Summary
Supramolecular nanostructures assembled by polymeric amphiphiles have been intensively studied during the last two decades. Such nanocarriers may be engineered to possess on-demand bio-responsitivity for the prevention, diagnosis, and treatment of human diseases. The successful development of several nanoassembly-based polymer therapeutics further encouraged scientists to develop nano-vehicles to achieve controlled release, enhanced efficacy, improved specificity and reduced toxicity. Different from the abundant existing literatures on the hydrophobically or electrostatically driven self-assemblies and their therapeutic applications, this article reviews host-guest interaction mediated nanoassemblies, especially those constructed using cyclodextrins as the host entities. The excellent biocompatibility, complexation capacity, and chemical-sensitivity of cyclodextrin make cyclodextrin-containing polymers attractive to construct host-guest nanoassemblies. Such nanocarriers may be advantageous also because of the broad availability of cyclodextrins, their flexibility for structure/property modulation and their chemical-responsive characteristics.
Keywords: Host-guest interaction, Cyclodextrin, Supramolecular chemistry, Self-assembly, Nano-assembly, Micelle, Vesicle, Drug delivery, Gene therapy, Medical imaging
Introduction
Self-assembled polymer assemblies have been widely studied during the last decade, due to their scientific significance and broad applications. Among their diverse applications, therapeutic delivery for tumor therapy has been intensively investigated in recent years[1–9]. The application of polymer assemblies for drug delivery dates back to 1980s when Ringsdorf et al. first reported the self-assembly of micelles and their solubilizaiton effect on a hydrophobic dye[10]. Subsequent in-depth and systematic studies by a few groups led to the clinical trials of several advanced formulations based on polymeric assemblies [11–13]. Non-covalent interactions such as hydrophobic force enables amphiphilic copolymers self-assemble into multi-morphological supramolecular structures, including spherical micelles, polymersomes, wormlike micelles, multicompartment micelles, toroidal assemblies, cylindrical micelles and helical superstructures [14–21]. Advances in this field suggested the assembling interactions can also be electrostatic, hydrogen-bonding, metal-ligand coordination and stereocomplexation [22–26]. Recently host-guest recognitions have been found to be powerful for fabricating nano-assemblies with advanced functions[27, 28].
According to the definition by Cram et al., host-guest interactions involve a complementary stereoelectronic arrangement of binding sites in host and guest. The host component is defined as an organic molecule or ion whose binding sites converge in the complex, while the guest component’s binding sites diverge in the complex [29]. The intermolecular forces of host-guest systems vary in strength from strong electrostatic interactions, through hydrogen bonding, cation-π, π-π stacking, and metal-ligand binding, to weak van der Waals attractions, and solvent reorganization [30]. The pioneering work in this field was accomplished by Charles Pedersen, Donald Cram and Jean-Marie Lehn[30–32]. Since then, a broad spectrum of organic compounds have been designed and synthesized as molecular hosts, including crown ethers, cavitands, spherands, cryptands, carcerands, calixarenes, cryptophanes, cyclodextrins, steroid clathrates, dendrimers, cucurbiturils and metalloporphyrin[30–39]. These host molecules have been broadly employed to construct multi-functionalized structures and supramolecular devices at all scales. Cyclodextrins (CDs), due to their extremely low toxicity, excellent biocompatibility and good complexation capacity with a variety of lipophilic molecules, are the most widely employed host units to construct assemblies for biomedical applications.
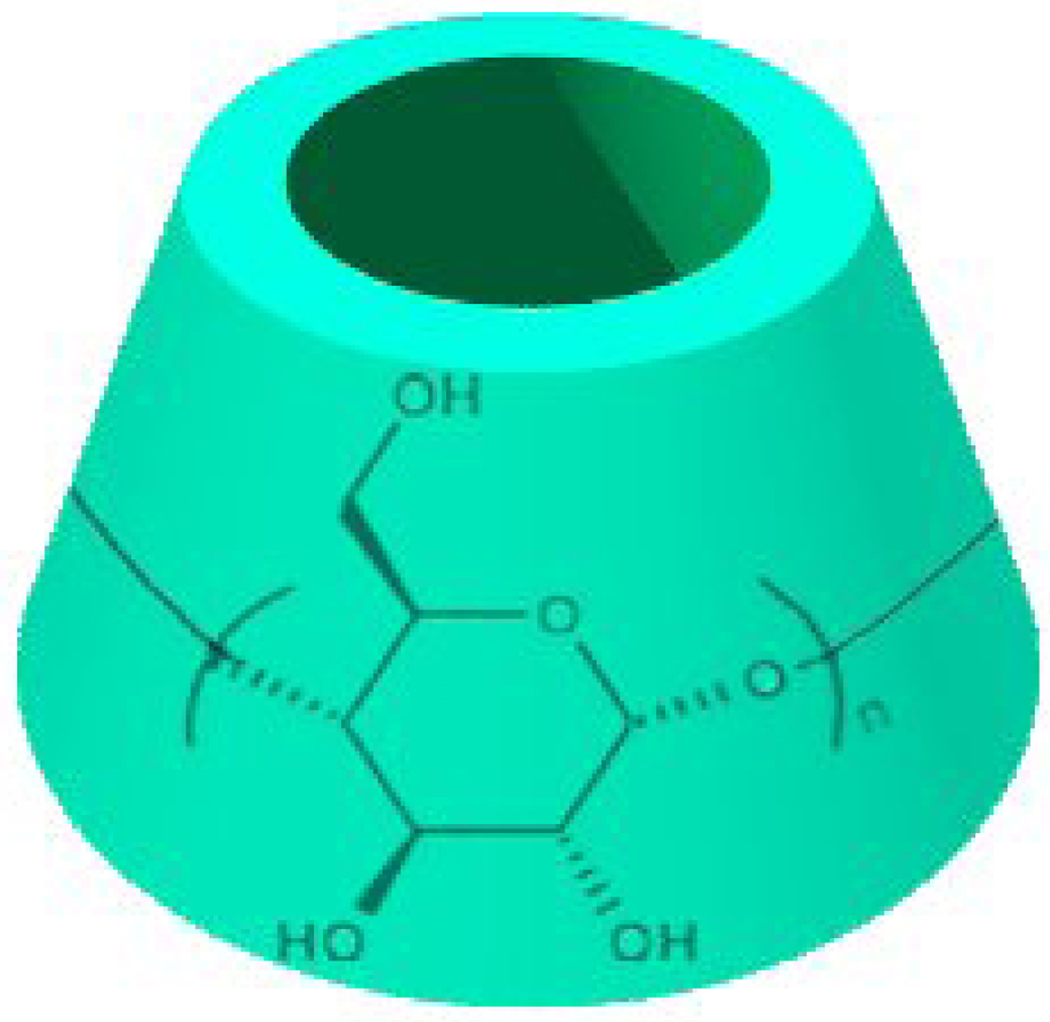
CDs are a family of naturally occurring oligosaccharides constructed from α(1→4)-linked glucose units. The most common members are α-, β-, and γ-CD, which contain 6, 7, and 8 units, respectively. CDs are truncated cone-shaped molecules with a hollow, tapered cavity of 0.79 nm in depth (Fig. 1). The top and bottom diameters of the cavity are 0.47 and 0.53 nm for α-CD, 0.60 and 0.65 nm for β-CD, and 0.75 and 0.83 nm for γ-CD, respectively[40]. Since their discovery by Villiers in 1891[41], these cyclic carbohydrates have been of great interest for scientists in multiple fields[27, 40, 42–48]. Whereas CDs are the mostly extensively studied ones among all potential hosts, the supramolecular structures and potential biomedical applications of CD-based polymers were not extensively investigated until recently. This article reviews the state-of-the-art achievements on host-guest polymer assemblies originated from CD polymers, with emphasis on their construction and applications in the pharmaceutics and biomedicine.
Figure 1.
Chemical structure of cyclodextrins (CDs). n=6, 7, and 8 represents α-, β-, and γ-CD, respectively.
