Abstract
DNA microarrays are used to profile changes in gene expression between samples in a high-throughput manner, but measurements of genes with low expression levels can be problematic with standard microarray substrates. In this work, we expand the detection capabilities of a standard microarray experiment using a photonic crystal (PC) surface that enhances fluorescence observed from microarray spots. This PC is inexpensively and uniformly fabricated using a nanoreplica molding technique, with very little variation in its optical properties within- and between-devices. By using standard protocols to process glass microarray substrates in parallel with PCs, we evaluated the impact of this substrate on a one-color microarray experiment comparing gene expression in two developmental stages of Glycine max. The PCs enhanced the signal-to-noise ratio observed from microarray spots by 1 order of magnitude, significantly increasing the number of genes detected above substrate fluorescence noise. PC substrates more than double the number of genes classified as differentially expressed, detecting changes in expression even for low expression genes. This approach increases the dynamic range of a surface-bound fluorescence-based assay to reliably quantify small quantities of DNA that would be impossible with standard substrates.
The DNA microarray is a valuable tool for high-throughput quantification of gene expression, allowing a large number of candidate genes to be examined for differential expression simultaneously without extensive prior knowledge of gene functions. Eukaryotic gene expression is typically characterized by a large number of genes expressed at very low levels and a decreasing number of genes expressed at high levels.1,2 Often the noise present in DNA microarray experiments is high enough that only a small fraction of genes assayed can be detected by fluorescence measurements. While sample variation and nonspecific binding play an important role in this experimental noise, microarray substrate fluorescence can contribute to noise as well. This may explain the poor performance of microarrays in detecting genes with low expression levels relative to other methods.3,4 To overcome the difficulties of quantifying the abundance of low expression genes, substrates that enhance the fluorescence observed from microarray spots can be used to achieve better assay performance.
Researchers have utilized various nanostructured metal substratres to achieve increased intensity from common microarray dyes, with signal enhancements ranging from 1 to 2 orders of magnitude. These methods include the growth of metal island films on a substrate5,6 or the deposition of nanoparticles fabricated by spray pyrolysis7 to produce optical resonances to which microarray dye excitation and/or emission can couple. However, the practical impact of these enhancement methods is unclear, because previous work has not characterized the signal-to-noise ratio (SNR) enhancement for a large number of spots over many substrates in a conventional gene expression microarray experiment. One potential obstacle in achieving this result is the need for an inexpensive nanoscale fabrication method achieving high-throughput and good uniformity over large areas; previous reports of microarray dye enhancement have not utilized photolithography to generate consistent patterns and thus are subject to a random arrangement of structures. Another potential obstacle is integration of these substrates with the existing commercial equipment used to fabricate and scan DNA microarrays, as previous literature has not explicitly demonstrated nanostructures over areas as large as conventional microscope slides. We addressed these issues by designing a nanostructured photonic crystal (PC) substrate capable of enhancing Cyanine-5 (Cy-5) fluorescence in a commercial microarray scanner and fabricating it by nanoreplica molding to fit standard microscope slides.
The PC substrates are composed of a subwavelength, periodic SiO2 surface structure coated with a high refractive index dielectric layer of TiO2, creating a periodic modulation in refractive index along the device surface. The periodic modulation gives rise to optical resonances8 that can be used to achieve fluorescence enhancement. These resonances can be used to generate strong optical near-fields at the device surface when spectrally aligned to the excitation wavelength9 and to spatially alter the fluorescence emission pattern to maximize light collection.10 The overall effect of these phenomena is to amplify the fluorescent signal from molecules within approximately 100 nm of the PC surface. While the first demonstrations of PC enhanced fluorescence for microarrays required expensive lithographic procedures for each device and yielded modest enhancement factors of approximately 6× signal enhancement in a commercial scanner,11,12 inexpensive and uniform fabrication over large areas in a nanoreplica molding process currently used to make commercial label-free biosensors13 has since been employed to make these structures.
Recently, PCs have been engineered by our group to enhance the common microarray dye Cyanine-5 (Cy-5) by more than 1 order of magnitude when scanned in a commercial microarray scanner.14 This work details the application of this PC design to a microarray experiment assessing differential expression between Glycine max cotyledons and trifoliates, which represent tissues from two distinct developmental stages in the soybean plant. Multiple PCs exhibiting highly uniform optical characteristics over the area of entire microscope slides were fabricated by nanoreplica molding. These PCs were processed using published protocols in parallel with commercial microarray substrates. By enhancing fluorescence, a larger number of genes can be detected above noise on the PC compared with glass substrates. This effect more than doubles the number of genes identified as differentially expressed between the trifoliate and cotyledon tissues, demonstrating that enhanced fluorescence offers practical benefit to a DNA microarray experiment.
