Abstract
In the Drosophila eye, neighboring ommatidia are separated by inter-ommatidial cells (IOCs). How this ommatidial spacing emerges during eye development is not clear. Here we demonstrate that four adhesion molecules of the Irre cell recognition module (IRM) family play a redundant role in maintaining separation of ommatidia. The four IRM proteins are divided into two groups: Kirre and Rst are expressed in IOCs, and Hbs and Sns in primary pigment cells (1°s). Kirre binds Hbs and Sns in vivo and in vitro. Reducing activity of either Rst or Kirre alone had minimal effects on ommatidial spacing, but reducing both together led to direct ommatidium:ommatidium contact. A similar phenotype was also observed when reducing both Hbs and Sns. Consistent with the role of these factors in sorting ommatidia, mis-expression of Hbs plus Sns within a single IOC led to complete separation of the cell from neighboring ommatidia. Our results indicate mutual preferential adhesion between ommatidia and IOCs mediated by four IRM proteins is both necessary and sufficient to maintain separation of ommatidia.
Keywords: cell adhesion, preferential adhesion, Drosophila, eye, pattern formation
Introduction
Organization of cells into a specific spatial configuration requires selective cell adhesion. During development, homotypic adhesion has been shown to promote cell aggregation. For example, positioning of the oocyte within the Drosophila ovary and aggregation of blastomeres in the mouse embryo are controlled by cadherins that act through homophilic interactions (De Vries et al., 2004; Godt and Tepass, 1998; Gonzalez-Reyes and St Johnston, 1998). A similar mechanism in the Drosophila eye acts locally to regulate aggregation of support (‘cone’) cells into a four-cell cluster (Hayashi and Carthew, 2004). On the other hand, heterotypic adhesion prevents cell aggregation. As an example, sorting of inter-ommatidial cells (IOCs) from multiple rows into a single line within the Drosophila eye is regulated by preferential adhesion, a situation in which overall adhesion between ‘unlike’ cells is stronger than that between ‘like’ cells (Bao and Cagan, 2005). Preferential adhesion of IOCs to ommatidia is mediated by Hibris (Hbs) and Roughest (Rst) through heterophilic interactions (Bao and Cagan, 2005).
Hbs and Rst are adhesion molecules of the Irre cell recognition module (IRM) family conserved from C. elegans to flies and humans (Fischbach et al., 2009). IRM proteins are involved in multiple processes during animal development. For example, the C. elegans IRM adhesion molecules SYG-1 and SYG-2 are essential for axon guidance (Shen and Bargmann, 2003; Shen et al., 2004). In vertebrates, Nephrin and Neph1 are homologs of Hbs/SYG-2 and Rst/SYG-1, respectively, and Nephrin and Neph1 are required for kidney and muscle development (Sohn et al., 2009; Tryggvason et al., 2006). In humans, mutations in Nephrin lead to Congenital Nephrotic Syndrome characterized by heavy proteinuria at birth (Tryggvason et al., 2006). In Drosophila, there are four IRM proteins identified to date: Rst, Hbs, Sticks and stones (Sns), and Kin of irre (Kirre, also known as Duf). These proteins are involved in multiple developmental processes such as myoblast fusion, axon guidance and cell sorting (Artero et al., 2001; Bour et al., 2000; Dworak et al., 2001; Ruiz-Gomez et al., 2000; Strunkelnberg et al., 2001). In addition, Rst, the founding member of the IRM family, is known to be essential for correct spacing of olfactory sensory organs in Drosophila (Venugopala Reddy et al., 1999) although the mechanism is unknown. Recently, it has been shown that two IRM adhesion molecules Sns and Kirre are involved in formation of a slit diaphragm-like structure in the Drosophila nephrocyte (Weavers et al., 2009; Zhuang et al., 2009), supporting the functional conservation of IRM proteins across species.
In the Drosophila eye, ommatidia are separated from each other by secondary and tertiary pigment cells—referred to here as inter-ommatidial cells (IOCs)—and mechanosensory bristles (Figure 1A-A’). Ommatidia represent the unit eyes of the fly. Within each ommatidium, eight photoreceptor neurons are capped by six glial-like support cells: four cone cells and two primary pigment cells (1°s). Separation of ommatidia is essential for fly vision, as the IOCs optically insulate each ommatidium (Johannsen, 1924). Cells in the eye derive from a common pool of precursor cells of epithelial origin (Garcia-Bellido and Merriam, 1969). The initial spacing pattern of ommatidia is established when the first cell type emerges in the third instar larva. This early step of patterning requires cell signaling mediated by EGFR (Baker and Rubin, 1989; Baker and Yu, 1997; Baonza et al., 2001; Spencer et al., 1998; Tio et al., 1994). The remaining photoreceptor neurons, cone cells and 1°s are subsequently and sequentially recruited into each ommatidial cluster. Once the ommatidial clusters are established, IOCs re-arrange from multiple rows between ommatidia into a one-cell wide hexagonal lattice (Figure 1A).
Figure 1.
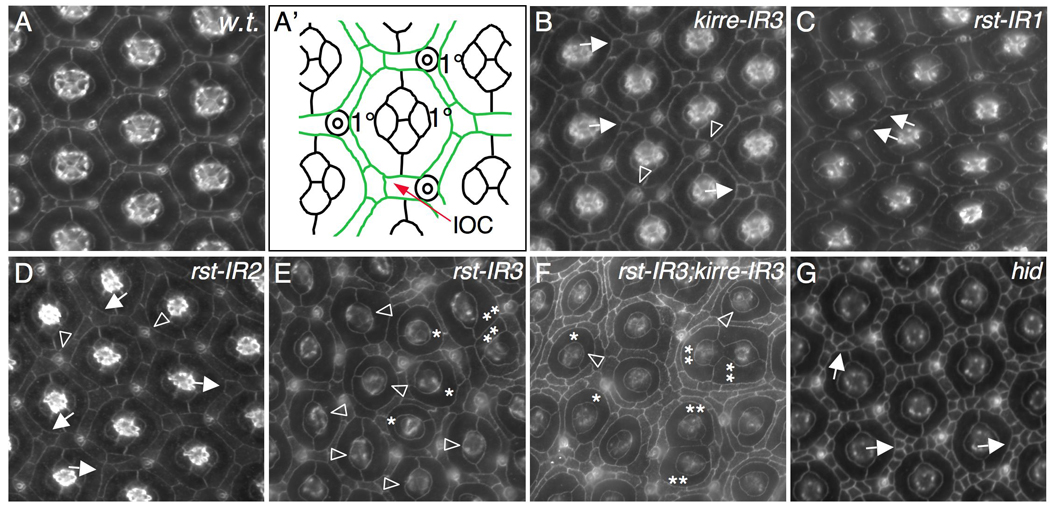
Kirre and Rst act redundantly in patterning ommatidia. Eyes at 42 h APF were stained with an antibody against either Armadillo (A–D) or E-cadherin (E–G). (A-A’) A wild type eye. Tracing of an ommatidium is shown in (A’); inter-ommatidial cells (IOCs) are pseudo-colored in green. IR1 and IR2 indicate expression of one and two copies of indicated RNAi transgenes, respectively. IR3 refers to expression of a single copy of a transgene together with Dicer-2. Expression of all transgenes is controlled by GMR-Gal4 in this figure. (B) Strong reduction of Kirre by kirre-RNAi (kirre-IR3). Single cells failed to be selected in vertices (arrows). Occasionally bristle groups were mis-positioned (arrowhead). (C) Mild reduction of Rst by expressing a single copy of rst-RNAi (rst-IR1). An extra cell is highlighted by an arrow. (D) Expression of two copies of rst-RNAi (rst-IR2). A single cell was not selected in the vertex (arrows). Cells were often found surrounding a bristle group (arrowheads). (E) Strong reduction of Rst by rst-RNAi (rst-IR3). Defects in cone cells (arrowheads) and 1°s (asterisks) are indicated. Two ommatidia in direct contact are highlighted by double asterisks. (F) Strong reduction of both Rst and Kirre. Defects in cone cells (arrowheads) and 1°s (asterisks) are indicted. Contacting ommatidia are highlighted by double asterisks. IOCs completely failed to sort into single line. (G) Ommatidial patterning in the pupal eye does not require cell death. In hid mutants, cell death was strongly blocked and cells failed to sort into single line (arrows). However, ommatidia were separated as in wild type.
There are two conundrums regarding this morphogenetic process. First, IOCs are known to have a lower affinity to each other than to 1°s (Bao and Cagan, 2005). Despite their expected low affinity, IOCs remain contiguous to maintain separation of ommatidia. Second, IOCs are constantly changing positions as revealed by live imaging (Larson et al., 2008). Nevertheless, ommatidia are rarely found to be in direct contact during development and are not found in contact in the adult. How IOCs maintain IOC-IOC contacts to separate ommatidia during eye development is not clear.