Host-guest nano-assemblies by β-CD containing hydrophilic polymers and hydrophobic guests
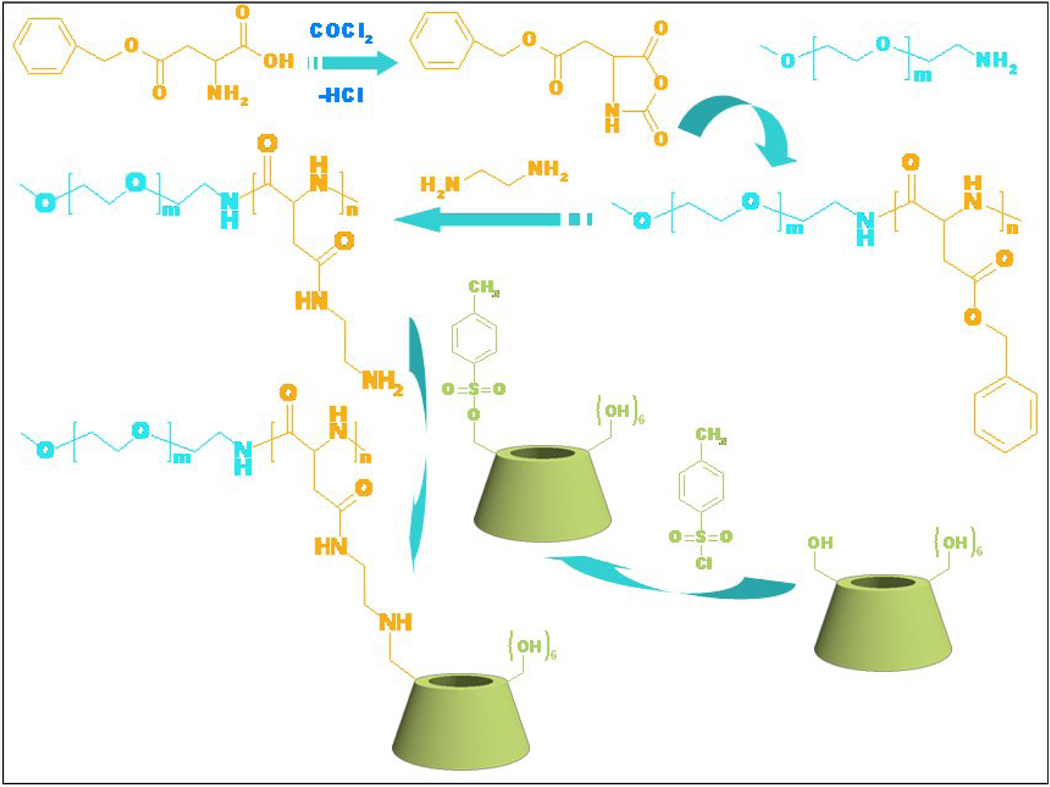
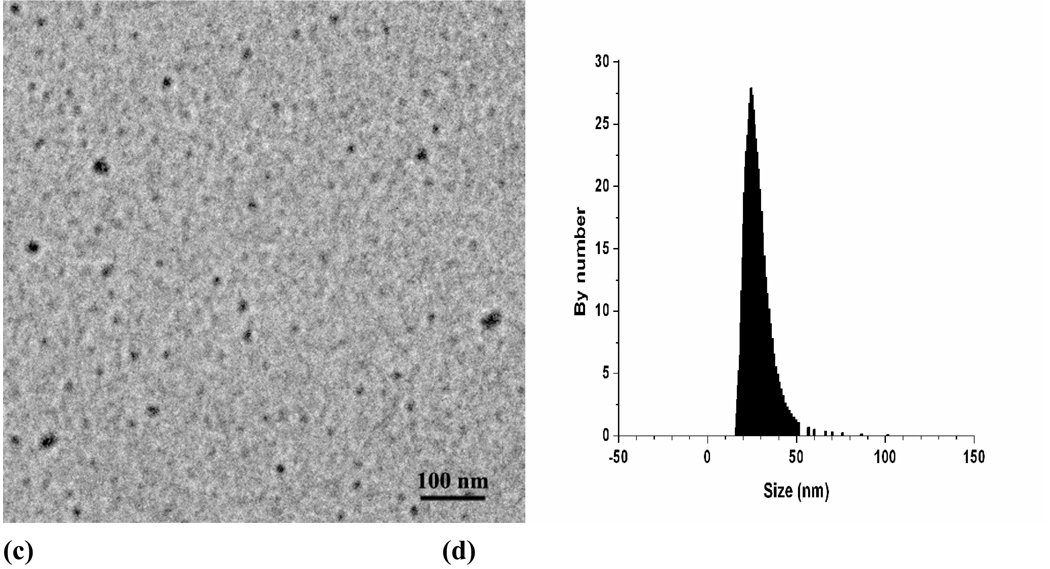
The host-guest interactions of CDs and hydrophobic entities have enabled the CDs and their derivatives to be the most widely employed host molecules functioning as delivery carriers and/or adjuvants for various hydrophobic drugs[45, 49–52]. With the aim to construct core-shell structured nano-assemblies as versatile delivery carriers, a diblock hydrophilic copolymer characterized by tandem alignment of one polyethylene glycol (PEG) block and one block carrying β-CD units on the side chain was employed [53]. The mentioned copolymer, abbreviated as PEG-b-PCD, could be synthesized by a macromolecular substitution reaction as illustrated in Fig. 2. Through optimized reaction conditions, β-CD can be highly efficiently conjugated to form the desired copolymer structure, as confirmed by 1H and 13C NMR spectra. The assembling of PEG-b-PCD in the presence of a guest molecule was initially studied by fluorescence measurements using pyrene as a probe. The significant excimer bands of pyrene molecules in the aqueous solutions of PEG-b-PCD suggested the potential formation of nano-assemblies. Straightforwardly, pyrene-mediated self-assembly was observed by atomic force microscopy (AFM) and transmission electron microscopy (TEM), which indicated the formation of spherical nanoparticles (Fig. 3a–c). The mean size determined by dynamic light scattering (DLS) was about 27 nm (Fig. 3d). A similar guest encapsulation concomitant with nanoparticle formation was found for another hydrophobic molecule, coumarin 102. These results suggested that the copolymer PEG-b-PCD may serve as delivery carrier for hydrophobic drugs. Indeed, detailed encapsulation studies using various drugs from ibuprofen (IBU), indomethacin (IND), dexamethasone (DMS), to rapamycin (RAP) revealed that lipophilic therapeutics of various structures can be efficiently loaded despite their different binding energies with β-CD[54]. This result also indicated that the inclusion capability of PEG-b-PCD copolymer is enhanced compared with free β-CD counterpart. The complexation interactions from multiple β-CD units in one or several polymer chains may have synergistically enhanced solubilization effect on guest molecules. Importantly, the payload in PEG-b-PCD assemblies could be released as proved by in vitro release experiments. The release behaviors of different drugs are mainly associated with their physicochemical properties such as solubility and molecular volume.
Figure 2.
Synthesis of β-CD containing copolymer PEG-b-PCD.
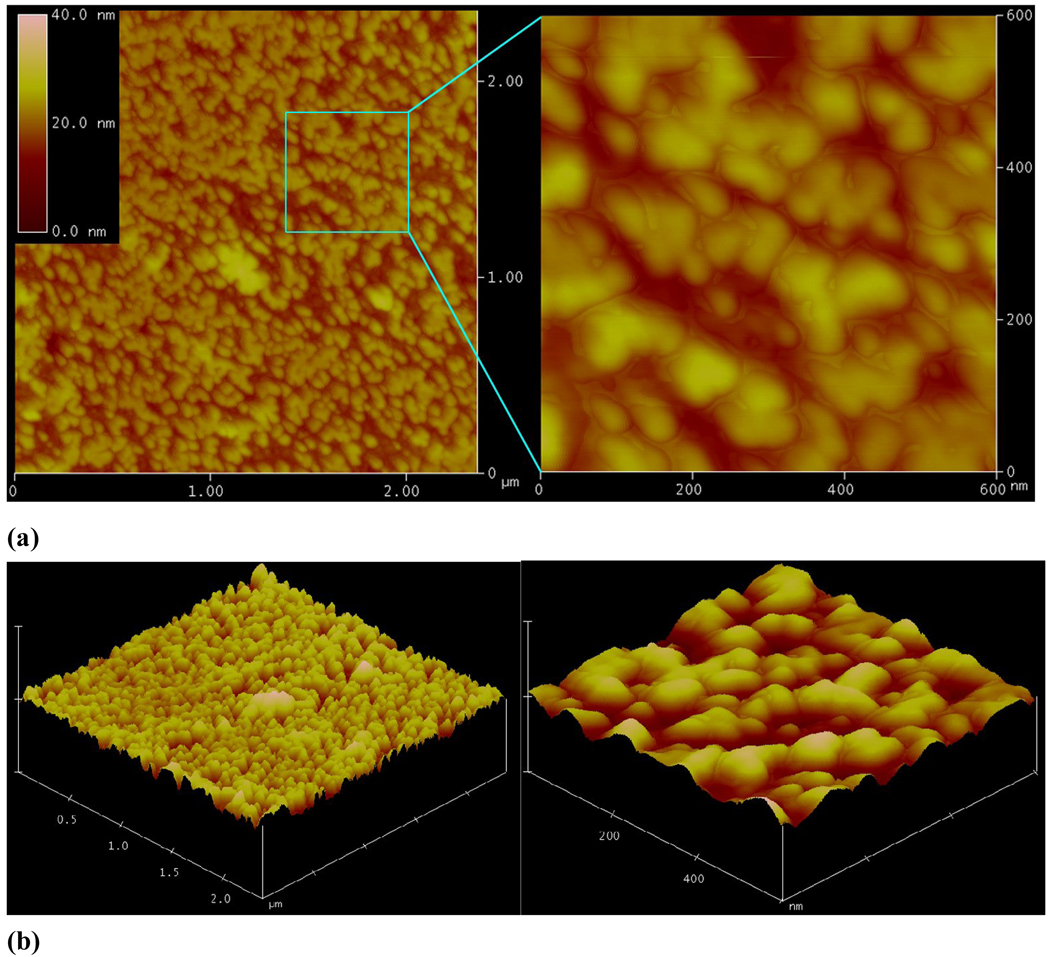
Figure 3.
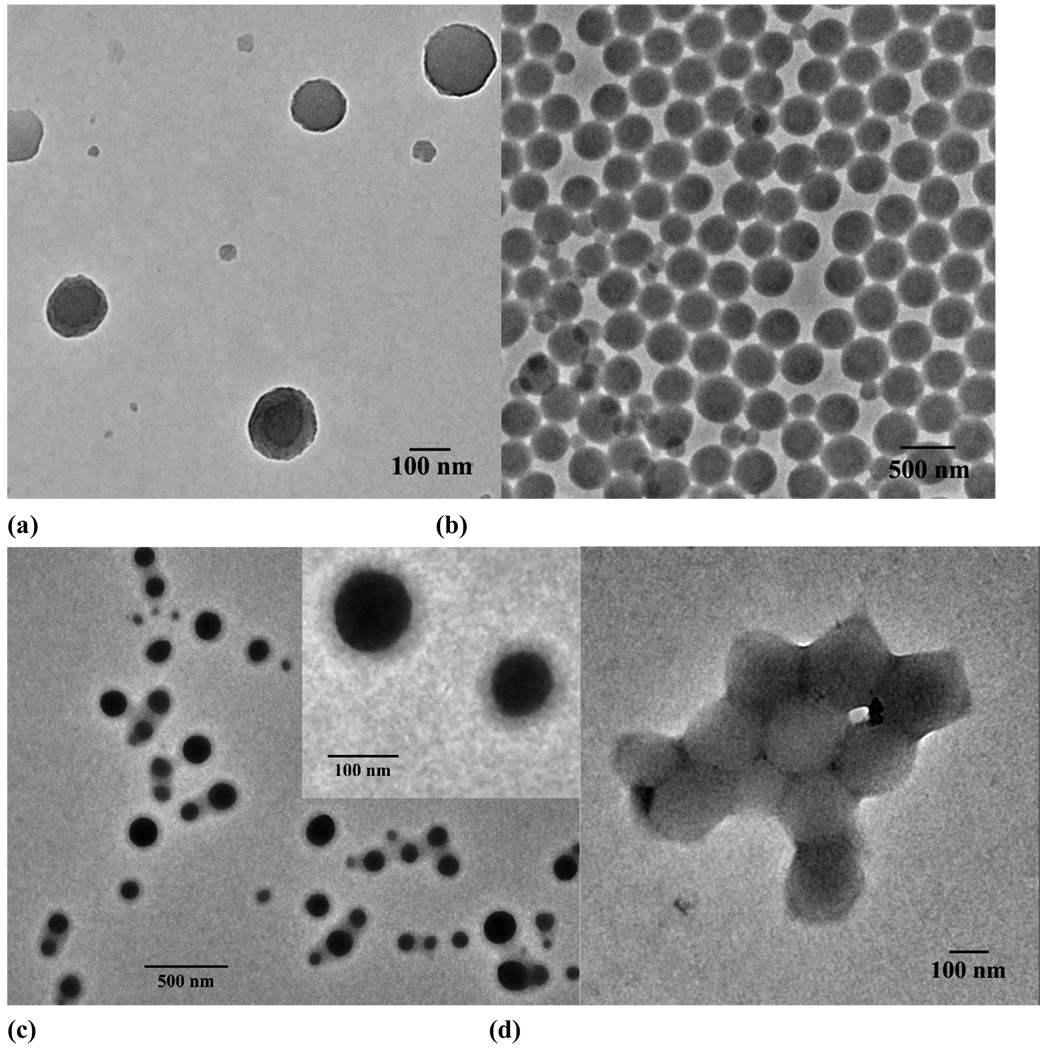
Pyrene mediated self-assembly of PEG-b-PCD. Tapping-mode atomic force microscopy (AFM) images: (a) Height and (b) 3D images. (c) TEM image and (d) size distribution by DLS. Total concentration was 2 mg/ml
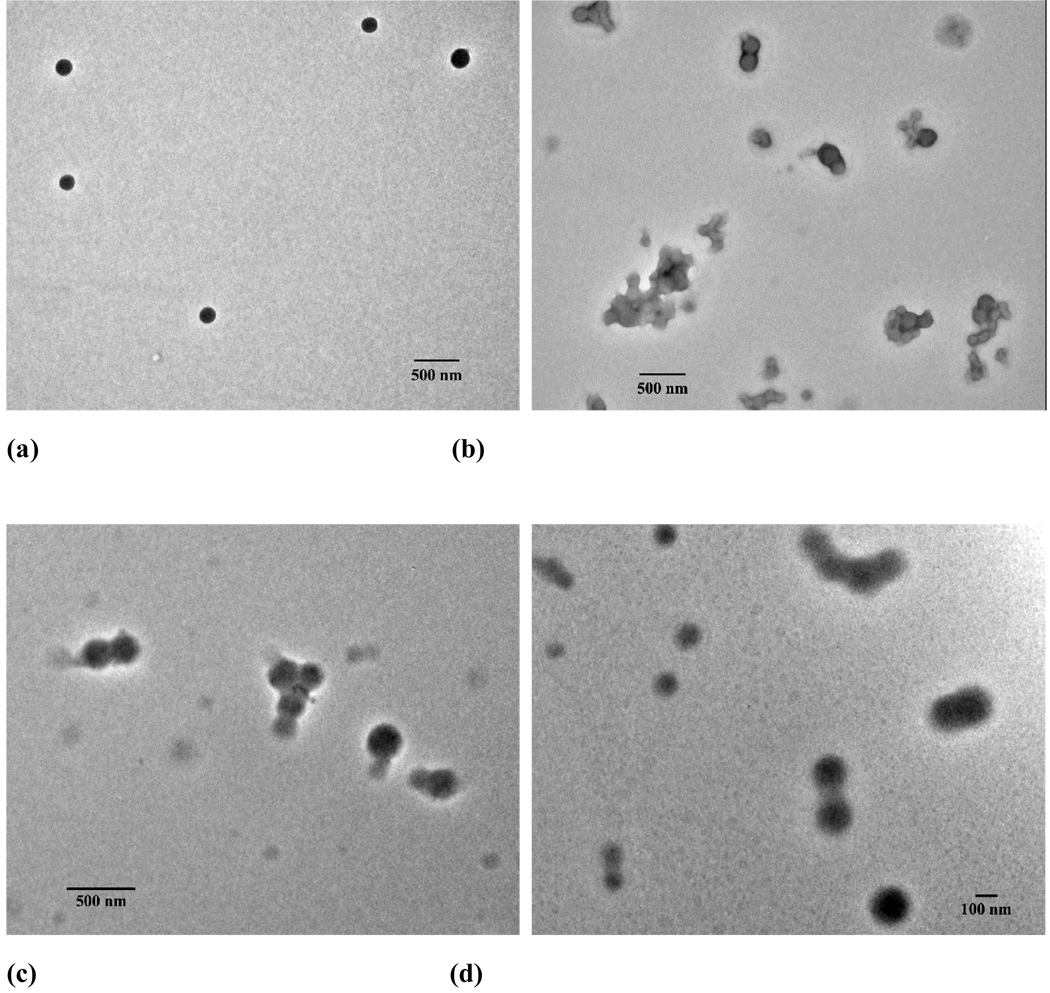
Besides small molecules, hydrophobic polymers were also found to be able to assist the assembly of PEG-b-PCD to form well-defined nanospheres[53]. Using poly(β-benzyl L-aspartate) (PBLA) with a Mn of 2000 as the model guest polymer, assemblies were successfully prepared by dialysis procedure. Spherical assemblies were constructed as observed by TEM (Fig. 4a–b) and SEM. The particle size was controlled by the content of PBLA, with the size increased from 118.7 to 209.2 nm when the feed ratio increased from 1:20 to 8:20. The core-shell structure was characterized by fluorescence and 1H NMR measurements. The rigid core was mainly composed of PBLA chains, while palisade PEG chains constituted the hydrophilic shell, which endows the particles with colloidal stability. A direct TEM observation by staining using phosphotungstic acid (PTA) revealed that the shell thickness was consistent with the chain length of PEG shell (Fig. 4c). Freeze-drying and subsequent reconstitution study demonstrated that the PBLA/PEG-b-PCD assemblies can be well-dispersed after lyophilization. In addition, long-term storage only slightly affected the size and morphology of these types of assembles. While the molecular weight of PBLA almost has no effect on the topology of the resultant nanoparticles, it does influence the particle size. For the assembling formulations with the same feed ratio, smaller size was observed for assemblies based on higher molecular weight PBLA[55]. Also, other hydrophobic polymers such as poly(D, L-lactide) (PDLLA) can function as the polymeric guest to direct the assembling of PEG-b-PCD (Fig. 4d).
