissues. Electrophoresis. 2002;23:1677–1681. doi: 10.1002/1522-2683(200206)23:11<1677::AID-ELPS1677>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 14.Pogribny I, Yi P, James SJ. A sensitive new method for rapid detection of abnormal methylation patterns in global DNA and within CpG islands. Biochem Biophys Res Commun. 1999;262:624–628. doi: 10.1006/bbrc.1999.1187. [DOI] [PubMed] [Google Scholar]
- 15.Vázquez-Chantada M, Ariz U, Varela-Rey M, Embade N, Martínez N, Fernández D, et al. Evidence for an LKB1/AMPK/eNOS cascade regulated by HGF, S-adenosymehtionine and nitric oxide in hepatocyte proliferation. Hepatology. 2009;49:608–617. doi: 10.1002/hep.22660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martínez-Chantar ML, Latasa MU, Varela-Rey M, Lu SC, García-Trevijano ER, Mato JM, et al. L-methionine availability regulates expression of the methionine adenosyltransferase 2A gene in human hepatocarcinoma cells: role of S-adenosylmethionine. J Biol Chem. 2003;278:19885–19890. doi: 10.1074/jbc.M211554200. [DOI] [PubMed] [Google Scholar]
- 17.Fulco M, Sartorelli V. Comparing and contrasting the roles of AMPK and SIRT1 in metabolic tissues. Cell Cycle. 2008;7:3669–3679. doi: 10.4161/cc.7.23.7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mato JM, Lu SC. Role of S-adenosyl-L-methionine in liver health and injury. Hepatology. 2007;45:1306–1312. doi: 10.1002/hep.21650. [DOI] [PubMed] [Google Scholar]
- 19.Lu SC, Alvarez L, Huang ZZ, Chen L, An W, Corrales FJ, et al. Methionine adenosyltransferase 1A knockout mice are predisposed to liver injury and exhibit increased expression of genes involved in proliferation. Proc Natl Acad Sci USA. 2001;98:5560–5565. doi: 10.1073/pnas.091016398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koonen DP, Jacobs RI, Febbraio M, Young ME, Soltys CL, Ong H, et al. Increased hepatic CD36 expression contributes to dyslipidemia associated with diet-induced obesity. Diabetes. 2007;56:2863–2871. doi: 10.2337/db07-0907. [DOI] [PubMed] [Google Scholar]
- 21.Chang BHJ, Li L, Paul A, Taniguchi S, Nannegari V, Heird W, et al. Protection against fatty liver but normal adipogenesis in mice lacking adipose differentiation-related protein. Mol Cell Biol. 2006;26:1063–1076. doi: 10.1128/MCB.26.3.1063-1076.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uno K, Kalagiri H, Yamada T, Ishogaki Y, Ogihara T, Imai J, et al. Neuronal pathway from the liver modulates energy expenditure and systemic insulin sensitivity. Science. 2006;312:1656–1659. doi: 10.1126/science.1126010. [DOI] [PubMed] [Google Scholar]
- 23.Leone TC, Weinheimer CJ, Kelly DP. A critical role for the peroxisome proliferators-activated receptor α (PPARα) in the cellular fasting response: the PPARα-null mouse as a model of fatty acids oxidation disorders. Proc Natl Acad USA. 1999;96:7473–7478. doi: 10.1073/pnas.96.13.7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martínez-Chantar ML, Vázquez-Chantada M, Garnacho M, Latasa MU, Varela-Rey M, Dotor J, et al. S-adenosylmethionine regulates cytoplasmic HuR via AMP-activated kinase. Gastroenterology. 2006;131:223–232. doi: 10.1053/j.gastro.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 25.Vázquez-Chantada M, Ariz U, Varela-Rey M, Embade N, Martínez-López N, Fernández-Ramos D, et al. Evidence for LKB1/AMP-activated protein kinase/endothelial nitric oxide synthase cascade regulated by hepatocyte growth factor, S-adenosylmethionine, an nitric oxide in hepatocyte proliferation. Hepatology. 2009;49:608–617. doi: 10.1002/hep.22660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschöp MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci USA. 2008;105:9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purushotham A, Schung TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009;9:327–338. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast Sir2 and human SIRT1. J Biol Chem. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 29.Fulco M, Cen Y, Zhao P, Hoffman EP, McBumey MW, Sauve AA, et al. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell. 2008;14:661–673. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lieber CS. Alcoholic fatty liver: Its pathogenesis and mechanism of progression to inflammation and fibrosis. Alcohol. 2004;34:9–19. doi: 10.1016/j.alcohol.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 31.Martínez-Chantar ML, Corrales FJ, Martínez-Cruz LA, García-Trevijano ER, Huang ZZ, Chen L, et al. Spontaneous oxidative stress and liver tumors in mice lacking methionine adenosyltransferase 1A. FASEB J. 2002;16:1292–1294. doi: 10.1096/fj.02-0078fje. [DOI] [PubMed] [Google Scholar]
- 32.Lathe R. Steroid and sterol 7-hydroxylation: ancient pathways. Steroids. 2002;67:967–977. doi: 10.1016/s0039-128x(02)00044-2. [DOI] [PubMed] [Google Scholar]
- 33.Leclerq IA, Farrell GC, Field J, Bell DR, Gonzalez FJ, Robertson GR. CYP2E1 and CYP4A as microsomal catalysts of lipid peroxides in murine nonalcoholic steatohepatitis. J Ciln Inv. 2000;105:1067–1075. doi: 10.1172/JCI8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caro AA, Cederbaum AI. Inhibition of CYP2E1 catalytic activity in vitro by S-adenosyl-L-methionine. Biochem Pharmacol. 2005;69:1081–1083. doi: 10.1016/j.bcp.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 35.Ungerstedt JS, Blömback M, Söderström T. Nicotinamide is a potent inhibitor of proinflammatory cytokines. Clin Exp Immunol. 2003;131:48–52. doi: 10.1046/j.1365-2249.2003.02031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calvisi DF, Ladu S, Gorden A, Farina M, Conner EA, Lee JS, et al. Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterology. 2006;130:1117–1128. doi: 10.1053/j.gastro.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 37.Wobser H, Dorm C, Weiss TS, Amann T, Bollheimer C, Bütther, et al. Lipid accumulation in hepatocytes induces fibrogenic activationnof hepatic stellate cells. Cell Res. 2009;19:996–1005. doi: 10.1038/cr.2009.73. [DOI] [PubMed] [Google Scholar]
- 38.Jou J, Choi SS, Diehl AM. Mechanisms of disease progression in nonalcoholic fatty liver disease. Semin Liver Dis. 2008;28:370–379. doi: 10.1055/s-0028-1091981. [DOI] [PubMed] [Google Scholar]
- 39.Green RG. Subclinical pellagra. Its diagnostic and treatment. Schizophrenia. 1970;2:70–79. [Google Scholar]
- 40.Vague P, Picq R, Bernal M, Lassmann-Vague V, Vialettes B. Effect of nicotinamide treatment on the residual insulin secretion in type 1 (insulin-dependent) diabetic patients. Diabetologia. 1989;32:316–321. doi: 10.1007/BF00265549. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.