Abstract
Cell polarity is involved in many biological functions such as development, wound healing and immune responses. In human neutrophils, polarization is characterized by the translocation of distinct sets of signaling molecules to opposite ends of the cell and the rapid rearrangement of cytoskeleton to initiate migration. While many image-based studies have described cellular morphology and the intensity level of polarity signaling molecules, systematic characterization of the spatial distribution of polarity signaling molecules has been lacking. Here we designed a collection of analytical features to quantify spatial phenotypes of polarity molecules. We compared our features to commonly used polarity readouts and found that they captured additional aspects of the polarization dynamics that were not contained in the existing features. Our work provides a starting point to identify informative features for the study of neutrophil polarization.
Index Terms: cell polarity, neutrophils, fluorescence imaging, image processing, feature extraction
1. INTRODUCTION
Cell polarity is essential for many biological processes implicated in development, axon guidance, tissue repairs and innate immunity. Leucocytes such as neutrophils can respond to external cues by initiating a cascade of signaling events that involve fast production of actin networks, change of cell shape and the development of polarity [1–6]. Polarized neutrophils are characterized by an actin-enriched leading edge that enables protrusion at the front and a myosin-based trailing edge that facilitates detachment at the back (Figure 1a). Coordination between the dynamic reorganization of signaling molecules and cytoskeletal changes allows neutrophils to move toward the invading pathogens, a process known as chemotaxis, and eliminate them from the host system. Recent advances in time-lapse video microscopy and fluorescence microscopy have allowed observation of the intracellular location of signaling proteins in chemotactic human neutrophils [1–6]. A large body of work has been devoted to devising numerical features to describe cellular phenotypes such as morphology, and the expression level, subcellular localization (e.g. nuclear, cytosolic or membrane regions), and texture of fluorescently stained proteins [7–12]. However, these features are not specifically designed to capture polarity phenotypes, such as the width, depth, location and size of actin and myosin-enriched regions, which could help us understand the development of front and back during polarization. Quantitative characterization of spatial polarity remains a