Abstract
This study includes 24 reports of chromosome counts in Tripleurospermum Sch. Bip. (20 taxa), Anthemis L. (1 species) and Matricaria L. (3 taxa) belonging to the tribe Anthemideae of the family Asteraceae from Turkey. Chromosome numbers of these taxa are 2n = 2x = 18, 3x = 27 and 4x = 36. Nine counts are new reports, one is not consistent with previous report, and the remainder confirm earlier information. Statistically significant differences depending on ploidy level, stomatal length or environmental factors such as altitude were determined in Tripleurospermum. Several systematic and evolutionary aspects of the genera are discussed in the light of chromosomal data.
Keywords: Anthemis, Cytotaxonomy, Matricaria, Polyploidy, Stomatal length, Tripleurospermum, Turkey
Introduction
The Asteraceae is one of the largest plant families (Bremer 1994). Its many genera and species, its worldwide distribution and the fact that it comprises many useful plants have made it the subject of large numbers of karyological studies (Watanabe 2009 and references therein). The Anthemideae is a medium-sized tribe in the family (Vallès et al. 2005), comprising 111 genera with c. 1,800 species, distributed worldwide (extratropical) but with main concentrations in Central Asia, the Mediterranean region and southern Africa (Oberprieler et al. 2007). It is generally accepted as a relatively natural assemblage.
The principal taxonomic problems within the tribe are almost entirely relationships between genera, but also circumscription of genera, especially within subtribal groups such as the Artemiisinae, Chrysantheminae and Tanacetinae (Bremer and Humphries 1993). A complete convenient classification has not yet been achieved. The circumscription of Anthemideae remains relatively unchanged since early work (Oberprieler et al. 2007). The tribe was monographed on the basis of mostly morphological phylogeny (Bremer and Humphries 1993), but there is little congruence with any of the previous classifications (Oberprieler et al. 2007). To complicate matters further, molecular phylogenies for the tribe as a whole (Watson et al. 2000) are largely incongruent with either morphological phylogeny (Bremer and Humphries 1993) or the previously proposed classifications (Oberprieler et al. 2007).
The genera Tripleurospermum Sch. Bip., Anthemis L. and Matricaria L. belong to the tribe Anthemideae and comprise c. 40, 175 and six species, respectively, which are distributed mainly in Europe, temperate Asia, North America and North Africa (Oberprieler et al. 2007). It is also difficult to determine the exact number of species without a monographical treatment of the whole genus Tripleurospermum, because its species have often been referred to genera including Pyrethrum Zinn., Chrysanthemum L., Matricaria L. and Chamaemelum Mill. (Pobedimova 1995).
Problems of taxonomy in these genera cannot be ignored. Species of Tripleurospermum, Anthemis and Matricaria are similar to each other in their morphological characteristics and are also similar to many other Anthemideae genera in their habit. Therefore, they have been confused both taxonomically and nomenclaturally with each other and with other Anthemideae genera (Jeffrey 1979; Xifreda 1985; Kerguélen et al. 1987; Pobedimova 1995; Applequist 2002; Hansen and Christensen 2009).
The taxonomy of these genera continues to be subject to much confusion, mainly because of the approaches to species delimitation that have been employed, resulting in varying numbers of recognized species. Initially, Tripleurospermum was assigned to the genus Matricaria, but later was recognized as a separate genus on the basis of the different structure of its achenes and the occurrence of a tetrasporic embryo sac (Harling 1951). However, Rauschert (1974) and Kay (1976) misapplied the name Matricaria to refer exclusively to the species of Tripleurospermum (Bremer and Humphries 1993). In addition, Oberprieler (2001) showed that Tripleurospermum is closely related to Anthemis in the strict sense and not to Matricaria. This is supported by the tetrasporic embryo sac shared by Tripleurospermum and Anthemis (versus monosporic in Matricaria and other Anthemideae genera).
Most cytological studies conducted in the genera have concentrated on the chromosome count, with little work having been focussed on detailed karyological criteria for taxonomic purposes (Mitsuoka and Ehrendorfer 1972; Inceer and Beyazoglu 2004; Garcia et al. 2005). In spite of the very large number of chromosome counts performed in Matricaria, to our knowledge, no chromosomal data on Turkish populations have been reported in the literature. Chromosomal data on Tripleurospermum and Anthemis are also scarce. The aim of the present study was to document the chromosome numbers of Tripleurospermum, Anthemis and Matricaria in Turkey, which is regarded as one of their main centres of diversification and speciation. We also discuss some systematic and evolutionary aspects of Tripleurospermum in the light of the chromosomal data. Finally, we have tested the possibility of the existence of any relationship between ploidy level and stomatal length or environmental factors in Tripleurospermum.
Materials and methods
Plant materials
Materials for chromosome count study include 24 taxa of Tripleurospermum, Anthemis and Matricaria. Vouchers are deposited in the herbarium at the Karadeniz Technical University, Department of Biology (KTUB) and H. Inceer collections. All taxa were directly collected from native populations.
Chromosome counts
Root tip meristerms obtained directly from natural populations, cultivated plants or germinated achenes were used for chromosome analysis. Samples were pretreated with 0.05% colchicine for 2–5 h. They were then fixed in absolute ethanol–glacial acetic acid (3:1) for at least 24 h at 4°C, hydrolysed in 1 N HCl at 60°C for 10 min and then rinsed in tap water for 2–3 min (Inceer and Hayirlioglu-Ayaz 2007). Staining was carried out in 1% aqueous aceto-orcein for 12–18 h at room temperature, and squashes were made in 45% acetic acid. The best metaphase plates were photographed using a Leica DM 4000 microscope and a Leica DFC 490 digital camera.
To assess the existence of published chromosome counts in the taxa studied, we used the common indexes of plant chromosome numbers cited by Inceer and Beyazoglu (2004), as well as the online chromosome number databases, Index to Plant Chromosome Numbers (Missouri Botanical Garden, http://motot.mobot.org/W3T/Search/ipcn.html) and Index to Chromosome Numbers in the Asteraceae (Watanabe 2009; http://www.lib.kobe-u.ac.jp/infolib/meta_pub/G0000003asteraceae_e).