MATERIALS AND METHODS
Photonic Crystal Fabrication and Characterization
The PCs used for this work were designed by simulation software employing Rigorous Coupled-Wave Analysis (DiffractMOD, RSoft Design Group, Inc.) to align optical resonances to the excitation (632.8 nm) and emission wavelengths (670-710 nm) of Cy-5. As described in previous work,14 the period of the structure, grating depth, and thickness of the high refractive index TiO2 layer were manipulated given the known refractive indices of PC materials to achieve optical resonances overlapping both excitation and emission wavelength ranges. These PCs were then fabricated by nanoreplica molding13 to create six distinct devices for microarray experiments. The silicon “master” for the molding process consisted of a 360 nm period one-dimensional grating structure with a 60 nm grating depth and 50% duty cycle, patterned on an 8 in. silicon wafer by deep-UV lithography. After immersion in 2% dichlorodimethylsilane (PlusOne Repel-Silane ES, GE Healthcare) to promote clean release, a UV-curable liquid polymer (Gelest, Inc.) with index of refraction npolymer = 1.46 was dispensed on a sheet of polyethylene terephthalate (PET), and the grating pattern was transferred with a roller. After curing the polymer under a high-intensity ultraviolet lamp (Xenon) for 90 s through the transparent PET sheet, 300 nm of SiO2 (nSiO2 = 1.46) and 160 nm of TiO2 (nTiO2 = 2.35) were added to the grating structure by sputter coating. The completed PCs were cut into 1 in. × 3 in. sections and adhered to glass microscope slides with an optically clear adhesive (3M).
The PCs were initially profiled for surface characteristics by atomic force microscopy (Dimension 3000, Digital Instruments) to compare the actual dimensions with the PC design. The PCs were then optically characterized by passing broadband light in the visible spectrum from a tungsten halogen lamp (Ocean Optics) through a polarizer and collimator before transmission through the device, which was aligned on a rotational stage to be perpendicular to the direction of incident light.15 PCs were illuminated with both transverse magnetic (incident electric field perpendicular to grating lines) and transverse electric (incident electric field parallel to grating lines) polarizations to characterize the two distinct resonances. Light transmitted through the PCs was collected by an optical fiber and measured using a UV-visible light spectrometer (Ocean Optics).
Slide Preparation
PCs were cleaned by O2 plasma treatment and incubated with 3-glycidoxypropyltrimethoxysilane at 185 mTorr overnight for surface functionalization. The control slides for microarray experiments were commercially available silanized glass slides (Corning GAPS II). Oligonucleotides were printed on the slides using a Genetix QArray2 robot. A set of previously annotated 192 70-mer oligonucleotides derived from publicly available soybean EST and mRNA sequences was spotted on the slides with 40 repeats per sequence per slide. These 192 oligonucleotides are a subset of a larger 19 200 set of oligonucleotides detailed in previous work.16 Each of six spotted PCs was matched with a spotted glass control slide to receive identical experimental treatments.
Microarray Sample Preparation and Hybridization
Sample RNA was extracted using previously published protocols.16 Cotyledon RNA was extracted from freeze-dried soybean cultivar Williams seeds with fresh weight between 100-200 mg. Cotyledon RNA was purified using a Qiagen RNeasy kit. Trifoliate RNA was extracted from freeze-dried rolled-up trifoliates of soybean cultivar Williams from leaves between 0.5 and 1.5 in. in length. Sample RNA was labeled with Cy-5 by reverse transcription. Three replicate slides were hybridized for each of the two tissue samples, with an identical number of glass slides processed in parallel with the PCs. Slides were blocked prior to hybridization with bovine serum albumin to prevent nonspecific binding, hybridized at 42 °C overnight, and washed as described previously.16
Data Collection
Slides were scanned with a Tecan LS Reloaded scanner with a transverse magnetic polarized laser (λ = 632.8 nm) at normal incidence and an emission filter spanning 670–710 nm. All slides were scanned at 10 μm resolution. Initial scans were at equal photomultiplier tube (PMT) gain to compare fluorescence intensities at equal measurement conditions, but afterward, gains were adjusted for each slide such that spots with the largest fluorescence intensities did not saturate the scanner PMT. Fluorescence images were analyzed using GenePix Pro 6.0 to compute spot and local background intensities as well as their standard deviations for each spot.
Signal-to-Noise Ratio Analysis
Fluorescence data was analyzed to calculate signal-to-noise ratios (SNRs) for each spot at identical scan conditions, where the SNR is the local background-subtracted spot intensity divided by the standard deviation of the local background pixels. For each of the six PCs and six glass slides, within-slide repeats (40 per probe sequence) were averaged to generate a SNR value for each of the 192 sequences probed in the experiment. A SNR enhancement factor was calculated by dividing the PC SNR value for each gene by the glass SNR value for the same gene. The proportion of detected genes was determined by calculating the percentage of genes on each slide with a SNR > 3.
Differential Expression Analysis
Expression data was analyzed using the Linear Models for Microarray Data (LIMMA) package in R. Data was background corrected using the normalized plus exponential convolution model with an offset of one.17 Quantile normalization was used to normalize between arrays. Log-transformed Cyanine-5 intensities were condensed by averaging within-slide repeats and then fit to a linear model. Empirical Bayes moderated t-statistics were calculated to assess differential expression between the trifoliate and cotyledon samples, with p-values adjusted by the Benjamini and Hochberg method to control the false discovery rate.18 The significance level for testing was set to α = 0.05.
