SUMMARY
Reprogramming of human somatic cells uses readily accessible tissue, such as skin or blood, to generate embryonic-like induced pluripotent stem cells (iPSCs). This procedure has been applied to somatic cells from patients who are classified into a disease group, thus creating ‘disease-specific’ iPSCs. Here we examine the challenges and assumptions in creating a disease model from a single cell of the patient. Both the kinetics of disease onset and progression as well as the spatial localization of disease in the patient’s body are challenges to model construction. New tools in genetic modification, reprogramming, biomaterials, and animal models can be used to address these challenges.
Unleashing the powerful tools of modern cell biology to dissect mechanisms of human disease requires large quantities of cells and tissues from specific sets of patients. Human pluripotent stem cells have the potential to generate all tissues in the body (Lowry et al., 2008; Park et al., 2008a; Reubinoff et al., 2000; Takahashi et al., 2007; Thomson et al., 1998; Yu et al., 2007) and therefore provide researchers in the lab critical access to patient-derived biomaterial, which constitute the principal input for such studies of disease. However, there are many technical challenges in generating and manipulating human pluripotent cells before they can be thought to be faithful models of specific diseases. This review will focus on the use of a rising class of pluripotent cells, “reprogrammed” human induced pluripotent stem cells (hiPSCs), to model human disease pathogenesis.
Modeling human diseases “in a dish” is firmly rooted in human embryonic stem cell (hESC) biology. In 1998, Thomson and colleagues derived hESC lines by culturing human blastocysts in a cocktail of growth factors and supporting mouse feeder cells (Thomson et al., 1998). These hESC lines were immediately heralded as foundational for cell replacement therapy and for modeling human diseases (Gearhart, 1998). Both ‘forward’ and ‘reverse’ genetics approaches have been utilized with hESCs to elucidate mechanisms of disease. In the reverse approach, it is possible to test the effects of pre-defined gene mutations in cells through use of preimplantation genetic diagnosis (PGD). By performing PGD on embryos, researchers were able to prospectively identify embryos with particular genetic disorders, and then derive ‘disease-specific’ hESCs for cystic fibrosis (Mateizel et al., 2006; Pickering et al., 2005), Huntington’s disease (Mateizel et al., 2006), Fragile X syndrome (Eiges et al., 2007), and Turner’s syndrome (Urbach and Benvenisty, 2009). Studies using such reverse genetics approaches are limited, because PGD embryos are only available for a very restricted number of human diseases. The forward genetics approach starts with a mutagenesis step, typically using known gene loci correlated with specific disease, followed by the identification of a disease phenotype in hESCs or their derivatives. In the case of Lesch-Nyhan disease, the hprt1 gene was mutated in hESCs through homologous recombination. The resulting hESCs showed an absence in hprt1 activity and produced more uric acid than unmodified “wild-type” cells (Urbach et al., 2004). These ‘Lesch-Nyhan specific’ hESC lines can be used to further define the molecular mechanisms of the disease and to screen for drugs that rescue hprt1 activity. Generating mutant hESC lines as disease models has been pursued in many laboratories, however these studies have faced challenges because of the inefficient methods to genetically modify hESCs (Giudice and Trounson, 2008). Of course, in the cases for which known disease-associated genetic loci are unknown, and those for which no obvious disease phenotype could be screened in hESCs, the forward reverse approach is also not tenable for disease model generation.
Concurrent to the development of ‘disease-specific’ hESC lines, a new technique of deriving human pluripotent stem cells has rapidly evolved since 2007. hiPSCs were first generated through viral transduction of four transcription factors into previously banked human fibroblasts (Lowry et al., 2008; Park et al., 2008a; Takahashi et al., 2007; Yu et al., 2007). These techniques have now been applied to blood or skin samples harvested from patients diagnosed with specific diseases (Dimos et al., 2008; Ebert et al., 2009; Hotta et al., 2009; Maehr et al., 2009; Park et al., 2008b; Ye et al., 2009; Soldner et al, 2009), however, thus far only a handful of reports have observed a disease phenotype in vitro (Ebert et al., 2009; Lee et al., 2009; Raya et al., 2009; Ye et al., 2009).
Recent work with rodents has tested the developmental potential of iPSCs and their potential for the treatment of diseases. Differentiation of mouse iPSCs can be directed in vitro into cardiovascular (Kuzmenkin et al., 2009; Narazaki et al., 2008; Schenke-Layland et al., 2008), hematopoietic (Hanna et al., 2007; Schenke-Layland et al., 2008; Xu et al., 2009), neural (Wernig et al., 2008), and hepatic progenitor cells (Cantz et al., 2008), and recently, mouse iPSCs passed the most stringent test of pluripotency by generating full-term adult mice in tetraploid complementation assays (Boland et al., 2009; Kang et al., 2009; Zhao et al., 2009). Further, mouse iPSCs obtained from adult fibroblasts can be used to restore physiological function of diseased tissues in vivo, as demonstrated by using iPSC-derived hematopoietic cells in a humanized mouse model of sickle cell anemia (Hanna et al., 2007). Also, endothelial/endothelial progenitor cells derived from mouse iPSCs injected directly into the liver of irradiated hemophilia A mice extended their survival for more than 3 months and rescued depleted Plasma FVIII levels (Xu et al., 2009). Finally, functional dopamine neurons could be generated from reprogrammed mouse fibroblasts, and transplantation of these neurons, like mESC-derived neurons, could restore dopamine function when grafted into Parkinsonian rats (Wernig et al., 2008). These studies establish that iPSCs have vast potential to generate a variety of functional cell types and can be used to modify the course of disease in rodents.
Although animal models continue to produce key insights into disease mechanisms, these systems have limitations that could be potentially overcome by human cellular models of disease. Many transgenic murine models of congenital and acquired diseases do not faithfully mirror the respective human pathophysiology. For example, mice carrying the same genetic deficiencies as Fanconi anemia patients do not develop the spontaneous bone marrow failure that is the hallmark of the human disease (Chen et al., 1996). Differences in tissue composition, anatomy, and physiology between animals and humans all may underlie these observations. For instance, the heart rates of humans and mice differ by ten fold (Davies and Morris, 1993; Sothern and Gruber, 1994), and arrhythmias can have very different consequences in these two species. Another challenge in using murine disease models can arise from the differences in colinearity of the human and mouse genomes and the lack of conservation of gene order. Mice engineered to be trisomic for those sections of the mouse genome that are orthologous to the human Down syndrome critical region failed to recapitulate human cranial abnormalities (Nelson and Gibbs, 2004) or neurodegeneration (Reeves et al., 1995) commonly associated with Down syndrome. Further, only a relatively small subset of age-regulated gene expression changes are conserved from mouse to man (Loerch et al., 2008). Lastly, the inbred genetic background of mice can also influence the phenotype resulting from the disease-associated mutations. Humans are of mixed genetic background and this complexity results in phenotypical variations of genetically defined diseases. To overcome these drawbacks of using animals to model human diseases, new hiPSCs have been generated and explored for disease modeling in a few cases, as described below.
Current strategies of establishing hiPS cellular models of disease
The establishment of disease models through patient-specific reprogramming involves two steps: first, derivation of hiPSCs from somatic cells of a patient and second, differentiating the hiPSCs into cell types affected by the patient’s disease. Below we illustrate common principles of this approach with diseases that have strong genetic etiologies.
Deriving hiPSCs
Typically, cells are harvested from a patient through a biopsy or blood sample. Harvested samples include adipose adult stem cells from lipoaspiration (Sun et al., 2009), the CD34+ fraction of blood samples (Ye et al., 2009), both fibroblasts and keratinocytes from skin samples (Aasen et al., 2008; Carey et al., 2009), and keratinocytes from plugged hair (Aasen et al., 2008). In addition, frozen banked tissues or cell lines, such as fetal brain cortices (Hester et al., 2009) or cord blood (Giorgetti et al., 2009; Haase et al., 2009), can be reprogrammed. Choosing which patient donor tissue(s) to reprogram depends on the type of disease and on the expected pattern of disease progression. For example, in myeloid proliferative disorders, a heterozygous JAK2-V617F genotype is observed in 100% of colony-forming erythroid progenitors in their CD34+ cells, and these cells were chosen as the source of cells for reprogramming (Ye et al., 2009). Any contaminating skin or other blood cells present in the patient’s sample giving rise to an iPSC line would not contain the JAK2-V617F genotype that correlated with the disease. Hence, the iPSC lines generated in this study were genotyped to ensure that they carried the mutation. In contrast, the genetic mutation correlated with diseases such as spinal muscular atrophy type I (SMA) is present in all cells though the only cell types affected are motor neurons (Ebert et al., 2009). Therefore, for this class of diseases, readily accessible skin biopsies can be used as donor cells for hiPSC line derivation.