Figure 4.
Self-assembly of PEG-b-PCD in the presence of hydrophobic polymers. (a) PBLA: PEG-b-PCD = 1:20, (b) PBLA: PEG-b-PCD = 8:20, (c) PBLA/PEG-b-PCD assemblies staining with PTA, and (d) PDLLA: PEG-b-PCD = 2:20.
Pioneered by Kataoka and Kabanov et al.[56, 57], polyelectrolyte complex (PEC) assemblies have attracted great attention because of their usefulness for the delivery of biomacromolecules such as proteins and polynucleotides. In addition to nanoparticles with hydrophobic microdomains, polyelectrolyte complex assemblies can be fabricated based on host-guest interactions. For this purpose, adamantane carboxylic acid (ADCA) was selected as a guest molecule because the strong inclusion interaction of the adamantyl (Ada) group with β-CD has been well demonstrated[58]. Taking advantage of host-guest interactions between PEG-b-PCD and Ada, a pseudo-polyelectrolyte copolymer with one negatively charged block was prepared. Further electrostatic interaction of this supramolecular polyelectrolyte with polyethylenimine (PEI) resulted in PEC-like assemblies. Nearly spherical assemblies with a mean size of 100.7 nm were observed by TEM. These results demonstrate that PEG-b-PCD copolymers may potentially function as delivery vectors for water-soluble biomacromolecules such as proteins, therapeutic DNA and siRNA.
For all the above mentioned nano-assemblies, the host-guest interactions between β-CD units in PEG-b-PCD copolymer and hydrophobic group in guest compounds are responsible for the self-assembly process. Hydrophobic guest mediated self-assembly has also been observed for other β-CD containing polymers. Gref et al. synthesized β-CD polymers (polyβCD). These polymers can assemble into nano-assemblies of about 200 nm in the presence of hydrophobically modified dextrans [59]. Further tailoring of the physicochemical properties of nano-assmblies such as de-assembling and swelling can be achieved by changing the hydrophobic groups conjugated onto the dextrans[60]. These nanogels can be loaded with hydrophobic compounds, substantiating their potential applications in pharmaceutics as well as cosmetic field[61]. Jiang et al. synthesized a linear homopolymer (PGMA-CD) with flanking β-CDs by free radical polymerization of a β-CD containing acrylate monomer[28]. As evidenced by DLS, TEM and AFM, micellar assemblies (100–300 nm) can form through the host-guest complexation between PGMA-CD and an Ada containing hydrophobic polymer (PtBA-Ada). The micellar surface can be conveniently modified using functional compounds via host-guest interactions. Moreover, shell cross-linking and subsequent core removal resulted in the formation of hollow nano-capsules composed of β-CD containing polymers. These nano-assemblies with β-CD host units are useful for delivering either hydrophobic or hydrophilic compounds. Through a similar procedure, micelles with a biodegradable core of poly(ε-caprolactone) (PCL) were assembled [62]. By a one-step polycondensation of β-CD in the presence of both epichlorohydrin and choline chloride, Xiao and coworkers synthesized a cationic polyβCD[63]. These polymers can load antifungal agents (butylparaben and triclosan) into their lipophilic cavities, as confirmed by 1H-1H gCOSY spectrum. The antibacterial activities against E. coli ATCC 11229 of butylparaben in host-guest assemblies were substantially enhanced. These assembled nano-vehicles are therefore applicable to control bacterial infections.