challenge, largely due to the difficulty of translating visually interpretable patterns into accurate mathematical quantities. In this work, we designed a collection of intuitive, geometry-based features and showed that they captured visually distinct spatial polarity phenotypes. We found that the dynamic trends of our proposed features were consistent with previous biological observations and provided additional information on the dynamics of neutrophil polarization that were not contained within commonly used polarity readouts.
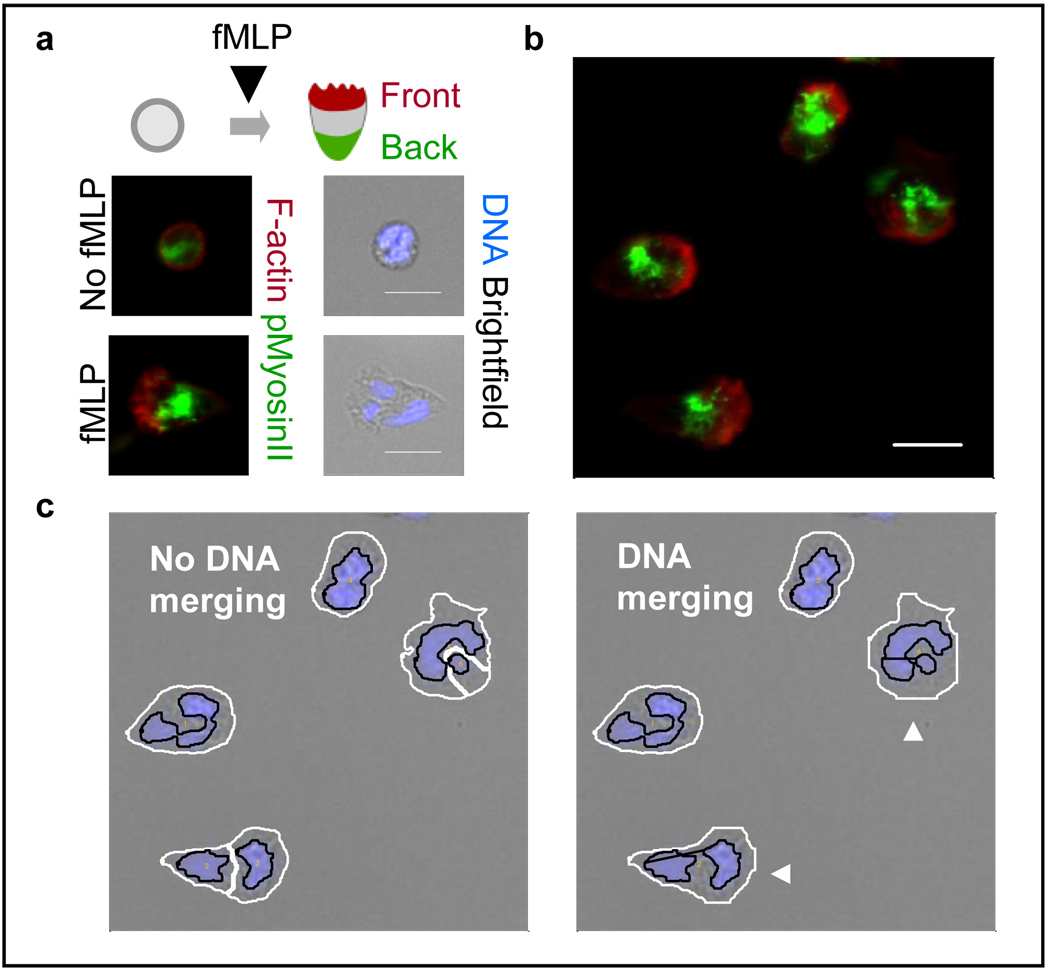
Figure 1. Fluorescence image processing allows visualization of polarity phenotypes in fMLP-stimulated human neutrophils.
a. fMLP induces morphological changes and formation of spatial polarity of F-actin and pMyosinII. b. Neutrophils show polarized morphology and spatial distribution of signaling molecules (10nM fMLP, 3min after stimulation). The protruding front is enriched in F-actin (red) while the contractile back is enriched in pMyosinII (green). Scale bars: 10µm. c. Customized image segmentation algorithm prevents error due to multilobal nuclear morphology. Shown are DNA (blue) and brightfield channels of the same cells as in part b. White/black lines: cellular/nuclear boundaries. Arrowheads: cells correctly segmented after DNA merging.
2. EXPERIMENTAL SETUP
2.1. Polarization assay and image processing
Human neutrophils were isolated from a single healthy donor’s blood [13] and seeded onto 96-well glass bottom imaging plates. The chemoattractant fMLP, a synthetic peptide that mimics the activity of bacterially-derived peptides, was then added to uniformly stimulate cells. We fixed cells at 0, 1, 3 and 5min after stimulation, respectively, to build a snapshot of the polarization process. We fluorescently stained two polarity signaling molecules, filamentous actin (F-actin) for the front and phosphorylated myosin II (pMyosinII) for the back, and also the cellular DNA for cell identification (Figure 1). Images were acquired using BD Bioimager 855 with 40× objective lens. The rolling-ball background subtraction [14] was applied to remove uneven background illumination in the images.
For cell segmentation, we noticed that human neutrophils had multilobal nuclei that could occupy disjoint regions in a 2D plane, thus causing segmentation errors. We developed a modified version of a previously reported watershed-based segmentation algorithm [12] that merged adjacent cellular regions if their nuclei were not excessively far apart and if both the coalesced DNA and cellular areas were below a certain physiological threshold (Figure 1c).