High-Throughput RNA Sequencing and Analysis
The mRNAs were also subjected to high-throughput sequencing (RNA-seq) performed at the Keck Center of the University of Illinois using the Illumina Genome Analyzer II resulting in 10–18 million total reads for leaf trifoliate and the immature cotyledon mRNA samples, respectively. After processing, the RNA-seq reads are all 70 bases in length. The sequence reads were aligned using Bowtie19 to the approximately 78 700 predicted Glyma gene models available at Phytozome 5.0 (http://www.phytozome.net) for the recently sequenced soybean genome.20 The Bowtie parameters allowed three mismatches to each Glyma model and allowed up to 25 gene model matches to detect repetitive gene models. Normalization of RNA-seq data as RPKM (reads per kilobase of gene model per million mapped reads) was calculated as shown in previous work.21 Assignment of the 70-mer oligos to Glyma gene models was also performed using Bowtie with the same parameters. Most of the 70-mer oligos on the arrays matched only one or a few Glyma models.
RESULTS
Characterization of Photonic Crystal Substrates
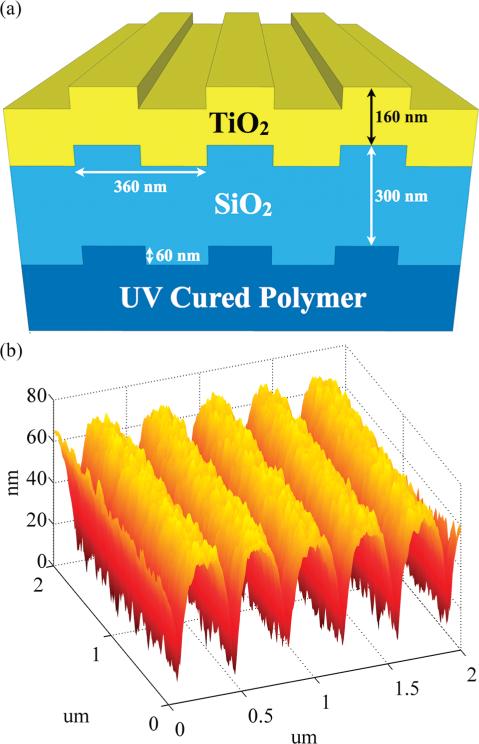
A schematic of the nanoreplica-molded one-dimensional PC design optimized in previous work14 to enhance fluorescence from Cy-5 (with period Λ = 360 nm and grating step height h = 60 nm) appears in Figure 1a. A representative atomic force micrograph detailing the surface structure appears in Figure 1b, with a measured period of Λ = 366 nm and a measured height of h = 50 nm showing good agreement with the expected dimensions.
Figure 1.
(a) Schematic of PC design dimensions. (b) Atomic force micrograph of completed PC structure (after TiO2 deposition), with a measured period of 366 nm and height of 50 nm.
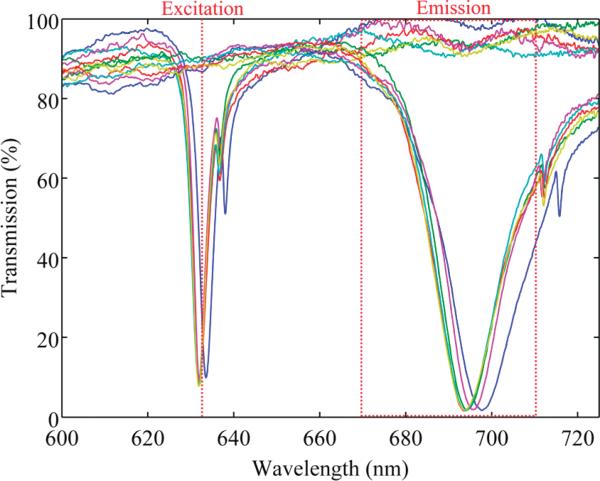
Enhancement of Cy-5 was achieved by aligning a narrow PC resonance (full-width at half-maximum, fwhm = 4 nm) with the laser excitation wavelength of 632.8 nm and engineering a second broad resonance (fwhm = 20 nm) to overlap the emission filter wavelengths of 670–710 nm (Figure 2). A more narrow excitation resonance increases the magnitude of the enhanced optical near fields at the device surface,22 while the broad extraction resonance maximizes the spectrum of emitted light redirected toward the detection optics.10 Good spectral uniformity of the narrow excitation resonance over large areas of the PC is required to ensure precise overlap of the resonance with a narrowband excitation source regardless of the location of a microarray spot on the PC. The nanoreplica molding fabrication process achieves this uniformity throughout individual microscope slide-sized PCs, with a maximum observed resonance wavelength standard deviation of σwithin-PC = 0.239 nm over 6 distinct PCs. Figure 2 illustrates that excellent between-device uniformity is achieved as well; the standard deviation in mean resonance wavelength between the 6 PCs used in this work is σbetween-PC = 0.691 nm, making both the within-device variation and between-device variation in resonance wavelength significantly smaller than the spectral width of the resonance.
Figure 2.
Optical transmission measurements from all six PCs (each represented by a colored solid line) used in this study, obtained by illuminating the devices with polarized, collimated white light. Resonances with narrow spectral features are excited when the PCs are illuminated with transverse magnetic polarized light and overlap the excitation wavelength of 632.8 nm (dotted line). Resonances with broader spectral features are excited when the PCs are illuminated with transverse electric polarized light and overlap the emission filter wavelengths of 670–710 nm (dotted box).