Temperature-sensitive assemblies via host-guest interactions
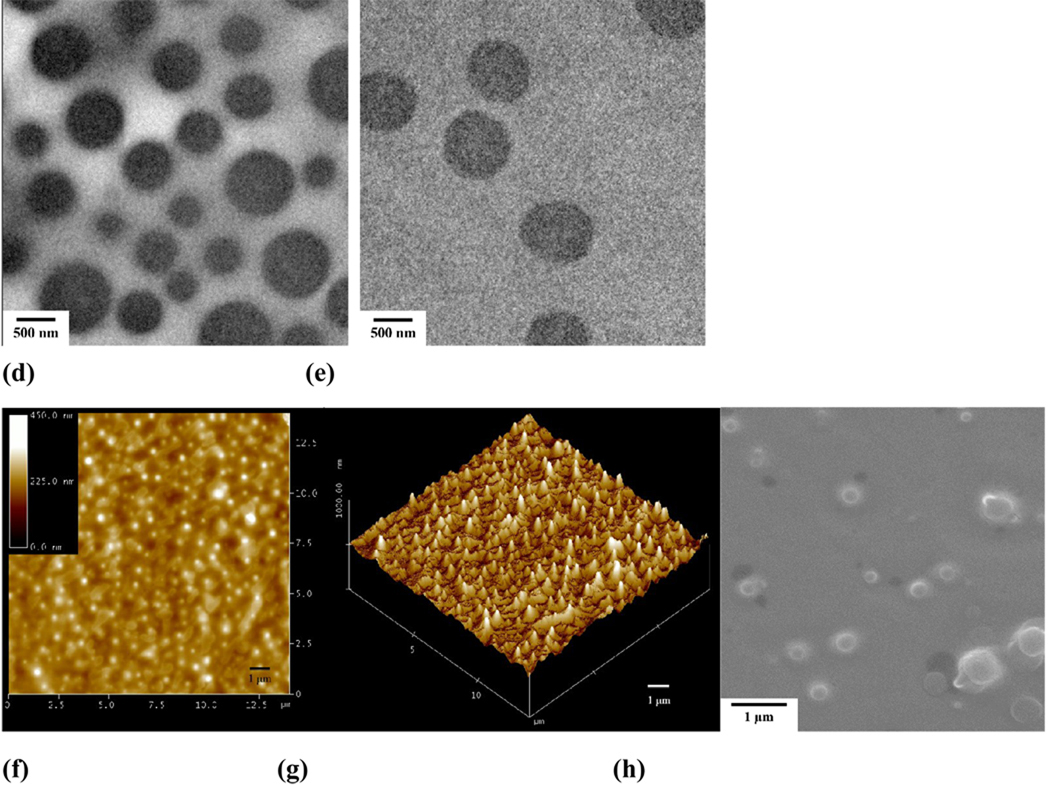
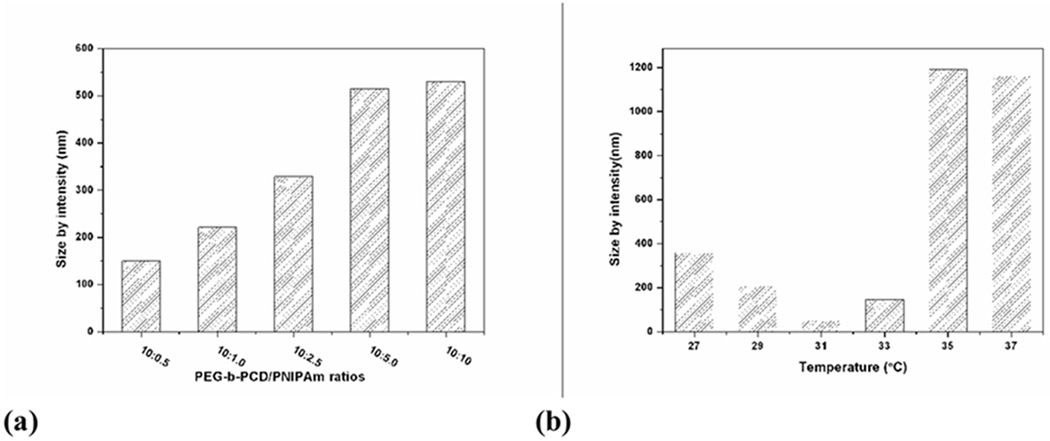
Temperature-responsive assemblies are of particular interest because of their potential applications such as in encapsulation, controlled delivery, gene transfection, imaging, and intelligent switches[64–69]. To construct thermosensitive polymer assemblies, graft or block copolymers with poly(N-isopropylacrylamide) (PNIPAm) as a responsive segment/block are frequently utilized[70]. Subsequent dissolution or dialysis results in the formation of assemblies with temperature-responsive outer shells. When PNIPAm, a well-known thermosensitive polymer was employed as guest polymer, novel temperature-responsive nano-assemblies could be fabricated by a spontaneous assembly process driving by the host-guest interactions between PNIPAm and PEG-b-PCD [71]. Morphological characterization by TEM, AFM and SEM showed the formation of nearly spherical assemblies. The size of the assemblies increased with the increase in the feed ratio of PNIPAm/PEG-b-PCD (Fig. 5a–e and Fig. 6a). 1H NMR and DLS characterizations revealed the thermosensitivity of assembled nanoparticles (Fig. 6b), which is related to the temperature-induced chain shrinkage and subsequent inter-particle aggregation.
Figure 5.
Spontaneous assembly of PNIPAm/PEG-b-PCD. TEM images of assemblies based on PNIPAm and PEG-b-PCD of various weight ratios: (a) 0.5 : 10; (b) 1 : 10; (c) 2.5 : 10; (d) 5 : 10; (e) 10 : 10. Assemblies with PNIPAm/PEG-b-PCD (2.5 : 10): (f) height AFM image, (g) 3D AFM image, and (h) SEM image.
Figure 6.
(a) Average particle size of assemblies based on PNIPAm and PEG-b-PCD of various weight ratios, DLS measurement was performed at 25°C; (b) Temperature-dependent particle size of assemblies with PNIPAm/PEG-b-PCD ratio of 2.5 : 10.
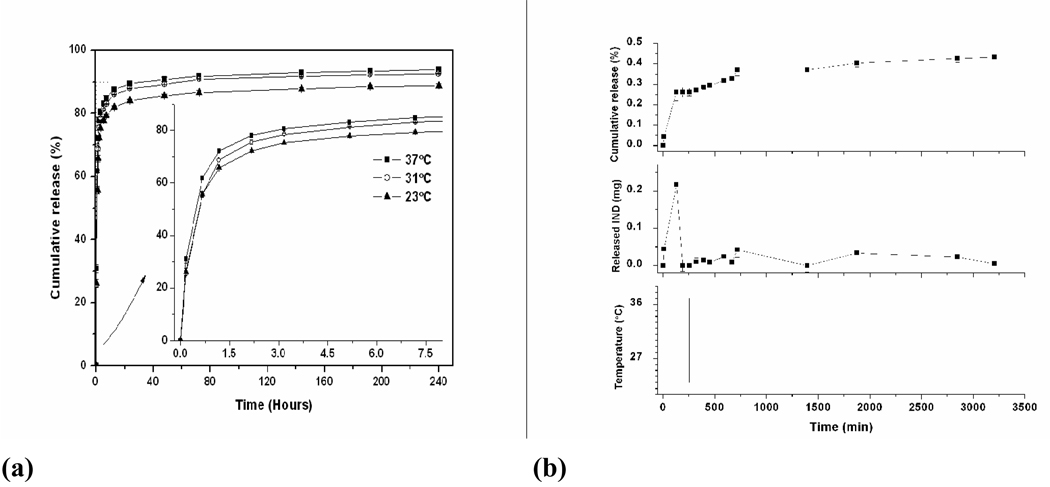
To substantiate the possible application of PNIPAm/PEG-b-PCD assemblies for the controlled drug delivery, indomethacin (IND) was selected as a model drug. A high efficient drug loading was achieved when dialysis was performed to prepare IND-containing assemblies. In vitro release in PBS (0.01 M, pH 7.4) at three different temperatures suggested the drug release could be accelerated by increasing temperature (Fig. 7a). These temperature-dependent release profiles should be related to the thermosensitivity of PNIPAm. In addition, the IND release from PNIPAm/PEG-b-PCD assemblies could be reversibly switched between on and off states in response to temperature through the LCST (Fig. 7b). The release of IND was selectively accelerated by heating the solution above the LCST, while it was decelerated to a certain degree at a temperature below the LCST. The IND release rate was accordingly changed upon temperature switching. Whereas similar release profiles have been reported for polymeric micelles with PNIPAm as shells[72, 73], the aforementioned study demonstrated that temperature modulated release profile can also be achieved through the guest-host assemblies with PNIPAm chains as the cores.
Figure 7.
Temperature-responsive in vitro release behaviors of IND from PNIPAm/PEG-b-PCD (5:10) assemblies. (a) Cumulative release curves at various temperatures, IND loading was 21.2 wt%. (b) temperature-switched release profiles, IND loading was 9.3 wt%.
Chemical-responsive nano-platforms for drug delivery
Stimulus-responsive assemblies have emerged as promising carriers that may enhance the therapeutic efficacy and minimize side effects[74]. Compared with other systems, chemical-sensitive assemblies can more conveniently achieve their responsive function by utilizing the biological or chemical signals that are specific to cells, tissues, organs, or disease sites. Alternatively, the administration of another compound can trigger the release of payload to achieve its therapeutic effect.
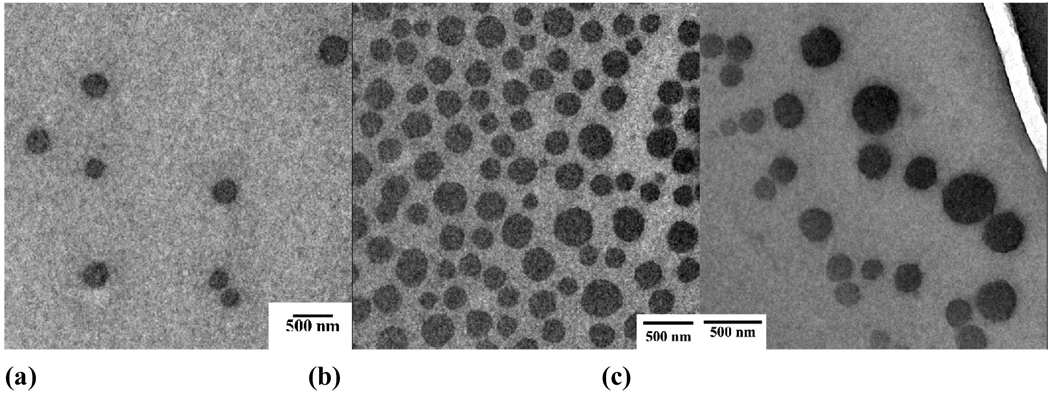
Due to the reversibility of the host-guest interactions between the β-CD and hydrophobic guests, the supramolecular systems based on such non-covalent interactions frequently display chemical sensitivity. Nanocarriers with chemical-sensitivity can be assembled by PEG-b-PCD copolymers in the presence of hydrophobic compounds (either polymers or small molecules). The effect of small molecule stimulants on the PBLA/PEG-b-PCD assemblies was observed by TEM. Small molecules including potassium iodide, benzyl alcohol and cetyltriethylammnonium bromide, were selected as the stimulants. In the presence of a small amount of stimulants, interparticle aggregates were formed (Fig. 8). This chemical-triggered aggregation of the assemblies is likely to be associated with the deshelling effect resulting from the competition between small stimulants and PBLA to complex with β-CD units. This unique characteristic might facilitate the intracellular trafficking of assemblies by weakening the hydration effect of the PEG shells.
Figure 8.
Small molecule stimulants triggered morphology evolution of assemblies based on PBLA/PEG-b-PCD (6:15): (a) control, (b) potassium iodide (0.05 M), (c) benzyl alcohol (0.05 M), (d) cetyltrimethylammonium bromide.