Stomatal measurements
Stomatal measurements were carried out on the lower (abaxial) epidermis of many of the fresh or dried leaves. Five slides were prepared for each taxon, and seven stomatal lengths were measured on each slide in random fields of view. Paradermal sections of abaxial epidermis were stained in hematoxylin for 10 min and mounted in glycerine. Stomata were measured under ×40 magnification using a light microscope with an ocular scale. Drawings were made using a lucida camera.
Statistical analysis
Statistical analysis was carried out to evaluate the relationships among the variables of ploidy level, stomatal length and altitude in Tripleurospermum using SPSS version 15.
Nomenclature
We follow herein the nomenclature adopted by Grierson (1975), Enayet Hossain (1975), Davis (1988), Oberprieler and Vogt (2006) and Inceer and Hayirlioglu-Ayaz (2008). Investigated taxa are listed by their genera, in alphabetical order.
Results and discussion
Chromosome counts
Many of our data represent new counts, whereas others confirm unique or scarce previous reports, or represent counts that differ from those cited previously. Sources and counts of the plant material studied here were as follows:
Tripleurospermum Sch. Bip.
T. baytopianum E. Hossain
A1 (E) Tekirdağ: Between Sarköy and Gölcük, slopes, meadows and roadsides, 250 m a.s.l., 17.iv.2008, Inceer 508. 2n = 18 (Fig. 1).
Figs. 1–7.
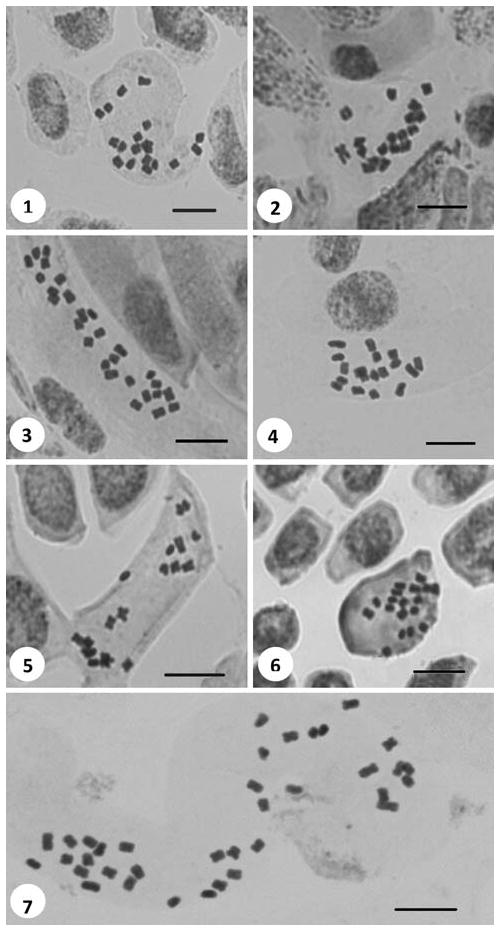
Somatic metaphases in Tripleurospermum. 1 T. baytopianum (2n = 18). 2 T. conoclinium (2n = 18). 3 T. corymbosum (2n = 27). 4 T. disciforme (2n = 18). 5 T. elongatum (2n = 18). 6 T. fissurale (2n = 18). 7 T. heterolepis (2n = 36). Scale bars 10 μm
To our knowledge, this is the first report of the chromosome number of this endemic species. It is only known from one locality in the literature (Enayet Hossain 1975).
T. conoclinium (Boiss. & Bal.) Hayek
B2 Izmir: Boz Dağ, cultivated fields, meadows and pastures, 1,178 m a.s.l., 07.iv.2008, Inceer 478. 2n = 18 (Fig. 2).
According to our data, this is the first report of the chromosome number of this species. Firstly, this species was determined in Turkish flora, and Enayet Hossain (1975) assigned it as endemic. Later, it was collected from the Island of Lesvos (Bazos and Yannitsaros 2004).
T. corymbosum E. Hossain
B9 Ağrı: Suluçem (Musun), Balık Gölü, meadows, cultivated fields, 2,098 m a.s.l., 11.vii.2008, Inceer 612. 2n = 27 (Fig. 3).
To our knowledge, this is the first report of the chromosome number of this species, which is endemic to Turkey.
T. disciforme (C. A. Meyer) Sch. Bip.
B2 Izmir: Boz Dağ, roadsides, water meadows, 1,021 m a.s.l., 06.vii.2008, Inceer 592. 2n = 18 (Fig. 4).
This is the first report from Turkish populations. Our diploid count agrees with many previous reports on material from other areas (Watanabe 2009 and references therein). It confirms that the diploid level predominates in this species, although the tetraploid level has also been reported in Iranian plants (2n = 36; Chehregani and Mehanfar 2008).
T. elongatum (Fisch. & Mey.) Bornm.
A9 Kars (Ardahan): Between Ardahan and Göle, roadsides, open meadows, stream sides, 1,800 m a.s.l., 18.vii.2007, Inceer 423. 2n = 18 (Fig. 5).
To our knowledge, this is the third chromosome count on this species. It confirms the chromosome number given by Magulaev (1992) in plants from the northern Caucasus and by Inceer and Beyazoglu (2004) from north-east Anatolia (different population).
T. fissurale (Sosn.) E. Hossain
A8 Artvin: Yusufeli, between Ispir and Yusufeli, near Kozahora, roadsides, rocky slopes, 617 m a.s.l., 04.vi.2007, Inceer 351. 2n = 18 (Fig. 6).
According to our data, this is the first report of the chromosome number of this species, which is endemic to Turkey.