Signal-to-Noise Ratio Analysis
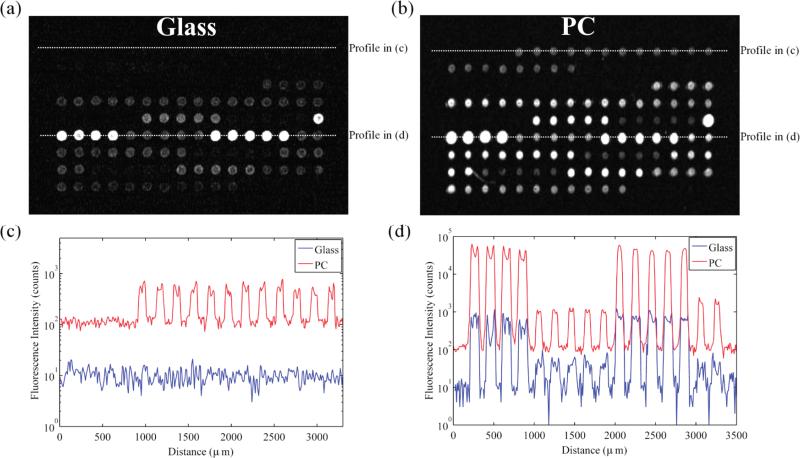
After pairing each PC with a control glass slide and printing a set of 192 oligonucleotides and negative controls on the slides,16 a one-color DNA microarray protocol was simultaneously run on each glass–PC pair. Three pairs of slides were hybridized with Cy-5 labeled cotyledon RNA (extracted from seeds), and three pairs were hybridized with Cy-5 labeled trifoliate RNA (extracted from leaves) from Glycine max cultivar Williams, representing two distinct tissues and stages of development. After averaging the 40 duplicates of the 192 genes on each slide, a ratio of averaged PC SNR to averaged glass SNR was generated for each PC-glass slide pair, resulting in a median SNR enhancement across all slides of 10.6×. The effect of this SNR enhancement on the raw fluorescence data is observed in Figure 3, which shows line profiles of identical probes for a microarray grid on both a PC and its control. Considerable enhancement is observed for spots of varying expression levels (Figure 3c, d), with low expression genes being much easier to discriminate from noise on the PC.
Figure 3.
Fluorescence images at identical gains of a single identical microarray grid on glass (a) and PC (b), with brightness and contrast adjustment to make the maximum number of spots visible on both images. For comparison of spot intensities, line profiles of identical locations on the grid for glass and PC are illustrated on the same plots. Lower expression genes appear in (c) and higher expression genes appear in (d).
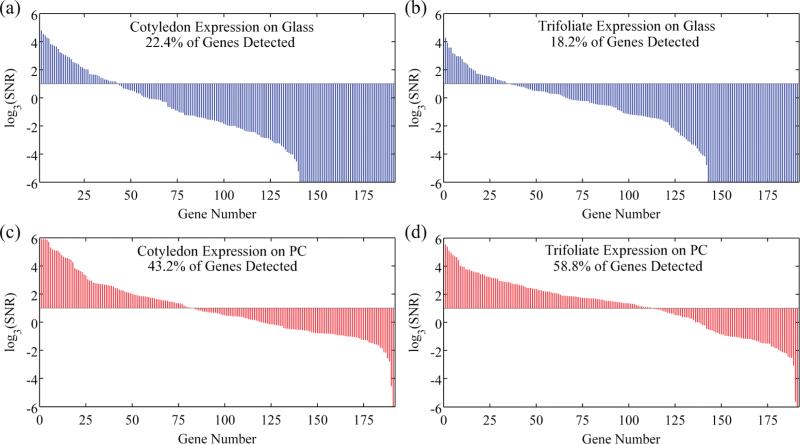
To explore the relevance of this SNR enhancement on gene expression measurements, additional fluorescence scans of PCs and glass slides were performed after gain adjustment to utilize the full dynamic range of the scanner photomultiplier tube (PMT). Because the PCs demonstrate fluorescence enhancement, they were scanned at lower PMT gain values, resulting in lower noise levels. A detection threshold of SNR = 3 was applied to determine the proportion of spots that could be detected on each slide. Across all cotyledon samples, 25.0% of spots could be detected on the glass slides, while 46.3% of the spots were detected on the PCs. A more dramatic increase in the number of detected spots was observed across all trifoliate samples, with 14.7% of spots on the glass slides being detected as compared to 49.0% of the spots on the PC. The number of genes that could be detected above noise thus almost doubled for the cotyledon sample and more than tripled for the trifoliate sample, as illustrated in plots of SNR for each gene for representative slides in Figure 4. SNR values (averaged across duplicate spots) are graphed for each gene in decreasing expression order for a single slide pair hybridized to a cotyledon sample (Figure 4a, c) and a single slide pair hybridized to a trifoliate sample (Figure 4b, d). As expected, negative control spots on both the PCs and the glass slides had SNRs below the detection threshold.
Figure 4.
Logarithmic plots of duplicate-averaged SNR values for the 192 probed genes on selected glass–PC slide pairs for each tissue. Genes are organized in decreasing expression order for each chip, and an SNR detection threshold of 3 appears as the cutoff line in each graph. SNR expression profiles for cotyledon RNA appear in (a) and (c) for a glass slide and its paired PC, respectively. Trifoliate RNA expression profiles are plotted in (b) and (d) for a glass slide and its paired PC, respectively. Negative control spots on all slides appeared below the detection threshold.