Here we show that IRM adhesion molecules function redundantly in maintaining separation of ommatidia. Based on their homology to the mammalian proteins Neph1 and Nephrin, the four IRM adhesion molecules can be divided into two groups: Kirre and Rst represent the Neph1 group, and Hbs and Sns the Nephrin group. We show the two groups of IRM adhesion molecules are expressed in complementary cell types. Each of these two groups forms stronger inter-group interactions than intra-group interactions. Our results demonstrate mutual preferential adhesion between ommatidia and IOCs play a role in maintaining ommatidial spacing.
Materials and Methods
Drosophila genetics
RNA interference (RNAi) flies UAS-kirre-IR and UAS-sns-IR were generated for this work. y w hsFLP, UAS-nlsGFP and Act5C>y+>Gal4 UAS-GFP were provided by Bloomington Stock Center. Other flies used: UAS-rst-IR and UAS-hbs-IR (Bao and Cagan, 2005), UAS-rst (Reiter et al., 1996), UAS-kirre/duf (Ruiz-Gomez et al., 2000; Strunkelnberg et al., 2001), UAS-sns (gift of Susan Abmayr), UAS-hbs (Dworak et al., 2001), rP298-lacZ (Nose et al., 1998), Gal4–54 (gift of Liqun Luo), l(3)k05017/ Df(3L)WR+X1 for the hid null mutant (gift of Nick Baker) and hsFLP MKRS (gift of Matthew Freeman). For the purpose of testing specificity of RNAi constructs, independent UAS-hbs-IR and UAS-rst-IR RNAi lines provided by the Vienna Drosophila RNAi Center (VDRC) were also used.
Construction of transgenes for RNAi
UAS-kirre-IR and UAS-sns-IR containing an inverted repeat (IR) for RNAi were generated following the strategy as previously described (Bao and Cagan, 2006). A 500-bp cDNA fragment of kirre (2505-2004) and 523-bp cDNA fragment of sns (1987–2509) were selected as target sequences. Transgenic lines were established by standard P-element mediated germline transformation. Multiple lines were isolated and all gave similar phenotypes. Except where noted, the following lines were used for this work: UAS-kirre [A202A1] on the third chromosome, UAS-kirre [D201A2] on the second chromosome and UAS-sns-IR [B103A1] on the third chromosome. Strong reduction of target genes was achieved by co-expressing a single copy of UAS-Dicer-2 (Dietzl et al., 2007).
Clonal analysis
Single-cell clones expressing a target gene were generated using a FLP-out technique (Basler and Struhl, 1994). To induce clones, pupae at 12 h APF were heat-shocked at 37°C in a water bath for 20 min.
Production of antibodies
The anti-Kirre antibody was raised in rats against the intracellular domain of Kirre/Duf (698–915 a.a.). The fusion protein containing 22 amino acids of an N-terminal His-Tag and the intracellular domain of Kirre was produced and purified from E. coli using the pET14b vector system (Novagen). Rats were immunized with 100 µg of purified protein mixed with Freund’s complete adjuvant and given six booster shots of 100 µg protein in Freund’s incomplete adjuvant at 2-week intervals thereafter.
The anti-Hbs antibody (AS14) was raised in rabbits using a peptide for the intracellular domain of Hbs (1083–1096 a.a.): AEPSNDDVYSKDDS. Immunization of rabbits was performed by GenScript Corp. (www.genscript.com).
Histochemistry
Immunostaining of the eye was performed as described (Bao and Cagan, 2005). Rat anti-Kirre (1:5000) and Rabbit anti-Hbs AS14 (1:2500) were generated for this work. Other primary antibodies used: mouse anti-Rst (Mab24A5.1, 1:100) (Schneider et al., 1995), rabbit anti-Sns (1:300) (Bour et al., 2000), rabbit anti-Echinoid (1:2500) (gift of Andrew Jarman) and rabbit anti-lacZ (1:2000; 5 Prime → 3 Prime). Rat anti-DE-cadherin (1:20) and mouse anti-Armadillo (1:10) were provided by Developmental Studies Hybridoma Bank at the University of Iowa. Secondary antibodies: Alexa 488 and Alexa 568 conjugated secondary antibodies (1: 5000; Molecular Probes); Cy5 conjugated secondary antibodies (1:1000; Jackson ImmunoResearch Laboratories). All images were taken using an epi-fluorescence microscope (Axioplan2, Carl Zeiss, Inc.). Images were minimally processed in Adobe Photoshop for cropping and contrast.
S2 cell culture and co-immunoprecipitation
S2 cell culture and co-immunoprecipitation were performed following the protocols as described previously (Bao and Cagan, 2005). Relevant proteins were immunoprecipitated using mouse anti-FLAG M2 agarose (Sigma). Primary antibodies used for western blot: mouse anti-flag M2 (1:3000; Sigma) and mouse anti-V5 (1:5000; Invitrogen). The secondary antibody used was goat anti-mouse IgG-HRP (1:5000; Cell Signaling).
To express proteins for co-immunoprecipitation, S2 cells were transiently transfected with appropriate plasmids. Proteins were expressed under the control of a tubulin promotor in CaSpeR4-tub, which was generated by removing EGFP from CaSpeR4-tub-EGFP (tubulin-EGFP) (Brennecke et al., 2003). The following constructs were used for cell culture: CaSpeR4-tub-kirre-3×flag, CaSpeR4-tub-hbs-3×flag, CaSpeR4-tub-rst-v5, CaSpeR4-tub-hbs-v5, CaSpeR4-tub-kirre-v5 and CaSpeR4-tub-sns-v5. To generate these constructs, a full length rst, hbs, kirre or sns cDNA was subcloned into pGEM-S1 (Bao and Cagan, 2006). The sequence encoding 3×Flag or V5 was then added in frame 3’ to each coding sequence. The whole sequence was then shuttled into CaSpeR4-tub.
Results
1. Separation of ommatidia requires Kirre and Rst
In an RNA-interference (RNAi)-based screen to uncover genes that functionally interact with rst we identified kirre, a gene known to act with rst in myoblast fusion (Ruiz-Gomez et al., 2000; Strunkelnberg et al., 2001). In the Drosophila eye, mild defects were observed when kirre alone was strongly reduced by RNAi (Figure 1B and supplemental Figure S1B-B’). Typically, bristle groups and IOCs were mis-positioned (Figure 1B) though ommatidia remained well separated as in wild type.
Reduction of rst activity, on the other hand, affected arrangement of IOCs, 1°s, and cone cells in a dose-dependent fashion. Moderate reduction of Rst by RNAi (rst-IR1 or rst-IR2) resulted in defects in positioning IOCs and bristle groups (Figure 1C–D; see also Bao and Cagan, 2005). Strong reduction of rst (rst-IR3) led to defects in organization of cone cells and 1°s as well as in IOCs (Figure 1E and Supplemental Figure S1G-G’). Although most IOCs sorted into single file, 19.1% of ommatidia were found in direct contact with a neighboring ommatidium (n=304 ommatidia; Figure 1E). A similar phenotype was also observed when independent RNAi lines (from the Vienna Drosophila RNAi Center) were utilized (Supplemental Figure S2A-A’), indicating that defects observed using these RNAi lines are due to depletion of Rst. Reducing both kirre and rst together significantly enhanced the patterning defects: 36.6% of ommatidia were found in direct contact with another ommatidium (n=424 ommatidia; Figure 1F). Further, IOCs showed little evidence of sorting into single file (Figure 1F). Therefore, Kirre and Rst act redundantly to regulate separation of ommatidia.
Rst is required for cell death in the pupal eye (Reiter et al., 1996; Wolff and Ready, 1991). Upon strong reduction of both Rst and Kirre, ectopic IOCs— arranged in multiple rows— were observed between ommatidia (Figure 1F). This enhanced phenotype indicates that Rst and Kirre together regulate cell death and IOC sorting as well as ommatidial patterning. To test whether abnormal ommatidial patterning following depletion of both Rst and Kirre is due to an indirect effect mediated by apoptosis, we used a hid null mutant (Grether et al., 1995; Yu et al., 2002). In genotypically hid mutant pupal eyes, cell death is completely blocked (Yu et al., 2002). However in hid mutant eyes ommatidia were arranged in a nearly perfect hexagonal array as in wild type (Figure 1G), indicating cell death is not required for patterning ommatidia.
2. Kirre is expressed in IOCs
To further understand the function of Kirre in separation of ommatidia we examined the sub-cellular localization of the Kirre protein. Rst is known to mainly localize at the border between IOCs and 1°s (Reiter et al., 1996). A low level of Rst protein was also found in intracellular vesicles within both 1°s and IOCs (Figure 2A). Using an antibody specific to Kirre, we found Kirre protein co-localized with Rst on the cell membrane as well as in vesicles (Figure 2A). The expression patterns of Rst and Kirre also matched nearly perfectly (Figure 2A).
Figure 2.