T. heterolepis (Freyn & Sint) Bornm.
A7 Gümüshane: Keçikaya Village, roadsides, 1,618 m a.s.l., 04.vii.2007, Inceer 382b. 2n = 36 (Fig. 7).
To our knowledge, this is the first report of the chromosome number of this species, which is endemic to Turkey.
T. hygrophilum (Bornm.) Bornm.
B1 Izmir: Yamanlar Dağı, near Pinus forest, open places, 887 m a.s.l., 15.iv.2007, Inceer 273. 2n = 18 (Fig. 8).
Figs. 8–14.
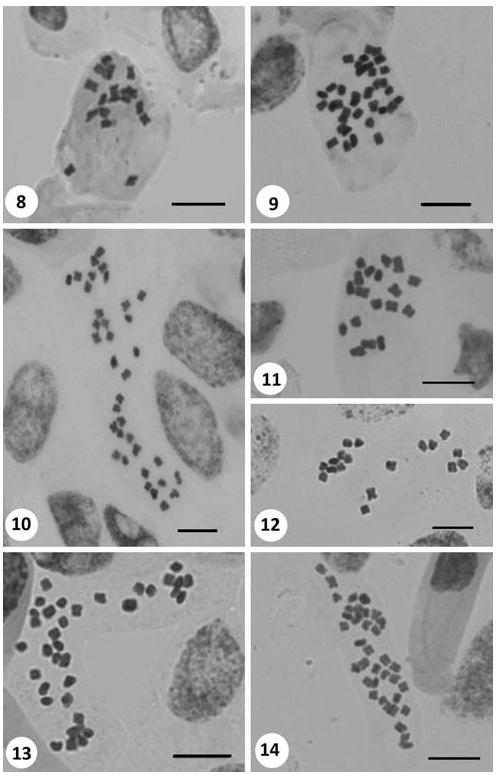
Somatic metaphases in Tripleurospermum. 8 T. hygrophilum (2n = 18). 9 T. inodorum (2n = 36). 10 T. kotschyi (2n = 36). 11 T. microcephalum (2n = 18). 12 T. parviflorum (2n = 18). 13 T. pichleri (2n = 36). 14 T. repens (2n = 36). Scale bars 10 μm
According to our data, this is the first report of the chromosome number of this species, which is endemic to Turkey.
T. inodorum (L.) Sch. Bip.
A9 Erzurum: Between Pasinler and Horasan, near Horasan, Köprü Village, roadsides, 1,600 m a.s.l., 11.vii.2008, Inceer 600. 2n = 36 (Fig. 9).
This is the first report from Turkish populations. Our count agrees with many others from different territories (Watanabe 2009 and references therein). Its chromosome number is shown to be variable between 18 and 36.
To our knowledge, the systematic position of T. inodorum is still a controversial problem. It has been confused both taxonomically and nomenclaturally with T. maritimum (L.) Koch and other Anthemideae genera (Applequist 2002).
T. kotschyi (Boiss.) E. Hossain
C5 Niğde: Ulukısla, Bolkar Mountains, near Karagöl, 2,600 m a.s.l., 29.vii.2008, Inceer 702. 2n = 36 (Fig. 10).
According to our data, this is the first report of the chromosome number of this species, which is endemic to Turkey.
T. microcephalum (Boiss.) Bornm.
B8 Mus: Fallow fields, banks, roadsides, 1,323 m a.s.l., 09.vii.2008, Inceer 594. 2n = 18 (Fig. 11).
To our knowledge, this is the first report of the chromosome number of this species.
T. parviflorum (Willd.) Pobed.
B2 Izmir: Boz Dağ, rocky places, cultivated fields, roadsides, 1,178 m a.s.l., 07.iv.2008, Inceer 479. 2n = 18 (Fig. 12).
To our knowledge, this is the third chromosome count on this species. It confirms the chromosome number given by Avetisian and Oganesian (1995) in plants from Armenia and by Inceer and Beyazoglu (2004) from north-east Anatolia.
T. pichleri (Boiss.) Bornm.
A2 Bursa: Uludağ, meadows, damp woods, near hotels, 1,828 m a.s.l., 11.vi.2008, Inceer 553. 2n = 36 (Fig. 13).
According to our data, this is the first report of the chromosome number of this species, which is endemic to Turkey.
T. repens (Freyn & Sint.) Bornm.
A7 Gümüshane: Gezge Village, meadows, 1,724 m a.s.l., 08.vii.2007, Inceer 383. 2n = 36 (Fig. 14).
To our knowledge, this is the second chromosome count on this species, which is endemic to Turkey. It confirms the chromosome number given by Inceer and Beyazoglu (2004) from Rize, north-east Anatolia.
T. rosellum (Boiss. & Orph.) Hayek var. album E. Hossain
A3 Bolu: Near Abant Lake, meadows, 1,331 m a.s.l., 12.vi.2008, Inceer 555. 2n = 18 (Fig. 15).
Figs. 15–20.