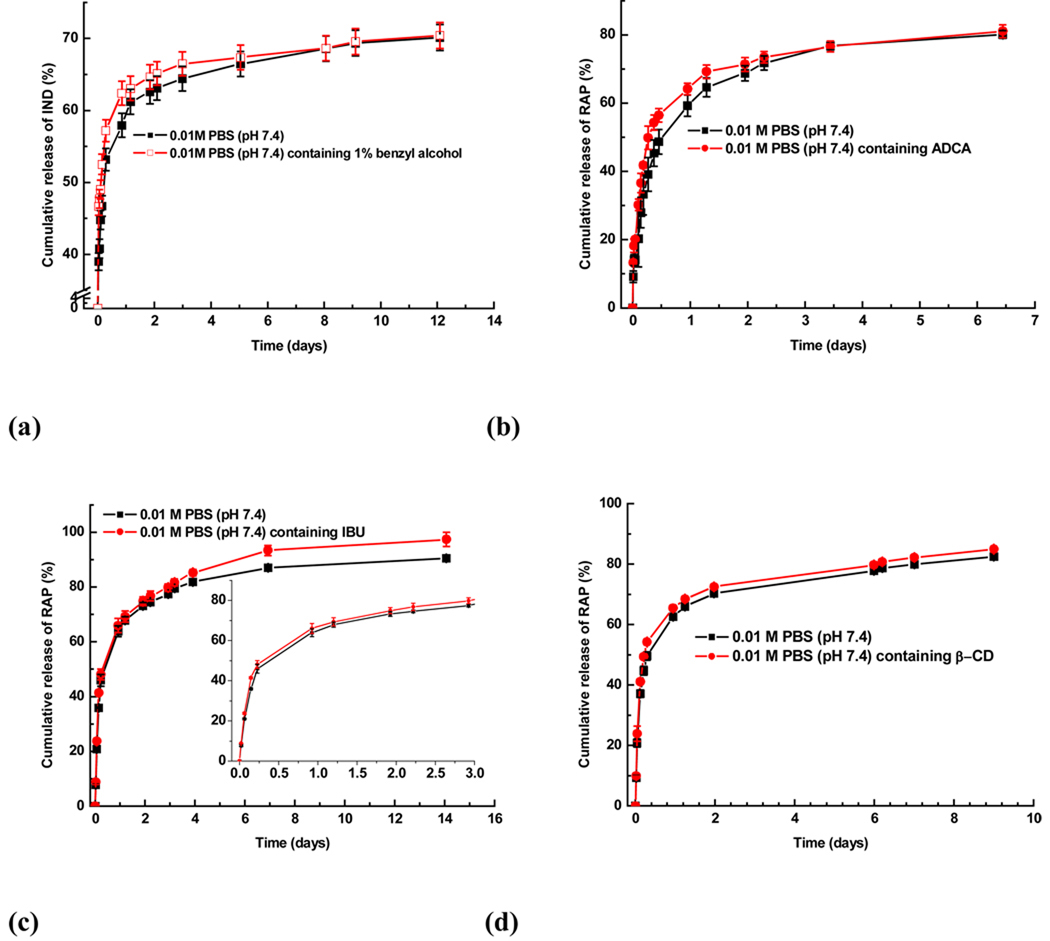
Stimulus-sensitive assemblies can also be formed by small molecules and the aforementioned PEG-b-PCD copolymers[54]. Initially, the chemical sensitivity was demonstrated by fluorescence experiments using pyrene as the probe. A decrease in the excimer intensity of pyrene/PEG-b-PCD system was observed when ADCA or β-CD was added to the aqueous solutions, which indicated the chemical-triggered release of pyrene from assemblies. Two hydrophobic drugs, i.e., IND and RAP, have been selected to substantiate the potential application of these chemical responsive assemblies. In vitro release studies were carried out in the presence of various chemical stimuli, including benzyl alcohol, ADCA, IBU and β-CD (Fig. 9). Independent of drug structure, the release rate in the initial stage could be accelerated by stimulants (benzyl alcohol and ADCA). In addition, an increase in the RAP release was also observed in the presence of another hydrophobic drug IBU. These results indicated that the release of payload from PEG-b-PCD based assemblies can be triggered by the competitive compounds against guest molecules. Free β-CD can serve as a competitor to the host polymer to spring and speed the release of cargo molecules (Fig. 9d).
Figure 9.
Chemical-responsive in vitro release profiles of either IND or RAP containing assemblies based on PEG-b-PCD.
Host-guest nano-carriers for gene delivery
Davis and coworkers synthesized a class of linear, cationic, β-CD-containing polymers (βCDPs) by copolymerizing difunctionalized β-CD monomers with other difunctionalized comonomers[75]. These polymers have low in vitro and in vivo toxicity. Polyplexes with size of <200 nm can be assembled by the condensation of pDNA using βCDPs. Importantly, in vitro transfection efficiency with βCDPs was comparable to those based on PEI (branched, 25 kDa) and Lipofectamine. The transfection activity and cell toxicity are critically related to the structure of this type of carbohydrate polycations[76], mainly the comonomer structure, charge center type as well as the cyclodextrin type and functionalization[77–79]. The colloidal stability in biological fluids can be further increased by surface decoration of the assembled polyplexes using adamantane conjugated PEG (PEG-AD), which is achieved through the host-guest interactions between β-CD and Ada group[80]. Furthermore, a targeting ligand can be introduced onto the PEG shell of assemblies containing gene segments, by using a transferrin-PEG-Ada conjugate [81]. In vivo transfection experiments in murine models and non-human primates as well as humans showed that the ligand functionalized nano-assemblies can efficiently deliver the gene payload to tumor tissues, and perform their therapeutic efficacy after intravenous injections[82–85]. These studies also confirmed the safety of these non-viral nanoparticles for systemic administrations.
Taking advantage of the host-guest interactions between β-CD units and Ada cationic derivatives, Burckbuchler et al. were able to construct a pseudo-polycation functioning as a vector for gene delivery[86]. Gel retardation, ζ-potential as well as surface enhanced Raman spectroscopy (SERS) experiments demonstrated the formation of polyplex assemblies by host-guest polycations and plasmid DNA (pDNA) encoding a luciferase gene. In addition, gel retardation and small angle neutron scattering (SANS) measurements suggested that the stability of assembled polyplexes was mainly related to the chemical structure of guest molecules. The poyplexes based on the polycation of Ada3 and Ada4 were relatively stable under higher NaCl concentrations. In vitro transfection was performed on HepG2 and KEK293 cells. The optimized polyplexes based on Ada4 exhibited a transfection efficiency in the same order of magnitude as that of DOTAP, a commercial transfection agent. Nevertheless, the former transfection was carried out in the presence of the fusogenic peptide JTS-1.
CDs based polypseudorotaxanes (CDPRs) are a family of polymers with CDs threading onto the polymer chains. Since their discoveries by several groups[87–89], there have been high interests in the development of functional polymer materials and delivery systems based on CDPRs [90–95]. Nevertheless, the applications of CDPR polymers for gene delivery only emerged as a new direction recently. Yui et al. synthesized a biocleavable polyrotaxane[96], in which dimethylaminoethyl-modified α-CDs were threaded onto a PEG chain capped with benzyloxycarbonyl tyrosine via disulfide linkages. AFM observation together with gel electrophoresis and ξ-potential measurements confirmed the formation of polyplexes as well as the successful condensation of pDNA. The cleavage of disulfide linkages under reducible conditions in cytosolic milieu facilitates the de-condensation and intracellular trafficking of pDNA cargo. In vitro transfection revealed that the assembled polyplexes can rapidly escape from endosome and deliver pDNA to the nucleus. On the other hand, Li et al. designed cationic CDPRs with multiple oligoethylenimine-grafted CDs threaded on a poly(ethylene oxide)-poly(propylene oxide)-poly(ethylene oxide) (PEO-PPO-PEO) chain [97]. These cationic polyrotaxanes with different oligoethylenimines can efficiently condense pDNA into nanoparticles, which was confirmed by agarose gel electrophoresis, particle size, and ξ-potential measurements. Importantly, these supramolecular polycations are less toxic than PEI (25 K) for cultured L929 and HEK 293 cells. Transfection studies using luciferase as a marker gene in HEK293 showed that the efficiency of assembled polyplexes was even comparable with those of PEI (25K). In addition, Liu et al. reported the condensation of calf thymus DNA by CDPRs with anthryl-modified-β-CDs threading onto polypropylene glycol (PPG)[98]. However, no cytotoxicity and transfection results have been reported for these CDPRs.
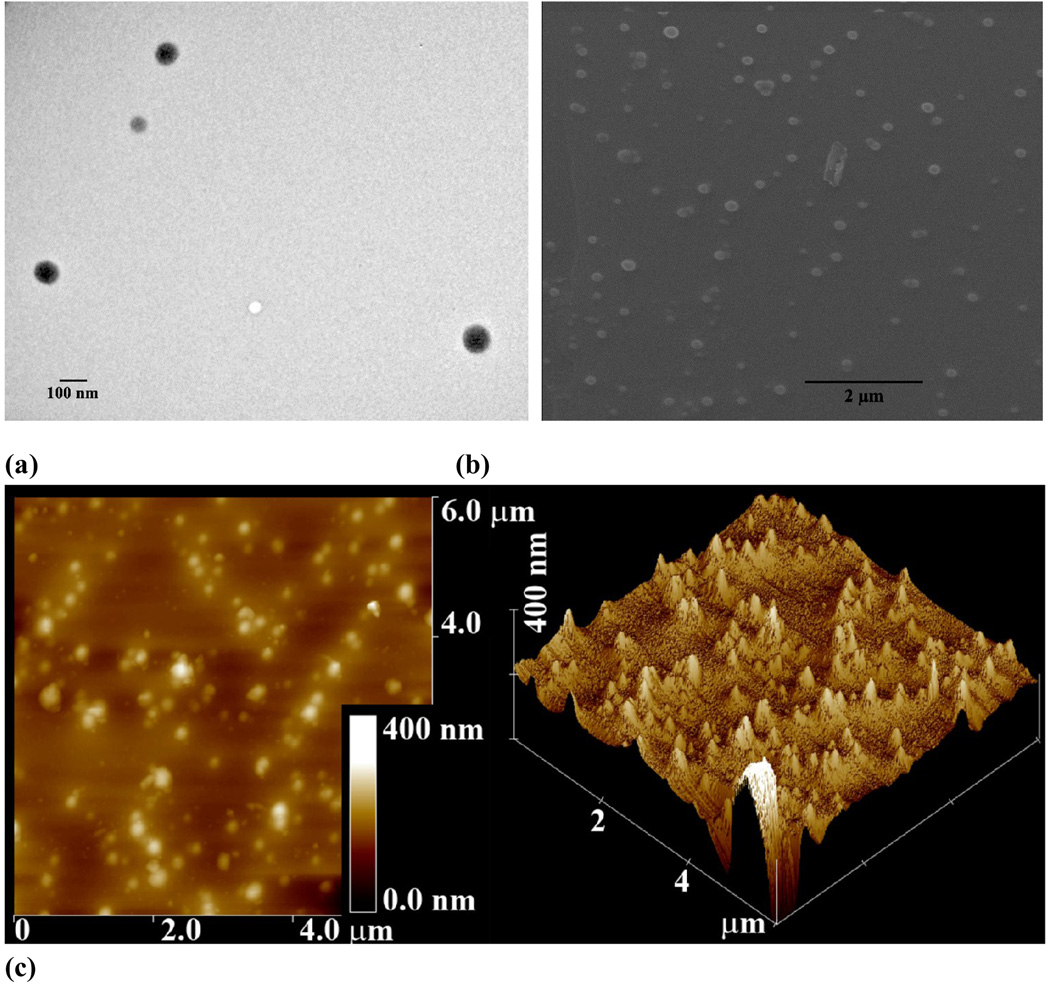
To develop a multifunctional polymer nanocarrier via host-guest interactions for simultaneous drug and gene delivery, β-CDs were conjugated onto branched polyethylenimine (PEI-CD) to synthesize a host polymer[99]. PBLA was used as a macromolecular guest for this host polymer to construct core-shell structured nano-assemblies. The spherical morphology of assemblies was confirmed by TEM, SEM and AFM observations (Fig. 10). The DLS analysis showed a mean size of 90.6 nm. The nature of the core of assemblies was characterized using fluorescence and 1H NMR spectroscopy. Together with the potentiometric titration and fluorescence anisotropy results, the core-shell structure, with rigid PBLA core surrounded by positively charged PEI segments, was proposed. TEM observation confirmed the core-shell structure of the nano-assemblies. Accordingly, the core composed of hydrophobic PBLA chain can accommodate hydrophobic drugs, while the cationic shell can condense pDNA.