Many types of cells are generated during the stochastic reprogramming process (Chan et al., 2009; Hanna et al., 2009), including transformed cells or “intermediates” (Chan et al., 2009; Mikkelsen et al., 2008; Sridharan et al., 2009), and any newly established cell lines must be extensively tested for pluripotency characteristics. For example, in somatic cells, endogenous alleles encoding transcription factors specific to ESCs, oct4 and nanog, are silenced through methylation, and demethylation of these alleles upon reprogramming is a key hallmark of fully reprogrammed iPSCs (Mikkelsen et al., 2008; Sridharan et al., 2009; Takahashi et al., 2007; Yu et al., 2007). Differentiation into all three germ layers using in vitro differentiation is also necessary (Ellis et al., 2009) and using teratoma assays is highly preferred (Daley et al., 2009). If mutant genes involved in the pathogenesis of the disease are expressed in the hiPSCs, transcriptional abnormalities in the somatic cells affected in the patient, such as aberrant splicing or reduced transcript levels, can be investigated in the established hiPSC lines (Ebert et al., 2009; Lee et al., 2009).
Differentiating hiPSCs to functional cells
Most disease phenotypes are only observed in lineage-committed or differentiated cells, and not in the ESCs or iPSCs. Thus, pertinent information on the pathogenesis of a disease may only be obtained from hiPSCs that have been differentiated in vitro to disease relevant differentiated cell types. Differentiation of hiPSCs into several cell types has already been achieved: neural progenitors (Chambers et al., 2009), motor neurons (Dimos et al., 2008; Ebert et al., 2009), dopaminergic neurons (Soldner et al., 2009), retinal cells (Osakada et al., 2009), hepatocytes (Sullivan et al., 2009), blood cells (Choi et al., 2009; Ye et al., 2009), adipocytes (Taura et al., 2009), endothelial cells (Choi et al., 2009; Sullivan et al., 2009), and fibroblasts (Hockemeyer et al., 2008; Maherali et al., 2008). During these differentiation protocols that can span multiple weeks, many cell types are generated, and transcription factor or surface marker expression is the typical approach used to assay the developmental stage of differentiation. The function of these differentiated cells, thus far, been assayed in only a few cases: reduced cell migration of neural crest hiPSC-derivatives (Lee et al., 2009) and erythropoiesis via colony formation assays of hematopoietic hiPSC-derivatives (Raya et al., 2009; Ye et al., 2009). Functional cellular and biochemical phenotypes, such as transcript splicing, were observed upon differentiation into specific cell types and were ultimately linked to of the known disease pathology to establish a ‘disease model’ (Ebert et al., 2009; Lee et al., 2009; Raya et al., 2009; Ye et al., 2009). Extending this experimental paradigm to diseases with either unknown or more complex, multicellular phenotypes, or to diseases involving cell types that have yet to be generated in vitro from hESCs or hiPSCs represents a major current limitation of the approach.
Technical challenges and emerging solutions
As introduced above, while the hiPSC approach to study pathogenesis has been attempted for a few monogenic diseases, disease modeling strategies to study more complex diseases face even greater challenges. A major concern to the study of human diseases in the cell culture dish is that it may be difficult or not practical to in vitro model diseases with a long-latency such as Alzheimer’s or Parkinson’s. In such cases, the dynamics of disease progression in the patient is likely to be vastly different from any phenotype developing in vitro in cells differentiated from patient-specific hiPSCs (Fig. 1a). One possibility to overcome this challenge would be the attempt to accelerate the appearance of pathological phenotypes in the cell culture dish by exposing the cells to environmental effects that may contribute to the disease, such as oxidative stress. Issues such as the kinetics of disease pathology have yet to be addressed with the strategies that have been published to date. A second major concern for the study of disease pathogenesis is that it may be difficult or impractical to model diseases in vitro with a single purified lineage-committed cell type. In current hiPSC modeling approaches possible interactions of the cell type that is affected in the patient with other cell types within a tissue or within the diseased patient’s body have yet to be systematically reconstructed (Fig. 1b), and in some cases differentiation protocols required to generate cell types of interest from pluripotent populations have not been established. Lastly, diseases with significant epigenetic components may be difficult to study in iPSCs, as the reprogramming process is expected to remove any epigenetic alterations associated with disease phenotypes. Hence, epigenetic alterations will not persist in the pluripotent iPSCs, an issue particularly relevant to sporadic and multifactorial disorders caused by a combination of genetic and environmental factors. Environmental factors, such as toxic metals and pesticides, general lifestyle and dietary habits have been associated with increased risk in some diseases and may affect the epigenome (Jaenisch and Bird, 2003). Thus, iPSCs from patients with sporadic diseases, which are caused predominantly by epigenetic alterations, may be of little value for mechanistic studies unless the epigenetic alterations also associate with unidentified genetic alterations.
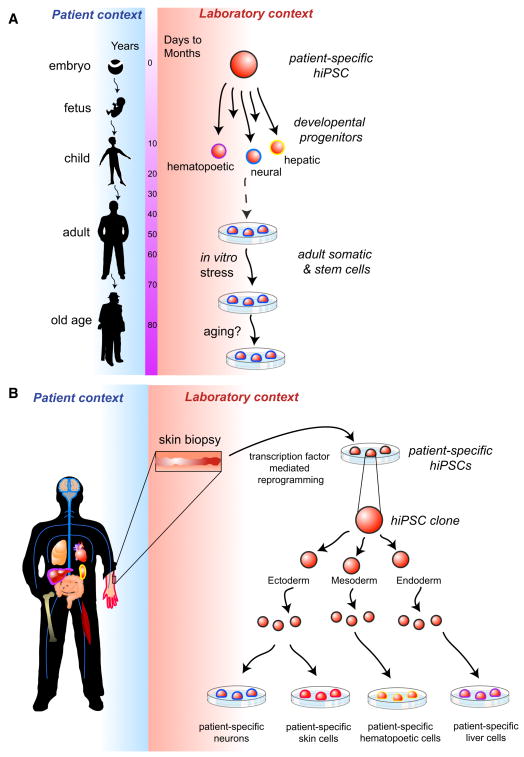
Figure 1. Technical challenges in generating cellular models of disease.
Recapitulating disease in the laboratory requires reconstruction of both the kinetics of disease development and pathology (a) as well as in the interaction of the principal diseased cell type with other cell types in the patient’s body (b). In (a), the dynamics of disease progression in the patient is likely to take years, while phenotypes developing in vitro in cells differentiated from patient-specific hiPSCs could be achieved in days to months. This acceleration could be achieved through in vitro stress, including exposure of the cells to environmental effects such as oxidative stress, or by promoting “ageing” in vitro. In (b), cells are typically harvested from a patient through a blood sample or biopsy (shown in this example, although any part of the body could be used). The harvested sample is reprogrammed to generate hiPSCs, and a hiPSC line is subsequently differentiated to produce specific cell types thought to be affected by the disease. Interaction of the principal diseased cell type with other cell types within a tissue or within the diseased patient’s body may need to be reconstructed in vitro for effective disease modeling.
Defining a disease-relevant phenotype will critically depend on the choice of ‘healthy wild-type’ control cells. A wide range of control cell lines could be used for comparison with a given patient-specific hiPSC line, including established hESC lines, or established hiPSC lines from healthy donors. To establish a general model of disease, a panel of lines derived from the same patient, as well as additional, unrelated patients suffering from the same disease should be compared to ensure that any observations are not specific to a given cell line, or a particular patient. For example, given the genetic background diversity that exists between unrelated individuals, the use of control cells lines derived from healthy siblings may be less likely to result in background-specific confounding results during experimental comparisons. In single gene diseases genetically-rescued hiPSC lines could represent an ideal isogenic control. In diseases with somatically acquired mutations, hiPSC lines isolated from unaffected cell types could be used as controls. For example in myeloid proliferative disorders that affect the hematopoietic system, hiPSC lines derived from the skin of the patient would serve as control lines for studies involving hiPSC lines derived from the diseased blood.