3. SPATIAL SIGNALING PHENOTYPES
3.1. Spatial tightness (ST)
Spatial tightness among a group of pixels measured their “spread” in space. We computed this quantity among the brightest pixels since we were interested in measuring the size of the F-actin/pMyosinII-enriched areas where active signaling occurred. We defined Tα as the set of pixels in the cellular region C whose intensity was in the top α% (α=10 in our study) and Dα, DC as the average pairwise pixel distances in Tα and C, respectively. The spatial tightness ST of a marker was defined as in (1):
| (1) |
3.2. Angular tightness (AT)
Angular tightness was designed to measure the lateral “width” of the area occupied by a group of pixels (e.g. Tα). To facilitate its computation, we first transformed the pixel intensity map into polar coordinates so that the cell center had radius ρ=0 while pixels on the cell contour had radius ρ=1 (Figure 2a). We determined the minimal aperture θα measured at the cell center (in radians) that covered 90% of Tα. The angular tightness was defined as:
| (2) |
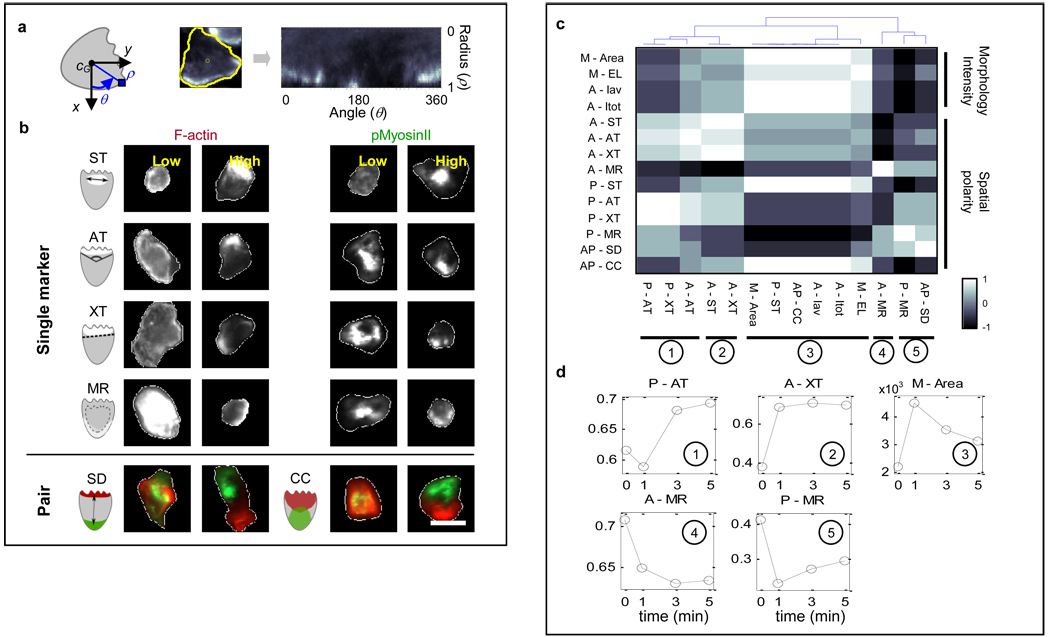
Figure 2. Computational image processing revealed distinct patterns of F-actin and pMyosinII spatial distribution during polarization.
a. Transformation of pixel intensity map into polar coordinates. b. Representative cells showing low/high values of the spatial polarity features. White contours: cell boundaries. White pixel areas denote high fluorescence intensity regions. Top panel: F-actin (left) and pMyosinII (right) features. Bottom panel: pairwise marker features. Scale bars: 10µm. c–d. The polarity features showed five distinct dynamic trends. An example of dynamic response was given for each case. Feature labels were prefixed by the following letters to represent different markers or feature categories: A for F-actin, P for pMyosinII, AP for F-actin and pMyosinII (pairwise) and M for morphology.
3.3. Axial tightness (XT)
Axial tightness was designed to quantify the “depth” of polarization of the active polarity signaling areas based on how tightly the brightest pixels could be partitioned to one “side” of the cell. We determined the axial tightness by searching for the linear bi-partition of the cell that could delimit 90% of Tα within the smallest area, Aα. By denoting A0 as the total cell area, axial tightness XT was defined as:
| (3) |
3.5. F-actin/pMyosinII separation distance (SD)
In addition to single-marker features, we also quantified the spatial separation between the F-actin and pMyosinII-enriched areas. This readout served to indicate to what extent the front and back signaling regions were segregated from each other. We defined SD as the Euclidean distance between the centers of F-actin (A) and pMyosinII (P) enriched regions (i.e. Tα(A), Tα(P)) normalized by the maximum distance between pixels in the cellular area, dmax:
| (4) |
3.6. F-actin/pMyosinII correlation coefficient (CC)
To measure the spatial co-variation of the intensity values between 2 channels, we rearranged the 2-D pixel intensity maps into 1-D vectors and computed their correlation coefficient. To reduce numerical noise due to pixels with low intensity values, in each channel we reset the intensity of a pixel to zero if its original value was in the bottom β% (β=25 in our study). We modified the correlation coefficient CC between the F-actin and pMyosinII intensity vectors (Iβ,A,I β,P) as in (5) so that its value ranged from 0 (perfect overlap) to 1 (no overlap) (“ ·” designates dot product):
| (5) |
We note that all the features considered here are defined to increase as cells display more spatial polarization. As can be seen in the representative images of neutrophils shown in Figure 2b, different values of our proposed features captured visually distinct patterns of spatial polarization of F-actin and pMyosinII.
