Abstract
Hox genes of the Abdominal-B (Abd-B) class regulate gonadal development in diverse metazoans. Here we have investigated the role of the Abd-B homolog egl-5 in C. elegans gonadal development. Previous work showed that egl-5 is required male-specifically in the gonad and that mutant gonads are highly dysgenic and possibly feminized. We have used sex-specific gonadal reporter genes to confirm that the gonads of egl-5 males are extensively feminized. Sex-specific expression of egl-5 requires the global sex determination gene tra-1 and the gonadal masculinizing gene fkh-6, but mutagenesis of a short male gonadal enhancer element in egl-5 suggested that this regulation is indirect. Ectopic expression of EGL-5 in hermaphrodites is sufficient to induce male gonadal gene expression, indicating that EGL-5 plays an instructive role in male gonadal fate determination. EGL-5 acts in parallel with a Wnt/β -catenin pathway to regulate male gonadal fates and can physically interact with the Wnt pathway transcription factor POP-1 and modulate activity of a POP-1 dependent reporter gene. We propose that EGL-5 imparts sex-specific function on POP-1 by recruiting it to male-specific gonadal target genes.
Keywords: gonad, Wnt, Hox, sexual differentiation
INTRODUCTION
Homeodomain-containing transcription factors (Hox proteins) were discovered on the basis of their remarkable role in specifying positional information along the body axis, a function widely conserved among metazoans (reviewed in Maeda and Karch, 2009; Pearson et al., 2005). Subsequently it became evident that Hox proteins regulate many other processes, including sexual development (Williams and Carroll, 2009). In Drosophila, for example, sex-specific expression of the Hox gene Scr is required for development of the male-specific sex comb and has influenced the morphological evolution of this structure (Barmina and Kopp, 2007).
Hox proteins of the Abdominal-B (ABD-B) family control varied aspects of sexual development. In Drosophila ABD-B acts in concert with the conserved sex determining transcription factor Doublesex (DSX) to direct sex-specific pigmentation of the abdomen and sexually dimorphic genital disc development, thereby helping to integrate sexual differentiation with spatial patterning (Sanchez et al., 2001; Williams et al., 2008). ABD-B also is required for male-specific gonadal development in Drosophila, specifying a group of male-specific gonadal cells (DeFalco et al., 2004). In C. elegans the Abd-B homolog egl-5 regulates positional identity of cells in both sexes and is required for several aspects of male sexual differentiation, mainly in the posterior of the animal. These include formation of male-specific sensory organs, sex muscle differentiation, and gonadal development (Chisholm, 1991; Ferreira et al., 1999; Ross and Zarkower, 2003).
Here we have further investigated the role of egl-5 in the male gonad. Gonadal development in C. elegans is highly sexually dimorphic. Somatic tissues of the gonad derive from two progenitor cells, Z1 and Z4 (hereafter referred to as somatic gonad progenitors, or SGPs), which form in both sexes during embryogenesis. During this period they associate with the germline progenitors Z2 and Z3 to form a four-cell gonadal primordium that is morphologically identical in the two sexes. During the first SGP division, which occurs in the first larval stage (L1), sexual dimorphism becomes evident, with a more highly asymmetric division of the SGPs in males than in hermaphrodites. The initial gonadal divisions during L1 serve to define the gonadal axes in both sexes and to establish the cell lineage precursors; subsequent development consists of further proliferation and differentiation of these lineages and gonadal leader cell migrations that elongate and shape the gonad as it grows. Hermaphrodites develop a symmetrical gonad in which a central uterus connects to two arms, each with a sheath surrounding mitotic and meiotic germ cells and a spermatheca to store sperm. The male gonad has a very different J-shaped structure consisting of a single arm with an acellular sheath containing mitotic and meiotic germ cells, a seminal vesicle (SV) that stores spermatids, and a vas deferens (VD) connecting the gonad to the cloaca.
Sex specificity in C. elegans gonadal development requires the global sex determination pathway, acting through the TRA-1/GLI transcription factor, which specifies gonadal sex during late embryogenesis and early L1 (Hodgkin, 1987; Mathies et al., 2004). Other genes mediate subsequent sexual differentiation of the gonad, these include the forkhead transcription factor, fkh-6 (Chang et al., 2004), the cyclin D homolog cyd-1 (Tilmann and Kimble, 2005), and the Wnt pathway components sys-1, pop-1, and lit-1 (Kalis et al., in press). Loss of each of these genes results in male gonads that are extensively feminized. While this clearly demonstrates a requirement for these genes in male gonadal differentiation none of these genes act sex-specifically in male gonadogenesis. This suggests that additional genes must confer sex-specificity on gonadal differentiation. egl-5 is a prime candidate for such a gene, as it is expressed sex-specifically in male gonads and egl-5 mutants have only male gonadal defects (Chisholm, 1991; Ferreira et al., 1999).
We recently identified egl-5 in a genome-wide RNAi screen as one of the genes whose depletion causes male gonadal feminization (Kalis et al., in press). Here we have further analyzed the role of egl-5 in gonad sex differentiation. We find that mutant males express reporter genes specific to most hermaphrodite gonadal cell types; this reveals much more extensive gonadal feminization than anticipated based on previous analysis of cellular morphology. Genetic epistasis tests and mutagenesis of an upstream gonadal regulatory element indicate that egl-5 expression is regulated indirectly by the global sex determination pathway and FKH-6. Importantly, ectopic EGL-5 was sufficient to masculinize the hermaphrodite gonad, suggesting that EGL-5 plays an instructive rather than a permissive role in male gonadal differentiation. Genetic analysis suggests that egl-5 functions in parallel with a Wnt/β-catenin pathway to modulate POP-1 transcriptional regulatory activity, and we found that EGL-5 and POP-1 can physically interact in yeast and in vitro. These genetic and physical interactions suggest that EGL-5 confers male-specific functions on the gonadal Wnt/β-catenin pathway, possibly by recruiting POP-1 to male-specific gonadal target genes.
MATERIALS AND METHODS
Worm culture and alleles
Nematodes were cultured as described (Stiernagle, 2006). All strains contain the high incidence of male mutation him-8 or him-5 except those carrying tra-1 alleles. Strains were maintained at 16 degrees C or 20 degrees C unless otherwise noted. All RNAi experiments were performed at 22 degrees C. The following alleles and arrays were used: Alleles- LGI: pop-1(q624); LGII: fkh-6(ez16, q641); LGIII: egl-5(n486, u202), tra-1(e1834, e1099); LGIV: him-8(e1489); LGV: him-5(e1490) Integrated arrays- LGI: tnIs5[lim-7::GFP] (Hall et al., 1999); LGV: qIs56[lag-2::GFP] (Siegfried and Kimble, 2002); LGX: ezIs1[K09C8.2::GFP] (Thoemke et al., 2005); LG unknown: syIs187[POPTOP] (Green et al., 2008); qIs90[ceh-22::venus] (Asahina et al., 2006); syIS50[cdh-3::GFP] (Pettitt et al., 1996); leIs8[pes-8::lacZ] (Hope, 1991); [nmy-2::PGL-1::mRFP1] (Wolke et al., 2007); bxIs12[egl-5::GFP] (Teng et al., 2004); bxIs13[egl-5::GFP] (Teng et al., 2004) Extrachromosomal arrays- pWC1 ezEx176[fkh-6::GFP] (Chang et al., 2004), leEx780[ZK813.3::GFP] (Chang et al., 2004), ezEx82[C49C3.12::GFP] (Thoemke et al., 2005), ezEx191[hs::EGL-5] (Jiang and Sternberg, 1998), syEx609[egl-5::GFP] (Teng et al., 2004) All egl-5::GFP promoters engineered for this paper detailed below.
Cloning of cis-regulatory regions of egl-5
All egl-5::GFP clones were constructed from PCR products cloned into the Sal I and Sph I restriction sites in the pes-10 minimal promoter, pPD107.94 (Fire Vector Kit). Genomic DNA was the template for pAKK4, which was used to make all subsequent clones. Site directed mutations were added by PCR amplification. All clones were confirmed by sequencing. For each injected clone 2–3 independent lines were examined. Clones are as follows, with nucleotides corresponding to cosmid C08C3: pAKK4(ezEx177): 23,448–26,295; pAKK8(ezEx178): 23,448–24,727; pAKK11(ezEx181): 23,904–24,727; pAKK33(ezEx202): 24,188–24,311. All mutated clones correspond to nucleotides 24,188–24,311 (As diagrammed in Fig. S2): pAKK26(ezEx195); pAKK28(ezEx200); pAKK30(ezEx198); pAKK27(ezEx196); pAKK29(ezEx197); pAKK32(ezEx201); pAKK34(ezEx203); pAKK35(ezEx204); pAKK36(ezEx205); pAKK37(ezEx206); pAKK38(ezEx207); pAKK39(ezEx208); pAKK40(ezEx216).