In the following we will highlight four technical challenges: 1) creation of reprogramming factor-free hiPSCs to minimize or eliminate genetic alterations in the derived iPSC lines; 2) gene targeting strategies to generate markers for differentiation and gene corrections; 3) establishing disease-relevant phenotypes in vitro; and 4) establishing disease-relevant phenotypes in vivo. In each of these areas, new tools are emerging that address these challenges and will make modeling with hiPSCs more tractable for complex diseases (Fig. 2). These challenges and emerging solutions are described below in the chronological order likely encountered by researchers in this field. First, only virus mediated reprogramming has been used thus far to generate hiPSCs that display some disease specific phenotypes from patients, and because the reprogramming vectors remained integrated in these disease-specific hiPSCs, it cannot be excluded that residual vector expression contributed to the observed phenotype.
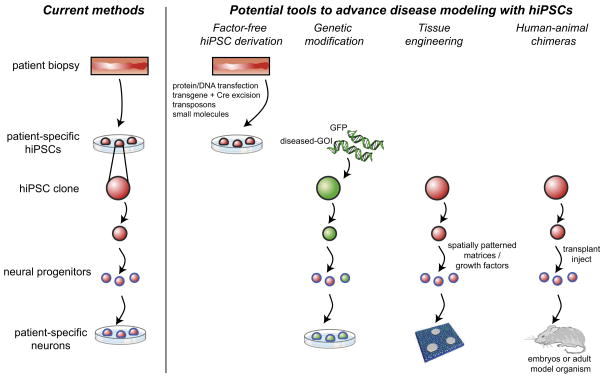
Figure 2. Emerging tools that may be used in disease modeling efforts.
Four emerging tools may address technical challenges in modeling diseases in patients using hiPSC technology. On the far left is a schematic of the current approach to disease modeling through reprogramming. New reprogramming strategies utilizing gene excision, DNA transfection, protein transfection, and small molecules could generate hiPSCs without integrated factors in the genome, termed “factor-free” hiPSCs. Advances in genetic modification allow for the tracking, accentuating, or accelerating pathological phenotypes through introduction of cell type–specific lineage reporters, as well as disruption, repair or overexpression of specific genes (e.g., GFP or disease-gene-of-interest, “disease-GOI”). Biomaterials can provide tailored microenvironmental stimuli, which include neighboring cells, extracellular matrix, soluble factors and physical forces, in order to reveal pathological mechanisms. Human-animal chimeras with human blood, neurons, and other tissues have been generated, and these tools could be used to interrogate the in vivo function of hiPSC derivatives.
Recently, novel derivation strategies have been devised to create ‘reprogramming factor-free’ hiPSCs. Second, gene targeting interventions aimed to disrupt, repair or overexpress genes in hiPSCs are integral in current strategies to mark and purify the differentiation stage of hiPSC derivatives and for genetic rescue of diseased cellular phenotypes. Current prospects for perturbing gene function in hiPSCs are described. Finally, traditional cell culture techniques with single differentiated hiPSC types may provide inadequate stresses or microenvironments to truly model the onset and/or progression of disease processes. Both biomaterials and animal-human chimeras are tools that overcome some of the limitations of traditional cell culture.
Strategies of deriving reprogramming factor-free hiPSCs
Residual expression of integrated copies of reprogramming factors in hiPSCs can affect the gene expression and potentially biological properties of the resulting iPSC derivatives. In the most salient example, use of c-Myc as a reprogramming factor led to high incidence of tumors in chimeras generated with mouse iPSCs and it is expected that this oncogene would function similarly in hiPSCs (Nakagawa et al., 2008). In a more systematic study through use of Cre-recombinase excisable viruses, hiPSCs were first derived through viral vector mediated transduction of reprogramming factors and subsequently followed by Cre-mediated excision of the vectors (Soldner et al., 2009). Such factor-free hiPSCs displayed a global gene expression profile that was more closely related to hESCs than to hiPSCs carrying the transgenes, consistent with the possibility that basal vector expression may affect the phenotype of the hiPSCs.
Various new methods have been developed to improve reprogramming technology to generate genetically unmodified or reprogramming “factor-free” hiPSCs (reviewed in (O’Malley et al., 2009)). At present, there is no clear optimal method, as each approach has strengths and disadvantages (Table 1). As noted above, the Cre-recombinase method efficiently reprograms cells, however viral elements flanking the loxP sites still remain after excision. Like the Cre-loxP recombination strategy, piggyBac transposition has achieved removal of exogenous reprogramming factors from genomic integration sites in iPSCs (Kaji et al., 2009; Woltjen et al., 2009). The piggyBac transposon/transposase system requires the inverted terminal repeats flanking a transgene and transient expression of the transposase enzyme to catalyse insertion or excision events. However, the identification of hiPSCs with minimal-copy vector insertions, integration site mapping, excision of the reprogramming cassettes and validation of factor-free clones can be a laborious process. Non-integrating strategies using episomes (Yu et al., 2009), adenoviral transfection (Stadtfeld et al., 2008), RNA viruses (Fusaki et al., 2009), or plasmid transfection (Gonzalez et al., 2009; Okita et al., 2008) are extremely inefficient. Though these approaches circumvent a few of these obstacles, it is difficult to completely exclude the possibility that vector subfragments integrated in the resulting iPSCs. Lastly, protein transfection can generate genetically-unmodified iPSCs, but at exceedingly low efficiencies (Kim et al., 2009; Zhou et al., 2009). A variety of small molecules could singly replace reprogramming factors (Huangfu et al., 2008; Ichida et al., 2009; Lyssiotis et al., 2009; Shi et al., 2008; Xu et al., 2008), but there has yet to be a demonstration of using only small molecules to reprogram somatic cells.
Table 1. Strategies for deriving “reprogramming factor-free” hiPSCs.
The first direct reprogramming strategies using viral transgenes encoding the transcription factors used in deriving hiPSCs generated hiPSCs with multiple integrated copies of viral transgenes. These transgenes may be reactivated during disease modeling, and new advances have generated iPSCs without integrated factors in the genome, termed “reprogramming factor-free” iPSCs.
| Strategy | Strengths | Potential Obstacles | References |
|---|---|---|---|
| Episomal vectors Adenoviral vectors Sendai viruses Transient transfection |
Use of non-integrating vector | Possibility of integrated vector subfragments; inefficient | (Fusaki et al., 2009;Gonzalez et al., 2009; Okita et al., 2008; Stadtfeld et al., 2008; Yu et al., 2009) |
| piggyBac transposon | Precise deletion possible | Excision may be inefficient and laborious | (Kaji et al., 2009; Woltjen et al., 2009) |
| Lentiviral vectors + Cre | Efficient reprogramming and vector deletion | Vector DNA external to the loxP sites remain integrated (viral promoters + LTRs) | (Chang et al., 2009; Soldner et al., 2009) |
| Arginine peptide tagged proteins | No genetic modification | Extremely low efficiency | (Kim et al., 2009; Zhou et al., 2009) |
| Small molecules | No genetic modification | Yet to be demonstrated: still requires at least one factor to be transduced | (Huangfu et al., 2008; Ichida et al., 2009; Lyssiotis et al., 2009; Shi et al., 2008) |
Genetic modification of hESCs and hiPSCs
Tracking, accentuating, or accelerating pathological phenotypes in the lab could greatly benefit from cell type–specific lineage reporters, as well as reliable tools to disrupt, repair or overexpress genes. First, cell type–specific lineage reporters would aid in the enrichment for specific cell types during in vitro differentiation, as differentiation techniques to generate specific somatic cell types affected by disease typically also produce progenitors and mixed cell cultures, which may interfere with in vitro assays of disease. Indeed, for many cell types of interest, efficient in vitro generation techniques have yet to be determined, and thus lineage-tracking tools will likely be required to achieve this important early step in the effort to model some human diseases. In addition, such reporters may facilitate the tracking of diseased cells in co-cultures and in chimeric animals after grafting or transplantation. Further, tools to disrupt, repair or overexpress genes could help isolate individual genetic components in complex disease models. Building up to a complex disease phenotype from combinations of single genetic modifications as well as rescuing phenotypes through gene modification would also be of interest. Lastly, overexpression of genes that stress or age cells might help to accentuate phenotypesand/or mimic the induction of disease onset in the laboratory context. hiPSCs provide an attractive pool of cells to modify since they indefinitely self-renew, although most methods could also be applied to hiPSC derivatives such as differentiated progenitors that can be easily expanded and banked.