4. VALIDATION OF SPATIAL FEATURES
4.1. Comparison of polarity features
For comparison, we computed several commonly used polarity readouts such as total/average cellular F-actin intensity (Itot, Iav), cell area and ellipticity (EL) [7]. We also computed the membrane-to-cytosol ratio (MR), a phenotype commonly used in studying cell signaling, defined here as the ratio between the total intensity of pixels with radius ρ>0.6 against pixels with ρ<0.6. We found that while some spatial features appeared redundant (e.g. AT and XT), spatial polarity did not necessarily correlate with morphology or intensity readouts. Polarization of F-actin/pMyosinII was not always associated with elongated shape or high fluorescence intensity and vice versa. For instance, some cells with low pMyosinII MR were elongated, while other cells with low F-actin MR were very bright in F-actin channel (Figure 2b). Our observation was consistent with previous reports that morphological changes and spatial polarity occurred at different times [5,6].
4.2. Correlation between dynamic responses
We next sought to identify the information content of different polarity features mentioned above in relationship to the process of polarization. To this end, we built a dynamic response vector for each feature f over the 4 observation time points, x(f) = [x1(f) … x4(f)]. The entries of x(f) were estimated by averaging the cell population median value over 6 repeated experiments. We used pairwise Spearman correlation coefficients between the vectors {x(f)} and identified five groups of features displaying distinct dynamic trends during polarization (Figure 2c). Commonly used readouts such as cell size, ellipticity and F-actin intensity were all found in the 3rd category, which was characterized by a stereotypical sharp-increase-gradual-decrease trend [1–6] (Figure 2d). In contrast, features in the 1st category (e.g. pMyosinII angular tightness, P-AT) first slightly decreased then rapidly increased after one minute. Furthermore, features in the 2nd category (e.g. A-ST, A-XT) increased until 3min after stimulation then maintained their level. The 4th and 5th categories of features such as the membrane-to-cytosol ratios of F-actin and pMyosinII (A-MR, P-MR) both showed decreasing trends, although with a slight nuance.
Visual examination of our image data indicated that unstimulated neutrophils had a thin layer of low level F-actin around the cell cortex. Upon stimulation, cell area expanded and strong F-actin production rapidly occurred near cellular membrane, while pMyosinII-enriched region translocated to the inner region of the cell. Subsequently, as F-actin polarized to one side of the cell, pMyosinII-enriched region gradually moved to the opposite side of the cell. This succession of events was previously observed in several studies [1–6] and consistently depicted by our polarity features. For instance, the dynamic trend of A-XT reflected that while cell area underwent a rapid expansion early on, the F-actin enriched area required longer time to focus and form the “front”. As for the membrane-to-cytosol ratio features A-MR and P-MR, the monotonic decrease of the F-actin readout described the fact that the F-actin network at the front became increasingly thicker in depth, which could be explained by the presence of a strong local positive feedback in the front signaling module that amplified F-actin production [1–3,5]. In contrast, the fast-decrease-slow-increase trend of P-MR reflected the facts that the spatial distribution of backness was constrained by F-actin and its establishment occurred gradually as the front signaling molecules concentrated in a narrow region [6]. In summary, our proposed features captured several dynamic trends of spatial redistribution of signaling molecules that were consistent with past observations. Our analysis identified informative phenotypes that could be added to the current polarity readouts to investigate neutrophil polarization.
5. CONCLUSION
In this work we implemented a collection of image-based features based on biological intuition to capture different spatial phenotypes of signaling molecules for frontness (F-actin) and backness (pMyosinII) in human neutrophils. These readouts are derived from the geometric patterns of pixel intensity and are designed to be robust to fluorescence intensity fluctuation during experiments. The proposed spatial features can provide additional information about polarization not contained within traditional morphology or intensity readouts. As new imaging probes are being created and analytical features made available, there is increasing need for determining which features (or categories of features) yield non-redundant biological information. Our work will provide a starting point for identifying information content within different polarity readouts and relating them to biological functions in neutrophils and other cell types.