Heat shock overexpression of EGL-5
To make EGL-5 over-expressing lines 20 ng/µl of pLG5 (hs::EGL-5) (Jiang and Sternberg, 1998), 120 ng/µl pBlueScript and 75 ng/µl of str-1::GFP (used as an injection marker) were injected into K09C8.2::GFP; him-8 or C49C3.12::GFP; him-8. To induce the expression of EGL-5, worms were synchronized by hypochlorite treatment and grown at 20 degrees C until the majority of animals were in the appropriate larval stage for heat shock. For heat shock, plates were wrapped in parafilm and submersed in a 30 degree C water bath for 1 hour, followed by at least 15 minutes in a 20 degree C water bath. Animals were assayed for GFP expression as young adults.
POPTOP analysis
Analysis of POPTOP reporter expression was performed as described previously (Kalis et al., in press), examining early to mid L3 males for mCherry expression. All images were captured at equivalent exposure levels.
FKH-6 binding site selection and gel mobility shift assay
Both the FKH-6 binding site selection and gel mobility shift assays were performed as previously described (Murphy et al., 2007). Briefly, FKH-6 binding sites were selected from random oligonucleotides by two rounds of immunoprecipitation followed by excision of the bound oligonucleotides from a gel mobility shift assay. The binding site consensus was derived from sequences of 61 selected and bound oligonucleotides.
Yeast two-hybrid assay
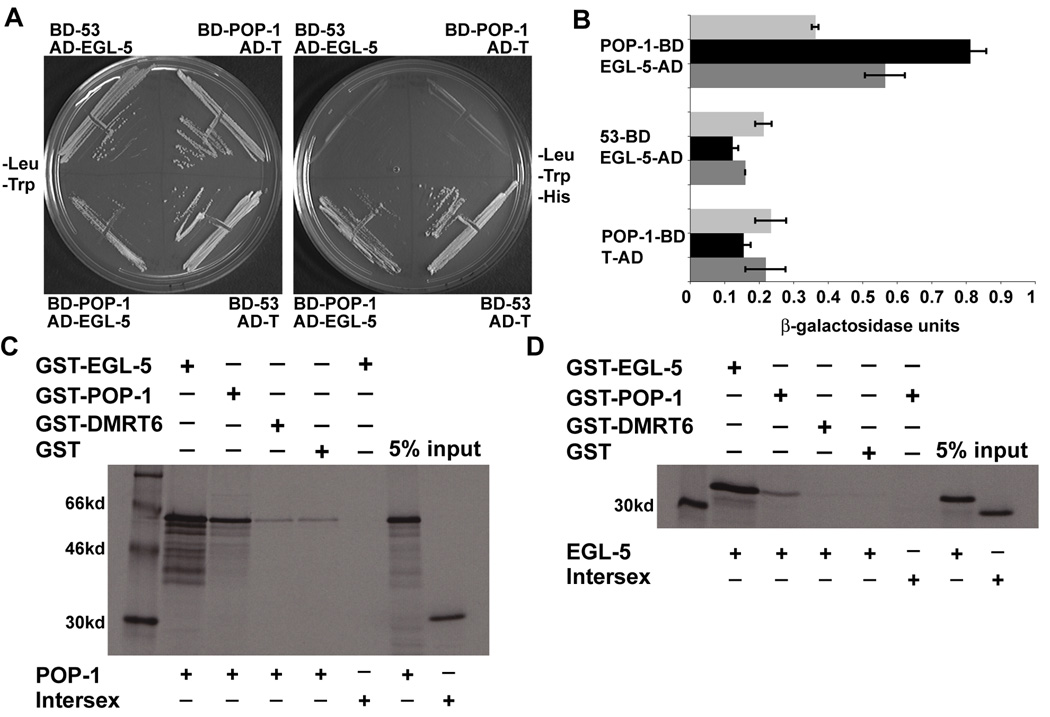
Full-length EGL-5 and POP-1 cDNAs were each cloned into the yeast two-hybrid vectors pGBKT7 BD and pGADT7 AD (Clontech Matchmaker Yeast Two-Hybrid System) and confirmed by sequencing. These plasmids were individually transformed into the yeast strain AH109, which contains four Gal4-responsive reporter genes. BD-EGL-5 strongly autoactivated and was not used further. BD-POP-1, AD-EGL-5, and positive controls BD-53 (murine p53) and AD-T (SV40 large T-antigen) were transformed pairwise into AH109 and selected on SD minimal media lacking leucine/tryptophan (transformation control), leucine/tryptophan/histidine or leucine/tryptophan/histidine/adenine. The positive interaction indicated by growth on plates lacking histidine was further confirmed using X-alpha Gal to assess activation of the MEL1 reporter (not shown) and X-beta Gal quantitative assay (Clontech Yeast Protocols Handbook) to assess activation of the lacZ reporter.
GST pulldown assay
Open reading frames for EGL-5, POP-1, and DMRT6 were cloned into pGEX20T (Huynh and Bardwell, 1998). BL21 cells expressing these GST fusion proteins were lysed with a French press and proteins purified on glutathione agarose (Sigma G4510). Open reading frames for EGL-5, POP-1 and, as a negative control, Drosophila Intersex (ISX), were also cloned into T7-plink (Dalton and Treisman, 1992). Radiolabeled proteins were prepared using TnTR quick-coupled transcription/translation system (Promega L1170) and S-35 methionine (PerkinElmer NEG-709A) according to the manufacturers’ instructions, with the addition of 20µM ZnSO4. Radiolabeled proteins were pre-cleared with unliganded glutathione beads in GST binding buffer (1X PBS, 100mM NaCl, 10% glycerol, 0.5% NP40, 2mM DTT, 0.5% non-fat dry milk, containing protease inhibitors) by rotation at room temperature for 30 minutes. After pre-clearing, radiolabeled proteins were combined with 2.5µg of the specific GST fusion proteins bound to glutathione agarose and incubated at room temperature with rotation for 30 minutes. After washing away unbound proteins, the bound proteins were resolved by SDS-PAGE and visualized by autoradiography. Input samples are equal to 5% of the total in each pulldown.
RESULTS
Extensive feminization of egl-5 mutant male gonads
Chisholm (1991) found that 5–10% of egl-5 mutant males have highly infolded gonadal tissue resembling that of the hermaphrodite uterus, suggesting possible gonadal feminization. In our RNAi screen we found that many egl-5(RNAi) males ectopically expressed a hermaphrodite-specific spermatheca or sheath GFP reporter, confirming that loss of EGL-5 can cause sexual transformation. To further investigate the degree and nature of sex reversal in egl-5 male gonads we examined expression of several sex-specific gonadal GFP reporters in two loss-of-function alleles of egl-5: the putative null allele u202 and the strong loss-of-function allele n486 (Chisholm, 1991; Wang et al., 1993).
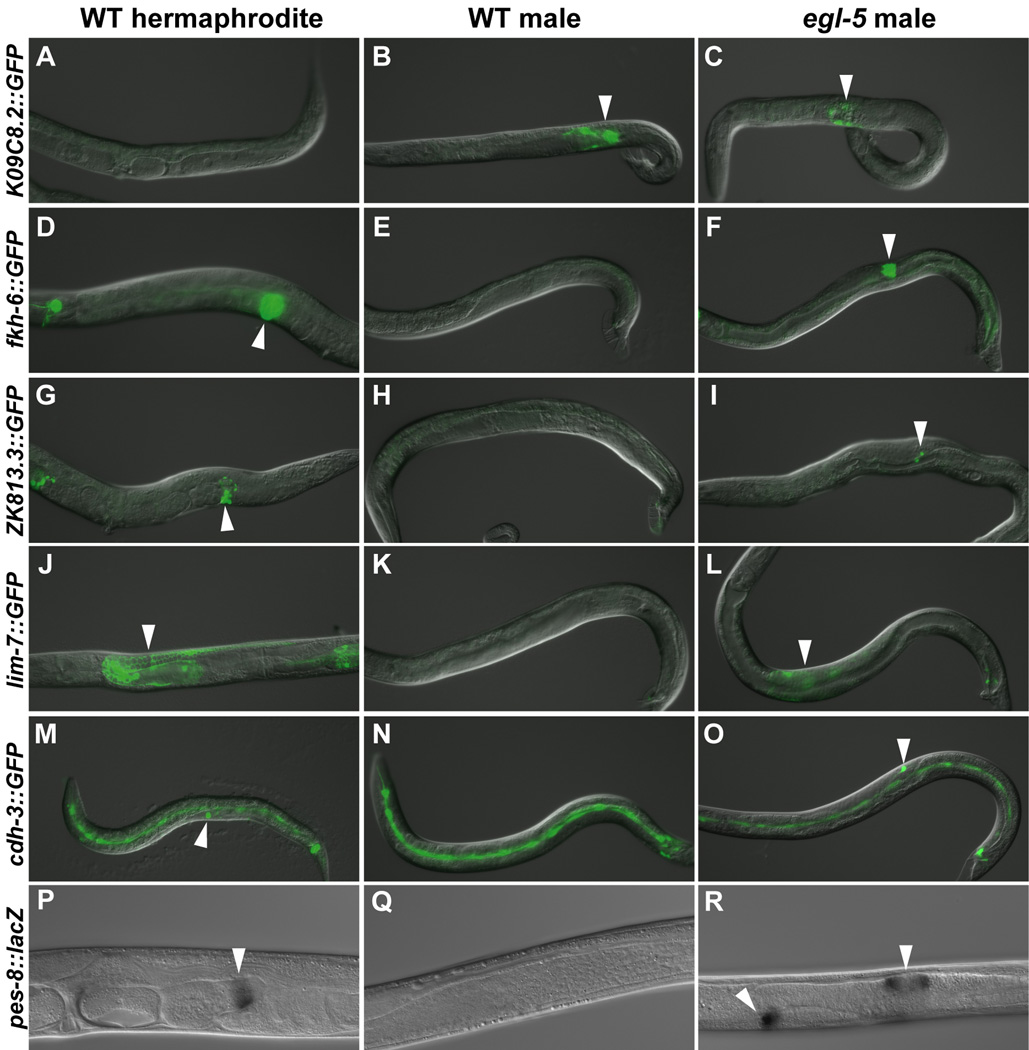
We first examined gonadal reporters that are expressed hermaphrodite-specifically in adults. Most egl-5 males expressed the spermatheca and sheath marker fkh-6::GFP, spermatheca marker ZK813.3::GFP, sheath marker lim-7::GFP, and uterine marker pes-8::lacZ (Fig. 1, Table S1). We therefore asked whether egl-5 mutants produce any male gonadal tissue, using the expression of the male-specific seminal vesicle and vas deferens reporter K09C8.2::GFP. All males retained K09C8.2::GFP expression, but the expression pattern was abnormal and often the number of expressing cells was reduced relative to controls (Fig. 1A–C, Table S1). We conclude that egl-5 male gonads contain cells with gene expression typical of most adult hermaphrodite gonadal cell types as well as male cell types and thus are extensively but incompletely sex-reversed.