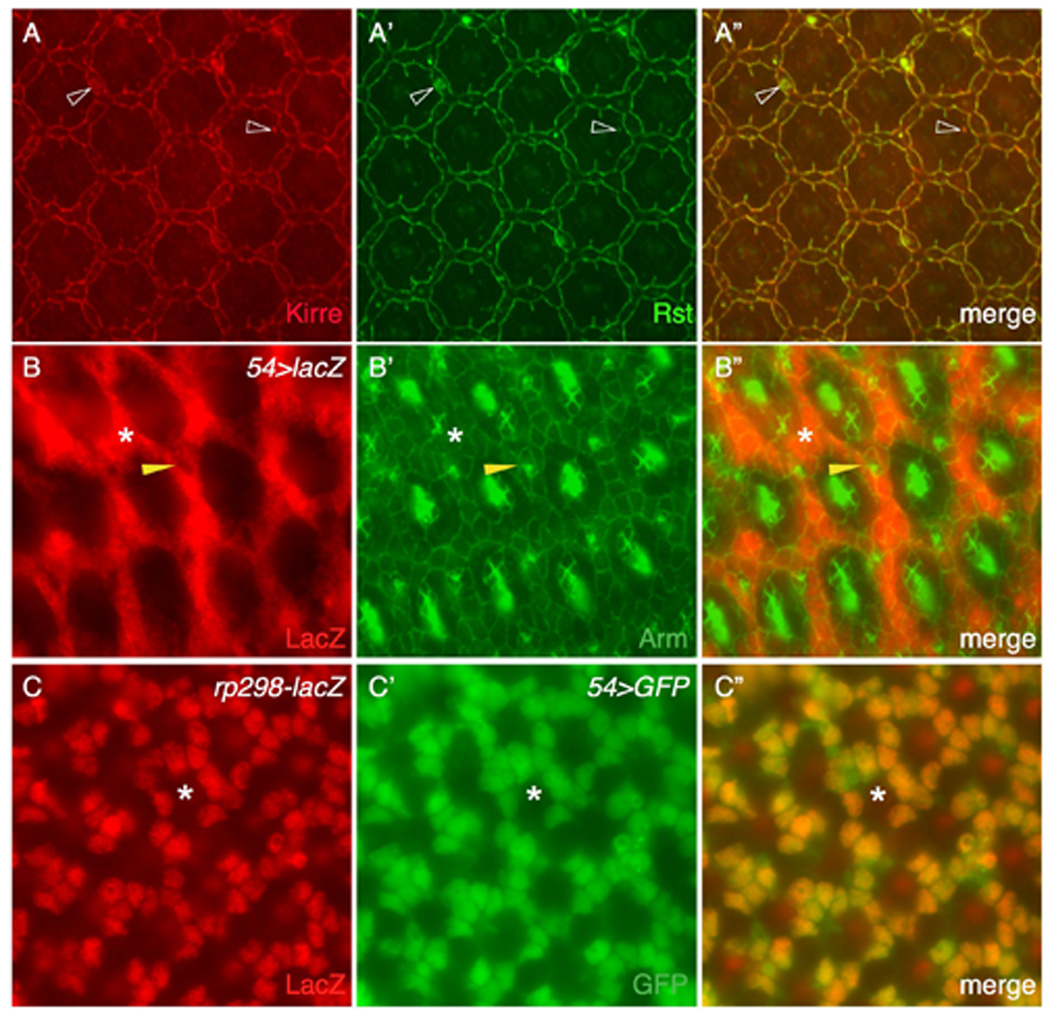
Kirre is expressed in IOCs. (A-A”) Kirre (red) co-localizes with Rst (green) on the surface. Kirre also co-localizes with Rst in all vesicles (open arrowheads). (B-B”) The Gal4–54; UAS-lacZ (54>lacZ) eye at 27 h APF was co-stained with anti-lacZ (red, left) and anti-Armadillo (green, middle) antibodies. Merged view is shown in the right panel. LacZ was detected exclusively in IOCs (asterisks). Arrowheads point to a bristle group from which lacZ staining was absent. (C-C”) kirre is transcribed in IOCs as revealed by an enhancer trap rP298-lacZ. The eye was stained with an anti-LacZ antibody (red) and marked by 54>GFP (green). Space for an ommatidium is indicated by an asterisk.
rst is mainly transcribed in IOCs at 27 h APF (Bao and Cagan, 2005). To assess the cell type in which kirre is transcribed we utilized the kirre reporter line rP298-lacZ, which expresses a nuclear LacZ readout of kirre expression (Nose et al., 1998). We marked IOCs with nuclear GFP driven by the IOC-specific driver 54-Gal4 (54>nlsGFP; Figure 2B). When co-expressed with 54>nlsGFP, rP298-lacZ was detected in IOCs (Figure 2C), indicating that kirre is transcribed in IOCs.
3. Separation of ommatidia requires Sns and Hbs
Sns is known to interact with Kirre during myoblast fusion while Hbs binds Rst during IOC sorting (Chen and Olson, 2001; Galletta et al., 2004; Bao and Cagan, 2005). To test whether Sns or Hbs is involved in maintaining separation of ommatidia, we reduced their activity during the stages of ommatidial assembly and IOC rearrangement. Mild defects including mis-positioned IOCs and bristles were observed when Sns alone was strongly reduced by RNAi (Figure 3A and supplemental Figure S1D-D’). Ommatidia were nonetheless well separated from each other as in wild type. Therefore, Sns is not by itself necessary for patterning ommatidia. Moderate reduction of Hbs by RNAi (hbs-IR1 or hbs-IR2) resulted in defects in IOCs and bristle group positioning (Figure 3B–C; see also Bao and Cagan, 2005). Strong reduction of hbs (hbs-IR3) led to defects in the organization of cone cells and 1°s as well as in IOCs, indicating that it has a prominent role in assembly each of these cell types (Figure 3D and supplemental Figure S1F-F’). Most IOCs sorted into single file and ommatidial spacing was not significantly affected. However, a few hbs-IR3 ommatidia— less than 4% (n=321 ommatidia)— were found in direct contact with each other (Figure 3D). Similar phenotypes were also observed when independent RNAi lines from VDRC were used (Supplemental Figure S2B-B’), suggesting defects seen in the eye using these RNAi lines are specifically due to depletion of Hbs.
Figure 3.
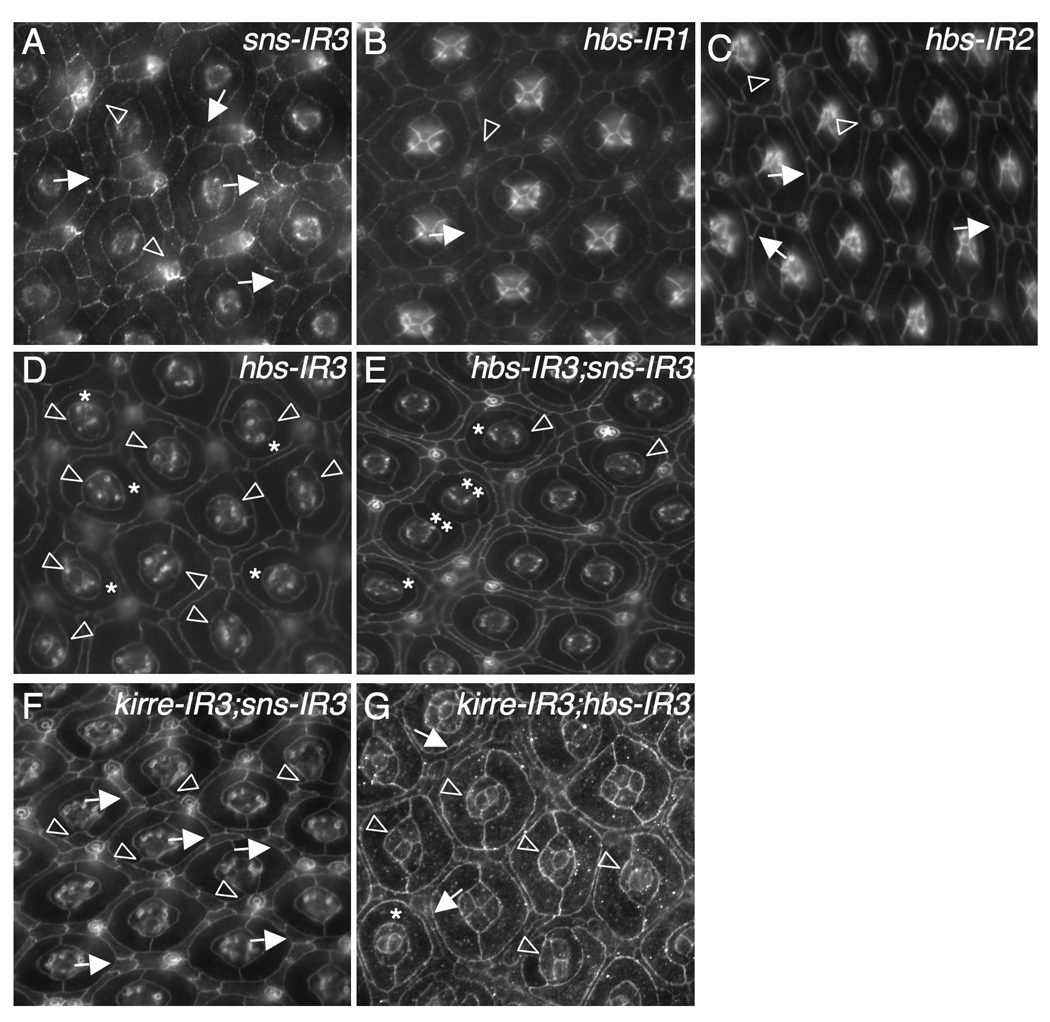
Sns and Hbs act redundantly in patterning ommatidia. Eyes at 42 h APF were stained with an antibody against either Armadillo (B–C), E-cadherin (A, D–F) or Echinoid (G). (A) Strong reduction of Sns by sns-RNAi (sns-IR3). Frequently, single cells failed to be selected at vertices (arrows). Occasionally, a cluster of cells was found surrounding a bristle group (arrowheads). (B) Mild reduction of Hbs by expressing a single copy of hbs-RNAi (hbs-IR1). A mis-positioned cell is highlighted by an arrow and a cluster of cells surrounding a bristle group indicated by an arrowhead. (C) Expression of two copies of hbs-RNAi (hbs-IR2). Single cells were not selected in vertices (arrows) and bristle groups misplaced (arrowheads). (D) Strong reduction of Hbs by hbs-RNAi (hbs-IR3). Defects in cone cells (arrowheads) and 1°s (asterisks) are highlighted. (E) Strong reduction of both Hbs and Sns. Ommatidia in direct contact are indicated by double asterisks. Defects in cone cells (arrowheads) and 1°s (asterisks) are highlighted. IOCs completely failed to sort into single file. (F) Strong reduction of both Kirre and Sns. Single cells failed to be selected within vertices (arrows). Extra cells (‘cone contact cells’) were commonly found in direct contact with cone cell quartets (arrowheads). (G) Strong reduction of both Kirre and Hbs. Frequently cone cells formed abnormal configurations (arrowheads). Occasionally IOCs failed to sort into single rows (arrows).