Differential Expression Analysis
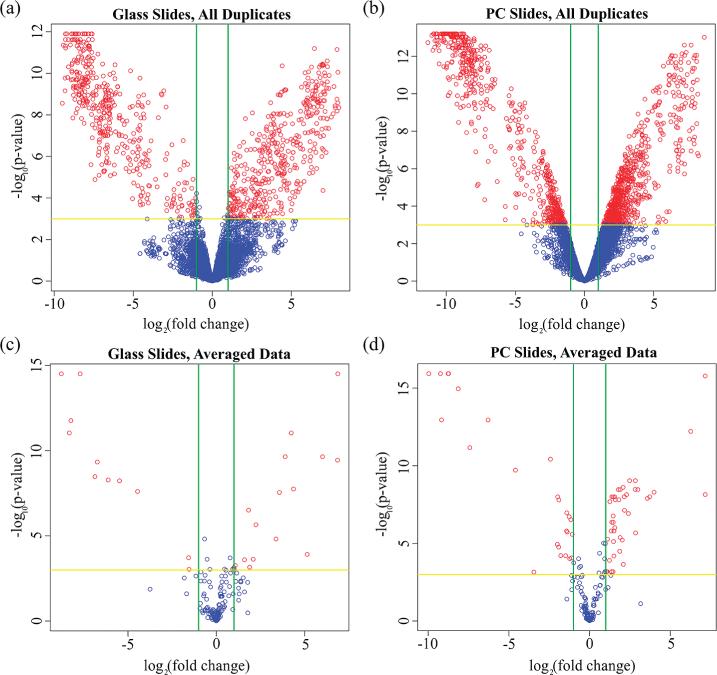
Background-corrected,17 normalized, log-transformed spot intensities were fit to a linear model using the LIMMA package in R, and empirical Bayes moderated t-statistics were calculated to assess differential expression in the trifoliate sample relative to the cotyledon sample. The analysis was carried out for glass slides and PCs separately to allow for comparison between the two substrates. Volcano plots (simultaneously illustrating the fold change and the adjusted p-value for each gene across glass slides or PCs) appear in Figure 5. Ideally, the plot should be a v-shape, since the p-value should decrease as the fold-change increases. However, this relationship is distorted by variation, since variation in fold-change measurements smears the curve horizontally and variation between samples leads to lower p-values (smearing the curve vertically). Figure 5a and b plot all 7680 spots without averaging of within-slide repeat spots in order to illustrate more clearly the effect of the PC on the experimental data. Red circles denote genes that have an adjusted p-value of less than 0.05 and a greater than 2-fold change. 1431 spots on the PCs fulfill these criteria, compared with 865 spots on the glass slides. The PC data more tightly conforms to the expected v-shape, suggesting there is less variation between within-slide repeats in fold-change values and p-values, particularly for spots with low fold-change value, compared to the glass slide. This lowered variation is expected to allow for discrimination of smaller changes in expression during statistical testing.
Figure 5.
Volcano plots detailing the relationship between fold-change and inverse p-value to assess differential expression between the trifoliate and cotyledon samples, with positive fold changes indicating increased trifoliate expression and negative fold changes indicating increased cotyledon expression. Green vertical lines represent the 2-fold change cutoff, and the yellow horizontal line denotes a p-value cutoff of 0.05. Genes meeting both thresholds are indicated by red spots. Unaveraged data representing all 7680 spots across all experimental slides (3 replicates per tissue) appear in (a) for the glass slides and (b) for the PCs, with 865 spots differentially expressed on glass slides and 1431 spots on the PCs. Averaging within-slide repeats condensed the data to 192 distinct genes and controls, which appear in (c) for the glass slides and (d) for the PCs. Of the 192 genes probed, 27 were classified as differentially expressed on the glass slides, while 68 met this classification on the PCs.
A similar analysis was carried out after averaging within-slide repeats, reducing the data set into 192 genes, with Figure 5c and d illustrating volcano plots for the averaged data set. On the glass slide, 27 genes fulfilled the criteria of statistically significant changes at an adjusted p-value less than 0.05 and a greater than 2-fold change, while on the PC, 68 genes fulfilled these criteria. Importantly, all 27 of the genes fulfilling these criteria on the glass slide were also identified as differentially expressed on the PCs (Table S-1 in the Supporting Information). The average measurement for these 27 genes are similar on both substrates, with measurements of 2350 counts (of fluorescence intensity) on the glass slides and 2880 counts on the PCs. However, an additional 41 genes were identified as differentially expressed on the PCs, suggesting that statistically significant changes in expression that were overwhelmed by noise on the glass slide could be identified on the PCs. Genes with a greater than 2-fold change as measured on the PCs (p < 0.05) but not the glass slides (Table S-2 in the Supporting Information) had an average expression level of 180 counts on the PCs, demonstrating lower expression levels than those genes classified as differentially expressed on the glass slides.