Tools to achieve expression of transgenes in hESCs or hiPSCs by random integration of vectors include retroviruses, lentiviruses, bacterial artificial chromosomes, synthetic gene delivery reagents, and a transposon/transposase system (Giudice and Trounson, 2008; Placantonakis et al., 2009). Viral gene transfer into hESCs can be inefficient, as adeno-associated virus and adenovirus vectors have been shown to transduce only 0.01–11% of undifferentiated hESCs (Smith-Arica et al., 2003). Lentiviral vectors are typically used instead for transgene expression, as these approaches achieve <40% transduction efficiency in hESCs (Xia et al., 2007). Recently, synthetic gene delivery approaches have been developed to rival viral delivery, as engineered polymers and cationic reagents have the ability to condense DNA into particles that facilitate cellular uptake and endosomal escape (Green et al., 2008). Finally, piggyBac transposition is host-factor independent and has recently been demonstrated to be functional in various hiPSCs (Kaji et al., 2009; Lacoste et al., 2009; Woltjen et al., 2009). Copy number and integration patterns of the transgenes are not easily controlled in these strategies.
Targeting specific endogenous genetic loci is a key technology to study gene function, as this strategy preserves the flanking genomic context of the target including important regulatory elements. Since the derivation of the first hESCs, only a few reports have described successful gene targeting by homologous recombination in hESCs (Costa et al., 2007; Davis et al., 2008; Irion et al., 2007; Zwaka and Thomson, 2003). These studies used both nonisogenic and isogenic constructs encoding a drug selectable cassette introduced into hESCs by electroporation or transfection with a cationic reagent. Isolation of correctly targeted clones involved drug selection and screening of clones through PCR or Southern analysis to check for proper integration of the vectors into the human genome. Recently, a technique, called ‘genome editing’, based on the introduction of DNA double-strand breaks by site-specific zinc-finger nucleases (ZFNs) to facilitate homologous recombination (Lombardo et al., 2007) has been used to target endogenous genes in hESCs and hiPSCs (Hockemeyer et al., 2009; Zou et al., 2009). A ZFN is generated by fusing the FokI nuclease domain to a DNA recognition domain composed of engineered zinc-finger motifs that specify the genomic DNA binding site for the chimeric protein. Upon binding of two such fusion proteins at adjacent genomic sites, the nuclease domains dimerize, become active and cut the genomic DNA. When a donor DNA that is homologous to the target on both sides of the double-strand break is provided, the genomic site can be repaired by homology-directed repair, allowing the incorporation of exogenous sequences placed between the homologous regions. To ensure the uniqueness of intended targets within the human genome, ZFNs containing multiple zinc fingers need to recognize composite sites of 20–50 bp. ZFNs were used to engineer several loci in hiPSCs: the disease-related pig-a locus (Zhou et al, 2009), the pitx3 locus, which is not expressed in hESCs, the oct4 locus to report on cell fate, and the aavs1 locus to be a ‘safe harbor’ for an inducible transgene (Hockemeyer et al., 2009). Though vector insertions into these four loci has been efficient, it is not clear as yet what fraction of genes can be targeted by this approach.
Towards tissue engineering with hiPSC derivatives to generate disease-relevant phenotypes
Cellular functions are influenced not only by cell-autonomous programs but also by microenvironmental stimuli, which include neighboring cells, extracellular matrix, soluble factors and physical forces. Engineered biomaterials and co-cultures may provide a powerful way to provide a richer context for studying disease relevant cell–cell interactions (Guilak et al., 2009). These contextual cues are particularly important for modeling non-cell autonomous pathology. In amyotrophic lateral sclerosis (ALS) for example, co-cultures of wild-type, hESC-derived motor neurons with mutant ALS astrocytes induced motor neuron death (Di Giorgio et al., 2008; Marchetto et al., 2008).
While full recapitulation of tissue architecture remains an elusive goal of tissue engineering, smaller functional units (10–100 μm) have been developed to study cellular responses to distinct local stimuli. Bhatia and colleagues used ‘soft lithography’ techniques to create micropatterned cell clusters, in which 500–μm-islands of human hepatic cells are surrounded by fibroblasts. These micropatterned cell cultures were then assessed for liver function through gene expression profiles, metabolism, secretion of liver-specific products and susceptibility to hepatotoxins (Khetani and Bhatia, 2008). Patterning approaches can also be applied in three-dimensional (3D) scaffolds, which have been generated from purified molecules such as collagen I, synthetic biomaterials, and from native extracellular matrices from which living cells were previously extracted (Yamada and Cukierman, 2007). Using hiPSC derivatives in combination with these and other advances in biomaterials such as microscale cell patterning and 3D tissue scaffolds could bridge the gap between traditional cell culture and animal models.
Generating disease-relevant phenotypes with human-animal chimeras
It may not be practical to in vitro model diseases with long latencies of onset and/or with complex pathophysiology. Thus, for some types of disease modeling in vivo approaches may be required. Chimeras provide long-term access to complex and changing environmental context for hiPSCs and are being currently being experimentally explored and optimized. A chimera is an organism in which tissues of genetically different constitution co-exist as a result of grafting, mutation, or some other process. Human-animal interspecific chimeras can be generated by grafting hiPSC-derived cells into embryos, fetuses, or adult animals (Behringer, 2007; Shultz et al., 2007). In several instances, xenografts created by transplantation of human cells into immune-privileged sites (e.g., anterior chamber of the eye or cheek pouch) has been used, however, the most widespread approaches utilize immunodeficient mice, such as the nude mouse, severe combined immunodeficiency (SCID) mouse, and NOG mouse. In this way animal chimeras engrafted with human tissues at orthotopic sites have been produced in efforts to generate ‘humanized’ animals (Friese et al., 2006).
Humanized mouse systems have recently had the most notable progress with hematopoietic, nervous system, and hepatic reconstitution with human adult stem cells or hESC derivatives (Behringer, 2007; Shultz et al., 2007). Adult human hematopoietic stem cells have been injected intravenously into irradiated adult or newborn recipients with significant engraftment (Ishikawa et al., 2005; Shultz et al., 2005). When undifferentiated hESCs were injected directly into the brain ventricles of fetal mice, human neurons and glia formed (Muotri et al., 2005), although it is not clear how differentiated cells were generated after injection and became incorporated into the brains of the host animal and why no hESC-derived teratomas formed. Human adult neural stem cells survive, migrate, and express differentiation markers for neurons and oligodendrocytes after long-term engraftment in spinal cord-injured NOD-SCID mice, and in the neonatal, the adult, or the injured rodent brain (Cummings et al., 2005; Guzman et al., 2007). Lastly, a hepatocyte-humanized mouse has been generated to exhibit human-type responses in a series of in vivo drug processing experiments and in the infection and propagation of hepatic viruses (Kneteman and Mercer, 2005). Currently, there have been no reports of using hiPSCs or their derivatives with such animal models, and these protocols will likely need to be refined to enable more robust engraftment and functionality of the transplanted human cells.
Outlook
Generating cellular models of disease is a large, long-term project that will likely take decades for the existing challenges to be addressed adequately for this approach to be applied to a wide range of diseases. However, progress has already been attained in several cases of modeling monogenic, cell autonomous diseases with developmental or early onset pathology (Table 2). The approach seems particularly tractable for ‘orphan-diseases’ where no animal model exists and where patients are few and far between. For more complex diseases we anticipate significant synergy among the four classes of emerging solutions consisting of reprogramming factor-free hiPSC derivation, genetic modification of hiPSCs, tissue engineering, and generating disease-relevant phenotypes with human-animal chimeras (Fig. 2). For example, tissue engineering approaches could generate splenic capsules from diabetes-specific hiPSCs that are implanted into humanized NOD mice to assay functional properties of the hiPSC derivatives. Further, reprogramming factor-free neural progenitors from idiopathic Parkinson’s disease hiPSC lines could be genetically modified to overexpress a mutant form of α-synuclein to accelerate late onset pathology. While disease modeling typically must be well developed before therapeutics can be identified and tested, several applications utilizing disease-specific human cell models have already been envisioned (Andersson and Lendahl, 2009; Colman and Dreesen, 2009; Daley and Scadden, 2008; Freund and Mummery, 2009; Rubin, 2008). These applications fall into three major categories: small molecule and protein therapeutic discovery, functional and chemical genomics to elucidate disease mechanisms, and cell replacement therapy.
Table 2. Salient motivating factors in choosing to model a disease with human iPSC technology.