Simultaneous depletion of both Hbs and Sns led to direct contact of 27.2% ommatidia (n=430 ommatidia) and complete failure of IOCs to sort into single file (Figure 3E). These phenotypes represent a strong enhancement of either knockdown alone. Further, these specific interactions between Hbs and Sns were also observed using weaker RNAi lines (data not shown). By contrast, strongly reducing either (i) Kirre plus Sns or (ii) Kirre plus Hbs led to only a slight phenotypic enhancement (Figure 3F–G). For example, when both Kirre and Sns were depleted, extra cells (‘cone contact cells’; Tanenbaum et al., 2000) were frequently found in direct contact with cone cell quartets. Depleting both Kirre and Hbs led to occasional IOCs in multiple rows between ommatidia (Figure 3G). In each case, ommatidia spacing was nonetheless nearly indistinguishable from single knockdown of either Sns or Hbs alone. Based on these results, we conclude that Hbs and Sns act redundantly to separate ommatidia.
4. Sns is expressed in 1°s
Both Sns and Hbs are expressed in the pupal eye (Fischbach et al., 2009). To further explore the mechanism by which Sns and Hbs act to separate ommatidia, we examined the sub-cellular localization of the Sns and Hbs proteins. Using a specific antibody for Hbs, we found Hbs protein was primarily localized at the border between IOCs and 1°s in a manner similar to Rst at 27 h APF (Figure 4A-A”). Also similar to Rst, low levels of Hbs were detected at 1°:1° as well as at cone:1° borders. Hbs protein was also found in vesicles in 1°s but not in IOCs; Hbs-containing vesicles co-localized with Rst in 1°s (Figure 4A-A”). Using a specific antibody for Sns (Bour et al., 2000), we found Sns co-localized with Kirre on the membrane and Sns and Kirre vesicles largely overlapped at 27 h APF (Figure 4B-B”). Further, both Sns and Kirre were found at the border between IOCs and bristle groups, consistent with previous reported localization (Fischbach et al., 2009).
Figure 4.
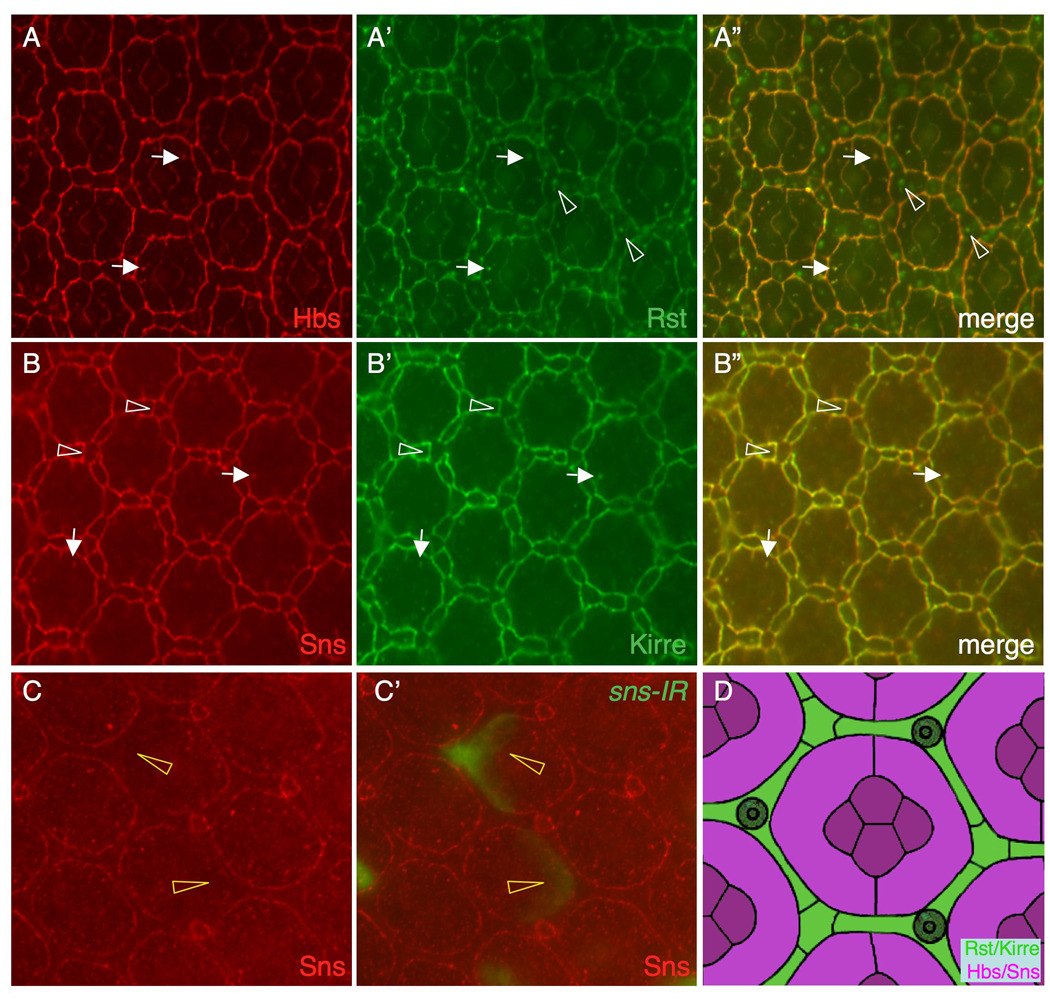
IRM proteins are expressed in complementary cell types. Eyes at 27 h APF are shown in A–B and an eye at 42 h APF in C. (A-A”) Hbs (red) and Rst (green) co-localize on the surface. Hbs also co-localizes with Rst in vesicles in 1°s (arrows). Rst is also found in vesicles in IOCs (arrowheads), where it does not co-localize with Hbs. A merged view is shown in A”. (B-B”) Sns (red) co-localizes with Kirre (green) on the cell surface. Sns and Kirre vesicles largely co-localize (arrows). Sns and Kirre were also found at the borders between IOCs and bristle groups (arrowheads). (C-C’) sns-RNAi (sns-IR3) was targeted to single cells (green, C’) and the eye was stained with an anti-Sns antibody (red). Sns is reduced at the border when sns-IR is targeted to 1°s (arrowheads). D) Schematic representation of expression domains of the IRM proteins. Hbs and Sns of the Nephrin group (magenta) are expressed 1°s and cone cells. Kirre and Rst of the Neph1 group are expressed in IOCs (green).
hbs is known to be transcribed in 1°s and cone cells in the pupal eye (Bao and Cagan, 2005). Levels of Sns protein on the surface membrane were reduced when Sns was knocked down in individual 1°s but not within IOCs (Figure 4C-C”), indicating that sns is transcribed within 1°s at this stage. Therefore, Sns and Hbs are expressed in a pattern complementary to Rst and Kirre, the former pair principally in 1°s while the latter in IOCs.