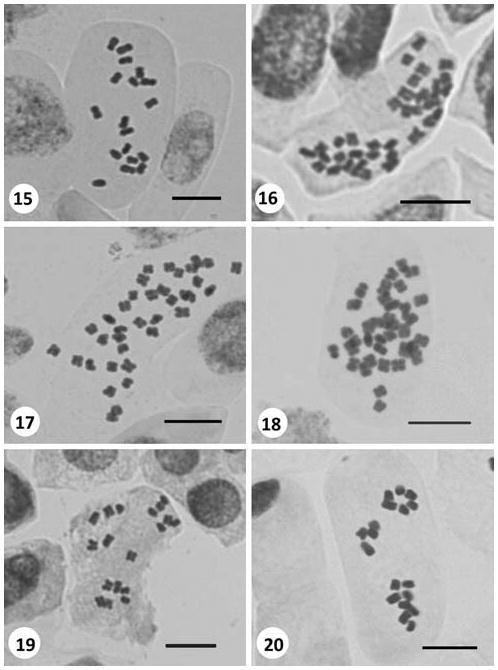
Somatic metaphases in Tripleurospermum. 15 T. rosellum var. album (2n = 18). 16 T. sevanense (2n = 36). 17 T. tempskyanum (2n = 36). 18 T. tenuifolium (2n = 36). 19 T. transcaucasicum (2n = 18). 20 T. ziganaense (2n = 18). Scale bars 10 μm
The single previous study on chromosome number in this taxon, which is endemic to Turkey, was 2n = 36 in north-east Anatolia population, Turkey (Inceer and Beyazoglu 2004). This is the second count and the first at the diploid level for this taxon. Both ploidy levels are based on x = 9. This taxon is thus represented by diploid and tetraploid cytotypes in Turkey. The cytotypes of T. rosellum var. album do not differ from each other with respect to plant morphology, but there may be physiological and ecological differences between them, comprising cytodemes. Similar results were found by Rottgardt (1956) in T. maritimum (L.) Koch, by Lankosz-Mróz (1976) in T. maritimum (L.) Koch ssp. inodorum Applequist from Poland and by Inceer and Beyazoglu (2004) in T. caucasicum (Willd.) Hayek from north-east Anatolia, Turkey.
T. sevanense (Manden.) Pobed.
C5 Niğde: Ulukısla, Bolkar Mountains, roadsides, 1,420 m a.s.l., 14.vii.2007, Inceer 396. 2n = 36 (Fig. 16).
Our chromosome count confirms the existence of the tetraploid cytotype of this species, reported by Inceer and Beyazoglu (2004), Garcia et al. (2005) and Inceer and Hayirlioglu-Ayaz (2007) on Turkish materials. Apart from this, one count on Armenian plants has reported the diploid level, 2n = 18 (Avetisian and Oganesian 1995).
T. tempskyanum (Freyn & Sint.) Hayek
A2 Bursa: Uludağ, near hotels, meadows, open places, 1,900 m a.s.l., 27.vi.2007, Inceer 361. 2n = 36 (Fig. 17).
Tripleurospermum tempskyanum was considered until now as endemic to Greece, being found in montane areas of north-west. This species is a new record for the flora of Turkey (unpublished data). Our tetraploid count confirms the chromosome number given by Constantinidis et al. (2002).
T. tenuifolium (Kit.) Freyn
A2 Bursa: Uludağ, near hotels, meadows, open places, 1,690 m a.s.l., 27.vi.2007, Inceer 353. 2n = 36 (Fig. 18).
This is the first report from Turkish populations. In the literature, chromosome numbers of this species are given as 2n = 2x = 18 (Jasiewicz and Mizianty 1975; Van Loon 1987; Strid and Tan 1991) and 2n = 4x = 36 (Avetisian and Oganesian 1995).
T. transcaucasicum (Manden.) Pobed.
A9 Kars (Ardahan): Between Ardahan and Göle, roadsides, step area, rocky places, 1,800 m a.s.l., 18.vii.2007, Inceer 422. 2n = 18 (Fig. 19).
To our knowledge, this is the third chromosome count on this species. It confirms the chromosome number given by Avetisian and Oganesian (1995) in plants from Armenia and Inceer and Beyazoglu (2004) from north-east Anatolia, Turkey.
T. ziganaense Inceer & Hayirlioglu-Ayaz
A7 Gümüshane: Zigana Dağı, between Zigana Pass and Torul, 1,300 m a.s.l., 22.vii.2008, Inceer 666. 2n = 18 (Fig. 20).
This is the first report of the chromosome number of this species. It is only known from one locality in the literature (Inceer and Hayirlioglu-Ayaz 2008). We only give the chromosome numbers of the topotype specimens.
Anthemis L.
A. macrotis (Rech. f.) Oberprieler & Vogt
C2 Muğla: Near Köyceğiz, roadsides, 11 m a.s.l., 10.iv.2008, Inceer 498. 2n = 18 (Fig. 21).
Figs. 21–24.
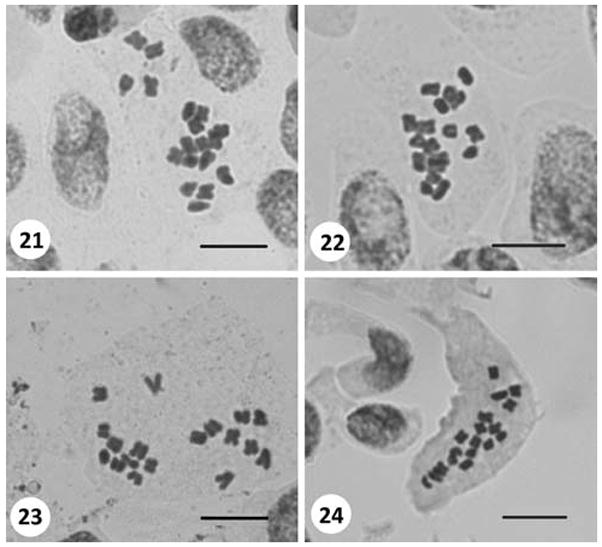
Somatic metaphases in Anthemis and Matricaria. 21 Anthemis macrotis (2n = 18). 22 Matricaria aurea (2n = 18). 23 M. chamomilla var. recutita (2n = 18). 24 M. matricarioides (2n = 18). Scale bars 10 μm
To our knowledge, this is second chromosome count of this endemic species. Our diploid count agrees with one previous report based on cultivated plants, which are not of known wild origin, from Edinburgh Royal Botanical Garden (Ratter and Milne 1973).
Matricaria L.
M. aurea (Loefl.) Sch. Bip.
C6: Gaziantep/Şanlıurfa: Between Nizip and Birecik, Dutlu, roadsides, near cultivated area, 440 m a.s.l., 08.v.2007, Inceer 322. 2n = 18 (Fig. 22).
According to our data, this is the first report from Turkish populations. Our diploid count agrees with many others from different territories (Watanabe 2009 and references therein). This number is shown to be very stable, because it is also the same in many other reports based on plants from very different regions.