Figure 10.
PBLA mediated self-assembly of PEI-CD into well-defined nanoparticles: (a) TEM, (b) SEM, and (c) AFM images.
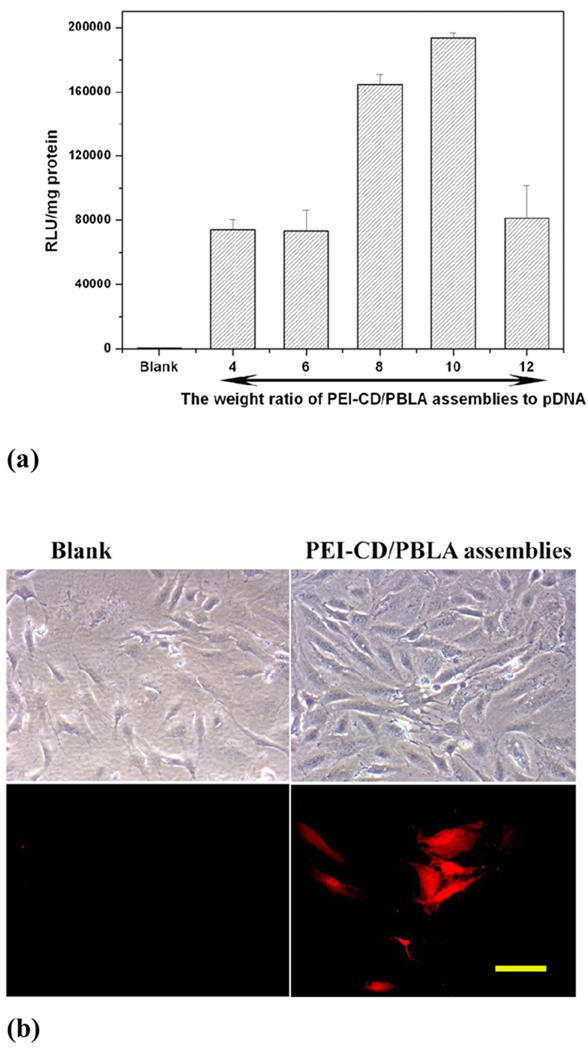
Dexamethasone (DMS), a potent steroidal anti-inflammatory drug can be efficiently loaded into and released from the PEI-CD/PBLA nanoparticles in a sustained manner. Gel retardation combined with EtBr exclusion assays demonstrated that the plasmid DNA (pDNA) could be complexed with PEI-CD/PBLA assemblies to form a condensed structure when the weight ratio was higher than 0.6. Cell culture experiments suggested that pDNA encoding red fluorescence protein can be transfected and expressed in osteoblast cells, using the polyplexes based on the assembled host-guest nanoparticles (Fig. 11). Interestingly, introducing DMS into the polyplexes can increase the transfection efficiency as well as dramatically decrease the cytotoxicity of assemblies. The decreased cytotoxicity was considered to be related to the anti-inflammatory and immunosuppressive activity of DMS that can inhibit proinflammatory cytokines such as tumor necrosis factor α (TNF-α) and antagonize the activation of the NF-κB pathway by direct and indirect mechanisms.
Figure 11.
(a) In vitro transfection of the luciferase gene into osteoblast cells by PBLA/PEI-CD nanoassemblies. Luciferase activity (RLU) was quantified using a luminometer 48 h after cells were transfected. Results were normalized to total cell protein. (b) Transfected cells were viewed by a fluorescence microscope 48 h after transfection. Excitation was performed with green light. Polyplexes with the weight ratio of PEI-CD/PBLA to pDNA of 8 were employed. The scale bar represents 100 µm.
Nanostructures based on other CD polymers via host-guest recognition
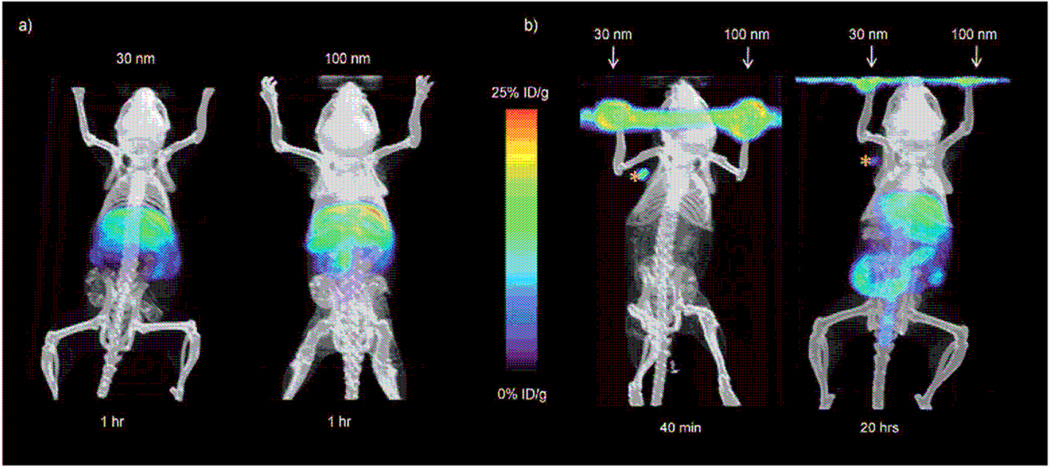
For biomedical imaging, Tseng et al. developed a convenient, flexible, and modular synthetic approach to prepare size-controllable supramolecular nanoparticles (SNPs) [100]. Three building polymers, i.e. Ada grafted polyamidoamine dendrimer (n-Ad-PAMAM), β-CD grafted branched polyethylenimine (CD-PEI), and Ada conjugated PEG (Ad-PEG), were synthesized. Molecular recognition between β-CD building blocks and Ada units in aqueous solutions led to assembled SNPs. Interestingly, the size of SNPs can be conveniently and reversibly modulated from 30 to 450 nm by changing the molar ratio of n-Ad-PEG/CD-PEI. The 64Cu-labeling of SNPs allowed the authors to explore the size-dependent biodistribution and lymph node drainage in mice models by microPET/CT (Fig. 12). Accordingly, SNPs thus assembled showed high potentials for therapeutic delivery and diagnostic imaging.
Figure 12.
MicroPET/CT studies at various times after injecting the mice with 30 and 100 nm 64Cu-labeled SNPs. a) In vivo biodistribution studies of the SNPs by systemically injecting the SNPs into mice through the tail veins. Left panel: 30 nm SNPs; right panel: 100 nm SNPs. b) Lymph node trafficking studies of SNPs using front footpad injections. The 30 and 100 nm SNPs were injected on different sides of footpads of a mouse. MicroPET/CT was carried out for 40 min immediately after injection (left) and 20 h after injection (right). From Ref.[100].
Shi et al. developed a method to construct double hydrophilic block copolymers via non-covalent supramolecular interactions. To this purpose, Ada terminated PNIPAm (Ad-PNIPAM70) and β-CD containing poly(4-vinylpyridine) (β-CD-P4VP30) were synthesized by the reversible addition-fragmentation chain transfer (RAFT) polymerization [101]. In aqueous solutions, a hydrophilic copolymer could be non-covalently formed by the host-guest inclusion between β-CD and Ada, as confirmed by 1H NOESY measurement. The pH and temperature dependent micellization was characterized by fluorescence measurement, DLS, static light scattering (SLS) and TEM. Typically, micelles with size less than 100 nm were assembled, with their morphology (either vesicles or spherical micelles) dependent on pH and temperature.
In addition to constructing 3D polymer nano-assemblies, host-guest interactions mediated assembly has also been demonstrated to be an efficient approach to assemble 2D nano-structures for drug and gene delivery. Most recently, Hammond et al. described a novel approach to fabricate layer-by-layer (LbL) multilayers for small-molecule delivery[102]. To prepare the LbL assemblies, poly(β-amino esters) were used as the degradable polycations, while poly(carboxymethyl-β-cyclodextrin) complexed with a small molecule served as the anionic supramolecular complex. Due to the inclusion capability of β-CD host to different hydrophobic groups, a large number of small-molecule therapeutics can be loaded into the nanoscaled coating by this approach. As shown by the authors, hydrophobic drugs such as ciprofloxacin, flurbiprofen, and diclofenac could be loaded and linear as well as programmable release kinetics can be achieved through hydrolytic top-down degradation. To fabricate a functionalized surface for gene delivery, Pun et al. synthesized β-CD containing PEI, which was then used to prepare pDNA containing polyplexes[103]. These nanoparticles (~100 nm) contain β-CD units on their surface, and therefore can be immobilized onto an Ada functionalized solid substrate via host-guest interactions between β-CD and Ada. The successful immobilization was confirmed by AFM analysis and quantification was performed via fluorescence assay. Despite the absence of biological evaluation, the study provided a specific and high-affinity loading method to functionalize solid surface for the delivery of biomacromolecules.
Concluding remarks
With the successful clinical application of assembled polymeric nano therapeutics for cancer therapy, supramolecular delivery systems are considered the most promising carriers for drug and gene delivery as well as other biomedical applications. This can be attributed to their high biocompatibility, prolonged circulation in bloodstream, broad payload spectrum from small molecules to biomacromolecules, unique size characteristics for tissue permeability and passive targeting as well as cellular endocytosis and organelle translocation, excellent formulation capability, easy tailoring/decoration for active targeting, and facile control of cargo release by the delicate molecular design. Introducing the host CDs into nanocarriers can further enhance the biocompatibility and lower the in vitro/vivo toxicity, and increase the convenience for additional surface functionalization. This has been well proven by the in-depth studies carried out by Davis’s group and other labs[6, 11]. Polymers with host CDs are able to assemble into nano-platforms with diverse characteristics. These polymers make the construction of highly complex superstructures and multifunctional nanosystems possible simply by a combinatorial strategy. For the core-shell structured nano-assemblies derived from the guest molecule mediated self-assembling of CD containing polymers, the core properties can be conveniently tailored by changing the guest component. The delivery profiles can be therefore modulated with great flexibility. Furthermore, chemical triggered biosensing, release or therapy can be easily achieved due to the reversibility of host-guest interactions.