5. Kirre binds both Hbs and Sns
To assess interactions between IRM adhesion molecules, we ectopically expressed Hbs or Sns within single cells in situ and assayed distribution of Rst and Kirre. Targeted ectopic Hbs was previously demonstrated to re-distribute Rst to adjacent membranes (Bao and Cagan, 2005). When Hbs was over-expressed in individual 1°s, ectopic Kirre was similarly recruited to the border (Figure 5A-A’), suggesting Kirre and Hbs function as a complex in trans. When a single copy of an sns transgene was over-expressed in 1°s, ectopic Kirre was found weakly at the border; two copies led to more robust recruitment of Kirre to the border (Figure 5B-B). In complementary experiments, over-expressing Kirre in single IOCs led to recruitment of ectopic Hbs and Sns to adjacent cell borders (Figure 5C–D). Similar results were obtained when Rst was over-expressed in single IOCs. Taken together, these results indicate that the four IRM adhesion molecules function pair-wise by forming complexes: Hbs/Sns in 1°s associate in trans with Rst/Kirre in IOCs.
Figure 5.
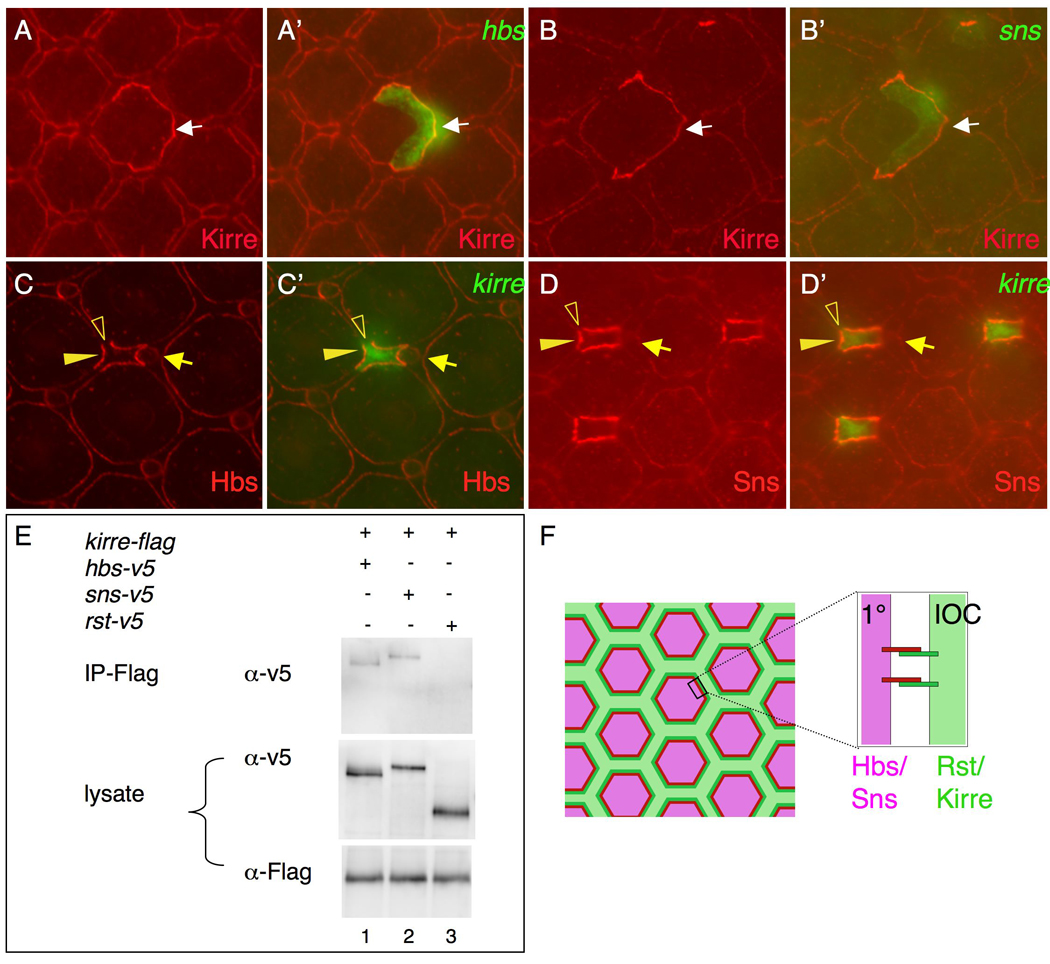
Kirre binds both Sns and Hbs. (A-A’) When Hbs (green) is over-expressed in a single 1°, ectopic Kirre (red) is recruited to the border (arrow). (B-B’) Upon over-expression of two copies of a sns transgene (green) in IOCs, ectopic Kirre (red) is attracted to the borders (arrow). (C-C’) When Kirre (green) is over-expressed in an IOC, ectopic Hbs is recruited to the border (arrowheads). Note ectopic Hbs is not found in between IOCs (open arrowheads). An arrow points to a bristle group. (D-D’) When Kirre (green) is over-expressed in IOCs, ectopic Sns is recruited to the border (arrowheads). Note ectopic Sns is not found in between IOCs (open arrowheads). An arrow points to a bristle group. (E) Both Sns and Hbs are co-immunoprecipitated with Kirre. Lane 1, Hbs+Kirre; Lane 2, Sns+Kirre; Lane 3, Rst+Kirre. Immunoprecitation was performed using an anti-Flag antibody.
To further assess direct protein-protein interactions we utilized cultured S2 cells. Direct interactions between Kirre and Sns and between Hbs and Rst have been reported in S2 cells (Bao and Cagan, 2005; Chen and Olson, 2001; Galletta et al., 2004). S2 cells were transfected with each construct separately; after co-culture, cells were lysed and co-immunoprecipitation was performed. Sns and Hbs were co-immunoprecipitated with Kirre (Figure 5E). In contrast, very little Rst was co-immunoprecipitated with Kirre (Figure 5E). Although we cannot exclude the possibility of intermediate factors in mediating interactions between IRM proteins, these data strongly suggest Kirre binds both Hbs and Sns in trans. Taken together, we conclude that these four IRM proteins form heterophilic interactions during ommatidial patterning (Figure 5F).
6. Hbs and Sns promote separation of cells from ommatidia
Our data identify all four IRM molecules as regulators of ommatidial patterning. We have previously demonstrated that Rst renders IOCs more adhesive to 1° than to other IOCs due to heterophilic interactions with Hbs (Bao and Cagan, 2005). This observation raises the possibility that Hbs and Sns similarly render ommatidia more adhesive to IOCs than to ommatidia. To test this possibility, we used FLP/FRT technology (Basler and Struhl, 1994) to mis-express Hbs and Sns within individual IOCs.
When a single copy of a sns transgene was expressed in an isolated IOC, the cell retained its normal contacts (Figure 6A). In contrast, expressing a single copy of an hbs transgene within an IOC led to partial segregation of the hbs-targeted IOC. Typically, the hbs-targeted IOC retained contact with one ommatidium and was flattened against it but was segregated away from the second ommatidium by neighboring IOCs, suggesting reduced adhesion between the hbs-targeted IOC and ommatidia (Figure 6B). Interestingly, the neighboring IOCs were part of the larger IOC hexagonal lattice, which established across the hbs-targeted IOC as if it were part of the adjacent ommatidium (Figure 6B). Neighboring IOCs established abnormally large interfaces with the hbs-targeted IOC, suggesting adhesion between the hbs-targeted IOC and neighboring IOCs was stronger than adhesion normally observed between IOCs (Figure 6B).
Figure 6.