M. chamomilla L. var. recutita (L.) Fiori
B1 Izmir: Odemis, between Izmir and Odemis, roadsides and cultived fields, 17 m a.s.l., 14.iv.2007, Inceer 258. 2n = 18 (Fig. 23).
This is the first report from Turkish populations. Our diploid count agrees with many others from different territories (Watanabe 2009 and references therein). This number is shown to be very stable, because it is also the same in many other reports based on plants from very different regions.
M. matricarioides (Less.) Porter ex Britton
A9 Kars (Ardahan): Between Ardahan and Göle, roadsides, 1,800 m a.s.l., 18.vii.2007, Inceer 420. 2n = 18 (Fig. 24).
To our knowledge, this is the first report from Turkish populations. Our diploid count agrees with many others from different territories (Watanabe 2009 and references therein). This number is shown to be very stable, because it is also the same in many other reports based on plants from very different regions.
Stomatal length
Stomatal frequency, guard cell length and stomatal plastid number have been used as morphological markers for identifying ploidy levels in many plant species (Bingham 1968; Santen and Casler 1986; Mishra 1997; Beck et al. 2003). It was noted that polyploids have larger stomata than diploids (Stebbins 1971; Seidler-Lozykowska 2003). A positive relationship was found between ploidy level and stomatal length in Tripleurospermum (r = 0.92, P = 0.01). The mean length of stomata varied from 24.87 to 29.69 μm in diploid plants and from 32.72 to 37.66 μm in tetraploids. On average, the tetraploids had a mean length of 35.00 ± 2.02 μm, which was found to be significantly longer (P < 0.01) than in diploid counterparts that had a mean length of 27.31 ± 1.99 μm (Table 1; Fig. 25). It was noted that, on examination of the diploid, triploid and tetraploid plants separately, there were significant differences (P < 0.01). The majority of the variation was associated with ploidy level.
Table 1.
Differences in stomatal length (mean ± standard deviation, SD) among ploidy levels in Tripleurospermum, as determined by Duncan’s multiple-range test from one-way analysis of variance (ANOVA); values with different letters are significant at P < 0.01
| Taxon | Chromosome number (2n) | Ploidy level | Stomatal length (μm) |
|---|---|---|---|
| T. baytopianum | 18 | 2x | 25.24 ± 1.89 |
| T. conoclinium | 18 | 2x | 28.56 ± 0.94 |
| T. corymbosum | 27 | 3x | 30.64 ± 1.44 |
| T. disciforme | 18 | 2x | 27.60 ± 1.14 |
| T. elongatum | 18 | 2x | 24.87 ± 1.57 |
| T. fissurale | 18 | 2x | 27.08 ± 1.34 |
| T. heterolepis | 36 | 4x | 35.92 ± 0.97 |
| T. hygrophilum | 18 | 2x | 26.48 ± 1.33 |
| T. inodorum | 36 | 4x | 32.72 ± 1.22 |
| T. kotschyi | 36 | 4x | 36.58 ± 1.81 |
| T. microcephalum | 18 | 2x | 25.28 ± 1.04 |
| T. parviflorum | 18 | 2x | 29.69 ± 0.25 |
| T. pichleri | 36 | 4x | 33.08 ± 0.46 |
| T. repens | 36 | 4x | 37.66 ± 0.47 |
| T. rosellum var. album | 18 | 2x | 28.32 ± 1.01 |
| T. sevanense | 36 | 4x | 34.46 ± 1.51 |
| T. tempskyanum | 36 | 4x | 35.72 ± 1.17 |
| T. tenuifolium | 36 | 4x | 33.88 ± 1.67 |
| T. transcaucasicum | 18 | 2x | 28.92 ± 1.39 |
| T. ziganaense | 18 | 2x | 28.25 ± 1.04 |
| Source of variation | Ploidy mean length (μm; among taxa) | ||
| Diploid taxa (2x) | 27.31 ± 1.99a | ||
| Triploid taxon (3x) | 30.64 ± 1.44b | ||
| Tetraploid taxa (4x) | 35.00 ± 2.02c |
Fig. 25.
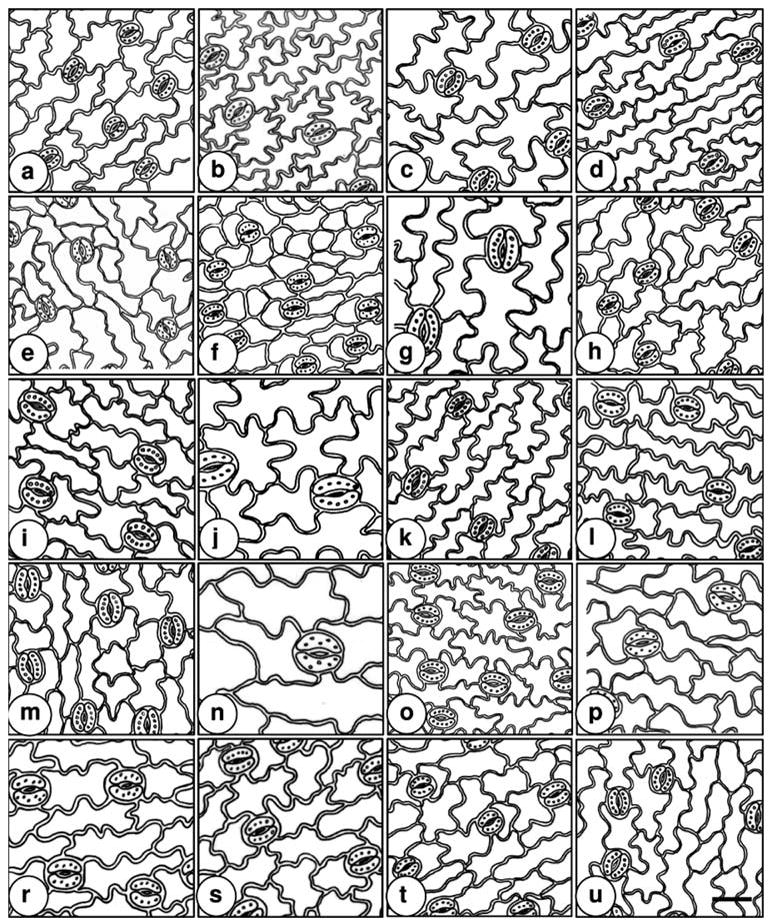
Drawings of paradermal sections in Tripleurospermum. a T. baytopianum; b T. conoclinium; c T. corymbosum; d T. disciforme; e T. elongatum; f T. fissurale; g T. heterolepis; h T. hygrophilum; i T. inodorum; j T. kotschyi; k T. microcephalum; l T. parviflorum; m T. pichleri; n T. repens; o T. rosellum var. album; p T. sevanense; r T. tempskyanum; s T. tenuifolium; t T. transcaucasicum; u T. ziganaense. Scale bar 50 μm
Conclusions