Multi-functionalized systems integrating multimodal imaging, simultaneous imaging and drug/gene delivery [104–107], and carriers armed with the modern biological tools at cellular and molecular levels[108], are even more exciting directions deserving our efforts in the coming years. These highly advanced systems hold great potentials to renovate current pharmaceutics and medicine, achieving ‘personalized medicine’. Due to their unique hosting capability and other desirable characters, cyclodextrins and their derivatives will remain the beloved building units for scientists to develop novel advanced delivery vehicles. However, biologically responsive materials and host molecules with more specificity need to be designed and synthesized or generated by modifying currently available materials. In addition, the development of novel host molecules suitable for biomedical applications is still necessitous to fabricate host-guest assemblies. The cucurbit[n]uril (n=5~10) family has emerged recently as new host compounds with unusual recognition properties such as high affinity, high selectivity, and tunable kinetics of association and dissociation [39, 109]. Nevertheless, few data about the use of cucurbit[n]uril family for constructing nano-assembled systems are available at this moment. Their safety and efficiency remain unclear for drug/gene delivery and other therapeutic applications. Intensive studies are mandatory to address these issues.
Acknowledgements
We gratefully acknowledge the financial support from the NIH (NIDCR DE015384 & DE017689: PXM). JXZ would like to thank the Start-up Funds from the Third Military Medical University.
Biographies

Dr. Jianxiang Zhang received his BS degree in polymer science and engineering from Xi’an Jiaotong University, PR China in 2001. In 2006, he obtained his PhD at Zhejiang University in polymer chemistry and physics. He was a postdoctoral fellow under the guidance of Professor Peter X Ma at the University of Michigan from 2006 to 2009. He took a faculty position in the Department of Pharmaceutics at the Third Military Medical University in 2009. His current research interests include biomaterials, self-assembly and controlled drug delivery. He has published over 30 peer-reviewed journal papers. He serves as a reviewer for 15 scientific journals, including Biomaterials, Chemical Communications, Journal of Materials Chemistry, Macromolecular Rapid Communications, Nanotechnology, Pharmaceutical Research and Soft Matter.

Dr. Peter X. Ma received his BS degree in Polymer Chemistry and Chemical Engineering from Tsinghua University (Beijing, China), and Ph.D. in Polymer Science and Engineering from Rutgers University. He then did his postdoctoral research at MIT and Harvard Medical School on Biomaterials and Tissue Engineering. In 1996, he joined the faculty of the University of Michigan, in the Department of Biologic and Materials Sciences (School of Dentistry), the Department of Biomedical Engineering, and the Macromolecular Science and Engineering Center (College of Engineering). He was elected to Fellow of the American Institute of Medical and Biological Engineering in 2006. He is currently the Richard H Kingery Endowed Collegiate Professor at the University of Michigan (Ann Arbor, MI).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Duncan R. Nat. Rev. Drug Discov. 2003;2:347–360. doi: 10.1038/nrd1088. [DOI] [PubMed] [Google Scholar]
- 2.Discher DE, Ortiz V, Srinivas G, Klein ML, Kim Y, David CA, Cai SS, Photos P, Ahmed F. Prog. Polym. Sci. 2007;32:838–857. doi: 10.1016/j.progpolymsci.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kabanov AV, Gendelman HE. Prog. Polym. Sci. 2007;32:1054–1082. doi: 10.1016/j.progpolymsci.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nat. Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 5.Torchilin VP. Pharm. Res. 2007;24:1–16. doi: 10.1007/s11095-006-9132-0. [DOI] [PubMed] [Google Scholar]
- 6.Davis ME, Chen Z, Shin DM. Nat. Rev. Drug Discov. 2008;7:771–782. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 7.Lee ES, Gao ZG, Bae YH. J. Control. Release. 2008;132:167–170. doi: 10.1016/j.jconrel.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bae Y, Kataoka K. Adv. Drug Deliv. Rev. 2009;61:768–784. doi: 10.1016/j.addr.2009.04.016. [DOI] [PubMed] [Google Scholar]
- 9.Wiradharma N, Zhang Y, Venkataraman S, Hedrick JL, Yang YY. Nano Today. 2009;4:302–317. [Google Scholar]
- 10.Pratten MK, Lloyd JB, Horpel G, Ringsdorf H. Makromol. Chem. 1985;186:725–733. [Google Scholar]
- 11.Heath JR, Davis ME. Annu. Rev. Med. 2008;59:251–265. doi: 10.1146/annurev.med.59.061506.185523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang L, Gu FX, Chan JM, Wang AZ, Langer R, Farokhzad OC. Clin. Pharmacol. Ther. 2008;83:761–769. doi: 10.1038/sj.clpt.6100400. [DOI] [PubMed] [Google Scholar]
- 13.Matsumura Y, Kataoka K. Cancer Sci. 2009;100:572–579. doi: 10.1111/j.1349-7006.2009.01103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Discher DE, Eisenberg A. Science. 2002;297:967–973. doi: 10.1126/science.1074972. [DOI] [PubMed] [Google Scholar]
- 15.Zhang LF, Eisenberg A. Science. 1995;268:1728–1731. doi: 10.1126/science.268.5218.1728. [DOI] [PubMed] [Google Scholar]
- 16.Won YY, Davis HT, Bates FS. Science. 1999;283:960–963. doi: 10.1126/science.283.5404.960. [DOI] [PubMed] [Google Scholar]
- 17.Li ZB, Kesselman E, Talmon Y, Hillmyer MA, Lodge TP. Science. 2004;306:98–101. doi: 10.1126/science.1103350. [DOI] [PubMed] [Google Scholar]
- 18.Pochan DJ, Chen ZY, Cui HG, Hales K, Qi K, Wooley KL. Science. 2004;306:94–97. doi: 10.1126/science.1102866. [DOI] [PubMed] [Google Scholar]
- 19.Cui HG, Chen Z, Zhong S, Wooley KL, Pochan DJ. Science. 2007;317:647–650. doi: 10.1126/science.1141768. [DOI] [PubMed] [Google Scholar]
- 20.Wang XS, Guerin G, Wang H, Wang Y, Manners I, Winnik MA. Science. 2007;317:644–647. doi: 10.1126/science.1141382. [DOI] [PubMed] [Google Scholar]
- 21.Cornelissen JJLM, Fischer M, Sommerdijk NAJM, Nolte RJM. Science. 1998;280:1427–1430. doi: 10.1126/science.280.5368.1427. [DOI] [PubMed] [Google Scholar]
- 22.Kabanov AV, Bronich TK, Kabanov VA, Yu K, Eisenberg A. J. Am. Chem. Soc. 1998;120:9941–9942. [Google Scholar]
- 23.Harada A, Kataoka K. Science. 1999;283:65–67. doi: 10.1126/science.283.5398.65. [DOI] [PubMed] [Google Scholar]
- 24.Nishiyama N, Kataoka K. J. Control. Release. 2001;74:83–94. doi: 10.1016/s0168-3659(01)00314-5. [DOI] [PubMed] [Google Scholar]
- 25.Chen DY, Jiang M. Acc. Chem. Res. 2005;38:494–502. doi: 10.1021/ar040113d. [DOI] [PubMed] [Google Scholar]
- 26.Goh TK, Tan JF, Guntari SN, Satoh K, Blencowe A, Kamigaito M, Qiao GG. Angew. Chem. Int. Ed. 2009;48:8707–8711. doi: 10.1002/anie.200903932. [DOI] [PubMed] [Google Scholar]
- 27.Davis ME, Brewster ME. Nat. Rev. Drug Discov. 2004;3:1023–1035. doi: 10.1038/nrd1576. [DOI] [PubMed] [Google Scholar]
- 28.Wang J, Jiang M. J. Am. Chem. Soc. 2006;128:3703–3708. doi: 10.1021/ja056775v. [DOI] [PubMed] [Google Scholar]
- 29.Kyba EP, Helgeson RC, Madan K, Gokel GW, Tarnowski TL, Moore SS, Cram DJ. J. Am. Chem. Soc. 1977;99:2564–2571. [Google Scholar]
- 30.Cram DJ. Angew. Chem. Int. Ed. 1988;27:1009–1112. [Google Scholar]
- 31.Lehn JM. Angew. Chem. Int. Ed. 1988;27:89–112. [Google Scholar]
- 32.Pedersen CJ. Angew. Chem. Int. Ed. 1988;27:1021–1027. [Google Scholar]
- 33.Boas U, Heegaard PMH. Chem. Soc. Rev. 2004;33:43–63. doi: 10.1039/b309043b. [DOI] [PubMed] [Google Scholar]
- 34.Bohmer V. Angew. Chem. Int. Ed. 1995;34:713–745. [Google Scholar]
- 35.Tashiro K, Aida T. Chem. Soc. Rev. 2007;36:189–197. doi: 10.1039/b614883m. [DOI] [PubMed] [Google Scholar]
- 36.Seel C, Vogtle F. Angew. Chem. Int. Ed. 1992;31:528–549. [Google Scholar]
- 37.Wenz G. Angew. Chem. Int. Ed. 1994;33:803–822. [Google Scholar]
- 38.Wallimann P, Marti T, Furer A, Diederich F. Chem. Rev. 1997;97:1567–1608. doi: 10.1021/cr960373b. [DOI] [PubMed] [Google Scholar]
- 39.Lagona J, Mukhopadhyay P, Chakrabarti S, Isaacs L. Angew. Chem. Int. Ed. 2005;44:4844–4870. doi: 10.1002/anie.200460675. [DOI] [PubMed] [Google Scholar]
- 40.Li S, Purdy WC. Chem. Rev. 1992;92:1457–1470. [Google Scholar]
- 41.Villiers A. Compt. Rend. Acad. Sci. 1891;112:536–538. [Google Scholar]
- 42.Saenger W. Angew. Chem. Int. Ed. 1980;19:344–362. [Google Scholar]
- 43.Villalonga R, Cao R, Fragoso A. Chem. Rev. 2007;107:3088–3116. doi: 10.1021/cr050253g. [DOI] [PubMed] [Google Scholar]
- 44.Takahashi K. Chem. Rev. 1998;98:2013–2033. [Google Scholar]