Fig. 1. Extensive feminization in the egl-5 male gonad.
Left column: wild type XX hermaphrodites. Middle column: wild type XO males. Right column: egl-5(u202 or n486) mutant XO males. All panels are overlaid DIC and fluorescence images except P–R, which are DIC alone. For each reporter, fluorescence images are identical exposures for all genotypes. All animals are young adults except M–O, which are L3 animals. (A–C) K09C8.2::GFP is expressed in the seminal vesicle and vas deferens of wild type males and in egl-5 mutants. (D–R) All reporters are expressed in a specific gonadal tissue in wild type hermaphrodites and ectopically expressed in egl-5 mutant males. (D–F) fkh-6::GFP is expressed in the spermatheca and sheath. (G–I) ZK813.3::GFP is expressed in the spermatheca. (J–L) lim-7::GFP is expressed in the sheath. (M–O) cdh-3::GFP is expressed in the anchor cell and rarely in egl-5 mutants. (P–R) pes-8::lacZ, X-gal staining is in the uterine valve.
Early gonadal development is relatively normal in egl-5 males
To determine when abnormal gonadal development of egl-5 mutant males begins, we examined the major early gonadal cell types. We first examined specification of the male-specific linker cell (LC) and male distal tip cells (mDTCs), based on expression of reporter genes and cell morphology. Most egl-5 mutants specified normal numbers of LCs and mDTCs as assayed by lag-2::GFP and ceh-22-GFP expression, respectively (Table S2). This is consistent with the normal appearance of the gonadal primordium reported in egl-5(n945) males (Chisholm, 1991). As a measure of early feminization we assayed formation of anchor cells, a hermaphrodite specific gonadal cell that forms during late L1 and later induces vulva formation (Kimble and Hirsh, 1979). Although egl-5(n486) male gonads occasionally contained a cell typical anchor cell morphology and expression of the anchor cell marker cdh-3::GFP, most mutant males did not (Fig. 1M–O, Table S1). Mutant males never formed a vulva (data not shown), presumably because the ectopic anchor cells were not properly located for vulval induction or were not functional. From these results we conclude that early gonadal development is essentially normal in egl-5 males and feminization occurs later.
egl-5 expression in the male gonad
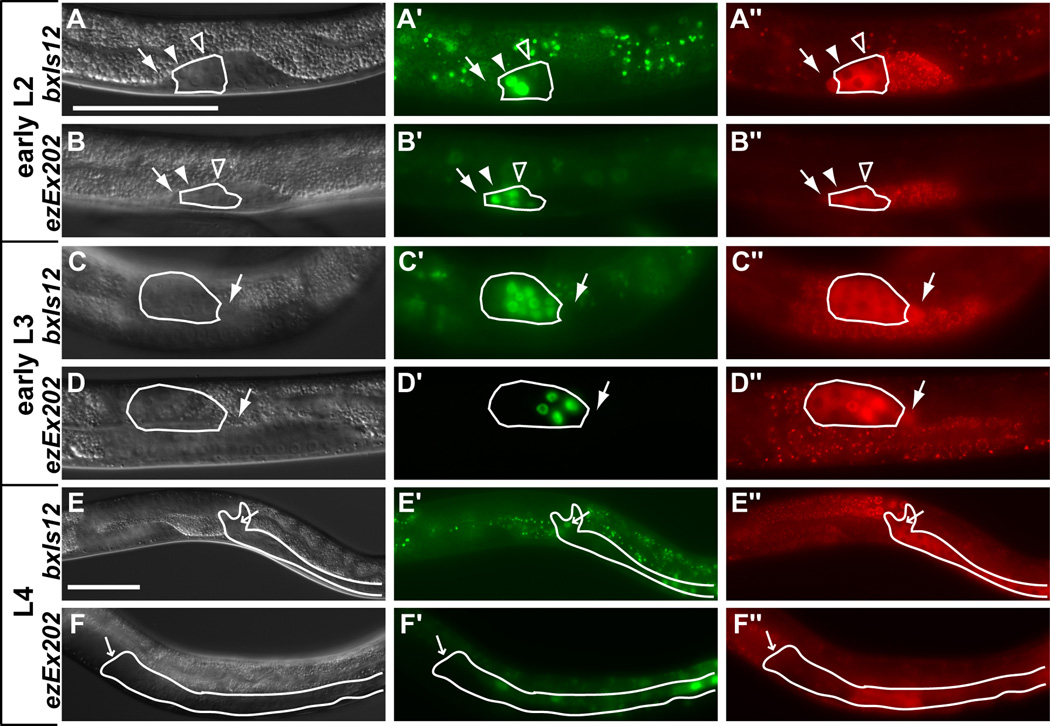
Previous antibody staining and reporter analysis found that egl-5 RNA and protein are expressed male-specifically in the gonad (Ferreira et al., 1999; Teng et al., 2004). We performed additional analysis of egl-5::GFP expression using the integrated reporter bxIs12, which contains 16 kb of egl-5 upstream sequences and has GFP fused in frame to exon 3 (Teng et al., 2004). We found that bxIs12 is expressed in most if not all somatic cells in the male gonad (VD and SV precursors and descendents) with the exception of the LC and mDTCs and their progenitors. Expression of bxIs12 begins during late L1, consistent with the relatively normal early gonadal development in egl-5 males, and by the L1 molt expression usually was visible in all seven vas deferens (VD) and seminal vesicle (SV) precursor cells clustered at the anterior end of the gonad (Fig. 2A). During most of larval development expression was much stronger in proximal than distal somatic cells (other than the LC). The timing and number of cells expressing bxIs12 is generally consistent with antibody staining. However, antibody staining was reported in both somatic and germ cells (Ferreira et al., 1999), whereas bxIs12 GFP is only expressed in somatic cells, based on lack of overlapping expression of the germ cell marker nmy-2::PGL-1::mRFP1 (Figs. 2 and S1). During anterior gonadal migration in L2 (Fig. S1C) and rapid somatic divisions of L3 most if not all of the somatic cells, which cluster at the proximal end, expressed GFP (Fig. 2C and Fig. S1E, G, I). During L4, gonadal expression decreased as morphogenesis was completed and persisted only in the seminal vesicle valve cells by the adult stage (Fig. 2E). From these results we conclude that bxIs12 contains the regulatory elements necessary for expression of egl-5 in the male somatic gonad.
Fig. 2. egl-5 gonadal expression.
Left column: DIC images. Middle column: (A, C, E) bxIs12 (egl-5::GFP with full regulatory region) expression (Teng et al., 2004), (B, D, F) exEx202 (pAKK33-123bp regulatory region required for egl-5 gonadal expression). Right column: germ cell marker nmy-2::PGL-1::mRFP1 (red punctate expression distinguishes germ cells from somatic cells (Wolke et al., 2007)). Gonadal regions containing VD and SV cells and descendents are outlined in white (green fluorescence outside white outlines is intestinal autofluorescence). White arrows indicate the LCs, filled arrowheads indicate the VD precursors, open arrowheads indicate the SV precursors, and small white arrows in rows E and F indicate the valve cells. (A, B) In early L2 GFP expression is present in the three vas deferens (VD) precursors (strong) and four seminal vesicle (SV) precursors (weaker) (all seven cells are not in focal plane). (C, D) In early L3 as the gonad reflexes to the posterior the VD and SV cells start dividing. GFP expression remains strong in VD daughters and proximal SV daughters and is much weaker in distal SV daughters. (E) In L4 as the VD and SV daughters terminally differentiate GFP expression is retained only in the four valve cells. (F) Expression is occasionally present in the valve cells of late L4 larvae and adults. Scale bars = 50µm.