The results listed above and in Table 1 confirm the existence of one basic chromosome number in the genera. All of the studied taxa have x = 9, the most common basic number in the tribe Anthemideae and the family Asteraceae (Fedorov 1969; Solbrig 1977; Schweizer and Ehrendorfer 1983; Oliva and Vallès 1994; Vallès and Siljak-Yakovlev 1997; Vallès et al. 2001, Inceer and Beyazoglu 2004; Vallès et al. 2005; Inceer and Hayirlioglu-Ayaz 2007). The ploidy levels range from 2n = 2x = 18, 2n = 3x = 27, to 2n = 4x = 36. To date, one ploidy level (2x) in Matricaria (Watanabe 2009 and references therein) and four ploidy levels (2x, 3x, 4x, 5x) in Tripleurospermum have been reported in the literature by various authors (Hüser 1930; Fedorov 1969; Frey et al. 1981; Razaq et al. 1994; Dobeš et al. 1997; Inceer and Beyazoglu 2004). The diploid and tetraploid levels are common than the triploid and penta-ploid levels in Tripleurospermum. The stability of ploidy level in Matricaria is noteworthy in Turkish populations. On the other hand, there are several arguments for a close relationship of Tripleurospermum and Anthemis in the light of embryological (Harling 1950, 1951, 1960), phytochemical (Greger 1977), cytological (Mitsuoka and Ehrendorfer 1972; Oberprieler 1998; Inceer and Hayirlioglu-Ayaz 2007) and molecular (Oberprieler 2001, 2005) studies, rather than Matricaria.
It is known that polyploids have wider geographical ranges, are more successful in colonizing newly available areas (Stebbins 1971; Soltis and Soltis 2000) and have greater tolerance to ecological conditions than their diploid counterparts (Hagerup 1933; Pandit et al. 2006). From our survey of the literature and chromosome counts of the 20 taxa, it is clear that the polyploid distribution patterns of Tripleurospermum follow these general trends. Ploidy level is positively correlated with stomatal length and altitude in the genus (Table 2). In addition, there is a positive relationship between stomatal length and altitude (Table 2). It is concluded that stomatal length can be used as a taxonomic criterion to determine diploid and polyploid species of Tripleurospermum.
Table 2.
Correlation coefficients among variables in Tripleurospermum
| Source of variation | 1 | 2 | 3 |
|---|---|---|---|
| Ploidy level (1) | 1.000 | 0.922* | 0.609* |
| Stomatal length (2) | – | 1.000 | 0.636* |
| Altitude (3) | – | – | 1.000 |
P = 0.01
Polyploidy is considered by many authors as an important evolutionary mechanism in plants (Bretagnolle et al. 1998; Soltis et al. 2004) and in some groups of the Anthemideae in particular (Solbrig 1977; Vallès et al. 2001, 2005; Inceer and Beyazoglu 2004; Guo et al. 2005; Inceer and Hayirlioglu-Ayaz 2007). This is supported by the present study, since nine populations belonging to Tripleurospermum are polyploid. Together with these counts, the rate of polyploid taxa of Tripleurospermum in Turkish flora has reached 53%, which would seem to suggest that polyploidy is important in the evolution and differentiation of its species. The high frequency of polyploids in the plants examined leads us to consider that Tripleurospermum is continously developing genetic and ecological evolutionary mechanisms leading to diversification and speciation, particularly in Anatolia. This is also supported by dispersal-vicariance analysis (DIVA) and molecular data from Oberprieler (2005). However, further investigations are needed to trace their genome donors and to determine the nature of polyploidy in the genus.
Acknowledgments
The authors thank Melahat Ozcan, Murat Bal and Dr. Faik Ahmet Ayaz for help during the development of this research; Dr. Robert H. Glew (School of Medicine, University of New Mexico) for his suggestions; anonymous reviewers for comments that improved the manuscript considerably; and the Scientific and Technical Research Council of Turkey (TUBITAK, TBAG Project No. 106T162) for financial support.