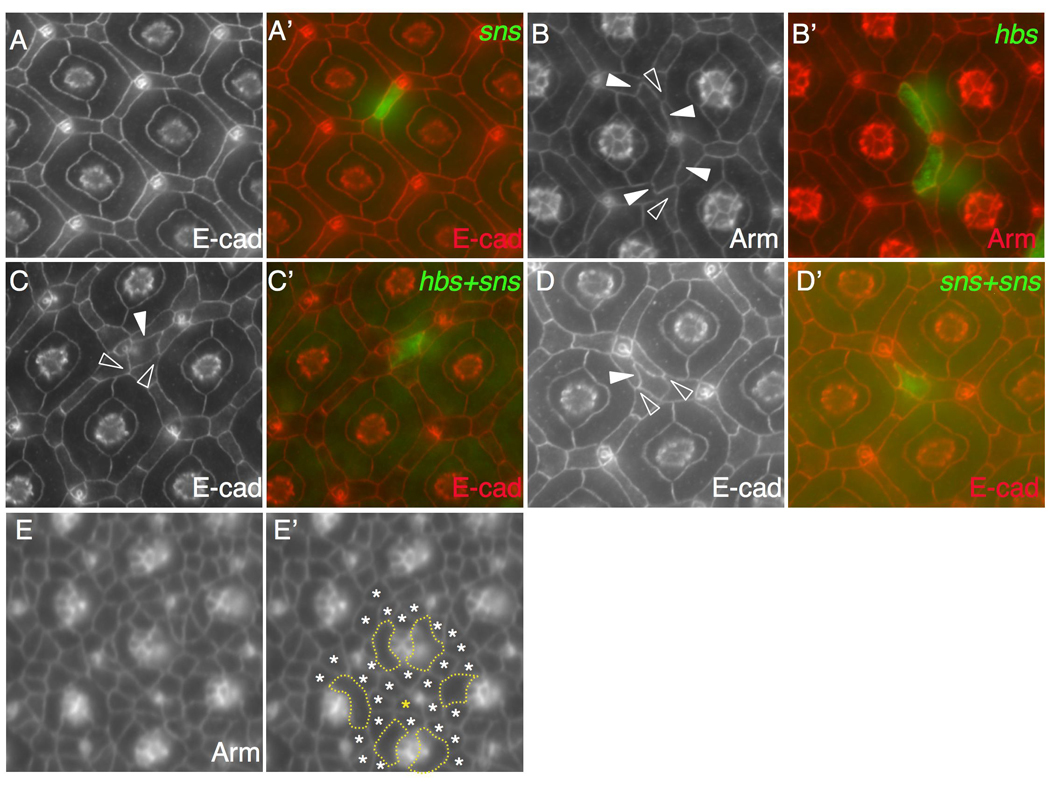
Sns and Hbs drive separation of cells from ommatidia. Eyes at 42 h APF were stained with an antibody against either Armadillo (Arm) or E-cadherin (E-cad) as indicated. Merged views in A–D are shown in A’-D’. (A-A’) When a single copy of sns (green) was expressed in a single cell, the cell retained its normal position. (B-B’) Upon expression of hbs (green), two cells were separated from one but retained contact with another ommatidium. Wild type IOCs maintained small interfaces with each other (open arrowhead) while target IOCs established larger interfaces with IOCs (arrowheads). (C-C’) When both hbs and sns were expressed in a single cell (green), the cell was fully separated from ommatidia. Wild type IOCs maintained small interfaces with each other (open arrowheads) but larger interfaces with the target IOC (arrowhead). (D-D’) When two copies of a sns transgene were expressed in a single cell (green), the cell was fully separated from ommatidia. Wild type IOCs maintained small interfaces with each other (open arrowheads) but larger interfaces with the target IOC (arrowhead). (E-E’) Pupal eye at 20 h APF. The eye was stained with an anti-Armadillo antibody. Emerging 1°s are outlined (E’). Most IOCs (white asterisks) are found in touch with at least one 1°. In the same area, an IOC (gold asterisk) not in touch with any 1° is highlighted.
Higher levels of hbs/sns expression, however, yielded a different outcome. Expressing both hbs and sns transgenes within a single IOC led to a newly emergent, striking phenotype: 41% of hbs/sns-targeted IOCs were fully separated from ommatidia with neighboring IOCs intercalated around all sides of the cell (n=48 clones, 5 eyes; Figure 6C). Surrounding IOCs maintained small interfaces with each other but larger interfaces with the hbs/sns-targeted cell, similar to the former’s behavior with bona fide ommatidia. These results suggest that expressing Sns plus Hbs is sufficient to guide cells into establishing a novel ommatidial-like niche that is separated from nearby ommatidia.
The ability of hbs/sns-targeted IOCs to separate from ommatidia raises two possibilities. The segregation effect could be a direct result of the higher protein levels provided by the two transgenes. Alternatively, it could reflect a newly emergent property when Hbs and Sns were expressed in the same cell. To distinguish between these possibilities, we expressed two copies of a sns transgene in single IOCs; recall that a single copy had no effect on patterning. Upon expression of the sns transgenes, 52% of target cells were fully separated from ommatidia (n=60 clones, 6 eyes; Figure 6D). This result indicates that segregation of IOCs is correlated with the amount of Sns/Hbs proteins expressed within the cell: low levels did not significantly alter the sorting behavior of the target cells (Figure 6A), moderate levels led to partial segregation of the target cells from nearby ommatidia (Figure 6B), and high levels led to fully segregated IOCs (Figure 6C–D). This effect on cell sorting was specific to Hbs and Sns since IOCs did not segregate from ommatidia upon over-expression of the classical adhesion molecules E-cadherin or N-cadherin (data not shown). Drosophila E- and N-cadherin mediate cell sorting through homophilic interactions in multiple tissues including the eye (Godt and Tepass, 1998; Gonzalez-Reyes and St Johnston, 1998; Hayashi and Carthew, 2004). Failure of E- and N-cadherin to direct segregation of IOCs highlights the importance of selective adhesion within the eye field to segregate cells. Taken together, we conclude that Sns and Hbs are sufficient to promote segregation of cells from ommatidia in a dose-dependent manner.
7. Separation of Sns/Hbs-expressing cells is an active process
Our data is consistent with a model in which high levels of Hbs or Sns can actively segregate a cell or cell cluster from neighboring ommatidia. This view assumes, however, that emerging IOCs are initially found next to ommatidia and later move into their new niche. The alternative is that most IOCs are already segregated away from ommatidia early, and Hbs/Sns act to maintain their positions. To distinguish between these two possibilities, we therefore examined the pupal eye at 20 h APF when 1°s are emerging (Figure 6E). At this early stage, 95.8% of IOCs (n=54 ommatidia, 4 eyes) were found in direct contact with at least one ommatidium (Figure 6E). We therefore conclude that most randomly generated hbs/sns-targeted cells were initially in direct contact with developing ommatidia and then were actively separated through the action of Hbs/Sns.
Discussion
In developing tissues, homotypic adhesion often drives cell aggregation. For instance, homotypic adhesion mediated by cadherins promotes aggregation of cone cells in the Drosophila eye, positioning of the Drosophila oocyte and aggregation of blastomeres in the mouse embryo (De Vries et al., 2004; Godt and Tepass, 1998; Gonzalez-Reyes and St Johnston, 1998; Hayashi and Carthew, 2004). Conversely, preferential adhesion mediated by heterophilic-interacting adhesion molecules promotes segregation of cells. In the Drosophila eye, for example, preferential adhesion mediated by IRM adhesion molecules promotes segregation of IOCs, leading to reduction of IOC:IOC contacts (Bao and Cagan, 2005). Nevertheless, IOCs do not fully separate from each other but rather they maintain a coherent hexagonal lattice, raising the question of how IOCs remain contiguous so that ommatidia are separated.
In this paper we show that preferential adhesion of ommatidia to IOCs plays a role in maintaining separation of ommatidia. In thermodynamics, adhesion strength corresponds to the work of adhesion (W) (Moore, 1972). We designate adhesion strength of ommatidia (specifically 1°s) and IOCs as W(o-o) and W(i-i), respectively, and adhesive strength between ommatidia and IOCs as W(o-i). Based on the preferential adhesion model (Bao and Cagan, 2005), IOCs have stronger adhesion to ommatidia than to themselves (W(o-i)> W(i-i)). In this paper we expand this initial finding and provide evidence that Hbs and Sns render ommatidia more adhesive to IOCs than to ommatidia (W(o-i)> W(o-o)).
Based on their homology to the mammalian proteins Neph1 and Nephrin, the four IRM adhesion molecules can be subdivided into two groups: Kirre and Rst in the Neph1 group, and Hbs and Sns in the Nephrin group. The IRM proteins of the Neph1 group are expressed in IOCs, and proteins of the Nephrin group in 1°s. Together with published observations (Bao and Cagan, 2005; Chen and Olson, 2001; Galletta et al., 2004), our results indicate that IRM proteins from one group form heterophilic interactions with proteins from another group (Figure 5E). Strength of interaction between proteins within the same group are much weaker (Figure 5E; and S. Bao and R. Cagan, unpublished). Therefore, IRM proteins prefer inter-group interactions to intra-group interactions. Rst has been shown to mediate preferential adhesion of IOCs to ommatidia through heterophilic interactions with Hbs (Bao and Cagan, 2005). The similarity between the two groups of IRM proteins with regards to preferred protein-protein interactions raises the possibility that Hbs and Sns mediate preferential adhesion of ommatidia to IOCs through heterophilic interactions with Kirre and Rst. Direct ommatidium:ommatidium contact upon reduction of either Kirre-plus-Rst or Hbs-plus-Sns supports this model (Figure 1, Figure 3).