For the disease categories listed in each row, the salient characteristics of each are highlighted in relation to the technical challenges of creating disease hiPSC models. This table is meant to be illustrative of different challenges facing several types of disease, and not an exhaustive list of hiPSC models. For example, in infectious diseases (e.g., HIV), a humanized mouse model with hiPSCs could explore human host factors that confer resistance. Three motivating factors for using hiPSC technology to model a disease are anticipated to be important for each class of diseases (denoted by green “+”). First, the genetics of the disease may be an important factor. On the one hand, if disease etiology is expected to involve many genetic lesions across the human genome, the hiPSC technology can provide a cell line which contains the appropriate disease-relevant combinations of these lesions. On the other hand, clear Mendelian genetics enables one to be certain that the genetic lesion is captured in the hiPS-derived cells. Second, if animal models could not reasonably be expected to recapitulate disease pathology or phenotype (most particularly for psychiatric/neurobehavioral disease), hiPSC-derived cells may be the best option to study the cellular changes involved in a particular diseases. Third, where there is well-characterized pathology in human diseases, phenotypes observed in hiPSC derivatives can be more easily related to those seen in patients. Two potential complicating factors are denoted by a red “+”. Non-cell autonomous pathology will likely be difficult to model with differentiated hiPSC cell types, and environmental stresses may be difficult to recapitulate experimentally. If disease-specific hiPSCs have been derived from particular patient groups, the references are listed on the right most column.
| Disease Categories |
Motivating Factors | Potential Complicating Factors | Existing hiPSC Line |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Multifactorial genetics |
Human- specific pathology |
Molecular & histopathology well- characterized |
Other | Non-cell autonomous pathology |
Environmental stresses |
Other | ||||
| Neurology | Neurodegenerative | Familial | ? | + | + | Mendelian genetics in many cases | + | ? | Early onset in some cases; Network dysfunction likely important | Huntington’s disease (Park et al., 2008b); dysautonomia (Lee et al., 2009); SMA type 1 (Ebert et al., 2009); ALS (Dimos et al., 2008) |
| Sporadic | + | + | + | Cellular pathology well- defined | + | + | Network dysfunction likely important | Parkinson’s disease (Park et al., 2008b; Soldner et al., 2009) | ||
| Neuro- developmental | + in some cases | + | + | Well-defined genetic lesions in many cases | + | ? | Complex phenotypes involving many parts of the body; Network dysfunction likely important | Rett Syndrome (Hotta et al., 2009); Down Syndrome (Park et al., 2008b) | ||
| Neurobehavioral/Psychological | + | + | ? | Opportunity to define cellular pathology | + | + | Network dysfunction likely important | None published | ||
| Hematology/Oncology | + | + | + | Well-defined genetic lesions in some cases; Correct known genetic defects readily via bone- marrow manipulation; Opportunity to distinguish epigenetic & genetic factors | ? | + in some cases | Assaying function of hematopoietic derivatives;iPSC state may erase important epigenetic alterations | Sickle-cell disease (Ye et al., 2009a); Fanconi anemia (Raya et al., 2009); Myeloproliferative disorders (Ye et al., 2009b) | ||
| Endocrinology | + | ? | + | Well-defined cellular pathology; sometimes well- defined genetic lesions | + | + | Autoimmune response likely important | Juvenile diabetes mellitus (Maehr et al., 2009; Park et al., 2008b) | ||
| Infectious diseases | + | + | ? | Amenable to reverse genetics to look for host susceptibility factors | ? | ? | Susceptibility could rely on specific aspects of physiology | None published | ||
Small molecule and protein therapeutic discovery
Recent hiPS cellular disease models already have lead to small molecule candidates for treating familial dysautonomia (Lee et al., 2009) and SMA type I (Ebert et al., 2009). Such cell-based screening efforts could also be applied to development of monoclonal antibodies or other protein therapeutics. Defining appropriate in vitro cell function to be used as the read-out in therapeutic screens will likely be linked to known indicators of disease amelioration observed in the clinic. In addition, hiPSC technology could help identify drugs that are only effective against diseased cells with particular genetic profiles and advance the detection of off-target drug toxicities. Lastly, this knowledge may help narrowing of target patient populations and even reduce the cost of therapeutic testing (Rubin, 2008).
Functional and chemical genomics to elucidate disease mechanisms
In addition to discovering new therapeutics, hiPS cellular disease models can be used in combination with new tools designed to systematically perturb cells in vitro in order to elucidate mechanisms of disease. Further, induction of such perturbations in hiPSCs or their derivatives could also inform the search for improved means of directing differentiation (Xu et al., 2008). Functional genomics approaches can perturb 102–104 of genes in parallel, as libraries of short interfering hairpins directed against all the genes in the human genome have been produced (Moffat et al., 2006). An alternative approach is the use of small organic molecules instead of genetic perturbations. This approach is referred to as “chemical genomics” and is used to illuminate the molecular mechanisms underlying biological processes by virtue of the capacity of small molecules to alter protein activity by binding to their target and inhibiting or activating their normal functions (Stockwell, 2004). Using either libraries of hairpins or small molecules, mechanisms of disease can be understood by gain-of-function screens, loss-of-function screens, or synthetic lethal screens (Grimm, 2004).
Recent studies involving stem cell differentiation (reviewed in (Xu et al., 2008)) and reprogramming (Ichida et al., 2009) provide proof-of-concept that complex biological mechanisms can be effectively understood through chemical and functional genomics approaches. hESCs have been treated with small molecule libraries to elucidate novel pathways that correlate with hESC self-renewal and differentiation activities (Desbordes et al., 2008), and similar methods will likely also be used with hiPSCs. Further, chemical and functional genomics screening has already identified genes and pathways important in enhancing hiPSC formation - TGFβ and ERK signaling using small molecules (Ichida et al., 2009) and p53 using smaller scale siRNA knockdowns (Zhao et al., 2008). Utilizing similar approaches with patient-specific hiPSCs could help elucidate unknown mechanisms of disease.
Cell transplantation into patients
Given the limitations of any in vitro or animal model of disease, it may be that some insight into human disease will only come after cell transplantation into diseased human tissue is attempted. Transplantation of hiPSC derivatives into focal, diseased lesions would likely be the first application of cell replacement therapy, although it is currently unclear whether fully differentiated cells or progenitor or stem cells would more easily reconstitute the tissues at the site(s) of disease. Challenges in using cell therapy with hiPSCs have been recently reviewed (Colman and Dreesen, 2009; Daley and Scadden, 2008; Freund and Mummery, 2009; Kiuru et al., 2009). In addition to the modeling considerations described herein, for the use of stem cell derivatives in human therapy, it will be particularly important to monitor cell karyotype to detect chromosomal abnormalities that could arise during prolonged cell culture (Spits et al., 2008). Karyotypic changes have been repeatedly reported for hESCs expanded in culture and might also be expected for hiPSCs, as these could cause tumorigenicity in addition to teratoma formation after transplantation of derivatives into patients. Lastly, factor-free reprogramming in fully defined, feeder-free culture conditions will probably be a regulatory requirement for this class of cell-based therapeutics.
Closing Comments
Disease models utilizing patient-specific hiPSCs will likely generate a wealth of information and data that could be combined with genetic analyses of disease. The combination of genetic and hiPSC trait information may allow early and more accurate prediction and diagnosis of disease and disease progression. Further, the redefinition of disease subtypes through such disease modeling is likely to provide many examples of differential response to therapy, and understanding of individual responses to drugs will have implications for their use and development by the pharmaceutical industry. Such work is envisioned, in one example, to proceed in collaboration with the “Personal Genomes Project.” Lastly, this research will require new social arrangements between patients, doctors, and bench researchers which may challenge existing institutional and state policy in regards to property, privacy, and human dignity involving such novel human cellular material and modeling data. These societal challenges are outside of the scope of this article, but will likely also guide the development of this research area insofar as it is conceived as an effort to respond directly to clinical need. Therefore, research at the bench is likely to be structured to take into account the legal, ethical, regulatory and economic environment that mediates basic research from clinical application. It will be important to maintain awareness of these extrinsic factors as this research area develops in order to produce the most robust science while also responding the urgency of developing new clinical tools.
By framing disease at the cellular level with human embryonic-like material, in contrast to the genetic level or the model organism level, researchers in this field face unique technical challenges. Emerging solutions involve several areas of biomedical research and are likely to be used to produce new molecular understanding of a wide range of diseases.