Sex-specific regulation of egl-5 in the somatic gonad
To investigate how egl-5 male-specific gonadal expression is established, we mapped the cis-regulatory elements responsible. First we examined reporters with 5’ truncations relative to bxIs12 GFP (Fig. S2A) (Teng et al., 2004). None had gonadal expression, indicating that at least one essential regulatory element is located distally, in sequences unique to bxIs12. Indeed, a reporter with the most distal 2.7 kb of bxIs12 fused to the pes-10 minimal promoter fully recapitulated the male-specific gonadal expression of the bxIs12 reporter. Further dissection of this interval yielded a 123 bp element that is sufficient for male-specific expression in the somatic gonad when fused to a pes-10 minimal promoter (pAKK33, ezEx202). The time of onset and the pattern of gonadal expression promoted by ezEx202 were very similar to that of bxIs12 during larval development but expression in the adult valve cells was reduced or absent, indicating that additional sequences are required for sustained gonadal expression of egl-5 in adults (Fig. 2 and Fig. S1).
Because the global sex determination pathway terminating in tra-1 is required for sex-specific differentiation of the gonad (Hodgkin, 1987; Mathies et al., 2004), male-specific expression of egl-5 presumably is regulated by tra-1, either directly or indirectly. To confirm this we analyzed tra-1(null) XX animals for expression of egl-5::GFP and found that these pseudomales express egl-5::GFP in the somatic gonad at the normal developmental times (Table 1). We also found that gonadal phenotypes of tra-1 egl-5 XX pseudomales are similar to those of egl-5 mutant XO males, confirming that egl-5 is required for male gonadal development in tra-1 mutants (Table 2).
Table 1.
fkh-6 and tra-1 regulate egl-5 reporter expression
| genotype | males with GFP in Z1/Z4 in L1 |
n | males with GFP in non DTC/LC somatic gonad cells in L3 |
n |
|---|---|---|---|---|
| egl-5::GFP(bxIs12) | 0% | 100 | 100% | 100 |
| egl-5::GFP(ezEx202) | 0% | 25 | 100% | 30 |
| fkh-6::GFP(ezEx176) | 50% | 8 | 0% | 30 |
| bxIs12; fkh-6(ez16) | 0% | 12 | 0% | 11 |
| bxIs12; tra-1(e1834)a | ND | ND | 67%b | 9 |
| ezEx202; tra-1(e1834)a | 0% | 13 | ND | ND |
| ezEx176; egl-5(n486) | 33% | 12 | 0% | 31 |
XX psuedomales scored
Lack of reporter expression in some tra-1(e1834) psuedomales is likely due to the partially penetrant somatic gonad abnormality found in tra-1(null) animals (Hodgkin, 1987).
Table 2.
Genetic interaction of egl-5 with fkh-6 and tra-1.
| genotype | 4–8 cell gonad |
not elongated gonad |
partially or abnormally elongated gonad |
WT | n |
|---|---|---|---|---|---|
| WT | 0 | 0 | 0 | 100 | 100 |
| egl-5(n486) | 0 | 4 | 96 | 0 | 172 |
| egl-5(u202) | 0 | 4 | 96 | 0 | 56 |
| tra-1(e1834) | 3 | 25 | 34 | 37 | 59 |
| fkh-6(q641)b | 0 | 100 | 0 | 0 | 198 |
| egl-5(n486) tra-1(e1834)a | 0 | 58 | 42 | 0 | 53 |
| fkh-6(q641); tra-1(e1099)ab | 90 | 10 | 0 | 0 | 42 |
| fkh-6(ez16); egl-5(n486) | 3 | 97 | 0 | 0 | 34 |
XX pseudomales scored
from Chang et al. 2004
fkh-6 is required for male gonadal development during the first division of the SGPs, whereas egl-5 is expressed later in L1. Based on this temporal order, it is likely that egl-5 acts downstream of fkh-6 in gonadal development. Consistent with this view, fkh-6 reporter expression was unaffected in egl-5 mutants but egl-5 reporter expression was severely reduced in fkh-6 mutants (Table 1). In addition, the gonadal phenotypes of fkh-6; egl-5 double mutants resembled those of fkh-6 single mutants (Table 2). We conclude that egl-5 functions downstream of both tra-1 and fkh-6 in the male gonad.
We found previously that fkh-6 and tra-1 are required redundantly for proliferation of male gonadal cells (Chang et al., 2004). We therefore tested whether egl-5 might function in this process downstream of either tra-1 or fkh-6. We assessed the number of somatic cells in gonads of egl-5; fkh-6 males and egl-5 tra-1 pseudomales. In both cases although gonadal development was highly abnormal, the number of somatic cells was not severely reduced (data not shown). From these results we conclude that egl-5 is critical for male gonadal fate specification but is dispensable for mitotic proliferation in the male gonad. These results further support the idea that male fate and proliferation are controlled by separable pathways (Chang et al., 2004) and indicate that egl-5 functions exclusively in the male fate pathway.
Temperature shift experiments with temperature sensitive alleles of tra-2 and fem-1 have shown that the global sex determination pathway normally specifies gonadal sex before the first division of the SGPs (Klass et al., 1976; Nelson et al., 1978). fkh-6 also acts early in gonadogenesis; it is expressed in the SGPs and seems to function during their first division. Although egl-5 is not expressed until several hours after the first SGP division either TRA-1 or FKH-6 might directly bind the 123 bp gonadal regulatory element and regulate egl-5 transcription. To investigate this possibility we searched the regulatory element for potential binding sites for either protein. The TRA-1 consensus DNA binding site has been determined previously and confirmed in vivo (Conradt and Horvitz, 1999; Yi et al., 2000; Zarkower and Hodgkin, 1993). To identify potential FKH-6 binding sites it was necessary to determine FKH-6 DNA binding specificity. To do so, we selected from a pool of random oligonucleotides those bound in vitro by FKH-6, identified a consensus putative binding motif contained in these oligonucleotides, and confirmed the specificity of FKH-6 for binding to the consensus motif in gel mobility shift assays (Fig. S3 and Methods).
The gonadal regulatory element contained three potential TRA-1 binding sites and two potential FKH-6 binding sites, and we mutated these individually and in combination. TRA-1 represses egl-5 expression and thus mutating TRA-1 binding sites would be expected to cause ectopic reporter expression in XX gonads. However, mutating the three potential TRA-1 sites singly or in combinations had no affect or caused the loss of reporter expression (Fig. S2B). We cannot exclude the possibility that one or more bona fide TRA-1 sites overlaps an essential positive regulatory element that also was altered. However, although chromatin immunoprecipitation detects strong binding of TRA-1 to its known regulatory elements in mab-3 and xol-1, we detected no association of TRA-1 with the egl-5 regulatory element in L1 or L3 (M. Berkseth and DZ, unpublished data). FKH-6 promotes activity of egl-5 but we found that mutating one or both of the candidate FKH-6 binding sites in the gonadal regulatory element had no effect on reporter expression (Fig. S1B). We conclude from these results that egl-5 is an indirect target of FKH-6 and TRA-1 regulation.
egl-5 has been shown to be autoregulated in some tissues (Li et al., 2009). To determine whether this also is the case in the gonad we crossed the ezEx202(egl-5::GFP) into egl-5(u202) mutants. egl-5 mutants males expressed ezEx202 at normal levels, suggesting it is not autoregulated in the gonad (89% n=47).
EGL-5 expression is sufficient to direct male gonadal fates in hermaphrodite gonads
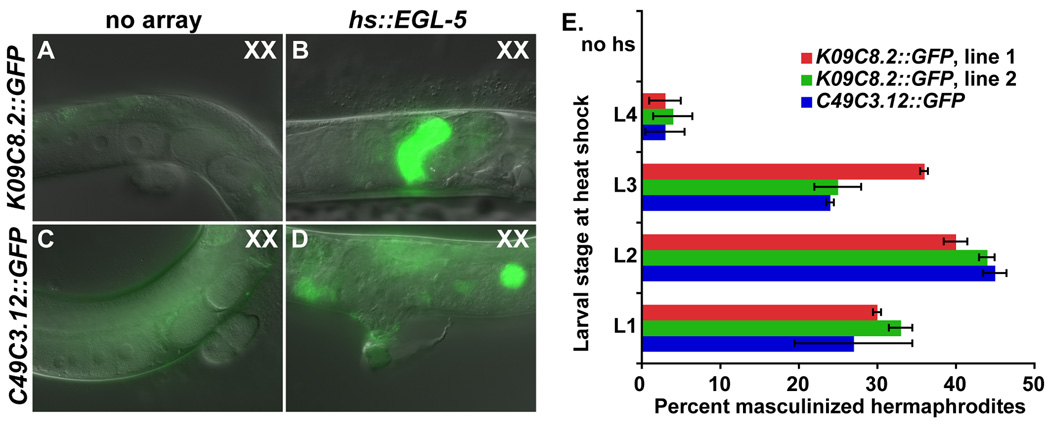
Analysis of egl-5 RNAi and loss-of-function mutations shows that egl-5 is necessary to execute many male cell fates and to prevent adoption of female fates in the gonads of XO animals. We next asked whether the mechanism by which EGL-5 acts in male fate determination is instructive or permissive by testing whether ectopic EGL-5 is sufficient to direct male differentiation in the XX gonad. We expressed EGL-5 from a heat shock promoter in XX animals carrying one of two male-specific gonadal GFP markers: the seminal vesicle marker K09C8.2::GFP or the vas deferens marker C49C3.12::GFP. In control XX animals lacking hs::EGL-5 neither reporter was expressed after heatshock (Fig. 3A, C). In contrast, XX animals ectopically expressing EGL-5 also expressed both male gonadal GFP markers, indicating that they were masculinized (Fig. 3B, D). We asked during which developmental stages EGL-5 expression can induce male gonadal cell fates by varying the stage in which heat shock was performed. The highest level of masculinization occurred when heat shock was performed during L2, but XX gonads could be masculinized even by expression starting in L3, indicating that sexual fates remain plastic in the gonad during this stage. We conclude from these results that egl-5 expression not only is necessary for determination of male gonadal cell fates in XO animals, but also is sufficient to promote male fates in the gonads of XX animals.