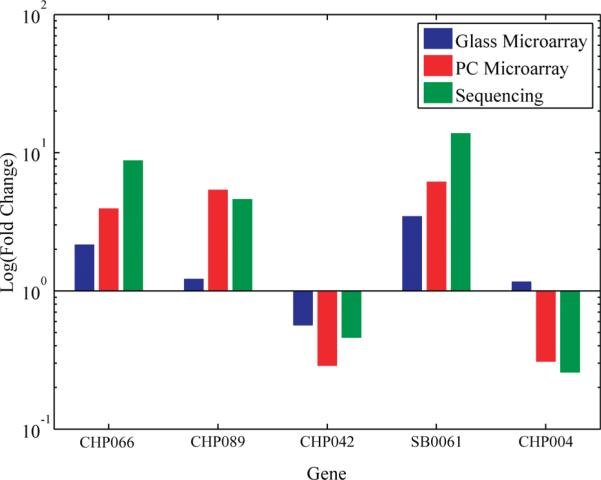
Validation of the microarray data by an independent method was obtained by high-throughput sequencing with the Illumina platform yielding 10–18 million total reads for leaf trifoliate and the immature cotyledon mRNA samples, respectively. High quality reads of 70 bases in length were mapped to 78 700 soybean gene models to obtain a quantitative view gene expression. Fold change values for 5 randomly selected genes from the 41 genes found to be differentially expressed on the PC but not the glass substrates (Table S-2 in the Supporting Information) are plotted in Figure 6, with a comparison of glass microarray, PC microarray, and sequencing data. The direction of differential expression of the genes represented by a majority of probes on the arrays was confirmed by the transcriptome sequencing data, although the absolute fold changes vary due to the fundamental differences between techniques. For example, Table S-3 in the Supporting Information shows agreement for 22 of 26 genes (one sequence was highly repetitive in the genome and thus could not be quantified) found to be differentially expressed on both PC and glass microarrays and Table S-4 in the Supporting Information shows agreement for 39 of 41 genes found to be differentially expressed only on PC microarrays. It is also apparent that the PC arrays detect genes with lower average RPKM values (average RPKM of 65.0 for both samples in Table S-4 in the Supporting Information) than detected reliably on the glass slides alone (average RPKM of 2160 for both samples in Table S-3 in the Supporting Information).
Figure 6.
Logarithmic plot of fold change comparisons between glass microarray, PC microarray, and sequencing results for five genes randomly selected from the list of genes found to be differentially expressed on the PCs but not on the glass slides. Fold change values for glass and PC microarrays were calculated by determining the ratio between average microarray expression level for trifoliate samples to average microarray expression level for cotyledon samples. A similar ratio was calculated using number of reads for sequencing data. The PC microarray results show similar directions and magnitudes of change compared to sequencing data for all 5 genes, while glass microarray data for CHP089 and CHP004 does not agree with the sequencing data.
As shown in Table S-1 in the Supporting Information (which lists genes with significant differences in expression as detected on both glass and PCs), a number of the genes found to be overexpressed in the seed cotyledons (with negative fold changes) include those that encode well-known soybean storage proteins (i.e., glycinin, lectin, Kunitz trypsin inhibitor, and the Bowman-Birk proteinase inhibitor) whose mRNA transcripts are abundant during seed embryogenesis.23,24 For example, RNA-seq transcriptomics data confirms some of these with very large RPKM values of up to 17 340 in the seed and no detectable transcripts in the leaves (Table S-3 in the Supporting Information). On the other hand, those genes encoding photosynthetic proteins as the Rubisco small chain precursor and chlorophyll a/b binding protein are overexpressed in the trifoliate leaves, as expected. As shown in Table S-2 in the Supporting Information, the additional genes detected as differentially expressed with significant p-values on the PCs represent various enzymes and transcription factors found to be expressed at lower levels by RNA transcriptome sequencing (Table S-4 in the Supporting Information), demonstrating the usefulness of the PCs to detect low expression transcripts.
DISCUSSION
By engineering PC resonances for compatibility with a commercial laser scanner, the benefits of enhanced fluorescence can be applied to a standard microarray experiment with no changes to the experimental protocol. While the initial photolithography process needed to fabricate the silicon mold of the grating has high costs, a single round of photolithography on silicon can be translated into thousands of devices that are fabricated uniformly over large areas. By fabricating the mold on an 8 in. wafer, there is a large degree of flexibility in fitting PCs to preferred labware formats such as microscope slides and microtiter plates. Because PCs were cut to fit standard microscope slides, they could be processed with existing protocols and scanned with commercially available equipment, allowing for convenient adoption of PC substrates in a standardized experiment. Not only does the nanoreplica molding process provide a convenient form factor for the substrates, it also enables the excellent level of optical uniformity required to ensure that every spot on the microarray experiences the same level of enhancement. This is key to ensure that the data obtained from PCs does not have a higher level of variation than the data obtained from glass slides.