Acknowledgments
We would like to thank Vikram Khurana, Frank Soldner, Julien Muffat, Ben Hurlbut, and members of the Jaenisch lab for helpful discussions. R.J. is supported by grants from the NIH: RO1-HDO45022, R37-CA084198, RO1-CA087869. K.S. is supported by the Society in Science: The Branco-Weiss fellowship. RJ is an advisor to Stemgent and a cofounder of Fate Therapeutics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aasen T, Raya A, Barrero MJ, Garreta E, Consiglio A, Gonzalez F, Vassena R, Bilić J, Pekarik V, Tiscornia G, et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nature Biotechnology. 2008;26:1276–1284. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- Andersson ER, Lendahl U. Regenerative medicine: a 2009 overview. J Intern Med. 2009;266:303–310. doi: 10.1111/j.1365-2796.2009.02157.x. [DOI] [PubMed] [Google Scholar]
- Behringer RR. Human-animal chimeras in biomedical research. Cell Stem Cell. 2007;1:259–262. doi: 10.1016/j.stem.2007.07.021. [DOI] [PubMed] [Google Scholar]
- Boland MJ, Hazen JL, Nazor KL, Rodriguez AR, Gifford W, Martin G, Kupriyanov S, Baldwin KK. Adult mice generated from induced pluripotent stem cells. Nature. 2009;461:91–94. doi: 10.1038/nature08310. [DOI] [PubMed] [Google Scholar]
- Cantz T, Bleidissel M, Stehling M, Schöler HR. In vitro differentiation of reprogrammed murine somatic cells into hepatic precursor cells. Biological Chemistry. 2008;389:889–896. doi: 10.1515/BC.2008.107. [DOI] [PubMed] [Google Scholar]
- Carey BW, Markoulaki S, Hanna J, Saha K, Gao Q, Mitalipova M, Jaenisch R. Reprogramming of murine and human somatic cells using a single polycistronic vector. Proc Natl Acad Sci USA. 2009;106:157–162. doi: 10.1073/pnas.0811426106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nature biotechnology. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan E, Ratanasirintrawoot S, Park I, Manos P, Loh Y, Huo H, Miller J, Hartung O, Rho J, Ince T, et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nature Biotechnology. 2009 doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- Chang CW, Lai YS, Pawlik KM, Liu K, Sun CW, Li C, Schoeb TR, Townes TM. Polycistronic lentiviral vector for “hit and run” reprogramming of adult skin fibroblasts to induced pluripotent stem cells. Stem Cells. 2009;27:1042–1049. doi: 10.1002/stem.39. [DOI] [PubMed] [Google Scholar]
- Chen M, Tomkins DJ, Auerbach W, McKerlie C, Youssoufian H, Liu L, Gan O, Carreau M, Auerbach A, Groves T, et al. Inactivation of Fac in mice produces inducible chromosomal instability and reduced fertility reminiscent of Fanconi anaemia. Nature Genetics. 1996;12:448–451. doi: 10.1038/ng0496-448. [DOI] [PubMed] [Google Scholar]
- Choi KD, Yu J, Smuga-Otto K, Salvagiotto G, Rehrauer W, Vodyanik M, Thomson J, Slukvin I. Hematopoietic and endothelial differentiation of human induced pluripotent stem cells. Stem Cells. 2009;27:559–567. doi: 10.1634/stemcells.2008-0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman A, Dreesen O. Pluripotent stem cells and disease modeling. Cell Stem Cell. 2009;5:244–247. doi: 10.1016/j.stem.2009.08.010. [DOI] [PubMed] [Google Scholar]
- Costa M, Dottori M, Sourris K, Jamshidi P, Hatzistavrou T, Davis R, Azzola L, Jackson S, Lim SM, Pera M, et al. A method for genetic modification of human embryonic stem cells using electroporation. Nat Protoc. 2007;2:792–796. doi: 10.1038/nprot.2007.105. [DOI] [PubMed] [Google Scholar]
- Cummings BJ, Uchida N, Tamaki SJ, Salazar DL, Hooshmand M, Summers R, Gage FH, Anderson AJ. Human neural stem cells differentiate and promote locomotor recovery in spinal cord-injured mice. Proc Natl Acad Sci USA. 2005;102:14069–14074. doi: 10.1073/pnas.0507063102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley GQ, Lensch MW, Jaenisch R, Meissner A, Plath K, Yamanaka S. Broader implications of defining standards for the pluripotency of iPSCs. Cell Stem Cell. 2009;4:200–201. doi: 10.1016/j.stem.2009.02.009. author reply 202. [DOI] [PubMed] [Google Scholar]
- Daley GQ, Scadden DT. Prospects for stem cell-based therapy. Cell. 2008;132:544–548. doi: 10.1016/j.cell.2008.02.009. [DOI] [PubMed] [Google Scholar]
- Davies B, Morris T. Physiological parameters in laboratory animals and humans. Pharm Res. 1993;10:1093–1095. doi: 10.1023/a:1018943613122. [DOI] [PubMed] [Google Scholar]
- Davis RP, Ng ES, Costa M, Mossman AK, Sourris K, Elefanty AG, Stanley EG. Targeting a GFP reporter gene to the MIXL1 locus of human embryonic stem cells identifies human primitive streak-like cells and enables isolation of primitive hematopoietic precursors. Blood. 2008;111:1876–1884. doi: 10.1182/blood-2007-06-093609. [DOI] [PubMed] [Google Scholar]
- Desbordes SC, Placantonakis DG, Ciro A, Socci ND, Lee G, Djaballah H, Studer L. High-throughput screening assay for the identification of compounds regulating self-renewal and differentiation in human embryonic stem cells. Cell Stem Cell. 2008;2:602–612. doi: 10.1016/j.stem.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giorgio FP, Boulting GL, Bobrowicz S, Eggan KC. Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying an ALS-causing mutation. Cell Stem Cell. 2008;3:637–648. doi: 10.1016/j.stem.2008.09.017. [DOI] [PubMed] [Google Scholar]
- Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, Croft GF, Saphier G, Leibel R, Goland R, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- Ebert AD, Yu J, Rose FF, Mattis VB, Lorson CL, Thomson JA, Svendsen CN. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiges R, Urbach A, Malcov M, Frumkin T, Schwartz T, Amit A, Yaron Y, Eden A, Yanuka O, Benvenisty N, et al. Developmental study of fragile X syndrome using human embryonic stem cells derived from preimplantation genetically diagnosed embryos. Cell Stem Cell. 2007;1:568–577. doi: 10.1016/j.stem.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Ellis J, Bruneau BG, Keller G, Lemischka IR, Nagy A, Rossant J, Srivastava D, Zandstra PW, Stanford WL. Alternative induced pluripotent stem cell characterization criteria for in vitro applications. Cell Stem Cell. 2009;4:198–199. doi: 10.1016/j.stem.2009.02.010. author reply 202. [DOI] [PubMed] [Google Scholar]
- Freund C, Mummery CL. Prospects for pluripotent stem cell-derived cardiomyocytes in cardiac cell therapy and as disease models. J Cell Biochem. 2009;107:592–599. doi: 10.1002/jcb.22164. [DOI] [PubMed] [Google Scholar]
- Friese MA, Jensen LT, Willcox N, Fugger L. Humanized mouse models for organ-specific autoimmune diseases. Curr Opin Immunol. 2006;18:704–709. doi: 10.1016/j.coi.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad, Ser B. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gearhart J. New potential for human embryonic stem cells. Science. 1998;282:1061–1062. doi: 10.1126/science.282.5391.1061. [DOI] [PubMed] [Google Scholar]
- Giorgetti A, Montserrat N, Aasen T, Gonzalez F, Rodríguez-Pizà I, Vassena R, Raya A, Boué S, Barrero MJ, Corbella BA, et al. Generation of induced pluripotent stem cells from human cord blood using OCT4 and SOX2. Cell Stem Cell. 2009;5:353–357. doi: 10.1016/j.stem.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudice A, Trounson A. Genetic modification of human embryonic stem cells for derivation of target cells. Cell Stem Cell. 2008;2:422–433. doi: 10.1016/j.stem.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Gonzalez F, Barragan Monasterio M, Tiscornia G, Montserrat Pulido N, Vassena R, Batlle Morera L, Rodriguez Piza I, Izpisua Belmonte JC. Generation of mouse-induced pluripotent stem cells by transient expression of a single nonviral polycistronic vector. Proc Natl Acad Sci USA. 2009;106:8918–8922. doi: 10.1073/pnas.0901471106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JJ, Zhou BY, Mitalipova MM, Beard C, Langer R, Jaenisch R, Anderson DG. Nanoparticles for Gene Transfer to Human Embryonic Stem Cell Colonies. Nano Letters. 2008;8:3126–3130. doi: 10.1021/nl8012665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm S. The art and design of genetic screens: mammalian culture cells. Nat Rev Genet. 2004;5:179–189. doi: 10.1038/nrg1291. [DOI] [PubMed] [Google Scholar]