Fig. 3. EGL-5 expression is sufficient to specify male cell fates in XX gonads.
K09C8.2::GFP is expressed in seminal vesicle of wild type males and C49C3.12::GFP in the vas deferens. (A, C) Hermaphrodites lacking the hs::egl-5 array express neither of the male-specific reporters (GFP in C is gut autofluorescence). (B, D) Hermaphrodites with the hs::egl-5 array express both male-specific reporters after heat shock. (E) Animals were heat shocked during the indicated larval stage and examined as young adults for ectopic expression of reporter genes in the hermaphrodite gonad. Lines 1 and 2 are two independently isolated extrachromosomal arrays of hs::egl-5. Error bars indicate the SEM.
EGL-5 and a Wnt/β-catenin pathway cooperatively promote male gonadal development
We found previously that a Wnt/β-catenin pathway is required for male gonadal fate specification during larval development (Kalis et al., in press) and a Wnt pathway has been shown to regulate egl-5 expression in some cell types (Jiang and Sternberg, 1998; Li et al., 2009; Maloof et al., 1999). We therefore investigated the relationship between the Wnt pathway and egl-5. The Wnt/β-catenin pathway functioning in gonadal development includes the TCF/LEF transcription factor POP-1 and the β-catenin co-activator SYS-1 (Miskowski et al., 2001; Siegfried et al., 2004; Siegfried and Kimble, 2002). To find out whether this pathway is required for egl-5::GFP expression we assayed expression of the bxIS12 and ezEx202 reporters in males depleted for sys-1 and found that they retained reporter expression [ezEx202 sys-1(RNAi) 100% with expression, n=55, bxIs12 sys-1(RNAi) 99% with expression, n=70]. We also mutated a potential binding site for POP-1 in ezEx202, and this had no affect on expression (Fig. S2B). We conclude that the gonadal Wnt pathway is not required for egl-5 expression.
To ask whether egl-5 is required for Wnt/β-catenin activity we used a reporter with seven TCF/LEF binding sites fused to a pes-10 minimal promoter driving mCherry coding sequences (the “POPTOP” reporter (Green et al., 2008)). Expression of this reporter is dependent upon POP-1 activity, as pop-1 hypomorphic mutants have severely reduced numbers of POPTOP positive cells (Fig. 4A). We found previously that developing XO gonads have more POPTOP expressing cells than XX gonads, but that the intensity of expression is higher in XX than XO gonadal cells (Kalis et al, in press). Normal numbers of gonadal cells expressing POPTOP were present in egl-5 mutant male gonads (Fig. 4A). Unexpectedly, however, POPTOP expression in the mutant cells was stronger than in cells of control males and the intensity resembled that normally seen in XX gonads (Fig. 4B–D). Thus EGL-5 is not required for Wnt pathway activity in the male gonad but instead EGL-5 somehow modulates POP-1 activity in this tissue. As a further test of the interaction between EGL-5 and the Wnt pathway we compared lim-7::GFP expression in egl-5 males to that in egl-5; sys-1(RNAi) males. There was no significant enhancement of the egl-5 phenotype when sys-1 was depleted (Table 3). These results are consistent with EGL-5 and POP-1 acting jointly to specify male gonadal fates.
Fig. 4. Wnt pathway activity in egl-5 mutants.
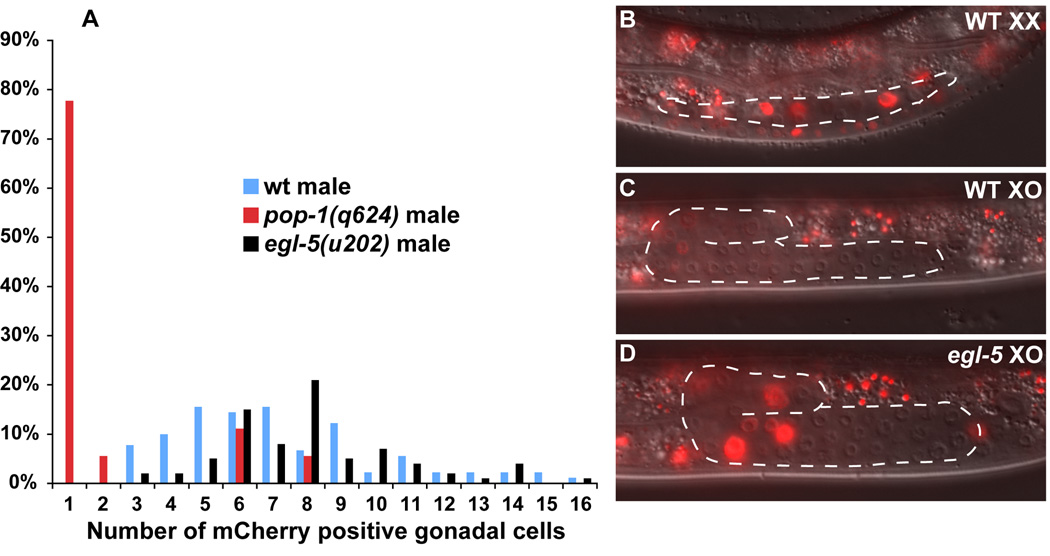
Wild type and pop-1 data are from Kalis et al., in press. All animals were scored in early to mid L3 for POPTOP expression. (A) Percent of animals with each number of gonadal cells expressing mCherry. The number of gonadal cells positive for POPTOP expression in egl-5 mutant males is the same as wild type males. (B) XX wild type hermaphrodite POPTOP expression. (C) XO wild type male POPTOP expression is less intense than that of hermaphrodites. (D) XO egl-5 POPTOP expression is present but at the higher level typical of hermaphrodites.
Table 3.
sys-1 fails to enhance egl-5 male gonadal feminization
| % GFP + males | n | SEM | |
|---|---|---|---|
| lim-7::GFP; control RNAi | 1 | 181 | 0.5 |
| lim-7::GFP; sys-1(RNAi) | 33 | 181 | 3.5 |
| lim-7::GFP; egl-5(u202); control RNAi | 71 | 213 | 3 |
| lim-7::GFP; egl-5(u202); sys-1(RNAi) | 69 | 208 | 4 |
The parallel function of EGL-5 and POP-1 in sexual fate specification suggests that they may regulate common transcriptional targets. We therefore tested whether the two proteins physically interact. First we performed a yeast two-hybrid experiment testing the interaction of full length EGL-5 and POP-1. We found that POP-1 fused to the Gal4 DNA binding domain weakly interacted with EGL-5 fused to the Gal4 transcriptional activation domain, activating three independent reporter genes (Fig. 5A, B, not shown). We confirmed this interaction by GST-pulldowns. GST-EGL-5 was able to pull down POP-1 as well as EGL-5. Likewise, GST-POP-1 was also able to pull down EGL-5 and POP-1. This suggests that POP-1 and EGL-5 can homodimerize and heterodimerize. POP-1 self-interaction has been observed previously in yeast two-hybrid assays (Li et al., 2004). As negative controls, we found that GST alone or GST-DMRT6 (an unrelated transcription factor) bound POP-1 and EGL-5 to a much lesser extent than GST-POP-1; also GST-EGL-5 and GST-POP-1 and GST-EGL-5 were unable to pull down the Drosophila Intersex protein (Fig. 5C, D). Collectively these data indicate that EGL-5 and POP-1 can physically interact and thus may act together in the male gonad to promote male fates.
Fig. 5. Physical interaction of EGL-5 and POP-1.
(A, B) Yeast two-hybrid interaction of EGL-5 and POP-1. (A) BD-POP-1 and AD-EGL-5 transformed yeast cells can grow on -Leu/-Trp/-His media like the positive control BD-53 and AD-T and unlike either negative control, BD-POP-1 and AD-T or BD-53 and AD-EGL-5. (B) Quantitative Beta-Galactosidase assay demonstrating BD-POP-1 and AD-EGL-5 have 3-fold higher Beta-Galactosidase activity than the negative controls. Error bars indicate standard deviation. (C, D) GST-pulldown of EGL-5 by POP-1 and POP-1 by EGL-5. (C) GST-EGL-5 can pull down POP-1 protein but not Intersex. GST-POP-1 can also pull down POP-1. GST-DMRT6 and GST alone pull down POP-1 is much weaker than either GST-EGL-5 or GST-POP-1. (D) GST-POP-1 can also pull down EGL-5 and not Intersex more that GST-DMRT6 or GST alone, however, not as strongly as GST-EGL-5.