The signal enhancement factor primarily used in previous work in this field is defined as spot intensity subtracted by the local background observed on the enhancement substrate divided by the same value observed on the glass slide or control substrate. The signal enhancement factor observed from Cy-5 spots with high expression genes in this microarray experiment was approximately 60×, which is identical to the enhancement demonstrated in previous work with this substrate.14 Thus, the PC signal enhancement compares favorably to experiments with metal island films that have yielded signal enhancement factors of 10–40×.5,6 However, the signal enhancement factor is not an ideal measurement to assess the practical utility of the substrate. This work has focused on SNR enhancement rather than signal enhancement because microarray data analysis programs use SNR values to classify spots as detected or not detected. It is possible to achieve good signal enhancement without achieving similar SNR enhancement if a substrate enhances fluorescence but has a large noise value thus voiding any advantages of fluorescence enhancement. Without knowledge of a substrate's impact on the SNR observed from spots, it is difficult to ascertain whether a substrate will benefit a target assay. The PC not only attains a large signal enhancement but it also achieves an SNR enhancement of approximately 10× (measured over all spots in the experiment), suggesting that the array can detect hybridization at concentrations 10× lower than can be detected on glass substrates.
The noise in a DNA microarray experiment arises primarily from the following sources: sample variation, nonspecific binding, instrumentation, and substrate fluorescence. Variation in the amount of nucleic acid sample captured is accounted for by hybridizing multiple arrays and figures prominently into tests of significance for differential expression experiments. Nonspecific binding is controlled largely by blocking and hybridization conditions and is assessed by evaluating negative control spots. The noise observed from instrumentation can be characterized by measurements of dark noise, but in this experiment, this represents only <5 counts relative to substrate noise levels more than 5× greater than this value. Substrate fluorescence is typically not manipulated because most microarray protocols have been optimized for a few common substrates. For genes with low expression levels, however, substrate fluorescence can be a significant contributor to noise. Changes in expression may not be large enough to overcome the noise in normal substrates, despite the fact these genes may be just as important as high expressors for cellular function.
Utilizing PCs as substrates amplifies the fluorophore intensity relative to the substrate fluorescence intensity and decreases the impact of substrate fluorescence on the measurements. Because the signal-to-noise ratio is enhanced by more than 1 order of magnitude on PC substrates, genes with expression levels that were lower than the noise floor on glass substrates can now be measured on PCs. This allows researchers to preserve the advantageous throughput of microarrays while increasing the sensitivity of their measurements. The practical effect of the PC is to improve the dynamic range of the expression measurements and allow for quantification of low expression genes. These low expression genes are not only detected, as evidenced by the increased number of genes above the SNR threshold, but also changes in the expression of these genes can be observed in the context of statistical testing. The direction of differential expression in these low expression genes is confirmed by sequencing data, which agreed with the microarray analysis for 39 of the 41 genes identified as differentially expressed only on PC microarrays. The capability of the PCs to measure low expression genes is reflected by the differences in average expression intensity between genes that were classified as differentially expressed only on the PCs (Table S-2 in the Supporting Information), with an average intensity of 148 counts for the seed sample genes and an average intensity of 212 counts for the leaf sample genes, compared to genes classified as differentially expressed on glass slides (Table S-1 in the Supporting Information), with an average intensity of 2930 counts for the seed sample genes and 2830 counts for the leaf sample genes. This finding is validated by the sequencing data as well. The 41 genes in Tables S-4 and S-2 in the Supporting Information corresponding to genes detected as differentially expressed on the PC slides had an average RPKM value of 56 in the leaf sample and 67 in the seed sample, whereas the 26 genes detected as differentially expressed on both PC and glass slides had much higher average RPKMs in both the leaf (341 PRKM) and seed (3965 RPKM) samples, respectively. Thus, both microarray and sequencing data suggest the PC can reliably quantify genes with expression levels at least 1 order of magnitude lower than measured with conventional glass microarrays. By expanding the dynamic range of the microarray experiment, the number of genes for which statistically significant changes in expression could be observed improved from 27 to 68 genes, or from 13 to 34% of the genes probed in the experiment. Because the gene expression follows a power law distribution, modest enhancements in the performance of the assay can dramatically increase the number of genes researchers are able to probe in this microarray format. This data thus suggests that the detection capabilities of microarray protocols currently used today can be greatly expanded by substitution of conventional substrates with enhanced fluorescence substrates such as PCs.
The increased SNRs provided by PCs may allow researchers to perform experiments that are currently problematic on glass slides. Because lower amounts of bound sample can be detected with the PC, sample sizes may be reduced to volumes that would be difficult to probe using normal glass substrates. This may be particularly helpful for profiling gene expression in smaller tissue samples or small populations of rare cells such as stem cells. Alternately, the reduction in experimental variation afforded by this substrate may allow researchers to confidently identify differentially expressed genes with fewer replicates, which may also prove useful with small sample sizes or rare cells. This approach is not limited to conventional DNA microarray experiments. Any surface-bound biomolecular assay can be performed on these PCs for improved performance, as is illustrated in previous work with immunoassays.25 This substrate can also potentially be adapted to improve reliability of novel technologies such as next-generation genomic sequencing platforms, since these instruments make extensive use of fluorescent molecules.