- Guilak F, Cohen DM, Estes BT, Gimble JM, Liedtke W, Chen CS. Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell. 2009;5:17–26. doi: 10.1016/j.stem.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman R, Uchida N, Bliss TM, He D, Christopherson KK, Stellwagen D, Capela A, Greve J, Malenka RC, Moseley ME, et al. Long-term monitoring of transplanted human neural stem cells in developmental and pathological contexts with MRI. Proc Natl Acad Sci USA. 2007;104:10211–10216. doi: 10.1073/pnas.0608519104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase A, Olmer R, Schwanke K, Wunderlich S, Merkert S, Hess C, Zweigerdt R, Gruh I, Meyer J, Wagner S, et al. Generation of induced pluripotent stem cells from human cord blood. Cell Stem Cell. 2009;5:434–441. doi: 10.1016/j.stem.2009.08.021. [DOI] [PubMed] [Google Scholar]
- Hanna J, Saha K, Pando B, van Zon J, Lengner C, Creyghton M, van Oudenaarden A, Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009 doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Wernig M, Markoulaki S, Sun CW, Meissner A, Cassady JP, Beard C, Brambrink T, Wu LC, Townes TM, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- Hester ME, Song S, Miranda CJ, Eagle A, Schwartz PH, Kaspar BK. Two factor reprogramming of human neural stem cells into pluripotency. PLoS ONE. 2009;4:e7044. doi: 10.1371/journal.pone.0007044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, Dekelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nature Biotechnology. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Cook EG, Gao Q, Mitalipova M, Jaenisch R. A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell. 2008;3:346–353. doi: 10.1016/j.stem.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta A, Cheung AYL, Farra N, Vijayaragavan K, Séguin CA, Draper JS, Pasceri P, Maksakova IA, Mager DL, Rossant J, et al. Isolation of human iPS cells using EOS lentiviral vectors to select for pluripotency. Nature Methods. 2009;6:370–376. doi: 10.1038/nmeth.1325. [DOI] [PubMed] [Google Scholar]
- Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nature biotechnology. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichida J, Blanchard J, Lam K, Son E, Chung J, Egli D, Loh K, Carter A, Di Giorgio F, Koszka K, et al. A Small-Molecule Inhibitor of Tgf-beta Signaling Replaces Sox2 in Reprogramming by Inducing Nanog. Cell Stem Cell. 2009 doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irion S, Luche H, Gadue P, Fehling HJ, Kennedy M, Keller G. Identification and targeting of the ROSA26 locus in human embryonic stem cells. Nature biotechnology. 2007;25:1477–1482. doi: 10.1038/nbt1362. [DOI] [PubMed] [Google Scholar]
- Ishikawa F, Yasukawa M, Lyons B, Yoshida S, Miyamoto T, Yoshimoto G, Watanabe T, Akashi K, Shultz LD, Harada M. Development of functional human blood and immune systems in NOD/SCID/IL2 receptor {gamma} chain(null) mice. Blood. 2005;106:1565–1573. doi: 10.1182/blood-2005-02-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature Genetics. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang L, Wang J, Zhang Y, Kou Z, Gao S. iPS cells can support full-term development of tetraploid blastocyst-complemented embryos. Cell Stem Cell. 2009;5:135–138. doi: 10.1016/j.stem.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Khetani SR, Bhatia SN. Microscale culture of human liver cells for drug development. Nature biotechnology. 2008;26:120–126. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiuru M, Boyer JL, O’Connor TP, Crystal RG. Genetic control of wayward pluripotent stem cells and their progeny after transplantation. Cell Stem Cell. 2009;4:289–300. doi: 10.1016/j.stem.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneteman NM, Mercer DF. Mice with chimeric human livers: who says supermodels have to be tall? Hepatology. 2005;41:703–706. doi: 10.1002/hep.20681. [DOI] [PubMed] [Google Scholar]
- Kuzmenkin A, Liang H, Xu G, Pfannkuche K, Eichhorn H, Fatima A, Luo H, Saric T, Wernig M, Jaenisch R, et al. Functional characterization of cardiomyocytes derived from murine induced pluripotent stem cells in vitro. The FASEB Journal. 2009:1–13. doi: 10.1096/fj.08-128546. [DOI] [PubMed] [Google Scholar]
- Lacoste A, Berenshteyn F, Brivanlou AH. An efficient and reversible transposable system for gene delivery and lineage-specific differentiation in human embryonic stem cells. Cell Stem Cell. 2009;5:332–342. doi: 10.1016/j.stem.2009.07.011. [DOI] [PubMed] [Google Scholar]
- Lee G, Papapetrou E, Kim H, Chambers S, Tomishima M, Fasano C, Ganat Y, Menon J, Shimizu F, Viale A, et al. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009 doi: 10.1038/nature08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loerch PM, Lu T, Dakin KA, Vann JM, Isaacs A, Geula C, Wang J, Pan Y, Gabuzda DH, Li C, et al. Evolution of the aging brain transcriptome and synaptic regulation. PLoS ONE. 2008;3:e3329. doi: 10.1371/journal.pone.0003329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, Ando D, Urnov FD, Galli C, Gregory PD, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nature biotechnology. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- Lowry WE, Richter L, Yachechko R, Pyle AD, Tchieu J, Sridharan R, Clark AT, Plath K. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci USA. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyssiotis CA, Foreman RK, Staerk J, Garcia M, Mathur D, Markoulaki S, Hanna J, Lairson LL, Charette BD, Bouchez LC, et al. Reprogramming of murine fibroblasts to induced pluripotent stem cells with chemical complementation of Klf4. Proc Natl Acad Sci USA. 2009;106:8912–8917. doi: 10.1073/pnas.0903860106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehr R, Chen S, Snitow M, Ludwig T, Yagasaki L, Goland R, Leibel RL, Melton DA. Generation of pluripotent stem cells from patients with type 1 diabetes. Proc Natl Acad Sci USA. 2009;106:15768–15773. doi: 10.1073/pnas.0906894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Ahfeldt T, Rigamonti A, Utikal J, Cowan C, Hochedlinger K. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell. 2008;3:340–345. doi: 10.1016/j.stem.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto MCN, Muotri AR, Mu Y, Smith AM, Cezar GG, Gage FH. Non-cell-autonomous effect of human SOD1 G37R astrocytes on motor neurons derived from human embryonic stem cells. Cell Stem Cell. 2008;3:649–657. doi: 10.1016/j.stem.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Mateizel I, De Temmerman N, Ullmann U, Cauffman G, Sermon K, Van de Velde H, De Rycke M, Degreef E, Devroey P, Liebaers I, et al. Derivation of human embryonic stem cell lines from embryos obtained after IVF and after PGD for monogenic disorders. Hum Reprod. 2006;21:503–511. doi: 10.1093/humrep/dei345. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Nakashima K, Toni N, Sandler VM, Gage FH. Development of functional human embryonic stem cell-derived neurons in mouse brain. Proc Natl Acad Sci USA. 2005;102:18644–18648. doi: 10.1073/pnas.0509315102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nature biotechnology. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Narazaki G, Uosaki H, Teranishi M, Okita K, Kim B, Matsuoka S, Yamanaka S, Yamashita JK. Directed and systematic differentiation of cardiovascular cells from mouse induced pluripotent stem cells. Circulation. 2008;118:498–506. doi: 10.1161/CIRCULATIONAHA.108.769562. [DOI] [PubMed] [Google Scholar]
- Nelson DL, Gibbs RA. Genetics. The critical region in trisomy 21. Science. 2004;306:619–621. doi: 10.1126/science.1105226. [DOI] [PubMed] [Google Scholar]
- O’Malley J, Woltjen K, Kaji K. New strategies to generate induced pluripotent stem cells. Current Opinion in Biotechnology. 2009 doi: 10.1016/j.copbio.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- Osakada F, Jin ZB, Hirami Y, Ikeda H, Danjyo T, Watanabe K, Sasai Y, Takahashi M. In vitro differentiation of retinal cells from human pluripotent stem cells by small-molecule induction. Journal of Cell Science. 2009;122:3169–3179. doi: 10.1242/jcs.050393. [DOI] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008a;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ. Disease-specific induced pluripotent stem cells. Cell. 2008b;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering SJ, Minger SL, Patel M, Taylor H, Black C, Burns CJ, Ekonomou A, Braude PR. Generation of a human embryonic stem cell line encoding the cystic fibrosis mutation deltaF508, using preimplantation genetic diagnosis. Reprod Biomed Online. 2005;10:390–397. doi: 10.1016/s1472-6483(10)61801-9. [DOI] [PubMed] [Google Scholar]