DISCUSSION
We have examined the role of the Hox transcription factor EGL-5 in specifying male gonadal cell fates and find that the gonads of egl-5 mutant males are extensively feminized, based on analysis of four hermaphrodite-specific gonadal reporters. This work confirms and extends the previous observation that a minority of egl-5 mutant males contain cells with uterine morphology. We find that ectopic EGL-5 expression is sufficient to activate male-specific reporters in the hermaphrodite gonad, suggesting that it plays an instructive rather than a permissive role in male fate specification. egl-5 is not expressed during the initial phase of gonadal development and it is not required when the global sex determination cascade is active; instead it promotes male sexual fate specification after the initial SGP divisions. Among genes whose loss is known to cause gonadal feminization (Chang et al., 2004; Kalis et al., in press; Tilmann and Kimble, 2005), egl-5 is unusual in being expressed male-specifically. Thus EGL-5 is a prime candidate to impart male specific function on other gonadal regulators. Indeed, genetic epistasis, reporter gene analysis, and physical interaction indicated that EGL-5 acts with POP-1 to promote male fates.
Male-specific gonadal expression of egl-5
The egl-5 gene has a large and complex regulatory region, which has been analyzed in detail and contains a number of distinct regulatory modules specifying tissue-, stage- and sex-specific expression in posterior muscle, neuronal, and hypodermal cells (Li et al., 2009; Teng et al., 2004; Zhang and Emmons, 2009). However, neither the regulatory sequences controlling gonadal expression of egl-5 nor the trans-acting factors involved have been investigated previously. We defined a short region that is necessary and sufficient for male-specific gonadal expression of reporter genes, but the trans-acting factors that directly regulate this element remain elusive. Sex-specific early gonadal development depends on the global sex determination pathway acting through tra-1 and also requires fkh-6, but both genes appear to regulate egl-5 transcription indirectly. We found that repression of egl-5 reporters in the XX gonad requires tra-1 and that the gonadal regulatory element contains several potential TRA-1 binding sites. However, mutagenesis of potential TRA-1 binding sites in the gonadal regulatory element did not support a direct function for TRA-1 in regulation of egl-5 transcription. Similarly, mutating potential FKH-6 binding sites did not eliminate expression of egl-5 reporters. We conclude that tra-1 and fkh-6 regulate egl-5 gonadal expression indirectly, via intermediary regulatory factors yet to be identified. Our determination of the preferred FKH-6 DNA binding site should help to find these regulatory factors.
egl-5 regulation of male gonadal cell fates
Loss of egl-5 caused male gonadal feminization similar to that caused by loss of Wnt/β-catenin pathway members, including sys-1 and pop-1. Additionally, loss of either egl-5 or Wnt/β-catenin signaling disrupts cell division asymmetry (Chisholm, 1991; Kalis et al., in press). These similar effects on gonadal development prompted us to investigate the relationship between egl-5 and the Wnt/β-catenin pathway. We found that the Wnt/β-catenin pathway is not required for egl-5 reporter expression and that egl-5 is not required for activation of a POP-1 dependent POPTOP reporter. However, in egl-5 mutant males POPTOP expression was much higher than in wild type, resembling the expression in wild type hermaphrodite gonads and suggesting that EGL-5 somehow modifies the activity of POP-1. The physical interaction we detected between EGL-5 and POP-1 in yeast and in vitro suggests that the effect of EGL-5 on 21 POP-1 activity involves direct contact between the two proteins. A model consistent with these results (Fig. 6) is that EGL-5 and POP-1 cooperatively regulate a group of male-specific transcriptional targets with recognition elements for both proteins; in the absence of EGL-5 more POP-1 is available to bind the POPTOP reporter, which lacks EGL-5 binding sites. The effect on POPTOP reporter expression potentially reflects small changes in the amount of available POP-1, as POP-1 transcriptional activity has been shown to be extremely sensitive to dosage and to the ratio of POP-1 to SYS-1 (Phillips and Kimble 2009). Gonads of egl-5; sys-1 double mutant males were not more severely feminized than those of egl-5 single mutants, a result also consistent with the two genes regulating common targets. Cooperative gene regulation with egl-5 may help explain how the Wnt/β-catenin pathway, which is required for gonadal development in both sexes, performs additional male-specific gonadal functions. It will be important to identify the gonadal targets of POP-1 and EGL-5.
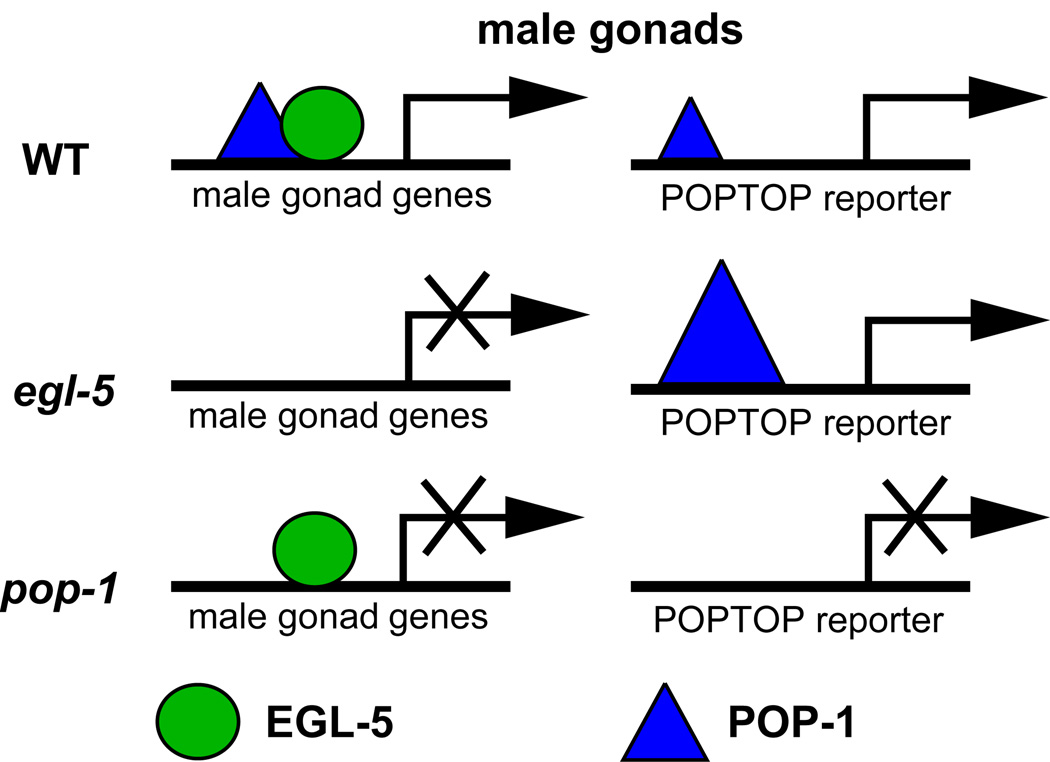
Fig. 6. Model for functional interaction of EGL-5 and POP-1 in the gonad.
Based on genetic and physical interaction, we suggest that EGL-5 helps recruit POP-1 to male gonadal target genes containing recognition sites for both proteins, imposing male-specific function on the gonadal Wnt/β-catenin pathway. In the absence of either EGL-5 or Wnt/β-catenin pathway members there is a failure to promote male specific cell fates because these shared targets are not activated. In this model the recruitment of POP-1 to male gonadal target genes by EGL-5 limits the amount of POP-1 available to activate the POPTOP reporter, accounting for the lower expression of this reporter in the male gonad. In egl-5 mutant males POP-1 is not recruited to male-specific target genes, allowing more efficient activation of the POPTOP reporter, which lacks EGL-5 recognition sites.
ABD-B homologs interact functionally with Wnt pathways in varied contexts, including the regulation of other aspects of C. elegans development, and these provide examples of how EGL-5 and POP-1 may interact to regulate target genes. In C. elegans T cell development POP-1 cooperatively activates expression of the Meis protein PSA-3 with the ABD-B homolog NOB-1, and POP-1 recruits the Hox cofactor CEH-20 to the nucleus to stimulate NOB-1 DNA binding activity (Arata et al., 2006). In Drosophila spiracle development ABD-B is involved in a complex tier of transcriptional regulation that includes co-regulation of target gene transcription together with Wnt and Hh signaling pathways, as well as regulation of the signaling pathways themselves (Merabet et al., 2005). In both cases Adb-B genes and Wnt signaling cooperate to induce the expression of genes required to establish cell or tissue identities in a manner analogous to the regulation we have observed in the male gonad. Combinatorial regulation by HOX proteins and Wnt pathways is likely to be a common means for multicellular organisms to deploy a limited number of cell signaling pathways in the specification and development of complex cell types and complex patterns (Bondos, 2006).