References
- Applequist WL. A reassesment of the nomenclature of Matricaria L. and Tripleurospermum Sch. Bip. (Asteraceae) Taxon. 2002;51:757–761. [Google Scholar]
- Avetisian VE, Oganesian ME, editors. Campanulaceae, Asteraceae. Koeltz Scientific Books; Havlickuv Brod: 1995. Flora Armenii 9. [Google Scholar]
- Bazos I, Yannitsaros A. Floristic reports from the island of Lesvos (Greece) I. Dicotyledones: Aceraceae to Guttiferae. Edinb J Bot. 2004;61(1):49–86. [Google Scholar]
- Beck SL, Dunlop RW, Fossey A. Stomatal length and frequency as a measure of ploidy level in black wattle, Acacia mearnsii (de Wild) Bot J Linn Soc. 2003;141:177–181. [Google Scholar]
- Bingham ET. Stomatal chloroplast in alfalfa at four ploidy levels. Crop Sci. 1968;8:509–511. [Google Scholar]
- Bremer K. Asteraceae cladistics and classification. Timber; Portland: 1994. [Google Scholar]
- Bremer K, Humphries CJ. Genetic monograph of the Asteraceae–Anthemideae. Bull Nat His Mus Lond (Bot) 1993;23:71–177. [Google Scholar]
- Bretagnolle F, Felber F, Calame FG, Küpfer P. La polyloiödie chez les plantes. Bot Helv. 1998;108:5–37. [Google Scholar]
- Chehregani A, Mehanfar N. New chromosome counts in the tribe Anthemideae (Asteraceae) from Iran. Cytologia. 2008;73(2):189–196. [Google Scholar]
- Constantinidis T, Bareka EP, Kamari G. Karyotaxonomy of Greek serpentine angiosperms. Bot J Linn Soc. 2002;139:109–124. [Google Scholar]
- Davis PH. Flora of Turkey and East Aegean Islands. Vol. 10. Edinburgh University Press; Edinburgh: 1988. [Google Scholar]
- Dobeš C, Hahn B, Morawetz W. Chromosomenzahlen zur Gefässpflanzen-Flora Österreichs. Linz Biol Beitr. 1997;29:5–43. [Google Scholar]
- Enayet Hossain ABM. Tripleurospermum Schultz Bip. In: Davis PH, editor. Flora of Turkey and the East Aegean Islands. Vol. 5. Edinburgh University Press; Edinburgh: 1975. pp. 295–311. [Google Scholar]
- Fedorov AA, editor. Chromosome numbers of flowering plants. Nauka; Leningrad: 1969. [Google Scholar]
- Frey L, Mizianty M, Mirek Z. Chromosome numbers of Polish vascular plants. Fragm Florist Geobot. 1981;27:581–590. [Google Scholar]
- Garcia S, Inceer H, Garnatje T, Vallès J. Genome size variation in some representatives of the genus Tripleurospermum. Biol Plant. 2005;49(3):381–387. [Google Scholar]
- Greger H. Anthemideae: chemical review. In: Heywood VH, Harborne JB, Turner BL, editors. The biology and chemistry of the Compositae. Academic Press; London: 1977. pp. 899–894. [Google Scholar]
- Grierson AJC. Matricaria L. In: Davis PH, editor. Flora of Turkey and the East Aegean Islands. Vol. 5. Edinburgh University Press; Edinburgh: 1975. pp. 293–295. [Google Scholar]
- Guo YP, Saukel J, Mittermayr R, Ehrendorfer F. AFLP analysis demonstrate genetic divergence, hybridization, and multiple polyploidization in the evolution of Achillea (Asteraceae–Anthemideae) New Phytol. 2005;166:273–290. doi: 10.1111/j.1469-8137.2005.01315.x. [DOI] [PubMed] [Google Scholar]
- Hagerup AC. Studies on polyploid ecotypes in Vaccinum uliginosum L. Hereditas. 1933;18:122–128. [Google Scholar]
- Hansen HV, Christensen KI. The common chamomilla and the scentless mayweed revisited. Taxon. 2009;58(1):261–264. [Google Scholar]
- Harling G. Embryological studies in the Compositae, part I, Anthemideae–Anthemidinae. Acta Horti Berg. 1950;15:135–168. [Google Scholar]
- Harling G. Embryological studies in the Compositae, part II, Anthemideae–Chrysantheminae. Acta Horti Berg. 1951;16:1–56. [Google Scholar]
- Harling G. Further embriyological and taxonomical studies in Anthemis L. and some related genera. Svensk Bot Tidskr. 1960;54:572–590. [Google Scholar]
- Hüser W. Untersuchungen über die anatomie und wasserökologie einiger Ostseestrandpflanzen. Planta. 1930;11:485–508. [Google Scholar]
- Inceer H, Beyazoglu O. Karyological studies in Tripleurospermum (Asteraceae, Anthemideae) from north-east Anatolia. Bot J Linn Soc. 2004;146:427–438. [Google Scholar]
- Inceer H, Hayirlioglu-Ayaz S. Chromosome numbers in the tribe Anthemideae (Asteraceae) from Turkey. Bot J Linn Soc. 2007;153:203–211. [Google Scholar]
- Inceer H, Hayirlioglu-Ayaz S. Tripleurospermum ziganaense (Asteraceae, Anthemideae), a new species from north-east Anatolia, Turkey. Bot J Linn Soc. 2008;158:696–700. [Google Scholar]
- Jasiewicz A, Mizianty M. Chromosome numbers of some Bulgarian plants. Fragm Florist Geobot. 1975;12:277–288. [Google Scholar]
- Jeffrey C. Note on the lectotypification of the names Cacalia L. Matricaria and Gnaphalium L. Taxon. 1979;28:349–351. [Google Scholar]
- Kay QON. Chamomilla S.F.Gray and Matricaria L. In: Tutin TG, Heywood VH, Burges NA, Moore DM, Valentine DH, Walters SM, Webb DA, editors. Flora Europaea. Vol. 4. Camridge University Press; Camridge: 1976. pp. 165–167. [Google Scholar]
- Kerguélen M, Bosc C, Lambinon J. Données taxonomiques nomenclaturales et chrologiques pour une révision de la flore de France. Lejeunia. 1987;120:1–264. [Google Scholar]
- Lankosz-Mróz M. Karyological investigations on Tripleurospermum maritimum (L.) Koch subsp. inodorum (L.) Hyl. ex Vaarama from Poland. Acta Biol Crac Ser Bot. 1976;19:93–105. [Google Scholar]