- Placantonakis DG, Tomishima MJ, Lafaille F, Desbordes SC, Jia F, Socci ND, Viale A, Lee H, Harrison N, Tabar V, et al. BAC transgenesis in human embryonic stem cells as a novel tool to define the human neural lineage. Stem Cells. 2009;27:521–532. doi: 10.1634/stemcells.2008-0884. [DOI] [PubMed] [Google Scholar]
- Raya A, Rodríguez-Pizà I, Guenechea G, Vassena R, Navarro S, Barrero MJ, Consiglio A, Castellà M, Río P, Sleep E, et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009;460:53–59. doi: 10.1038/nature08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves RH, Irving NG, Moran TH, Wohn A, Kitt C, Sisodia SS, Schmidt C, Bronson RT, Davisson MT. A mouse model for Down syndrome exhibits learning and behaviour deficits. Nature Genetics. 1995;11:177–184. doi: 10.1038/ng1095-177. [DOI] [PubMed] [Google Scholar]
- Reubinoff BE, Pera MF, Fong CY, Trounson A, Bongso A. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nature biotechnology. 2000;18:399–404. doi: 10.1038/74447. [DOI] [PubMed] [Google Scholar]
- Rubin LL. Stem cells and drug discovery: the beginning of a new era? Cell. 2008;132:549–552. doi: 10.1016/j.cell.2008.02.010. [DOI] [PubMed] [Google Scholar]
- Schenke-Layland K, Rhodes KE, Angelis E, Butylkova Y, Heydarkhan-Hagvall S, Gekas C, Zhang R, Goldhaber JI, Mikkola HK, Plath K, et al. Reprogrammed mouse fibroblasts differentiate into cells of the cardiovascular and hematopoietic lineages. Stem Cells. 2008;26:1537–1546. doi: 10.1634/stemcells.2008-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Do JT, Desponts C, Hahm HS, Schöler HR, Ding S. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;2:525–528. doi: 10.1016/j.stem.2008.05.011. [DOI] [PubMed] [Google Scholar]
- Shultz LD, Ishikawa F, Greiner DL. Humanized mice in translational biomedical research. Nat Rev Immunol. 2007;7:118–130. doi: 10.1038/nri2017. [DOI] [PubMed] [Google Scholar]
- Shultz LD, Lyons BL, Burzenski LM, Gott B, Chen X, Chaleff S, Kotb M, Gillies SD, King M, Mangada J, et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J Immunol. 2005;174:6477–6489. doi: 10.4049/jimmunol.174.10.6477. [DOI] [PubMed] [Google Scholar]
- Smith-Arica JR, Thomson AJ, Ansell R, Chiorini J, Davidson B, McWhir J. Infection efficiency of human and mouse embryonic stem cells using adenoviral and adeno-associated viral vectors. Cloning and Stem Cells. 2003;5:51–62. doi: 10.1089/153623003321512166. [DOI] [PubMed] [Google Scholar]
- Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, Hargus G, Blak A, Cooper O, Mitalipova M, et al. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sothern RB, Gruber SA. Further commentary: physiological parameters in laboratory animals and humans. Pharm Res. 1994;11:349–350. doi: 10.1023/a:1018992432608. [DOI] [PubMed] [Google Scholar]
- Spits C, Mateizel I, Geens M, Mertzanidou A, Staessen C, Vandeskelde Y, Van der Elst J, Liebaers I, Sermon K. Recurrent chromosomal abnormalities in human embryonic stem cells. Nature Biotechnology. 2008;26:1361–1363. doi: 10.1038/nbt.1510. [DOI] [PubMed] [Google Scholar]
- Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K. Role of the Murine Reprogramming Factors in the Induction of Pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockwell BR. Exploring biology with small organic molecules. Nature. 2004;432:846–854. doi: 10.1038/nature03196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan G, Hay D, Park I, Fletcher J, Hannoun Z, Payne C, Dalgetty D, Black J, Ross J, Samuel K, et al. Generation of functional human hepatic endoderm from human induced pluripotent stem cells. Hepatology. 2009 doi: 10.1002/hep.23335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Panetta NJ, Gupta DM, Wilson KD, Lee A, Jia F, Hu S, Cherry AM, Robbins RC, Longaker MT, et al. Feeder-free derivation of induced pluripotent stem cells from adult human adipose stem cells. Proc Natl Acad Sci USA. 2009;106:15720–15725. doi: 10.1073/pnas.0908450106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Taura D, Noguchi M, Sone M, Hosoda K, Mori E, Okada Y, Takahashi K, Homma K, Oyamada N, Inuzuka M, et al. Adipogenic differentiation of human induced pluripotent stem cells: comparison with that of human embryonic stem cells. FEBS Lett. 2009;583:1029–1033. doi: 10.1016/j.febslet.2009.02.031. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Urbach A, Benvenisty N. Studying early lethality of 45,XO (Turner’s syndrome) embryos using human embryonic stem cells. PLoS ONE. 2009;4:e4175. doi: 10.1371/journal.pone.0004175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbach A, Schuldiner M, Benvenisty N. Modeling for Lesch-Nyhan disease by gene targeting in human embryonic stem cells. Stem Cells. 2004;22:635–641. doi: 10.1634/stemcells.22-4-635. [DOI] [PubMed] [Google Scholar]
- Wernig M, Zhao JP, Pruszak J, Hedlund E, Fu D, Soldner F, Broccoli V, Constantine-Paton M, Isacson O, Jaenisch R. Neurons derived from reprogrammed fibroblasts functionally integrate into the fetal brain and improve symptoms of rats with Parkinson’s disease. Proc Natl Acad Sci USA. 2008;105:5856–5861. doi: 10.1073/pnas.0801677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hämäläinen R, Cowling R, Wang W, Liu P, Gertsenstein M, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Zhang Y, Zieth CR, Zhang SC. Transgenes delivered by lentiviral vector are suppressed in human embryonic stem cells in a promoter-dependent manner. Stem Cells and Development. 2007;16:167–176. doi: 10.1089/scd.2006.0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Alipio Z, Fink LM, Adcock DM, Yang J, Ward DC, Ma Y. Phenotypic correction of murine hemophilia A using an iPS cell-based therapy. Proc Natl Acad Sci USA. 2009;106:808–813. doi: 10.1073/pnas.0812090106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Shi Y, Ding S. A chemical approach to stem-cell biology and regenerative medicine. Nature. 2008;453:338–344. doi: 10.1038/nature07042. [DOI] [PubMed] [Google Scholar]
- Yamada KM, Cukierman E. Modeling tissue morphogenesis and cancer in 3D. Cell. 2007;130:601–610. doi: 10.1016/j.cell.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Ye L, Chang JC, Lin C, Sun X, Yu J, Kan YW. Induced pluripotent stem cells offer new approach to therapy in thalassemia and sickle cell anemia and option in prenatal diagnosis in genetic diseases. Proc Natl Acad Sci USA. 2009a;106:9826–9830. doi: 10.1073/pnas.0904689106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z, Zhan H, Mali P, Dowey S, Williams DM, Jang Y-Y, Dang CV, Spivak JL, Moliterno AR, Cheng L. Human induced pluripotent stem cells from blood cells of healthy donors and patients with acquired blood disorders. Blood. 2009b doi: 10.1182/blood-2009-04-217406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. Human Induced Pluripotent Stem Cells Free of Vector and Transgene Sequences. Science. 2009:1172482–1172482. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhao X-y, Li W, Lv Z, Liu L, Tong M, Hai T, Hao J, Guo C-l, Ma Q-w, Wang L, et al. iPS cells produce viable mice through tetraploid complementation. Nature. 2009;461:86–90. doi: 10.1038/nature08267. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Yin X, Qin H, Zhu F, Liu H, Yang W, Zhang Q, Xiang C, Hou P, Song Z, et al. Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell. 2008;3:475–479. doi: 10.1016/j.stem.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Zhou H, Wu S, Joo JY, Zhu S, Han DW, Lin T, Trauger S, Bien G, Yao S, Zhu Y, et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK, Chen G, Ye Z, Park IH, Daley GQ, et al. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaka TP, Thomson JA. Homologous recombination in human embryonic stem cells. Nature Biotechnology. 2003;21:319–321. doi: 10.1038/nbt788. [DOI] [PubMed] [Google Scholar]