Strong in vivo support for preferential adhesion between ommatidia and IOCs comes from mis-expression of Hbs and Sns in single IOCs. Here we refer to the Hbs/Sns-targeted IOCs as O’ cells: they retained most cell identity aspects of IOCs, and mimic ommatidia only inasmuch as they expressed Hbs/Sns. Accordingly, adhesion strength between the O’ cells and wild type IOCs is defined as W(o’-i), and adhesion strength between the O’ cells and ommatidia as W(o’-o). If adhesion between the O’ cell and ommatidia mediated by Hbs and Sns is stronger than adhesion between the O’ cell and IOCs, we expect the Hbs/Sns-targeted cells will remain attached to ommatidia (Figure 7). However, if Hbs/Sns-targeted O’ cells become more adhesive to IOCs than to ommatidia, these Hbs/Sns-targeted cells should separate away from all ommatidia (Figure 7). The observation that O’ cells do indeed sort away from ommatidia indicates that Hbs plus Sns render the O’ cell less adhesive to ommatidia than to IOCs (Figure 6C).
Figure 7.
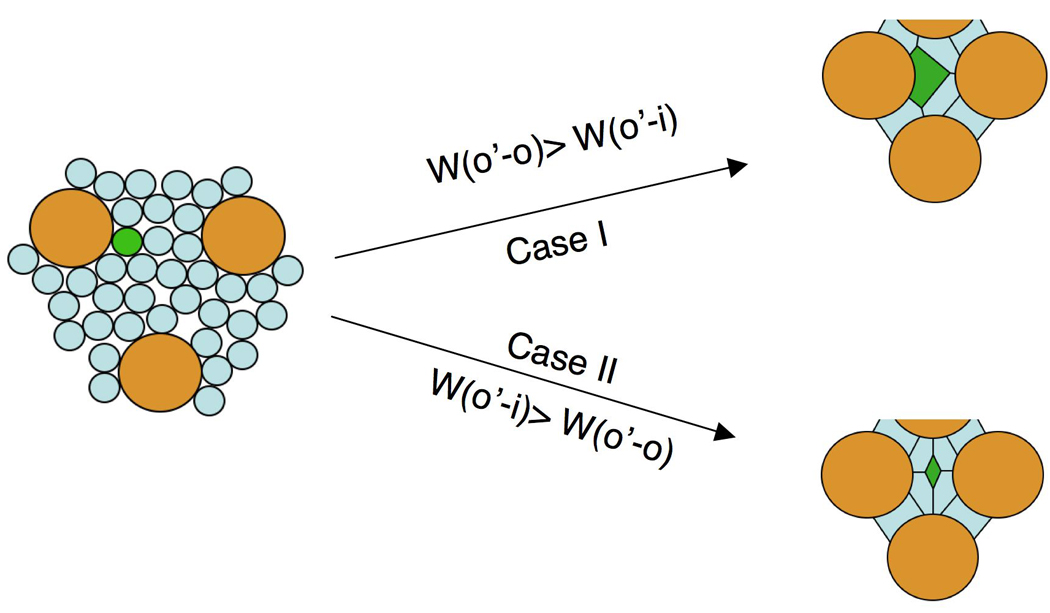
Hbs and Sns mediate preferential adhesion of ommatidia to IOCs. Ommatidia (o) are shaded in gold and IOCs (i) in light blue. A single IOC targeted with ectopic Hbs/Sns expression (O’ cell) is highlighted in green. If Hbs and Sns make the O’ cell more adhesive to ommatidia than to IOCs (W(o’-o) > W(o’-i)), upon mis-expression of Hbs/Sns, the O’ cell should remain attached to ommatidia (Case I). Conversely, if Hbs/Sns render the O’ cell less adhesive to ommatidia than to IOCs (W(o’-i) > W(o’-o)), the O’ cell should be detached from all ommatidia upon ectopic expression of Hbs/Sns (Case II).
These results strongly suggest expression of Hbs and Sns is sufficient to render ommatidia less adhesive to other ommatidia than to IOCs. As a result, ommatidia form preferential adhesion to IOCs. To take into account preferential adhesion of IOCs to ommatidia (Bao and Cagan, 2005), we propose an ommatidia/IOC mutual preferential adhesion model to explain both local cell sorting and global ommatidial patterning: (i) preferential adhesion of IOCs to ommatidia promotes reduced IOC/IOC contacts; (ii) preferential adhesion of ommatidia to IOCs minimizes ommatidium:ommatidium contacts; both types of preferential adhesion promote IOC:ommatidium contacts. Altering this mutual preferential adhesion by manipulating the strength of cell adhesion is sufficient to lead to changes in both local cell sorting and global ommatidial patterning.
Although our results highlight the importance of preferential adhesion in maintaining separation of ommatidia, we do not exclude the possibility that homotypic adhesion also contributes to this process. Rst has been shown to form homophilic interactions in cultured cells (Schneider et al., 1995) and Hbs can form homophilic interactions in vitro as well (S. Bao and R. Cagan, unpublished). To dissect the potential role of homotypic adhesion in maintaining separation of ommatidia, further work is needed to more precisely quantify differences in interactions among the IRM proteins in vivo.
Supplementary Material
Supplemental Figure S1. RNAi constructs strongly reduced target genes. Each RNAi construct was co-expressed with Dicer-2 driven by GMR-Gal4 (referred to as IR3). Eyes at 42 h APF were stained with specific antibodies and co-stained with junctional proteins as indicated. Wild type eyes were used as control (A, C, E and G). Staining for wild type and RNAi depleted eyes was performed in the same wells under the same condition.
Supplemental Figure S2 Specificity of RNAi constructs. Independent transgenic RNAi transgenes from the VDRC stock center were co-expressed with Dicer-2 driven by GMR-Gal4. Pupal eyes were stained with either an anti-Echinoid (A) or E-cadherin (B) antibody and co-stained with antibodies as indicated. (A-A’) Strong reduction of Rst using an RNAi transgene from VDRC led to patterning defects in a manner similar to using transgenes we generated for this work. Rst (red, A’) was undetectable. Defects in cone cells are indicated by arrowheads and contacting ommatidia highlighted by asterisks. (B-B’) Strong reduction of Hbs using an RNAi transgene from VDRC led to similar patterning defects to those by using RNAi transgenes generated for this work. Hbs (green, B’) was undetectable. Defects in cone cells are indicated by arrowheads and contacting ommatidia highlighted by asterisks.
Acknowledgements
We thank Susan Abmayr, Mary Baylies, Mar Ruiz-Gomez, Matthew Freeman, Stephen Cohen, Liqun Luo, Andrew Jarman, Akinao Nose and Nick Baker for flies, plasmids and antibodies. We also thank Claude Desplan, Elisabeth Knust and Richard Carthew for advices and comments on the manuscript. We are grateful to Gerit A. Linneweber for help in antibody production and Timothy Kang in imaging. We also thank the Bloomington Stock Center for flies and Developmental Studies of Hybridoma Bank, University of Iowa, for antibodies. This work is supported by the National Institute of Health (Grant R01-EY1149).
Abbreviations
- 1°
primary pigment cell
- Hbs
Hibris
- Kirre
Kin of irre
- Rst
Roughest
- Sns
Sticks and stones
- IOC
inter-ommatidial cell
- IRM
Irre cell Recognition Module
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare they have no competing financial interests.