- Magulaev AJ. Chromosome numbers in some species of vascular plants of the northern Caucasus flora. Bot Zhurn. 1992;77(10):88–90. [Google Scholar]
- Mishra M. Stomatal characteristic at different ploidy levels in Coffea L. Ann Bot. 1997;80:689–692. [Google Scholar]
- Mitsuoka S, Ehrendorfer F. Cytogenetics and evolution of Matricaria and related Genera (Asteraceae–Anthemideae) Österr Bot Zeitschr. 1972;120:155–200. [Google Scholar]
- Oberprieler C. The systematics of Anthemis L. (Compositae, Anthemideae) in W and C North Africa. Bocconea. 1998;9:1–328. [Google Scholar]
- Oberprieler C. Phylogenetic relationships in Anthemis L. (Compositae, Anthemideae) based on nrDNA ITS sequence variation. Taxon. 2001;50:745–762. [Google Scholar]
- Oberprieler C. Temporal and spatial diversification of Circum-Mediterranean Compositae–Anthemideae. Taxon. 2005;54(4):951–966. [Google Scholar]
- Oberprieler C, Vogt R. The taxonomic position of Matricaria macrotis (Compositae–Anthemideae) Willdenowia. 2006;36:329–338. [Google Scholar]
- Oberprieler C, Vogt R, Watson LE. XVI. Tribe Anthemidea Cass. In: Kadereit JW, Jeffrey C, Kubitzki K, editors. Flowering plants Eudicots. The families and genera of vascular plants. 1819. Vol. 8. Springer; Berlin: 2007. pp. 342–374. [Google Scholar]
- Oliva M, Vallès J. Karyological studies in some taxa of the genus Artemisia (Asteraceae) Can J Bot. 1994;72:1126–1135. [Google Scholar]
- Pandit MK, Tan HTW, Bisht MS. Polyploidy in invasive plant species of Singapore. Bot J Linn Soc. 2006;151:395–403. [Google Scholar]
- Pobedimova EG. Tripleurospermum Sch. Bip. In: Shiskin BK, Bobrov EG, editors. Flora USSR. XXVI. Bishen Singh Mahendra Pal Singh, Dehra Dun, India/Koertz Scientific Books; Koenigsten, Germany: 1995. pp. 181–213. [Google Scholar]
- Ratter JA, Milne C. Some angiosperm chromosome numbers. Notes Roy Bot Gard Edinb. 1973;32:429–438. [Google Scholar]
- Rauschert S. Nomenklatorische probleme in den gatung Matricaria L. Folia Geobot Phytotax. 1974;9:249–260. [Google Scholar]
- Razaq ZA, Vahidy AA, Ali Sl. Chromosome numbers in Compositae from Pakistan. Ann Missouri Bot Gard. 1994;8:800–808. [Google Scholar]
- Rottgardt K. Morphologische, cytologische und physiologische Untersuchungen von Ökotypen in Scleswig-Holstein. Beitr Biol Pflanzen. 1956;32:225–278. [Google Scholar]
- Santen EV, Casler EV. Evaluation of indirect ploidy indicators in Dactylis C. subspecies. Crop Sci. 1986;26:848–852. [Google Scholar]
- Schweizer D, Ehrendorfer F. Evolution of C-band patterns in Asteraceae–Anthemideae. Biol Zentr. 1983;102:637–655. [Google Scholar]
- Seidler-Lozykowska K. Determination of the ploidy level in chamomile [Chamomilla recutita (L.) Rausch.] strations rich in α-bisabolol. J Appl Genet. 2003;44(2):151–155. [PubMed] [Google Scholar]
- Solbrig OT. Chromosomal cytology and evolution in the family Compositae. In: Heywood VH, Harborn JB, Turner BL, editors. The biology and the chemistry of the Compositae I. Academic; London: 1977. pp. 269–281. [Google Scholar]
- Soltis PS, Soltis DE. The role of genetic and genomic attributes in the success of polyploids. Proc Natl Acad Sci U S A. 2000;97:7051–7057. doi: 10.1073/pnas.97.13.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS, Tate JA. Advanves in the study of polyploidy since plant speciation. New Phytol. 2004;161:173–191. [Google Scholar]
- Stebbins GL. Chromosomal evolution in higher plants. Edward Arnold; London: 1971. [Google Scholar]
- Strid A, Tan K. Matricaria L. In: Strid A, Tan K, editors. Mountain flora of Greece. Vol. 2. Edinburgh University Press; Edinburgh: 1991. pp. 450–452. [Google Scholar]
- Vallès J, Siljak-Yakovlev S. Cytogenetic studies in the genus Artemisia L.: flurochorome banded karyotypes of five taxa, including the Iberian endemic species Artemisia barrelieri Besser. Can J Bot. 1997;75:595–606. [Google Scholar]
- Vallès J, Torrell M, Garcia-Jacas N, Kampustina LA. New or rare chromosome counts in the genera Artemisia L and Mausolea Bunge (Asteraceae, Anthemidae) from Uzbekistan. Bot J Linn Soc. 2001;135:391–400. [Google Scholar]
- Vallès J, Garnatje T, Garcia S, Santz M, Korobrow A. Chromosome numbers in the tribes Anthemideae and Inuleae (Asteraceae) Bot J Linn Soc. 2005;148:77–85. [Google Scholar]
- Van Loon JCA. A cytotaxonomical atlas of the Balkan Flora. In: Löve A, Löve D, editors. Cytotaxonomical atlases. Vol. 4. J. Cramer; Berlin/Stuttgart: 1987. [Google Scholar]
- Watanabe W. Index to chromosome numbers in Asteraceae. 2009 http://www.lib.kobe-u.ac.jp/infolib/meta_pub/G0000003asteraceae_e, updated September 2009.
- Watson LE, Evans TM, Boluarte T. Molecular phylogeny and biogeography of tribe Anthemideae (Asteraceae), based on chloroplast gene ndhF. Mol Phyl Evol. 2000;15:59–69. doi: 10.1006/mpev.1999.0714. [DOI] [PubMed] [Google Scholar]
- Xifreda CC. Sonre el nombre cientifica correcta de la manzanilla (Matricaria recutita L., Asteraceae) Darwiniana. 1985;26:373–375. [Google Scholar]