- 45.Uekama K, Hirayama F, Irie T. Chem. Rev. 1998;98:2045–2076. doi: 10.1021/cr970025p. [DOI] [PubMed] [Google Scholar]
- 46.Breslow R, Dong SD. Chem. Rev. 1998;98:1997–2011. doi: 10.1021/cr970011j. [DOI] [PubMed] [Google Scholar]
- 47.Lipkowitz KB. Chem. Rev. 1998;98:1829–1873. doi: 10.1021/cr9700179. [DOI] [PubMed] [Google Scholar]
- 48.Singh M, Sharma R, Banerjee UC. Biotechnol. Adv. 2002;20:341–359. doi: 10.1016/s0734-9750(02)00020-4. [DOI] [PubMed] [Google Scholar]
- 49.Szente L, Szejtli J. Adv. Drug Deliv. Rev. 1999;36:17–28. doi: 10.1016/s0169-409x(98)00092-1. [DOI] [PubMed] [Google Scholar]
- 50.Brewster ME, Loftsson T. Adv. Drug Deliv. Rev. 2007;59:645–666. doi: 10.1016/j.addr.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 51.Carrier RL, Miller LA, Ahmed I. J. Control. Release. 2007;123:78–99. doi: 10.1016/j.jconrel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 52.Cal K, Centkowska K. Eur. J. Pharm. Biopharm. 2008;68:467–478. doi: 10.1016/j.ejpb.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 53.Zhang JX, Ma PX. Angew. Chem. Int. Ed. 2009;48:964–968. doi: 10.1002/anie.200804135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang JX, Ellsworth K, Ma PX. J. Control. Release. 2010;145:116–123. doi: 10.1016/j.jconrel.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang JX, Ma PX. 2010 To be submitted. [Google Scholar]
- 56.Harada A, Kataoka K. Macromolecules. 1995;28:5294–5299. [Google Scholar]
- 57.Kabanov AV, Bronich TK, Kabanov VA, Yu K, Eisenberg A. Macromolecules. 1996;29:6797–6802. [Google Scholar]
- 58.Rekharsky MV, Inoue Y. Chem. Rev. 1998;98:1875–1917. doi: 10.1021/cr970015o. [DOI] [PubMed] [Google Scholar]
- 59.Gref R, Amiel C, Molinard K, Daoud-Mahammed S, Sebille B, Gillet B, Beloeil JC, Ringard C, Rosilio V, Poupaert J, Couvreur P. J. Control. Release. 2006;111:316–324. doi: 10.1016/j.jconrel.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 60.Layre AM, Wintgens V, Gosselet NM, Dalmas F, Amiel C. Eur. Polym. J. 2009;45:3016–3026. [Google Scholar]
- 61.Daoud-Mahammed S, Couvreur P, Bouchemal K, Cheron M, Lebas G, Amiel C, Gref R. Biomacromolecules. 2009;10:547–554. doi: 10.1021/bm801206f. [DOI] [PubMed] [Google Scholar]
- 62.Ren SD, Chen DY, Jiang M. J. Polym. Sci. Part A: Polym. Chem. 2009;47:4267–4278. [Google Scholar]
- 63.Guan Y, Qian L, Xiao H. Macromol. Rapid Commun. 2007;28:2244–2248. [Google Scholar]
- 64.Kurisawa M, Yokoyama M, Okano T. J. Control. Release. 2000;69:127–137. doi: 10.1016/s0168-3659(00)00297-2. [DOI] [PubMed] [Google Scholar]
- 65.Arotçaréna M, Heise B, Ishaya S, Laschewsky A. J. Am. Chem. Soc. 2002;124:3787–3793. doi: 10.1021/ja012167d. [DOI] [PubMed] [Google Scholar]
- 66.Kabanov AV, Batrakova EV, Alakhov VY. J. Control. Release. 2002;82:189–212. doi: 10.1016/s0168-3659(02)00009-3. [DOI] [PubMed] [Google Scholar]
- 67.Rodriguez-Hernandez J, Checot F, Gnanou Y, Lecommandoux S. Prog. Polym. Sci. 2005;30:691–724. [Google Scholar]
- 68.Han HD, Choi MS, Hwang T, Song CK, Seong H, Kim TW, Choi HS, Shin BC. J. Pharm. Sci. 2006;95:1909–1917. doi: 10.1002/jps.20646. [DOI] [PubMed] [Google Scholar]
- 69.Li YY, Cheng H, Zhang ZG, Wang C, Zhu JL, Liang Y, Zhang KL, Cheng SX, Zhang XZ, Zhuo RX. ACS Nano. 2008;2:125–133. doi: 10.1021/nn700145v. [DOI] [PubMed] [Google Scholar]
- 70.Wei H, Cheng SX, Zhang XZ, Zhuo RX. Prog. Polym. Sci. 2009;34:893–910. [Google Scholar]
- 71.Zhang JX, Feng K, Cuddihy M, Kotov NA, Ma PX. Soft Matter. 2010;6:610–617. doi: 10.1039/c000898b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chung JE, Yokoyama M, Yamato M, Aoyagi T, Sakurai Y, Okano T. J. Control. Release. 1999;62:115–127. doi: 10.1016/s0168-3659(99)00029-2. [DOI] [PubMed] [Google Scholar]
- 73.Chung JE, Yokoyama M, Okano T. J. Control. Release. 2000;65:93–103. doi: 10.1016/s0168-3659(99)00242-4. [DOI] [PubMed] [Google Scholar]
- 74.Motornov M, Roiter Y, Tokarev I, Minko S. Prog. Polym. Sci. 2010;35:174–211. [Google Scholar]
- 75.Gonzalez H, Hwang SJ, Davis ME. Bioconjugate Chem. 1999;10:1068–1074. doi: 10.1021/bc990072j. [DOI] [PubMed] [Google Scholar]
- 76.Hwang SJ, Bellocq NC, Davis ME. Bioconjugate Chem. 2001;12:280–290. doi: 10.1021/bc0001084. [DOI] [PubMed] [Google Scholar]
- 77.Reineke TM, Davis ME. Bioconjugate Chem. 2003;14:247–254. doi: 10.1021/bc025592k. [DOI] [PubMed] [Google Scholar]
- 78.Reineke TM, Davis ME. Bioconjugate Chem. 2003;14:255–261. doi: 10.1021/bc025593c. [DOI] [PubMed] [Google Scholar]
- 79.Popielarski SR, Mishra S, Davis ME. Bioconjugate Chem. 2003;14:672–678. doi: 10.1021/bc034010b. [DOI] [PubMed] [Google Scholar]
- 80.Mishra S, Webster P, Davis ME. Eur. J. Cell Biol. 2004;83:97–111. doi: 10.1078/0171-9335-00363. [DOI] [PubMed] [Google Scholar]
- 81.Bartlett DW, Davis ME. Bioconjugate Chem. 2007;18:456–468. doi: 10.1021/bc0603539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pun SH, Tack F, Bellocq NC, Cheng JJ, Grubbs BH, Jensen GS, Davis ME, Brewster M, Janicot M, Janssens B, Floren W, Bakker A. Cancer Biol. Ther. 2004;3:641–650. doi: 10.4161/cbt.3.7.918. [DOI] [PubMed] [Google Scholar]
- 83.Hu-Lieskovan S, Heidel JD, Bartlett DW, Davis ME, Triche TJ. Cancer Res. 2005;65:8984–8992. doi: 10.1158/0008-5472.CAN-05-0565. [DOI] [PubMed] [Google Scholar]
- 84.Heidel JD, Yu ZP, Liu JYC, Rele SM, Liang YC, Zeidan RK, Kornbrust DJ, Davis ME. Proc. Natl. Acad. Sci. U. S. A. 2007;104:5715–5721. doi: 10.1073/pnas.0701458104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Davis ME, Zuckerman JE, Choi CHJ, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Burckbuchler V, Wintgens V, Leborgne C, Lecomte S, Leygue N, Scherman D, Kichler A, Amiel C. Bioconjugate Chem. 2008;19:2311–2320. doi: 10.1021/bc800070f. [DOI] [PubMed] [Google Scholar]
- 87.Ogata N, Sanui K, Wada J. J. Polym. Sci. Polym. Lett. Ed. 1976;14:459–462. [Google Scholar]
- 88.Harada A, Kamachi M. Macromolecules. 1990;23:2821–2823. [Google Scholar]
- 89.Wenz G, Keller B. Angew. Chem. Int. Ed. 1992;31:197–199. [Google Scholar]
- 90.Harada A, Hashidzume A, Takashima Y. Adv. Polym. Sci. 2006;201:1–43. [Google Scholar]
- 91.Huang FH, Gibson HW. Prog. Polym. Sci. 2005;30:982–1018. [Google Scholar]
- 92.Araki J, Ito K. Soft Matter. 2007;3:1456–1473. doi: 10.1039/b705688e. [DOI] [PubMed] [Google Scholar]
- 93.Loethen S, Kim JM, Thompson DH. Polym. Rev. 2007;47:383–418. [Google Scholar]
- 94.Li J, Loh XJ. Adv. Drug Deliv. Rev. 2008;60:1000–1017. doi: 10.1016/j.addr.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 95.Li J. Adv. Polym. Sci. 2009;222:79–112. [Google Scholar]
- 96.Ooya T, Choi HS, Yamashita A, Yui N, Sugaya Y, Kano A, Maruyama A, Akita H, Ito R, Kogure K, Harashima H. J. Am. Chem. Soc. 2006;128:3852–3853. doi: 10.1021/ja055868+. [DOI] [PubMed] [Google Scholar]
- 97.Li J, Yang C, Li HZ, Wang X, Goh SH, Ding JL, Wang DY, Leong KW. Adv. Mater. 2006;18:2969–2974. [Google Scholar]
- 98.Liu Y, Yu L, Chen Y, Zhao YL, Yang H. J. Am. Chem. Soc. 2007;129:10656–10657. doi: 10.1021/ja073882b. [DOI] [PubMed] [Google Scholar]
- 99.Zhang JX, Sun HL, Ma PX. ACS Nano. 2010;4:1049–1059. doi: 10.1021/nn901213a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang H, Wang ST, Su HL, Chen KJ, Armijo AL, Lin WY, Wang YJ, Sun J, Kamei KI, Czernin J, Radu CG, Tseng HR. Angew. Chem. Int. Ed. 2009;48:4344–4348. doi: 10.1002/anie.200900063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zeng JG, Shi KY, Zhang YY, Sun XH, Zhang BL. Chem. Commun. 2008:3753–3755. doi: 10.1039/b806858e. [DOI] [PubMed] [Google Scholar]
- 102.Smith RC, Riollano M, Leung A, Hammond PT. Angew. Chem. Int. Ed. 2009;48:8974–8977. doi: 10.1002/anie.200902782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Park IK, von Recum HA, Jiang SY, Pun SH. Langmuir. 2006;22:8478–8484. doi: 10.1021/la061757s. [DOI] [PubMed] [Google Scholar]
- 104.Torchilin VP. Adv. Drug Deliv. Rev. 2006;58:1532–1555. doi: 10.1016/j.addr.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 105.Suh WH, Suh YH, Stucky GD. Nano Today. 2009;4:27–36. [Google Scholar]
- 106.Park K, Lee S, Kang E, Kim K, Choi K, Kwon IC. Adv. Funct. Mater. 2009;19:1553–1566. [Google Scholar]
- 107.Kim J, Piao Y, Hyeon T. Chem. Soc. Rev. 2009;38:372–390. doi: 10.1039/b709883a. [DOI] [PubMed] [Google Scholar]
- 108.Hoffman AS. J. Control. Release. 2008;132:153–163. doi: 10.1016/j.jconrel.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 109.Lee JW, Samal S, Selvapalam N, Kim HJ, Kim K. Acc. Chem. Res. 2003;36:621–630. doi: 10.1021/ar020254k. [DOI] [PubMed] [Google Scholar]