ABD-B in sex-specific gonadal development: a theme with variations
Comparison of the role of EGL-5 in C. elegans gonadal development with that of ABD-B in Drosophila reveals that the two function analogously but not homologously. Both are required for proper execution of male but not female gonadal cell fates, but the mechanisms involved are quite different. ABD-B is required to specify “male-specific” somatic gonad precursors (msSGPs), which form in both sexes and survive only in males due to the action of the global sex determination pathway, acting through DSX (DeFalco et al., 2004). Thus ADB-B and the global sex determination pathway act jointly to specify msSGPs and retain them in males. By contrast, in C. elegans EGL-5 itself is expressed sex-specifically in the gonad due to regulation by the global sex determination pathway and helps drive gonadal cells toward male fates and away from female fates during gonadal differentiation. A similarity is that both ABD-B and EGL-5 can induce male gonadal cell fates when ectopically expressed [(DeFalco et al., 2004); this work]. Our results help illustrate how ABD-B proteins play a conserved role in male gonadal differentiation but intersect differently with the global sex determination pathway and likely act on distinct differentiation programs in different species.
Supplementary Material
Left column: bxIs12 (egl-5::GFP with full regulatory region) expression (Teng et al., 2004). Right column: exEx202 (pAKK33-123bp regulatory region required for egl-5 gonadal expression). All strains contain the germ cell marker nmy-2::PGL-1::mRFP1 (red punctate expression distinguishes germ cells from somatic cells (Wolke et al., 2007)). Gonadal regions containing VD and SV cells and descendents are outlined in white (green fluorescence outside white outlines is intestinal autofluorescence). White arrows indicate the LCs, filled arrowheads indicate the VD precursors, open arrowheads indicate the SV precursors, and small white arrows in rows E and F indicate the valve cells. (A, B) In early L2 GFP expression is present in the three vas deferens (VD) precursors (strong) and four seminal vesicle (SV) precursors (weaker) (all seven cells not in focus). (C, D) In late L2 the gonad has elongated anteriorly but the expression pattern remains in the same cells. (E, F) In early L3, as the gonad reflexes to the posterior, the VD and SV cells start dividing. GFP expression remains strong in VD daughters and proximal SV daughters and is much weaker in distal SV daughters. (G–J) GFP remains similar as the gonad continues migration through L3. (K) In L4, as the VD and SV daughters terminally differentiate, GFP expression is retained only in the four valve cells. (L) Expression is occasionally present in late L4 and adults in the valve cells. Scale bars = 50µm.
Extrachomosomal arrays designated “ezEx” contain egl-5 sequences fused to the Δpes-10 basal promoter (Fire Vector Kit) driving the expression of GFP. Plusses and minuses represent the presence or absence of male gonadal GFP fluorescence. (A) Heavy horizontal lines indicate egl-5 chromosomal regions contained within reporters. (B) Potential binding sites are indicated by colored horizontal bars above abridged pAKK33(ezEx202) sequence: FKH-6(blue), TRA-1(black), POP-1(red). Mutations in each reporter construct are indicated by nucleotides that differ from the abridged version of pAKK33.
The Forkhead transcription factor, FKH-6, consensus binding site was determined by in vitro selection from random oligonucleotides and differs from a second Forkhead transcription factor, PHA-4, binding site at six positions. In this gel mobility shift assay we show that a PHA-4 site (MYO-2) does not compete for FKH-6 binding, indicating that FKH-6 binds a specific sequence element that is distinct from the PHA-4 recognition sequence. Changing a FKH-6 site to conform to the PHA-4 consensus reduces binding of an unlabelled competitor, with the mut1 and mut4 changes having the greatest effect. All reactions were run on the same gel but gel image has lanes excised between lanes 3 and 4.
ACKNOWLEDGMENTS
We thank Drs. Scott Emmons, Ian Hope, Judith Kimble, Paul Sternberg, James Priess, and David Greenstein for strains and reagents, Kathleen Larson and Matthew Berkseth for experimental assistance, and Drs. Jennifer Ross Wolff and Mary Kroetz for critical reading of the manuscript. Many C. elegans strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resourses (NCRR). This work was supported by NIH grants GM53099 (DZ) and T32HD007480 (AKK).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Arata Y, Kouike H, Zhang Y, Herman MA, Okano H, Sawa H. Wnt signaling and a Hox protein cooperatively regulate psa-3/Meis to determine daughter cell fate after asymmetric cell division in C. elegans. Dev Cell. 2006;11:105–115. doi: 10.1016/j.devcel.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Asahina M, Valenta T, Silhankova M, Korinek V, Jindra M. Crosstalk between a nuclear receptor and beta-catenin signaling decides cell fates in the C. elegans somatic gonad. Dev Cell. 2006;11:203–211. doi: 10.1016/j.devcel.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Barmina O, Kopp A. Sex-specific expression of a HOX gene associated with rapid morphological evolution. Dev Biol. 2007;311:277–286. doi: 10.1016/j.ydbio.2007.07.030. [DOI] [PubMed] [Google Scholar]
- Bondos S. Variations on a theme: Hox and Wnt combinatorial regulation during animal development. Sci STKE. 2006;2006:pe38. doi: 10.1126/stke.3552006pe38. [DOI] [PubMed] [Google Scholar]
- Chang W, Tilmann C, Thoemke K, Markussen FH, Mathies LD, Kimble J, Zarkower D. A forkhead protein controls sexual identity of the C. elegans male somatic gonad. Development. 2004;131:1425–1436. doi: 10.1242/dev.01012. [DOI] [PubMed] [Google Scholar]
- Chisholm A. Control of cell fate in the tail region of C. elegans by the gene egl-5. Development. 1991;111:921–932. doi: 10.1242/dev.111.4.921. [DOI] [PubMed] [Google Scholar]
- Conradt B, Horvitz HR. The TRA-1A sex determination protein of C. elegans regulates sexually dimorphic cell deaths by repressing the egl-1 cell death activator gene. Cell. 1999;98:317–327. doi: 10.1016/s0092-8674(00)81961-3. [DOI] [PubMed] [Google Scholar]
- Dalton S, Treisman R. Characterization of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell. 1992;68:597–612. doi: 10.1016/0092-8674(92)90194-h. [DOI] [PubMed] [Google Scholar]
- DeFalco T, Le Bras S, Van Doren M. Abdominal-B is essential for proper sexually dimorphic development of the Drosophila gonad. Mech Dev. 2004;121:1323–1333. doi: 10.1016/j.mod.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Ferreira HB, Zhang Y, Zhao C, Emmons SW. Patterning of Caenorhabditis elegans posterior structures by the Abdominal-B homolog, egl-5. Dev Biol. 1999;207:215–228. doi: 10.1006/dbio.1998.9124. [DOI] [PubMed] [Google Scholar]
- Green JL, Inoue T, Sternberg PW. Opposing Wnt pathways orient cell polarity during organogenesis. Cell. 2008;134:646–656. doi: 10.1016/j.cell.2008.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DH, Winfrey VP, Blaeuer G, Hoffman LH, Furuta T, Rose KL, Hobert O, Greenstein D. Ultrastructural features of the adult hermaphrodite gonad of Caenorhabditis elegans: relations between the germ line and soma. Dev Biol. 1999;212:101–123. doi: 10.1006/dbio.1999.9356. [DOI] [PubMed] [Google Scholar]
- Hodgkin J. A genetic analysis of the sex-determining gene, tra-1, in the nematode Caenorhabditis elegans. Genes Dev. 1987;1:731–745. doi: 10.1101/gad.1.7.731. [DOI] [PubMed] [Google Scholar]
- Hope IA. 'Promoter trapping' in Caenorhabditis elegans. Development. 1991;113:399–408. doi: 10.1242/dev.113.2.399. [DOI] [PubMed] [Google Scholar]
- Huynh KD, Bardwell VJ. The BCL-6 POZ domain and other POZ domains interact with the co-repressors N-CoR and SMRT. Oncogene. 1998;17:2473–2484. doi: 10.1038/sj.onc.1202197. [DOI] [PubMed] [Google Scholar]
- Jiang LI, Sternberg PW. Interactions of EGF, Wnt and HOM-C genes specify the P12 neuroectoblast fate in C. elegans. Development. 1998;125:2337–2347. doi: 10.1242/dev.125.12.2337. [DOI] [PubMed] [Google Scholar]
- Kalis AK, Kroetz MB, Larson KM, Zarkower D. Functional Genomic Identification of Genes Required for Male Gonadal Differentiation in Caenorhabditis elegans. Genetics. doi: 10.1534/genetics.110.116038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J, Hirsh D. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev Biol. 1979;70:396–417. doi: 10.1016/0012-1606(79)90035-6. [DOI] [PubMed] [Google Scholar]
- Klass M, Wolf N, Hirsh D. Development of the male reproductive system and sexual transformation in the nematode Caenorhabditis elegans. Dev Biol. 1976;52:1–18. doi: 10.1016/0012-1606(76)90002-6. [DOI] [PubMed] [Google Scholar]
- Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, Goldberg DS, Li N, Martinez M, Rual JF, Lamesch P, Xu L, Tewari M, Wong SL, Zhang LV, Berriz GF, Jacotot L, Vaglio P, Reboul J, Hirozane-Kishikawa T, Li Q, Gabel HW, Elewa A, Baumgartner B, Rose DJ, Yu H, Bosak S, Sequerra R, Fraser A, Mango SE, Saxton WM, Strome S, Van Den Heuvel S, Piano F, Vandenhaute J, Sardet C, Gerstein M, Doucette-Stamm L, Gunsalus KC, Harper JW, Cusick ME, Roth FP, Hill DE, Vidal M. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Kulkarni RP, Hill RJ, Chamberlin HM. HOM-C genes, Wnt signaling and axial patterning in the C. elegans posterior ventral epidermis. Dev Biol. 2009;332:156–165. doi: 10.1016/j.ydbio.2009.05.567. [DOI] [PubMed] [Google Scholar]
- Maeda RK, Karch F. The bithorax complex of Drosophila an exceptional Hox cluster. Curr Top Dev Biol. 2009;88:1–33. doi: 10.1016/S0070-2153(09)88001-0. [DOI] [PubMed] [Google Scholar]
- Maloof JN, Whangbo J, Harris JM, Jongeward GD, Kenyon C. A Wnt signaling pathway controls hox gene expression and neuroblast migration in C. elegans. Development. 1999;126:37–49. doi: 10.1242/dev.126.1.37. [DOI] [PubMed] [Google Scholar]
- Mathies LD, Schvarzstein M, Morphy KM, Blelloch R, Spence AM, Kimble J. TRA-1/GLI controls development of somatic gonadal precursors in C. elegans. Development. 2004;131:4333–4343. doi: 10.1242/dev.01288. [DOI] [PubMed] [Google Scholar]
- Merabet S, Hombria JC, Hu N, Pradel J, Graba Y. Hox-controlled reorganisation of intrasegmental patterning cues underlies Drosophila posterior spiracle organogenesis. Development. 2005;132:3093–3102. doi: 10.1242/dev.01889. [DOI] [PubMed] [Google Scholar]
- Miskowski J, Li Y, Kimble J. The sys-1 gene and sexual dimorphism during gonadogenesis in Caenorhabditis elegans. Dev Biol. 2001;230:61–73. doi: 10.1006/dbio.2000.9998. [DOI] [PubMed] [Google Scholar]
- Murphy MW, Zarkower D, Bardwell VJ. Vertebrate DM domain proteins bind similar DNA sequences and can heterodimerize on DNA. BMC Mol Biol. 2007;8:58. doi: 10.1186/1471-2199-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson GA, Lew KK, Ward S. Intersex, a temperature-sensitive mutant of the nematode Caenorhabditis elegans. Dev Biol. 1978;66:386–409. doi: 10.1016/0012-1606(78)90247-6. [DOI] [PubMed] [Google Scholar]
- Pearson JC, Lemons D, McGinnis W. Modulating Hox gene functions during animal body patterning. Nat Rev Genet. 2005;6:893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- Pettitt J, Wood WB, Plasterk RH. cdh-3, a gene encoding a member of the cadherin superfamily, functions in epithelial cell morphogenesis in Caenorhabditis elegans. Development. 1996;122:4149–4157. doi: 10.1242/dev.122.12.4149. [DOI] [PubMed] [Google Scholar]
- Ross JM, Zarkower D. Polycomb group regulation of Hox gene expression in C. elegans. Dev Cell. 2003;4:891–901. doi: 10.1016/s1534-5807(03)00135-7. [DOI] [PubMed] [Google Scholar]
- Sanchez L, Gorfinkiel N, Guerrero I. Sex determination genes control the development of the Drosophila genital disc, modulating the response to Hedgehog, Wingless and Decapentaplegic signals. Development. 2001;128:1033–1043. doi: 10.1242/dev.128.7.1033. [DOI] [PubMed] [Google Scholar]
- Siegfried KR, Kidd AR, 3rd, Chesney MA, Kimble J. The sys-1 and sys-3 genes cooperate with Wnt signaling to establish the proximal-distal axis of the Caenorhabditis elegans gonad. Genetics. 2004;166:171–186. doi: 10.1534/genetics.166.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried KR, Kimble J. POP-1 controls axis formation during early gonadogenesis in C. elegans. Development. 2002;129:443–453. doi: 10.1242/dev.129.2.443. [DOI] [PubMed] [Google Scholar]
- Stiernagle T. Maintenance of C. elegans. WormBook. 2006:1–11. doi: 10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Y, Girard L, Ferreira HB, Sternberg PW, Emmons SW. Dissection of cis-regulatory elements in the C. elegans Hox gene egl-5 promoter. Dev Biol. 2004;276:476–492. doi: 10.1016/j.ydbio.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Thoemke K, Yi W, Ross JM, Kim S, Reinke V, Zarkower D. Genome-wide analysis of sex-enriched gene expression during C. elegans larval development. Dev Biol. 2005;284:500–508. doi: 10.1016/j.ydbio.2005.05.017. [DOI] [PubMed] [Google Scholar]
- Tilmann C, Kimble J. Cyclin D regulation of a sexually dimorphic asymmetric cell division. Dev Cell. 2005;9:489–499. doi: 10.1016/j.devcel.2005.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BB, Muller-Immergluck MM, Austin J, Robinson NT, Chisholm A, Kenyon C. A homeotic gene cluster patterns the anteroposterior body axis of C. elegans. Cell. 1993;74:29–42. doi: 10.1016/0092-8674(93)90292-x. [DOI] [PubMed] [Google Scholar]
- Williams TM, Carroll SB. Genetic and molecular insights into the development and evolution of sexual dimorphism. Nat Rev Genet. 2009;10:797–804. doi: 10.1038/nrg2687. [DOI] [PubMed] [Google Scholar]
- Williams TM, Selegue JE, Werner T, Gompel N, Kopp A, Carroll SB. The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell. 2008;134:610–623. doi: 10.1016/j.cell.2008.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolke U, Jezuit EA, Priess JR. Actin-dependent cytoplasmic streaming in C. elegans oogenesis. Development. 2007;134:2227–2236. doi: 10.1242/dev.004952. [DOI] [PubMed] [Google Scholar]
- Yi W, Ross JM, Zarkower D. Mab-3 is a direct tra-1 target gene regulating diverse aspects of C. elegans male sexual development and behavior. Development. 2000;127:4469–4480. doi: 10.1242/dev.127.20.4469. [DOI] [PubMed] [Google Scholar]
- Zarkower D, Hodgkin J. Zinc fingers in sex determination: only one of the two C. elegans Tra-1 proteins binds DNA in vitro. Nucleic Acids Res. 1993;21:3691–3698. doi: 10.1093/nar/21.16.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Emmons SW. Regulation of the Caenorhabditis elegans posterior Hox gene egl-5 by microRNA and the polycomb-like gene sop-2. Dev Dyn. 2009;238:595–603. doi: 10.1002/dvdy.21876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Left column: bxIs12 (egl-5::GFP with full regulatory region) expression (Teng et al., 2004). Right column: exEx202 (pAKK33-123bp regulatory region required for egl-5 gonadal expression). All strains contain the germ cell marker nmy-2::PGL-1::mRFP1 (red punctate expression distinguishes germ cells from somatic cells (Wolke et al., 2007)). Gonadal regions containing VD and SV cells and descendents are outlined in white (green fluorescence outside white outlines is intestinal autofluorescence). White arrows indicate the LCs, filled arrowheads indicate the VD precursors, open arrowheads indicate the SV precursors, and small white arrows in rows E and F indicate the valve cells. (A, B) In early L2 GFP expression is present in the three vas deferens (VD) precursors (strong) and four seminal vesicle (SV) precursors (weaker) (all seven cells not in focus). (C, D) In late L2 the gonad has elongated anteriorly but the expression pattern remains in the same cells. (E, F) In early L3, as the gonad reflexes to the posterior, the VD and SV cells start dividing. GFP expression remains strong in VD daughters and proximal SV daughters and is much weaker in distal SV daughters. (G–J) GFP remains similar as the gonad continues migration through L3. (K) In L4, as the VD and SV daughters terminally differentiate, GFP expression is retained only in the four valve cells. (L) Expression is occasionally present in late L4 and adults in the valve cells. Scale bars = 50µm.
Extrachomosomal arrays designated “ezEx” contain egl-5 sequences fused to the Δpes-10 basal promoter (Fire Vector Kit) driving the expression of GFP. Plusses and minuses represent the presence or absence of male gonadal GFP fluorescence. (A) Heavy horizontal lines indicate egl-5 chromosomal regions contained within reporters. (B) Potential binding sites are indicated by colored horizontal bars above abridged pAKK33(ezEx202) sequence: FKH-6(blue), TRA-1(black), POP-1(red). Mutations in each reporter construct are indicated by nucleotides that differ from the abridged version of pAKK33.
The Forkhead transcription factor, FKH-6, consensus binding site was determined by in vitro selection from random oligonucleotides and differs from a second Forkhead transcription factor, PHA-4, binding site at six positions. In this gel mobility shift assay we show that a PHA-4 site (MYO-2) does not compete for FKH-6 binding, indicating that FKH-6 binds a specific sequence element that is distinct from the PHA-4 recognition sequence. Changing a FKH-6 site to conform to the PHA-4 consensus reduces binding of an unlabelled competitor, with the mut1 and mut4 changes having the greatest effect. All reactions were run on the same gel but gel image has lanes excised between lanes 3 and 4.