References
- Artero RD, Castanon I, Baylies MK. The immunoglobulin-like protein Hibris functions as a dose-dependent regulator of myoblast fusion and is differentially controlled by Ras and Notch signaling. Development. 2001;128:4251–4264. doi: 10.1242/dev.128.21.4251. [DOI] [PubMed] [Google Scholar]
- Baker NE, Rubin GM. Effect on eye development of dominant mutations in Drosophila homologue of the EGF receptor. Nature. 1989;340:150–153. doi: 10.1038/340150a0. [DOI] [PubMed] [Google Scholar]
- Baker NE, Yu SY. Proneural function of neurogenic genes in the developing Drosophila eye. Curr Biol. 1997;7:122–132. doi: 10.1016/s0960-9822(06)00056-x. [DOI] [PubMed] [Google Scholar]
- Bao S, Cagan R. Preferential adhesion mediated by Hibris and Roughest regulates morphogenesis and patterning in the Drosophila eye. Dev Cell. 2005;8:925–935. doi: 10.1016/j.devcel.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Bao S, Cagan R. Fast cloning inverted repeats for RNA interference. Rna. 2006;12:2020–2024. doi: 10.1261/rna.258406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baonza A, Casci T, Freeman M. A primary role for the epidermal growth factor receptor in ommatidial spacing in the Drosophila eye. Curr Biol. 2001;11:396–404. doi: 10.1016/s0960-9822(01)00125-7. [DOI] [PubMed] [Google Scholar]
- Basler K, Struhl G. Compartment boundaries and the control of Drosophila limb pattern by hedgehog protein. Nature. 1994;368:208–214. doi: 10.1038/368208a0. [DOI] [PubMed] [Google Scholar]
- Bour BA, Chakravarti M, West JM, Abmayr SM. Drosophila SNS, a member of the immunoglobulin superfamily that is essential for myoblast fusion. Genes Dev. 2000;14:1498–1511. [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- Chen EH, Olson EN. Antisocial, an intracellular adaptor protein, is required for myoblast fusion in Drosophila. Dev Cell. 2001;1:705–715. doi: 10.1016/s1534-5807(01)00084-3. [DOI] [PubMed] [Google Scholar]
- De Vries WN, Evsikov AV, Haac BE, Fancher KS, Holbrook AE, Kemler R, Solter D, Knowles BB. Maternal beta-catenin and E-cadherin in mouse development. Development. 2004;131:4435–4445. doi: 10.1242/dev.01316. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Dworak HA, Charles MA, Pellerano LB, Sink H. Characterization of Drosophila hibris, a gene related to human nephrin. Development. 2001;128:4265–4276. doi: 10.1242/dev.128.21.4265. [DOI] [PubMed] [Google Scholar]
- Fischbach KF, Linneweber GA, Andlauer TF, Hertenstein A, Bonengel B, Chaudhary K. The irre cell recognition module (IRM) proteins. J Neurogenet. 2009;23:48–67. doi: 10.1080/01677060802471668. [DOI] [PubMed] [Google Scholar]
- Galletta BJ, Chakravarti M, Banerjee R, Abmayr SM. SNS: Adhesive properties, localization requirements and ectodomain dependence in S2 cells and embryonic myoblasts. Mech Dev. 2004;121:1455–1468. doi: 10.1016/j.mod.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Garcia-Bellido A, Merriam JR. Cell lineage of the imaginal discs in Drosophila gynandromorphs. J Exp Zool. 1969;170:61–75. doi: 10.1002/jez.1401700106. [DOI] [PubMed] [Google Scholar]
- Godt D, Tepass U. Drosophila oocyte localization is mediated by differential cadherin-based adhesion. Nature. 1998;395:387–391. doi: 10.1038/26493. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Reyes A, St Johnston D. The Drosophila AP axis is polarised by the cadherin-mediated positioning of the oocyte. Development. 1998;125:3635–3644. doi: 10.1242/dev.125.18.3635. [DOI] [PubMed] [Google Scholar]
- Grether ME, Abrams JM, Agapite J, White K, Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Carthew RW. Surface mechanics mediate pattern formation in the developing retina. Nature. 2004;431:647–652. doi: 10.1038/nature02952. [DOI] [PubMed] [Google Scholar]
- Johannsen OA. Eye structure in normal and eye-mutant Drosophilas. J Morph Physiol. 1924;39:337–349. [Google Scholar]
- Larson DE, Liberman Z, Cagan RL. Cellular behavior in the developing Drosophila pupal retina. Mech Dev. 2008;125:223–232. doi: 10.1016/j.mod.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore WJ. Physical Chemistry. 4th ed. Prentice Hall: 1972. [Google Scholar]
- Nose A, Isshiki T, Takeichi M. Regional specification of muscle progenitors in Drosophila: the role of the msh homeobox gene. Development. 1998;125:215–223. doi: 10.1242/dev.125.2.215. [DOI] [PubMed] [Google Scholar]
- Reiter C, Schimansky T, Nie Z, Fischbach KF. Reorganization of membrane contacts prior to apoptosis in the Drosophila retina: the role of the IrreC-rst protein. Development. 1996;122:1931–1940. doi: 10.1242/dev.122.6.1931. [DOI] [PubMed] [Google Scholar]
- Ruiz-Gomez M, Coutts N, Price A, Taylor MV, Bate M. Drosophila dumbfounded: a myoblast attractant essential for fusion. Cell. 2000;102:189–198. doi: 10.1016/s0092-8674(00)00024-6. [DOI] [PubMed] [Google Scholar]
- Schneider T, Reiter C, Eule E, Bader B, Lichte B, Nie Z, Schimansky T, Ramos RG, Fischbach KF. Restricted expression of the irreC-rst protein is required for normal axonal projections of columnar visual neurons. Neuron. 1995;15:259–271. doi: 10.1016/0896-6273(95)90032-2. [DOI] [PubMed] [Google Scholar]
- Shen K, Bargmann CI. The immunoglobulin superfamily protein SYG-1 determines the location of specific synapses in C. elegans. Cell. 2003;112:619–630. doi: 10.1016/s0092-8674(03)00113-2. [DOI] [PubMed] [Google Scholar]
- Shen K, Fetter RD, Bargmann CI. Synaptic specificity is generated by the synaptic guidepost protein SYG-2 and its receptor, SYG-1. Cell. 2004;116:869–881. doi: 10.1016/s0092-8674(04)00251-x. [DOI] [PubMed] [Google Scholar]
- Sohn RL, Huang P, Kawahara G, Mitchell M, Guyon J, Kalluri R, Kunkel LM, Gussoni E. A role for nephrin, a renal protein, in vertebrate skeletal muscle cell fusion. Proc Natl Acad Sci U S A. 2009;106:9274–9279. doi: 10.1073/pnas.0904398106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer SA, Powell PA, Miller DT, Cagan RL. Regulation of EGF receptor signaling establishes pattern across the developing Drosophila retina. Development. 1998;125:4777–4790. doi: 10.1242/dev.125.23.4777. [DOI] [PubMed] [Google Scholar]
- Strunkelnberg M, Bonengel B, Moda LM, Hertenstein A, de Couet HG, Ramos RG, Fischbach KF. rst and its paralogue kirre act redundantly during embryonic muscle development in Drosophila. Development. 2001;128:4229–4239. doi: 10.1242/dev.128.21.4229. [DOI] [PubMed] [Google Scholar]
- Tanenbaum S, Gorski S, Cagan RL. Genetic modifiers of the Drosophila irreC-rst locus: potential new regulators of retinal development and programmed cell death. Genetics. 2000;156:205–217. doi: 10.1093/genetics/156.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tio M, Ma C, Moses K. spitz, a Drosophila homolog of transforming growth factor-alpha, is required in the founding photoreceptor cells of the compound eye facets. Mech Dev. 1994;48:13–23. doi: 10.1016/0925-4773(94)90002-7. [DOI] [PubMed] [Google Scholar]
- Tryggvason K, Patrakka J, Wartiovaara J. Hereditary proteinuria syndromes and mechanisms of proteinuria. N Engl J Med. 2006;354:1387–1401. doi: 10.1056/NEJMra052131. [DOI] [PubMed] [Google Scholar]
- Venugopala Reddy G, Reiter C, Shanbhag S, Fischbach KF, Rodrigues V. Irregular chiasm-C-roughest, a member of the immunoglobulin superfamily, affects sense organ spacing on the Drosophila antenna by influencing the positioning of founder cells on the disc ectoderm. Dev Genes Evol. 1999;209:581–591. doi: 10.1007/s004270050292. [DOI] [PubMed] [Google Scholar]
- Weavers H, Prieto-Sanchez S, Grawe F, Garcia-Lopez A, Artero R, Wilsch-Brauninger M, Ruiz-Gomez M, Skaer H, Denholm B. The insect nephrocyte is a podocyte-like cell with a filtration slit diaphragm. Nature. 2009;457:322–326. doi: 10.1038/nature07526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff T, Ready DF. Cell death in normal and rough eye mutants of Drosophila. Development. 1991;113:825–839. doi: 10.1242/dev.113.3.825. [DOI] [PubMed] [Google Scholar]
- Yu SY, Yoo SJ, Yang L, Zapata C, Srinivasan A, Hay BA, Baker NE. A pathway of signals regulating effector and initiator caspases in the developing Drosophila eye. Development. 2002;129:3269–3278. doi: 10.1242/dev.129.13.3269. [DOI] [PubMed] [Google Scholar]
- Zhuang S, Shao H, Guo F, Trimble R, Pearce E, Abmayr SM. Sns and Kirre, the Drosophila orthologs of Nephrin and Neph1, direct adhesion, fusion and formation of a slit diaphragm-like structure in insect nephrocytes. Development. 2009;136:2335–2344. doi: 10.1242/dev.031609. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. RNAi constructs strongly reduced target genes. Each RNAi construct was co-expressed with Dicer-2 driven by GMR-Gal4 (referred to as IR3). Eyes at 42 h APF were stained with specific antibodies and co-stained with junctional proteins as indicated. Wild type eyes were used as control (A, C, E and G). Staining for wild type and RNAi depleted eyes was performed in the same wells under the same condition.
Supplemental Figure S2 Specificity of RNAi constructs. Independent transgenic RNAi transgenes from the VDRC stock center were co-expressed with Dicer-2 driven by GMR-Gal4. Pupal eyes were stained with either an anti-Echinoid (A) or E-cadherin (B) antibody and co-stained with antibodies as indicated. (A-A’) Strong reduction of Rst using an RNAi transgene from VDRC led to patterning defects in a manner similar to using transgenes we generated for this work. Rst (red, A’) was undetectable. Defects in cone cells are indicated by arrowheads and contacting ommatidia highlighted by asterisks. (B-B’) Strong reduction of Hbs using an RNAi transgene from VDRC led to similar patterning defects to those by using RNAi transgenes generated for this work. Hbs (green, B’) was undetectable. Defects in cone cells are indicated by arrowheads and contacting ommatidia highlighted by asterisks.