ACKNOWLEDGEMENTS
We thank members of the Altschuler and Wu labs for helpful discussions and readings of the manuscript. This research was supported by the National Institute of Health grants R01 GM081549 (L.F.W.) and R01 GM071794 (S.J.A.), the Welch Foundation I-1619 (S.J.A.) and I-1644 (L.F.W.).
Contributor Information
Chin-Jen Ku, Email: chin-jen.ku@utsouthwestern.edu.
Yanqin Wang, Email: yanqin.wang@utsouthwestern.edu.
Benjamin Pavie, Email: benjamin.pavie@utsouthwestern.edu.
Steven J. Altschuler, Email: steven.altschuler@utsouthwestern.edu.
Lani F. Wu, Email: lani.wu@utsouthwestern.edu.
REFERENCES
- 1.Xu J, et al. Divergent signals and cytoskeletal assemblies regulate self-organizing polarity in neutrophils. Cell. 2003;Vol. 114(no. 2):201–214. doi: 10.1016/s0092-8674(03)00555-5. [DOI] [PubMed] [Google Scholar]
- 2.Arrieumerlou C, Meyer T. A local coupling model and compass parameter for eukaryotic chemotaxis. Dev Cell. 2005;vol. 8(no. 2):215–227. doi: 10.1016/j.devcel.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Sasaki AT, et al. G protein–independent Ras/PI3K/F-actin circuit regulates basic cell motility. J Cell Biol. 2006;vol. 178(no. 2):185–191. doi: 10.1083/jcb.200611138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrew N, Insall RH. Chemotaxis in shallow gradients is mediated independently of PtdIns 3-kinase by biased choices between random protrusions. Nature Cell Biol. 2007;vol. 9:193–200. doi: 10.1038/ncb1536. [DOI] [PubMed] [Google Scholar]
- 5.Weiner OD, et al. Hem-1 complexes are essential for Rac activation, actin polymerization, and myosin regulation during neutrophil chemotaxis. PLoS Biol. 2006;vol. 4(2):e38. doi: 10.1371/journal.pbio.0040038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong K, et al. Neutrophil polarization: spatiotemporal dynamics of RhoA activity supports a self-organizing mechanism. Proc. Natl. Acad. Sci.USA. 2006;vol.103(no. 10):3639–3644. doi: 10.1073/pnas.0600092103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bakal C, et al. Quantitative morphological signatures define local signaling networks regulating cell morphology. Science. 2007;vol. 316:1753–1756. doi: 10.1126/science.1140324. [DOI] [PubMed] [Google Scholar]
- 8.Ponti A, et al. Two distinct actin networks drive the protrusion of migrating cells. Science. 2004;vol. 305:1782–1786. doi: 10.1126/science.1100533. [DOI] [PubMed] [Google Scholar]
- 9.Kasson PM, et al. Quantitative imaging of lymphocyte membrane protein reorganization and signaling. Biophysical J. 2005;vol. 88:579–589. doi: 10.1529/biophysj.104.048827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang L, Helmke BP. Spatiotemporal analysis of actin ruffling dynamics in living cells. Conf. Record of the 40th Asilomar Conf. on Signals, Systems and Computers; 2006. pp. 183–187. [Google Scholar]
- 11.Boland MV, Murphy RF. After sequencing: quantitative analysis of protein localization. IEEE Eng. Med. Biol. Mag. 1999;vol. 18:115–119. doi: 10.1109/51.790995. [DOI] [PubMed] [Google Scholar]
- 12.Loo LH, et al. Image-based multivariate profiling of drug responses from single cells. Nature Methods. 2007;vol. 4:445–453. doi: 10.1038/nmeth1032. [DOI] [PubMed] [Google Scholar]
- 13.Böyum A. Isolation of mononuclear cells and granulocytes from human blood. Scan. J. Clin. Lab. Invest. 1968;vol. 21:77–89. [PubMed] [Google Scholar]
- 14.Rasband WS. Bethesda, Maryland, USA: U.S. National Institutes of Health; ImageJ. 1997–2009 http://rsb.info.nih.gov/ij/