CONCLUSION
We have demonstrated that enhanced fluorescence is capable of significantly improving a DNA microarray that probes changes in gene expression between samples. By using a PC substrate with uniform optical characteristics over microscope slide-sized areas, the SNR from microarray spots was increased by an order of magnitude compared to commercial glass substrates. This SNR enhancement translated into a greater number of genes detected above the noise level and allowed for the detection of statistically signficant changes in low expression genes. After evaluating differential expression in soybean trifoliate tissue versus cotyledon tissue, more than twice as many genes were characterized as differentially expressed on the PCs compared to the glass slides, and many of these were validated by high-throughput mRNA sequencing data. Using a PC substrate for microarray experiments thus opens the possibility to interrogate the roles of genes that previously could not be reliably quantified in a high-throughput fashion.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the National Institutes of Health (grant no. GM086382A), the National Science Foundation (grant no. CBET 07-54122), SRU Biosystems, and the Illinois Soybean Association. Any opinions, findings, conclusions, or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Institutes of Health or the National Science Foundation. The authors thank Stephen Schulz, Brenda Hugh, Frank Jackson, and Kurt Albertson at SRU Biosystems for attaching photonic crystal substrates to microscope slides. The authors thank the staff at the Micro and Nanotechnology Laboratory at the University of Illinois at Urbana–Champaign. The authors also thank Sean Bloomfield for bioinformatics assistance and the staff of the Keck Center at the Biotechnology Center at University of Illinois for Illumina sequencing.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Tables S-1 and S-2 list genes found to have statistically significant changes in expression on glass and photonic crystal slides and Tables S-3 and S-4 show corresponding RNA transcript levels. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Kuznetsov VA, Knott GD, Bonner RF. Genetics. 2002;161:1321–1332. doi: 10.1093/genetics/161.3.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ueda HR, Hayashi S, Matsuyama S, Yomo T, Hashimoto S, Kay SA, Hogenesch JB, Iino M. Proc. Natl. Acad. Sci. U.S.A. 2004;101:3765–3769. doi: 10.1073/pnas.0306244101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuo WP, Liu F, Trimarchi J, Punzo C, Lombardi M, et al. Nat. Biotechnol. 2006;24:832–840. doi: 10.1038/nbt1217. [DOI] [PubMed] [Google Scholar]
- 4.t Hoen PAC, Ariyurek Y, Thygesen HH, Vreugdenhil E, Vossen RHAM, de Menezes RX, Boer JM, van Ommen G-JB, den Dunnen JT. Nucleic Acids Res. 2008;36:e141. doi: 10.1093/nar/gkn705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sabanyagam CR, Lakowicz JR. Nucleic Acids Res. 2007;35:e13. doi: 10.1093/nar/gkl1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moal EL, Leveque-Fort S, Potier M-C, Fort E. Nanotechnology. 2009;20:225502. doi: 10.1088/0957-4484/20/22/225502. [DOI] [PubMed] [Google Scholar]
- 7.Guo S-H, Tsai S-J, Kan H-C, Tsai D-H, Zachariah MR, Phaneuf RJ. Adv. Mater. 2008;20:1424–1428. [Google Scholar]
- 8.Rosenblatt D, Sharon A, Friesem AA. IEEE J. Quantum Electron. 1997;33:2038–2059. [Google Scholar]
- 9.Ganesh N, Mathias PC, Zhang W, Cunningham BT. J. Appl. Phys. 2008;103:083104. [Google Scholar]
- 10.Ganesh N, Block ID, Mathias PC, Zhang W, Chow E, Malyarchuk V, Cunningham BT. Opt. Express. 2008;16:21626–21640. doi: 10.1364/oe.16.021626. [DOI] [PubMed] [Google Scholar]
- 11.Neuschafer D, Budach W, Wanke C, Chibout S-D. Biosensors Bioelectron. 2003;18:489–497. doi: 10.1016/s0956-5663(02)00272-5. [DOI] [PubMed] [Google Scholar]
- 12.Budach W, Neuschafer D, Wanke C, Chibout S-D. Anal. Chem. 2003;75:2571–2577. doi: 10.1021/ac026390k. [DOI] [PubMed] [Google Scholar]
- 13.Cunningham B, Lin B, Qiu J, Li P, Pepper J, Hugh B. Sensors Actuators B. 2002;85:219–226. [Google Scholar]
- 14.Mathias PC, Wu H-Y, Cunningham BT. Appl. Phys. Lett. 2009;95:021111. doi: 10.1063/1.3184573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang F, Yen G, Cunningham BT. Opt. Express. 2010;18:11846–11858. doi: 10.1364/OE.18.011846. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez DO, Vodkin LO. BMC Genomics. 2007;8:468. doi: 10.1186/1471-2164-8-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, Smyth GK. Bioinformatics. 2007;23:2700–2707. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- 18.Benjamini Y, Hochberg Y. J. R. Stat. Soc. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 19.Langmead B, Trapnell C, Pop M, Salzberg SL. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, et al. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 21.Mortzazvi A, Williams BA, McCue K, Schaeffer L, Wold B. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 22.Mathias PC, Ganesh N, Zhang W, Cunningham BT. J. Appl. Phys. 2008;103:094320. [Google Scholar]
- 23.Thibaud-Nissen F, Shealy RT, Khanna A, Vodkin LO. Plant Physiol. 2003;132:118–136. doi: 10.1104/pp.103.019968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones SI, Gonzalez DO, Vodkin LO. BMC Genomics. 2010;11:136. doi: 10.1186/1471-2164-11-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mathias PC, Ganesh N, Cunningham BT. Anal. Chem. 2008;80:9013–9020. doi: 10.1021/ac801377k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.